Published online Sep 27, 2023. doi: 10.4240/wjgs.v15.i9.2032
This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Peer-review started: June 8, 2023
First decision: July 7, 2023
Revised: July 13, 2023
Accepted: August 4, 2023
Article in press: August 4, 2023
Published online: September 27, 2023
Processing time: 106 Days and 11.6 Hours
Early detection of colorectal cancer (CRC) is essential to reduce cancer-related morbidity and mortality. Stool DNA (sDNA) testing is an emerging method for early CRC detection. Syndecan-2 (SDC2) methylation is a potential biomarker for the sDNA testing. Aberrant DNA methylation is an early epigenetic event during tumorigenesis and can occur in the normal colonic mucosa during aging, which can compromise the sDNA test results.
To determine whether methylated SDC2 in sDNA normalizes after surgical resection of CRC.
In this prospective study, we enrolled 151 patients with CRC who underwent curative surgical resection between September 2016 and May 2020. Preoperative stool samples were collected from 123 patients and postoperative samples were collected from 122 patients. A total of 104 samples were collected from both preoperative and postoperative patients. Aberrant promoter methylation of SDC2 in sDNA was assessed using linear target enrichment quantitative methylation-specific real-time polymerase chain reaction. Clinicopathological parameters were analyzed using the results of SDC2 methylation.
Detection rates of SDC2 methylation in the preoperative and postoperative stool samples were 88.6% and 19.7%, respectively. Large tumor size (3 cm, P = 0.019) and advanced T stage (T3–T4, P = 0.033) were positively associated with the detection rate of SDC2 methylation before surgery. Female sex was associated with false positives after surgery (P = 0.030). Cycle threshold (CT) values were significantly decreased postoperatively compared with preoperative values (P < 0.001). The postoperative negative conversion rate for preoperatively methylated SDC2 was 79.3% (73/92).
Our results suggested that the SDC2 methylation test for sDNA has acceptable sensitivity and specificity. However, small size and early T stage tumors are associated with a low detection rate of SDC2 methylation. As the cycle threshold values significantly decreased after surgery, SDC2 methylation test for sDNA might have a diagnostic value for CRC.
Core Tip: This prospective study evaluated the detection of syndecan-2 (SDC2) methylation in preoperative and postoperative stool DNA samples of colorectal cancer (CRC) patients. The study demonstrated that the SDC2 methylation test showed high sensitivity (88.6%) for detecting CRC before surgery, indicating its potential as a non-invasive diagnostic tool. Postoperatively, the detection rate decreased to 19.7%, suggesting the normalization of SDC2 methylation after surgical resection. The study highlights the diagnostic value of SDC2 methylation in preoperative and postoperative stool samples, supporting its role as a non-invasive screening tool for CRC.
- Citation: Song JH, Oh TJ, An S, Lee KH, Kim JY, Kim JS. Comparative detection of syndecan-2 methylation in preoperative and postoperative stool DNA in patients with colorectal cancer. World J Gastrointest Surg 2023; 15(9): 2032-2041
- URL: https://www.wjgnet.com/1948-9366/full/v15/i9/2032.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v15.i9.2032
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and second leading cause of cancer-related deaths worldwide[1]. According to the 2019 Korean statistics, the prevalence of CRC was the third highest, following thyroid and lung cancer. From 2015 to 2019, the 5-year survival rate of CRC patients in Korea was 74.3%[2]. Patients with metastatic CRC have a 5-year survival rate of < 10%, whereas those with early detection of CRC have a 5-year survival rate of > 90%[3]. Therefore, the early detection of CRC is essential to reduce cancer-related morbidity and mortality.
Most patients with early-stage CRC are mostly asymptomatic. There are several screening tools for detecting asymptomatic CRC. The fecal occult blood test and fecal immunochemical test are non-invasive, inexpensive, and convenient methods. However, these tests have low sensitivity and specificity[4]. Computed tomography colonography can be used as an alternative examination for colonoscopy, which non-invasively examines the entire colon beyond the obstructive. However, there are concerns about radiation hazards and the disadvantage of detecting small polyps < 5mm in size. Colonoscopy is the most accurate method with the advantage of being able to perform procedures such as biopsy and polypectomy. In the NordICC trial, a prospective, multinational, and randomized controlled trial investigating the effectiveness of colonoscopy on CRC incidence and mortality, colonoscopy as a screening tool showed good compliance, a high rate of adenoma yield, and adequate performance[5]. However, colonoscopy is an invasive procedure that requires mechanical bowel preparation and carries the risk of complications, such as perforation or electrolyte imbalance. Therefore, there is a need to develop a non-invasive and highly accurate CRC screening tool, especially for those reluctant to undergo colonoscopy.
CRC carcinogenesis is associated with the accumulation of genetic and epigenetic alterations[6]. Aberrant DNA methylation is one of the most common molecular alterations involved in CRC[7]. Because exfoliated cells from tumors are present in stool samples from CRC patients, detection of aberrant methylation is emerging as a non-invasive diagnostic tool for CRC[8]. Several stool DNA (sDNA)-based methylation markers are available for the early detection of CRC[9,10]. Among these, syndecan-2 (SDC2) has been reported as a potential biomarker for sDNA testing[11-13]. Aberrant DNA methylation is an early epigenetic event during tumorigenesis that can occur in the normal colonic mucosa during aging[14], which may compromise the sDNA test results. If preoperatively methylated SDC2 (meSDC2) normalizes after the surgical resection of CRC, it may have diagnostic value and may be used for postoperative surveillance[15]. Therefore, we conducted a prospective observational study to compare SDC2 methylation in the sDNA of patients with CRC before and after surgery.
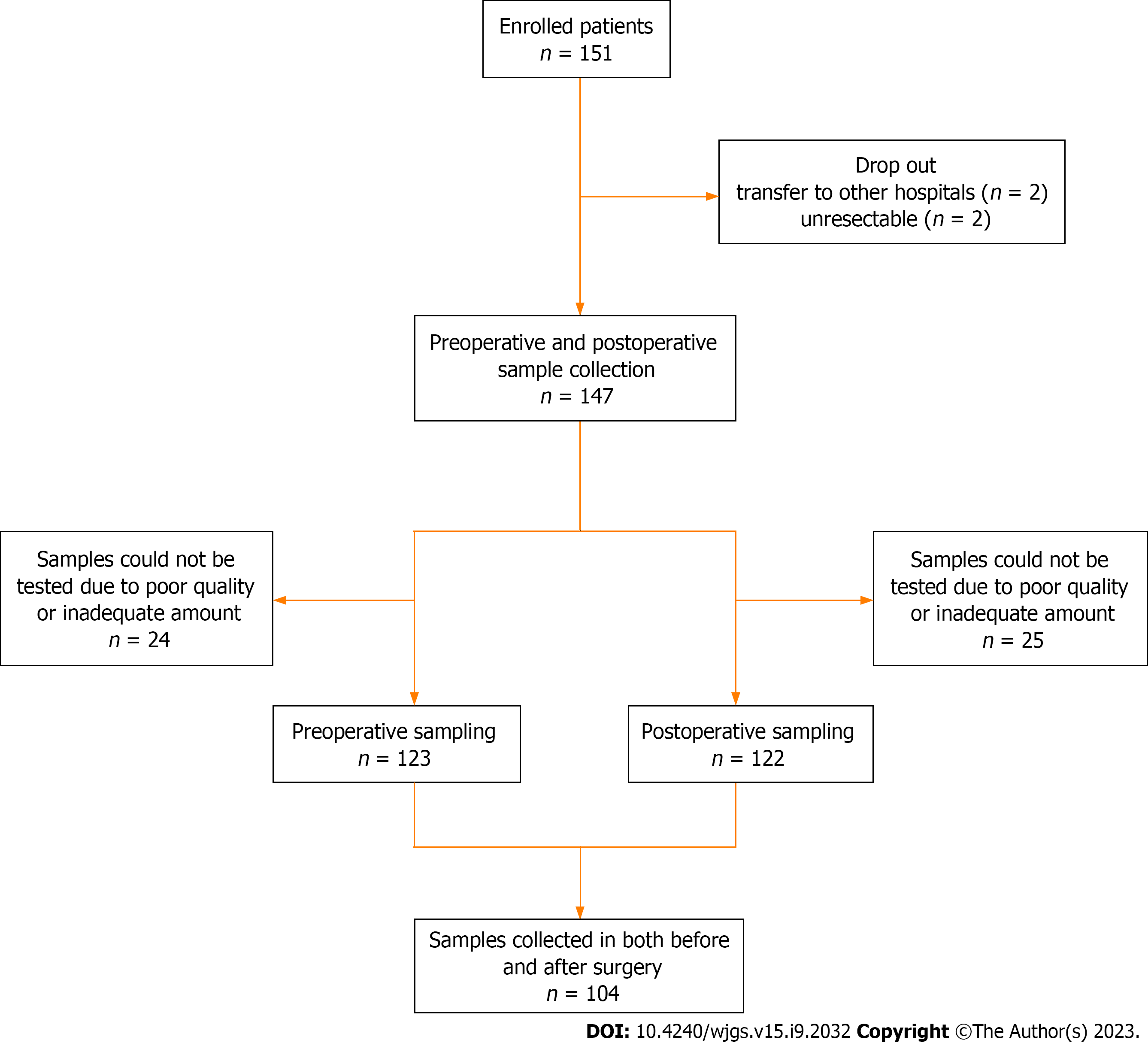
In this prospective study, we enrolled 151 patients diagnosed with CRC who underwent surgical resection with curative intent between September 2016 and May 2020 at the Department of Surgery, Chungnam National University Hospital (South Korea). This observational study was approved by the Institutional Review Board of our institute (No. 2016-05-018). The exclusion criteria were as follows: (1) Patients under 18 or over 80 years of age; (2) Difficulty in obtaining informed consent due to mental health issues; (3) Tumor complications such as bleeding, perforation, or obstruction; and (4) Palliative operation. Among the 151 enrolled patients, four dropped out. Two patients were transferred to other hospitals, and two were unresectable. Clinical data were collected for 147 patients. Stool samples were collected from 123 patients preoperatively and 122 patients postoperatively, excluding those due to poor sample quality and inadequate sample amount. A total of 104 samples were collected from both preoperative and postoperative patients. Figure 1 shows the flowchart of the enrolled patients.
For each subject, at least 2 g of voided stool sample was collected in 20 mL of preservative buffer (Genomictree, Inc., Daejeon, South Korea) using a disposable spatula from four to five different locations. The samples were collected from the patients before and after definitive surgery. Inadequate stool specimens were not included in the methylation analysis (e.g., diarrhea or loose stools).
sDNA was isolated using the GT Nucleic Acid PREP Kit II (Genomictree, Inc., Daejeon, South Korea) according to the manufacturer’s instructions. The Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific, MA, United States) was used to determine the DNA concentration. Briefly, all stool samples were weighed and homogenized in a preservative buffer using a multiple vortex mixer (MIULAB, Hangzhou, China). After homogenization, 1–2 g of each stool sample was used for DNA isolation.
Each two µg of stool-derived genomic DNA was chemically modified with sodium bisulfite using the EZ DNA Methylation-Gold Kit (ZYMO Research, CA, United States) according to the manufacturer’s instructions. Bisulfite-converted DNA was purified using a Zymo-Spin IC column (ZYMO Research) and eluted with 10 µL of distilled water. Bisulfite-converted DNA was either used immediately for methylation analysis or stored at -20 ºC until use.
The meSDC2 linear target enrichment (LTE)-quantitative methylation-specific real-time polymerase chain reaction (qMSP) assay was performed in duplicate reactions for each sample as described by Han et al[13]. A highly sensitive two-step meSDC2 LTE-qMSP assay was used to measure SDC2 methylation in sDNA. First, LTE was used to enrich meSDC2 target DNA and control COL2A1 DNA from the bisulfite-modified DNA. The region of the COL2A1 gene lacking CpG dinucleotides was used as a control to estimate the amount of amplifiable template and the adequacy of bisulfite conversion. The LTE reaction mixture (20 µL) containing 2.0 µg of bisulfite-converted sDNA, 50 nmol/L each of SDC2 methylation-specific antisense (5′-AAAGATTCGGCGACCACCGAACGACTCAAACTCGAAAACTCG-3’) and COL2A1 gene-specific antisense primers (5′-AAAGATTCGGCGACCACCGACTAICCCAAAAAAACCCAATCCTA-3′) attached to a 5’ universal sequence (5’-AAAGATTCGGCGACCACCGA-3’), and 4 µL of 5× AptaTaq polymerase chain reaction (PCR) master mix (Roche Diagnostics, Basel, Switzerland) was prepared. The thermal cycling conditions were as follows: 95 °C for 5 min, followed by 35 cycles of 95 °C for 15 s and 60 °C for 60 s. Next, the reaction mixture volume was scaled up to 40 µL, containing 8 µL of 5× AptaTaq PCR master mix, 250 nmol/L SDC2 methylation-specific sense primer (5′-GTAGAAATTAATAAGTGAGAGGGC-3′), 125 nmol/L SDC2 probe (5′-FAM-TTCGGGGCGTAGTTGCGGGCGG-3′), 125 nmol/L COL2A1 sense primer (5′GTAATGTTAGGAGTATTTTGTGGITA-3′), 62.5 nmol/L COL2A1 probe (5′-Cy5-AGAAGAAGGGAGGGGTGTTAGGAGAGG-3′), and 250 nmol/L universal sequence primer. Thermal cycling conditions were as follows: 95 °C for 5 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Heating and cooling rates were 20 °C/s and 15 °C/s, respectively. For each run, bisulfite-converted methylated (HCT116) and unmethylated genomic DNA (whole-genome amplified human lymphocyte DNA) were used as methylation controls. Non-template and non-template bisulfite-converted controls were also included in the study.
PCR for SDC2 and COL2A1 was performed in a single tube. meSDC2 LTE-qMSP was performed using Rotor-Gene Q real-time PCR instrument (Qiagen, Hilden, Germany). The cycle threshold (CT) value was calculated using the Rotor-Gene Q software. Lower CT values indicated higher levels of SDC2 methylation. For PCR analysis, SDC2 methylation was detected if the CT was less than 40 cycles. It was not detected if CT was not measurable. Samples were categorized as positive if at least one of the two reactions showed detectable SDC2, and they were considered negative if SDC2 methylation was not measurable in both reactions. The test results were acceptable only when the CT value of COL2A1 was < 31. If COL2A1 was not detected, or the CT value was > 31, the test was repeated. Neither the personnel involved in laboratory work nor data analysis of the SDC2 methylation results was informed of colonoscopic findings or pathology outcomes as reference standards. Oh TJ and An S are employees and shareholders of Genomictree, Inc.
Statistical analysis of the data was performed using IBM SPSS Statistics 26 (SPSS Inc., Chicago, IL, United States). Chi-square and/or Fisher’s exact tests were used for categorical variables. Detection rates were evaluated dichotomously as ‘0’ for methylation-negative and ‘1’ for methylation-positive.
The CT values before and after surgery according to tumor node metastasis (TNM) stage were compared using analysis of variance (ANOVA). A paired t-test was performed to compare CT values before and after surgery for each patient. A P value < 0.05 was considered statistically significant.
In addition, we retrieved literatures by Reference Citation Analysis to supplement the latest cutting-edge research results.
Patient characteristics are described in Table 1. In total, 97 male (66%) and 50 female (34%) patients were included in the preoperative or postoperative sampling. The mean age was 62.3. Total colonoscopy was performed before surgery in 133 patients (90.5%), and remnant advanced adenoma (RAA) was present in 18 patients (12.2%). Among the 18 patients with RAA, five remained SDC2 methylation-positive after surgery. The recurrence rate was 6.8% in all patients.
| Patients with preoperative or postoperative stool sampling, n = 147 (%) | Patients with preoperative and postoperative stool sampling, n = 104 (%) | |
| Sex | ||
| Male | 97 (66.0) | 66 (63.5) |
| Female | 50 (34.0) | 38 (36.5) |
| Age, yr | ||
| Mean (range) | 62.3 (35-80) | 62.8 (35-80) |
| Tumor location | ||
| Right colon | 45 (30.6) | 29 (27.9) |
| Left colon | 62 (42.2) | 51 (49.0) |
| Rectum | 40 (27.2) | 24 (23.1) |
| Histology | ||
| WD | 6 (4.1) | 4 (3.8) |
| MD | 132 (89.8) | 94 (90.4) |
| PD | 9 (3.4) | 5 (4.8) |
| Mucinous | 4 (2.7) | 1 (1.0) |
| Tumor size, cm | ||
| Mean (range) | 3.7 (0.0-15.0) | 3.5 (0.0-9.0) |
| T stage | ||
| Tis | 5 (3.4) | 4 (3.8) |
| T1 | 22 (15.0) | 15 (14.4) |
| T2 | 27 (18.4) | 19 (18.3) |
| T3 | 74 (50.3) | 52 (50.0) |
| T4 | 19 (12.9) | 14 (13.5) |
| N stage | ||
| N0 | 93 (63.3) | 66 (63.5) |
| N+ | 54 (36.7) | 38 (36.5) |
| TCF before surgery | ||
| Not done | 14 (9.5) | 9 (8.7) |
| Done | 133 (90.5) | 95 (91.3) |
| RAA | ||
| Not present | 129 (87.8) | 90 (86.5) |
| Present | 18 (12.2) | 14 (13.5) |
| Recurrence | ||
| No | 137 (93.2) | 98 (94.2) |
| Yes | 10 (6.8) | 6 (5.8) |
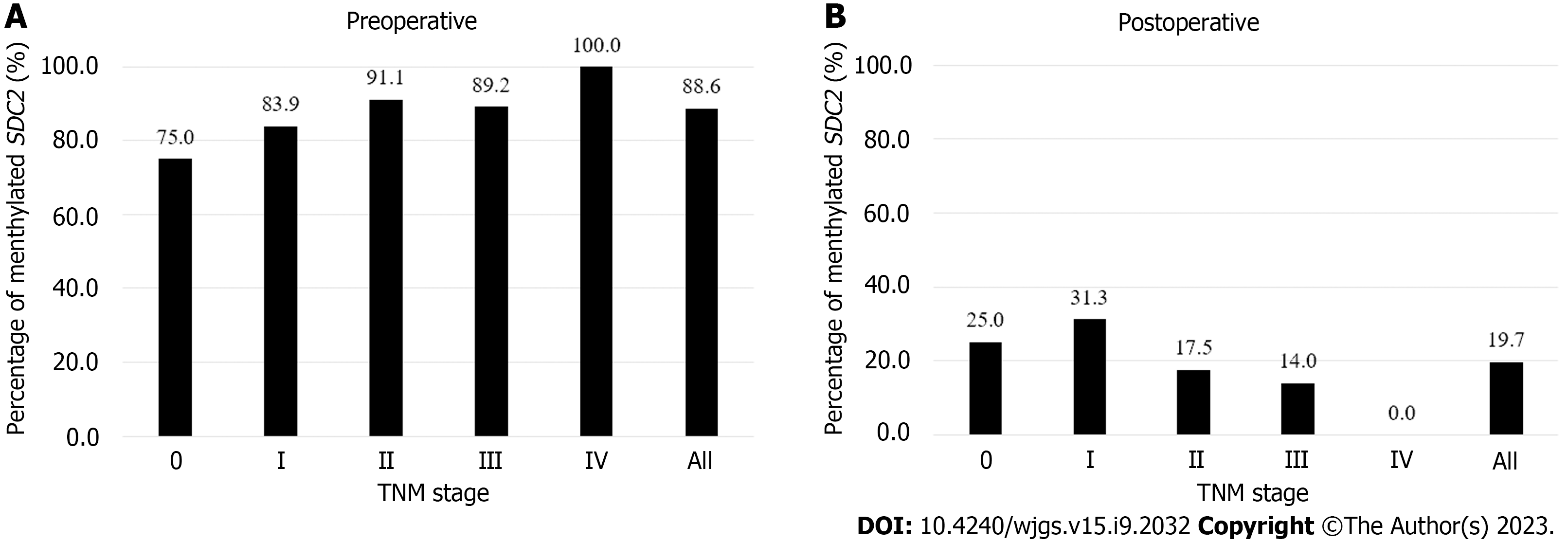
Figure 2 shows the detection rates of SDC2 methylation according to TNM stage. The positivity rate of the SDC2 methylation test before surgery was 88.6% (109/123), and the false negative rate was 11.4% (14/123). The positivity rates for stages 0, I, II, III, and IV were 75% (3/4), 83.9% (26/31), 91.1% (41/45), 89.2% (33/37), and 100% (6/6), respectively (Figure 2A). The detection rate of SDC2 methylation after surgery was 19.7% (24/122). The detection rates for stages 0, I, II, III, and IV were 25% (1/4), 31.2% (10/32), 17.5% (7/40), 14.0 (6/43), and 0% (0/3), respectively (Figure 2B). The demographic analysis of patients according to the preoperative and postoperative SDC2 methylation results is presented in Table 2. Large tumor size (3 cm, P = 0.019) and advanced T stage (T3–T4, P = 0.033) were associated with positivity of SDC2 methylation before surgery. Additionally, female patients showed more false positives after surgery (P = 0.030).
| Characteristics | Preoperative (n = 123) | Postoperative (n = 122) | ||||
| Negative, n = 14 (%) | Positive, n = 109 (%) | P value | Negative, n = 98 (%) | Positive, n = 24 (%) | P value | |
| Sex | 0.375 | 0.030 | ||||
| Male | 11 (78.6) | 69 (63.3) | 68 (69.4) | 11 (45.8) | ||
| Female | 3 (21.4) | 40 (36.7) | 30 (30.6) | 13 (54.2) | ||
| Age, yr | 0.512 | 0.845 | ||||
| < 65 | 9 (64.3) | 60 (55.0) | 55 (56.1) | 14 (58.3) | ||
| ≥ 65 | 5 (35.7) | 49 (45.0) | 43 (43.9) | 10 (41.7) | ||
| Location | 0.664 | 0.079 | ||||
| Right colon | 5 (35.7) | 30 (27.5) | 34 (34.7) | 3 (12.5) | ||
| Left colon | 7 (50.0) | 51 (46.8) | 42 (42.9) | 12 (50.0) | ||
| Rectum | 2 (14.3) | 28 (25.7) | 22 (22.4) | 9 (37.5) | ||
| Histology | 0.226 | 1.000 | ||||
| WD/MD | 12 (85.7) | 103 (94.5) | 92 (93.9) | 23 (95.8) | ||
| PD/Mucinous | 2 (14.3) | 6 (5.5) | 6 (6.1) | 1 (4.2) | ||
| Tumor size, cm | 0.019 | 0.145 | ||||
| < 3 | 10 (71.4) | 42 (38.5) | 41 (41.8) | 14 (58.3) | ||
| ≥ 3 | 4 (28.6) | 67 (61.5) | 57 (58.2) | 10 (41.7) | ||
| T stage | 0.033 | 0.137 | ||||
| Tis-T2 | 9 (64.3) | 34 (31.2) | 33 (33.7) | 12 (50.0) | ||
| T3-T4 | 5 (35.7) | 75 (68.8) | 65 (66.3) | 12 (50.0) | ||
| N stage | 0.769 | 0.152 | ||||
| N0 | 10 (71.4) | 70 (64.2) | 58 (59.2) | 18 (75.0) | ||
| N+ | 4 (28.6) | 39 (35.8) | 40 (40.8) | 6 (25.0) | ||
| TNM stage | 0.769 | 0.152 | ||||
| 0-II | 10 (71.4) | 70 (64.2) | 58 (59.2) | 18 (75.0) | ||
| III-IV | 4 (28.6) | 39 (35.8) | 40 (40.8) | 6 (25.0) | ||
| TCF before surgery | 0.627 | 0.685 | ||||
| Not done | 2 (14.3) | 10 (9.2) | 9 (9.2) | 1 (4.2) | ||
| Done | 12 (85.7) | 99 (90.8) | 89 (90.8) | 23 (95.8) | ||
| RAA | 0.692 | 0.172 | ||||
| Not present | 13 (92.9) | 94 (86.2) | 88 (89.8) | 19 (79.2) | ||
| Present | 1 (7.1) | 15 (13.8) | 10 (10.2) | 5 (20.8) | ||
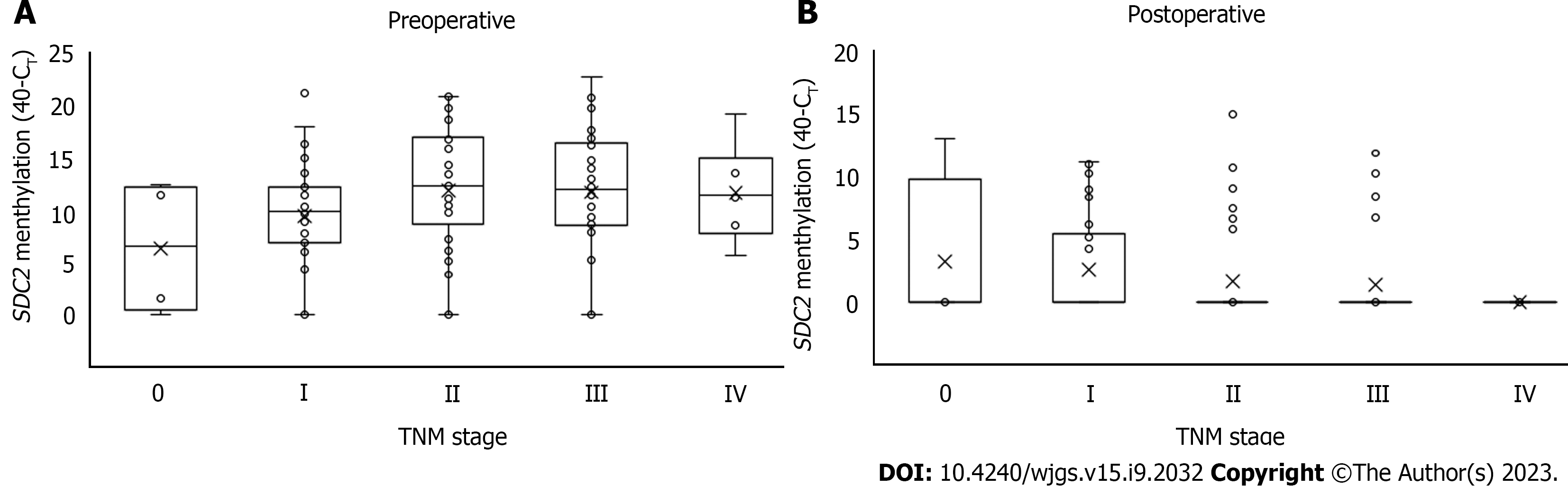
The distribution of CT values in preoperative and postoperative SDC2 methylation according to TNM stage is shown in Figure 3. There were no significant differences in the CT values of SDC2 methylation preoperatively (P = 0.901) or postoperatively (P = 0.332) according to TNM stage.
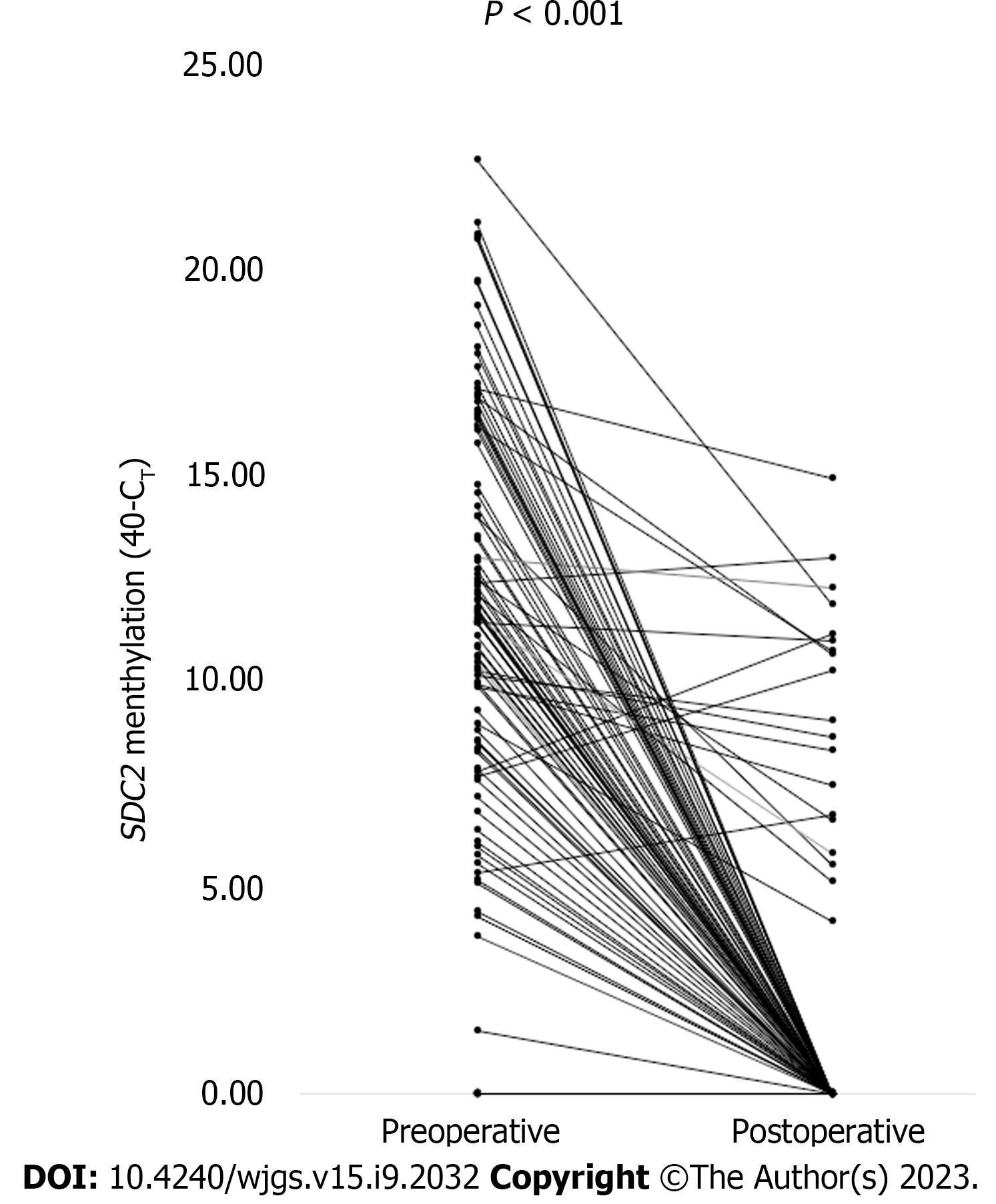
Figure 4 shows the CT values of meSDC2 for each patient before and after surgery. Among the 104 patients from whom stool samples were obtained before and after surgery, 92 patients showed positive results for preoperative SDC2 methylation. The postoperative negative conversion rate for preoperatively meSDC2 was 79.3% (73/92). The CT values of SDC2 methylation significantly decreased postoperatively compared to the preoperative values (P < 0.001).
Various screening tools are used for the early detection of CRC to reduce cancer-related morbidity and mortality. Among the screening tools, colonoscopy is the most accurate method and is considered the gold standard with high sensitivity and specificity. However, colonoscopy is an invasive procedure that requires mechanical bowel preparation and carries the risk of complications, such as perforation or electrolyte imbalance. Hence, it is not yet useful as a mass-screening test[16]. Recently, non-invasive and highly accurate tests using sDNA methylation have been reported[6,7,9,10]. Among the several known sDNA biomarkers, SDC2 was found to be the most accurate single gene[17]. Therefore, SDC2 methylation in sDNA has been proposed as a non-invasive mass-screening tool for the early detection of CRC.
Recently, Wang et al[18] reported a meta-analysis that evaluated the diagnostic performance of the SDC2 methylation test for detecting CRC. Twelve studies were included in the meta-analysis, and all articles were retrospective studies. Among them, seven studies measured meSDC2 in sDNA, and five studies measured it in blood. The overall sensitivity was 80% and the specificity was 95%; and the sensitivity and specificity of sDNA test were 83% and 94%, respectively. In the present study, the detection rates of SDC2 methylation in preoperative and postoperative were 88.6% and 19.7%, respectively. These values represent a sensitivity of 88.6% (109/123, 95%CI: 82%–96%) and specificity of 80.3% (98/122, 95%CI: 72%–87%), and the results were comparable to those of several previous studies.
Zhao et al[19] reported the detection rate of meSDC2 in stool samples from 94 patients with CRC and 124 normal healthy individuals. There were no significant differences in the detection rates of meSDC2 based on the age, sex, or stage. As the tumor size increased, the positive detection rate of meSDC2 also increased (P < 0.05). Similarly, we found that a large preoperative tumor size and advanced T stage were associated with high detection rates of SDC2 methylation. Females showed more false positive results after surgery. This result was thought to be due to the small sample size. The combination of SDC2 with other biomarkers may improve CRC detection rates[9,19].
Several studies have reported that SDC2 methylation is not related to the clinical stage[11,13,18]. The present study also showed similar results. There was no association between the detection rate of SDC2 methylation and clinical stage. Although there was no statistically significant difference, the detection rate of SDC2 methylation according to stage showed a gradual increase. Moreover, Oh et al[12] found that the sensitivity of SDC2 methylation test tended to gradually increase with an increase in the stage. Further studies are needed to validate whether clinical stage affects the detection rate of meSDC2.
Few studies have compared sDNA test results before and after surgery. Kisiel et al[15] evaluated sDNA markers (NDRG4 and BMP3) in 22 patients with CRC before and after surgery. They demonstrated that methylated sDNA markers present in patients normalized following surgical resection. Nishioka et al[20] compared the preoperative and postoperative sDNA levels of 54 patients with CRC who underwent surgical resection. Aberrant methylation of sDNA markers (CDH4 and GATA5) was detected in 23 (42.3%) preoperative stool samples from patients with CRC. Methylated alleles of these genes were not found in the postoperative sDNA. To the best of our knowledge, no studies have compared meSDC2 levels before and after surgery. We compared the preoperative and postoperative CT values of meSDC2 in 92 patients who tested positive for SDC2 methylation before surgery. The CT values of SDC2 methylation in sDNA were significantly decreased postoperatively compared to the preoperative values. These results indicate that SDC2 methylation test has diagnostic value and may be used for surveillance.
In our study, twenty-four (19.7%) patients with CRC remained positive for meSDC2 in the sDNA after surgical resection. Among these patients, there was no recurrence after surgery during the median follow-up period of 46 (1–67) months. Since molecular-level methylation precedes phenotypic tumorigenesis, these patients require follow-up with concern for possible recurrence. However, six patients who experienced recurrence after surgery showed negative results for SDC2 methylation. The correlation between postoperative SDC2 methylation and recurrence was not statistically significant (P = 0.597). Therefore, based on these results, the present study seemed unlikely to show the usefulness of meSDC2 in the sDNA as surveillance after CRC surgery. In addition, the liver and peritoneum are the most common sites for CRC metastasis[21], with rare occurrences in other parts of the body[22]. These recurrent cases of metastasis are difficult to detect with a sDNA test, making it difficult to use stool tests as a surveillance tool.
Similar to stool DNA testing, dysbiosis in the microbiota can be used as a test for early detection of CRC by obtaining fecal samples[23]. Several microbial species, such as F. nucleatum, B. fragilis, and F. Prausnitzii, are known to act as a driving force in the occurrence of CRC. Hence, detecting these microbial species in stool samples can aid in the early identification of CRC[24]. Non-invasive fecal biomarkers, such as aberrant methylation of sDNA tests or dysbiosis in the microbiota, can be expected to make important contributions to the early diagnosis and therapeutic implications of CRC in the future.
The present study had several limitations. First, selection bias may have existed because this study was conducted at a single institution with a small sample size. Second, it is insufficient to determine its value as a surveillance tool because SDC2 methylation was only compared before and after surgery, without long-term follow-up. Therefore, multicenter prospective studies with long-term follow-up are necessary to assess the feasibility of surveillance.
We demonstrated that the SDC2 methylation test in sDNA has acceptable sensitivity and specificity. However, small-size and early T stage tumors are associated with a low detection rate of SDC2 methylation. As the CT values significantly decrease after surgery, SDC2 methylation of the sDNA test exhibits diagnostic value for CRC.
Colorectal cancer (CRC) is a significant cause of morbidity and mortality worldwide, emphasizing the need for early detection. Stool DNA (sDNA) testing is a promising non-invasive method for CRC detection, and syndecan-2 (SDC2) methylation has been identified as a potential biomarker for this test.
The study aimed to investigate whether SDC2 methylation in sDNA normalizes after surgical resection of CRC, which could have implications for the diagnostic value and postoperative surveillance of SDC2 methylation.
The study aimed to compare the detection rates of SDC2 methylation in preoperative and postoperative stool samples of CRC patients and assess the association between SDC2 methylation and clinicopathological parameters. The study also sought to evaluate the change in SDC2 methylation levels before and after surgery.
A prospective study enrolled 151 CRC patients who underwent surgical resection. Stool samples were collected before and after surgery, and SDC2 methylation in sDNA was assessed using a quantitative methylation-specific real-time polymerase chain reaction. The association between SDC2 methylation and clinicopathological parameters was analyzed.
The detection rate of SDC2 methylation was significantly higher in preoperative stool samples (88.6%) compared to postoperative samples (19.7%). Large tumor size and advanced T stage were associated with higher detection rates before surgery, while female sex was associated with false positives after surgery. The cycle threshold (CT) values significantly decreased after surgery, indicating a normalization of SDC2 methylation. The postoperative negative conversion rate for preoperatively methylated SDC2 was 79.3%.
The study findings suggest that the SDC2 methylation test in sDNA has acceptable sensitivity and specificity for CRC detection. However, the detection rate is lower for small-size and early T stage tumors. The significant decrease in CT values after surgery indicates the diagnostic value of SDC2 methylation testing for CRC.
Further research is needed to validate the findings and assess the long-term utility of SDC2 methylation testing as a surveillance tool for postoperative CRC patients. Multicenter prospective studies with extended follow-up periods are warranted to evaluate the feasibility and effectiveness of SDC2 methylation testing in clinical practice.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Surgery
Country/Territory of origin: South Korea
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kisiel JB, United States; Lucke-Wold B, United States S-Editor: Zhang H L-Editor: A P-Editor: Cai YX
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. |
| 2. | Korean statistics 2019. [cited 2023 Jun 8]. Korean Statistical Information Service [Internet]. Available from: https://kosis.kr/. |
| 3. | Dashwood RH. Early detection and prevention of colorectal cancer (review). Oncol Rep. 1999;6:277-281. |
| 4. | Morikawa T, Kato J, Yamaji Y, Wada R, Mitsushima T, Shiratori Y. A comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population. Gastroenterology. 2005;129:422-428. |
| 5. | Bretthauer M, Kaminski MF, Løberg M, Zauber AG, Regula J, Kuipers EJ, Hernán MA, McFadden E, Sunde A, Kalager M, Dekker E, Lansdorp-Vogelaar I, Garborg K, Rupinski M, Spaander MC, Bugajski M, Høie O, Stefansson T, Hoff G, Adami HO; Nordic-European Initiative on Colorectal Cancer (NordICC) Study Group. Population-Based Colonoscopy Screening for Colorectal Cancer: A Randomized Clinical Trial. JAMA Intern Med. 2016;176:894-902. |
| 6. | Berger BM, Ahlquist DA. Stool DNA screening for colorectal neoplasia: biological and technical basis for high detection rates. Pathology. 2012;44:80-88. |
| 7. | Kadiyska T, Nossikoff A. Stool DNA methylation assays in colorectal cancer screening. World J Gastroenterol. 2015;21:10057-10061. |
| 8. | Vedeld HM, Goel A, Lind GE. Epigenetic biomarkers in gastrointestinal cancers: The current state and clinical perspectives. Semin Cancer Biol. 2018;51:36-49. |
| 9. | Chen J, Sun H, Tang W, Zhou L, Xie X, Qu Z, Chen M, Wang S, Yang T, Dai Y, Wang Y, Gao T, Zhou Q, Song Z, Liao M, Liu W. DNA methylation biomarkers in stool for early screening of colorectal cancer. J Cancer. 2019;10:5264-5271. |
| 10. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287-1297. |
| 11. | Niu F, Wen J, Fu X, Li C, Zhao R, Wu S, Yu H, Liu X, Zhao X, Liu S, Wang X, Wang J, Zou H. Stool DNA Test of Methylated Syndecan-2 for the Early Detection of Colorectal Neoplasia. Cancer Epidemiol Biomarkers Prev. 2017;26:1411-1419. |
| 12. | Oh TJ, Oh HI, Seo YY, Jeong D, Kim C, Kang HW, Han YD, Chung HC, Kim NK, An S. Feasibility of quantifying SDC2 methylation in stool DNA for early detection of colorectal cancer. Clin Epigenetics. 2017;9:126. |
| 13. | Han YD, Oh TJ, Chung TH, Jang HW, Kim YN, An S, Kim NK. Early detection of colorectal cancer based on presence of methylated syndecan-2 (SDC2) in stool DNA. Clin Epigenetics. 2019;11:51. |
| 14. | Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681-8686. |
| 15. | Kisiel JB, Yab TC, Taylor WR, Mahoney DW, Ahlquist DA. Stool methylated DNA markers decrease following colorectal cancer resection--implications for surveillance. Dig Dis Sci. 2014;59:1764-1767. |
| 16. | Ransohoff DF. How much does colonoscopy reduce colon cancer mortality? Ann Intern Med. 2009;150:50-52. |
| 17. | Gachabayov M, Lebovics E, Rojas A, Felsenreich DM, Latifi R, Bergamaschi R. Performance evaluation of stool DNA methylation tests in colorectal cancer screening: a systematic review and meta-analysis. Colorectal Dis. 2021;23:1030-1042. |
| 18. | Wang L, Liu Y, Zhang D, Xiong X, Hao T, Zhong L, Zhao Y. Diagnostic accuracy of DNA-based SDC2 methylation test in colorectal cancer screening: a meta-analysis. BMC Gastroenterol. 2022;22:314. |
| 19. | Zhao G, Liu X, Liu Y, Li H, Ma Y, Li S, Zhu Y, Miao J, Xiong S, Fei S, Zheng M. Aberrant DNA Methylation of SEPT9 and SDC2 in Stool Specimens as an Integrated Biomarker for Colorectal Cancer Early Detection. Front Genet. 2020;11:643. |
| 20. | Nishioka Y, Ueki T, Hokazono K, Nagayoshi K, Tanaka M. Comparative detection of aberrantly methylated DNA in preoperative and postoperative stool from patients with colorectal cancers. Int J Biol Markers. 2015;30:e81-e87. |
| 21. | Pretzsch E, Bösch F, Neumann J, Ganschow P, Bazhin A, Guba M, Werner J, Angele M. Mechanisms of Metastasis in Colorectal Cancer and Metastatic Organotropism: Hematogenous versus Peritoneal Spread. J Oncol. 2019;2019:7407190. |
| 22. | Lucke-Wold B, Bonasso PC, Cassim R. Primary colon adenocarcinoma with metastatic disease to the rectum followed by the left axilla. Am Med Stud Res J. 2017;4:57-61. |
| 23. | Villéger R, Lopès A, Veziant J, Gagnière J, Barnich N, Billard E, Boucher D, Bonnet M. Microbial markers in colorectal cancer detection and/or prognosis. World J Gastroenterol. 2018;24:2327-2347. |
| 24. | Abdulla MH, Agarwal D, Singh JK, Traiki TB, Pandey MK, Ahmad R, Srivastava SK. Association of the microbiome with colorectal cancer development (Review). Int J Oncol. 2021;58. |