Copyright
©The Author(s) 2021.
World J Diabetes. Jul 15, 2021; 12(7): 1116-1130
Published online Jul 15, 2021. doi: 10.4239/wjd.v12.i7.1116
Published online Jul 15, 2021. doi: 10.4239/wjd.v12.i7.1116
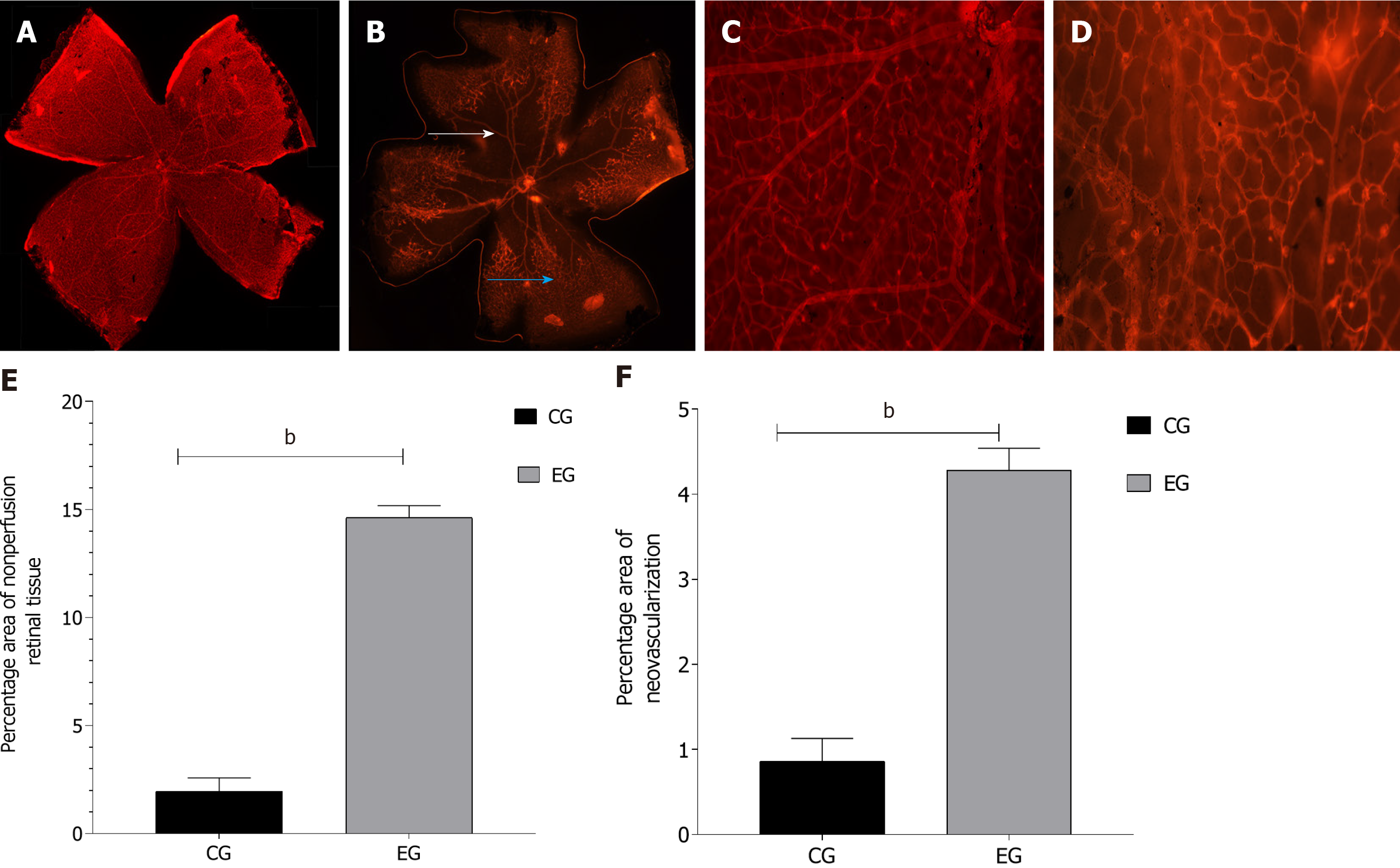
Figure 1 Evaluation of retinal neovascularization.
A: Retina patch morphology of control group (CG); B: Retina tissue morphology of experimental group (EG) (white arrow indicates no perfusion area, blue arrow indicates neovascularization); C: Retina local enlarged map of CG; D: Retina local enlarged map of EG; E: Retina no perfusion area statistical map; F: Retina neovascularization cluster area statistical map. bP < 0.01.
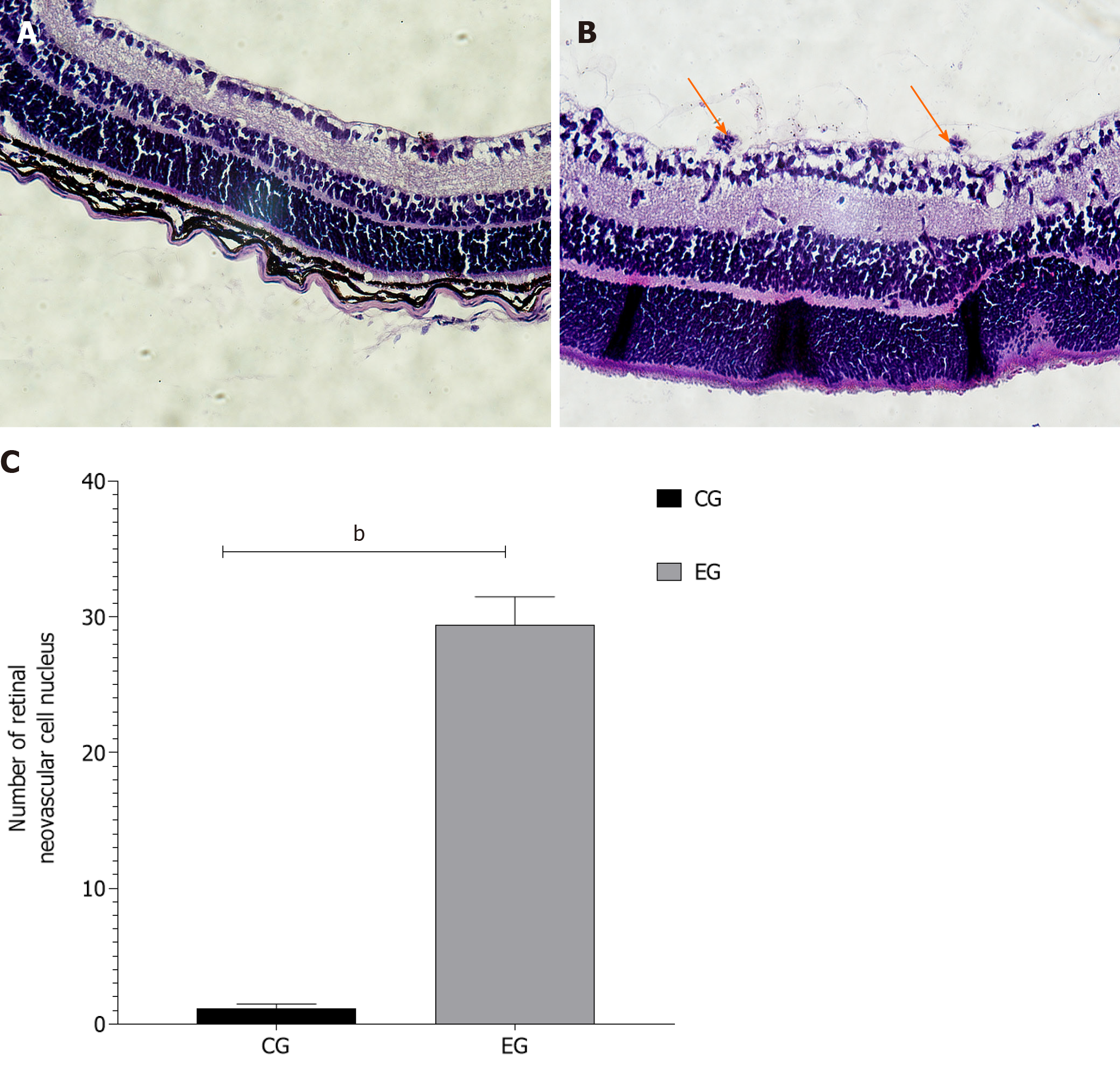
Figure 2 Quantitative analysis of vascular endothelial cells.
A: Hematoxylin and eosin (HE) staining of paraffin-embedded retina sections in the control group (CG) (200 ×); B: HE staining of paraffin-embedded retina sections in the experimental group (EG) (200 ×, orange arrow indicates that endothelial cell nucleus breaks through the inner limiting membrane); C: Number of nuclei breaking through the inner limiting membrane in the two groups. bP < 0.01.
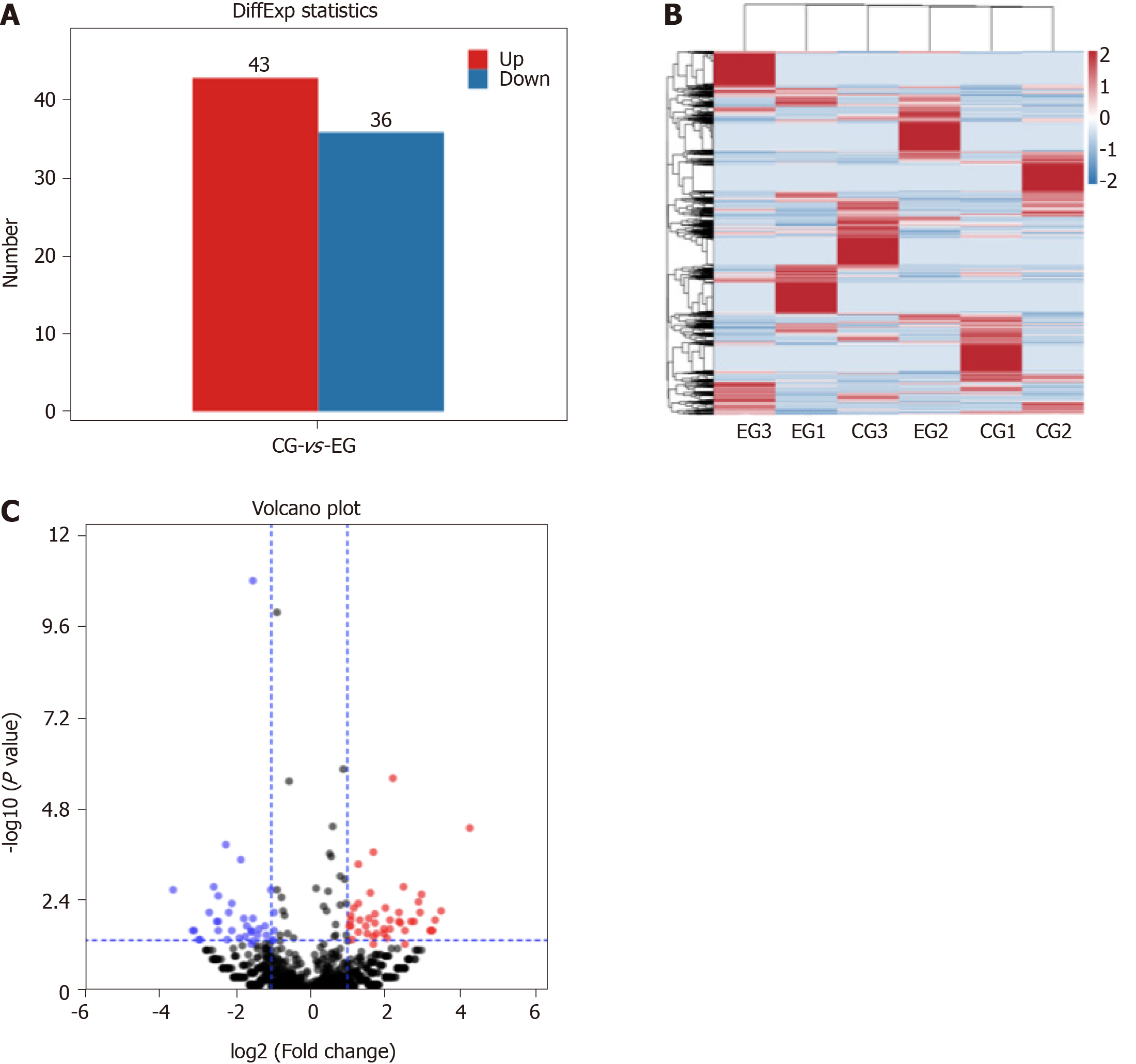
Figure 3 Screening results of differentially expressed genes.
A: Number of differentially expressed piRNAs between experimental group (EG) and control group (CG); B: Clustering analysis of differentially expressed piRNAs; C: Volcano map of differentially expressed piRNAs.
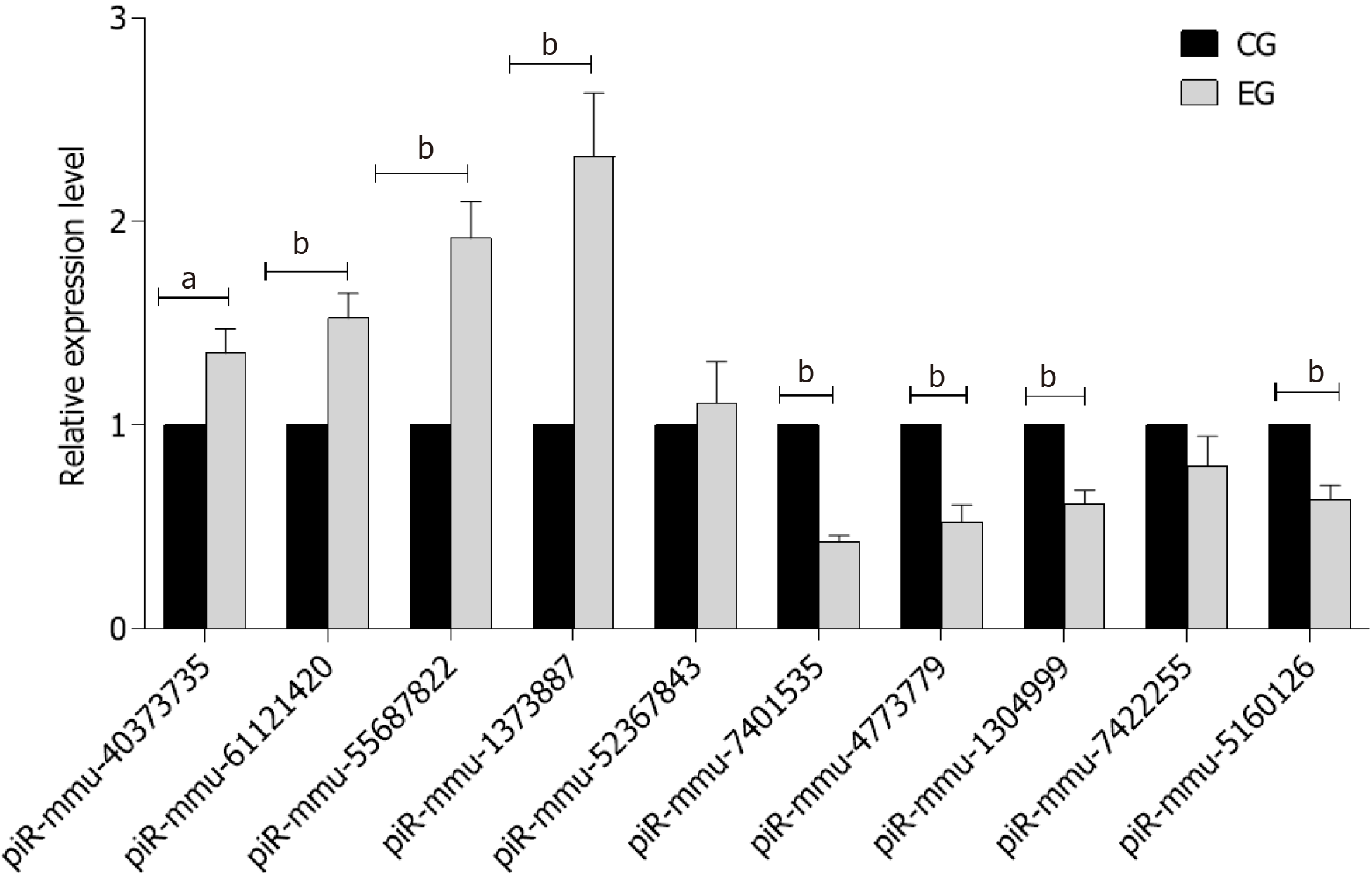
Figure 4 Statistics of ten piRNA validation results.
aP < 0.05; bP < 0.01. CG: Control group; EG: Experimental group.
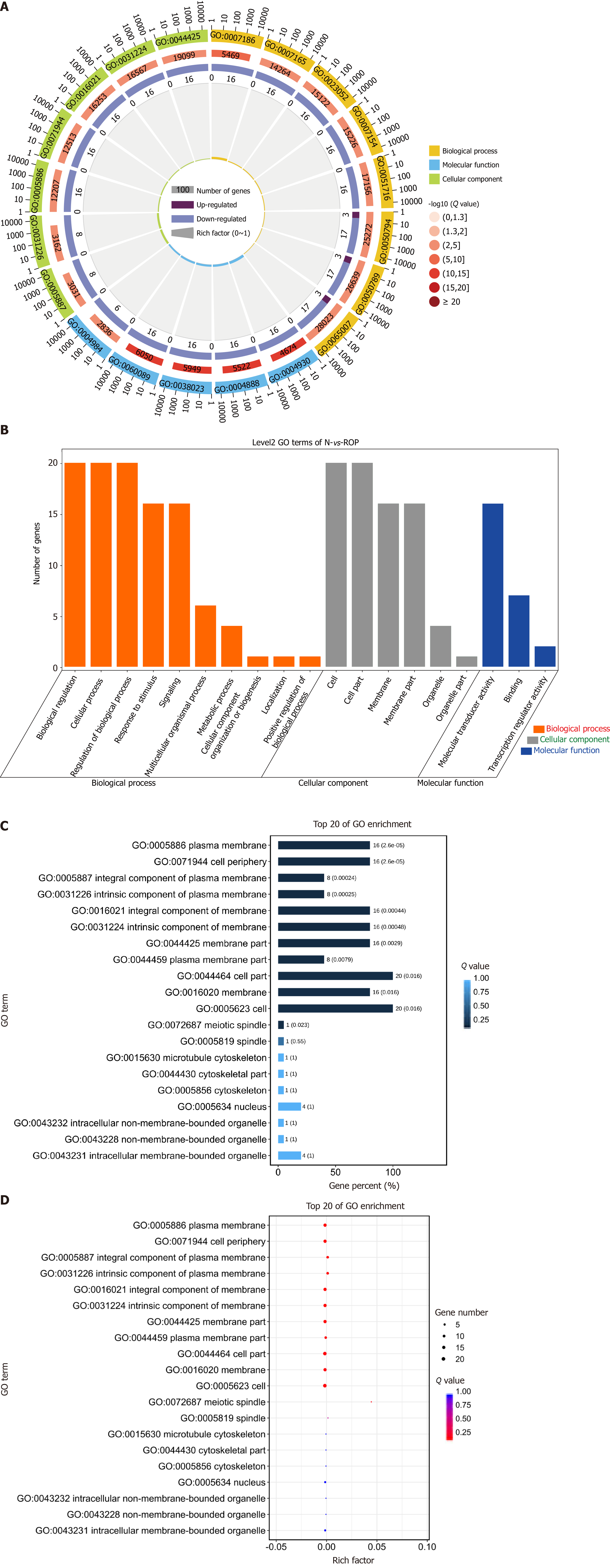
Figure 5 Gene Ontology analysis results.
A: Gene Ontology (GO) enrichment circle diagram (first circle: top 20 GO terms, the coordinate scale of gene number is presented outside the circle. Second circle: number and Q value of the GO term in the background gene. Third circle: number of GO term piRNA target genes. Fourth circle: rich factor value of each GO term piRNA); B: Bar graph of GO enrichment (abscissa shows the level 2 GO term, whereas the ordinate shows the number of genes in the term); C: Bar graph of GO enrichment (a darker color, results in a smaller Q value; the value in the column is the number and Q value of the GO term); D: GO enrichment bubble chart (size indicates the quantity; a redder color leads to a smaller Q value).
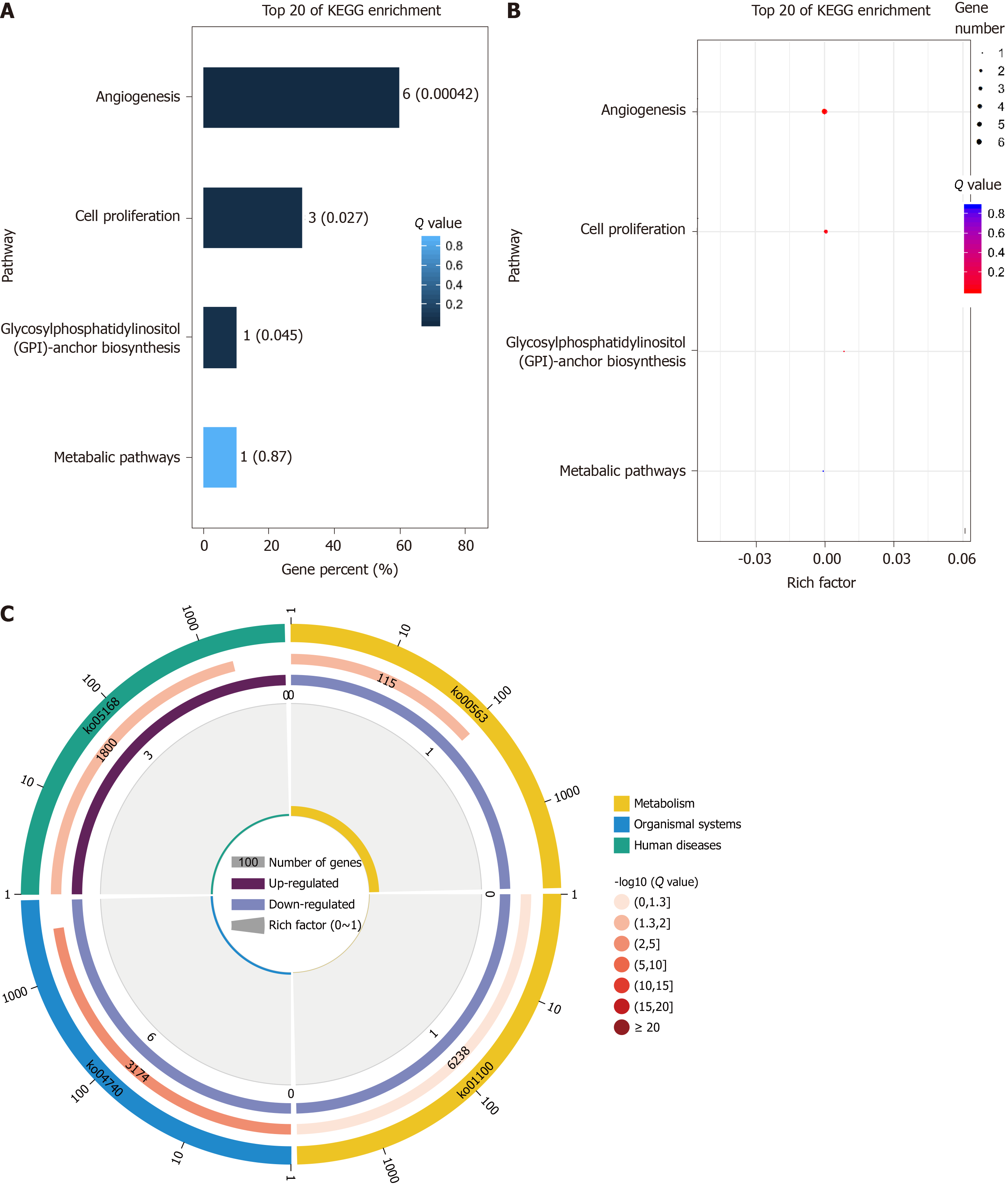
Figure 6 KO analysis results.
A: KO enrichment bar chart (top 20 pathways with the lowest Q value were used to draw the chart. The ordinate represents the pathways, whereas the abscissa shows the percentage of the number of pathways in all piRNA target genes; a darker color correlates with a smaller the Q value. The value in the column is the number and Q value of the pathways); B: KO enrichment bubble chart [top 20 pathways with the lowest Q value were used to plot. The ordinate shows the pathway, whereas the abscissa shows the enrichment factor (the number of piRNA target genes in the pathway divided by all numbers). The size indicates the number; a redder color indicates a smaller Q value]; C: KO enrichment circle diagram [first circle: top 20 enriched pathways; outside the circle is the coordinate scale of gene number, and different colors represent different classes. Second circle: number and Q value of the pathway in the background gene; the more the number of genes, a longer bar and smaller Q value leads to a redder color. Third circle: number of piRNA target genes in the pathway. Fourth circle: rich factor value of each pathway (number of piRNA target genes in the pathway divided by all numbers); background grid line, each grid represents 0.1].
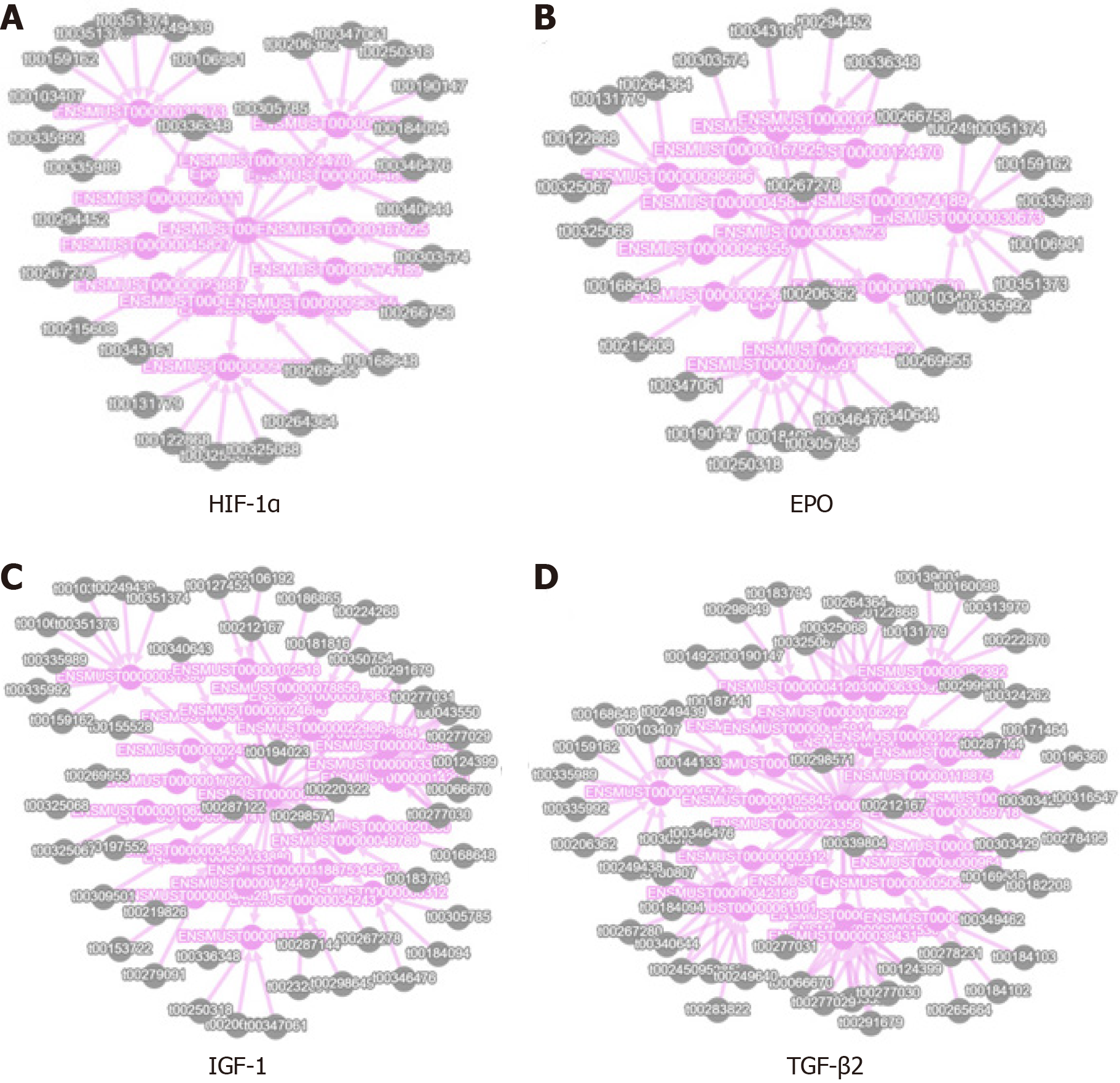
Figure 7 Protein interaction network analysis.
A-D: Stand for differential expression results of piRNA and EPO (B), HIF-1α (A), IGF1 (C), TGF-β2 (D) which are associated with retinal neovascularization.
- Citation: Yu Y, Ren KM, Chen XL. Expression and role of P-element-induced wimpy testis-interacting RNA in diabetic-retinopathy in mice. World J Diabetes 2021; 12(7): 1116-1130
- URL: https://www.wjgnet.com/1948-9358/full/v12/i7/1116.htm
- DOI: https://dx.doi.org/10.4239/wjd.v12.i7.1116