Published online Oct 15, 2015. doi: 10.4251/wjgo.v7.i10.250
Peer-review started: May 8, 2015
First decision: July 17, 2015
Revised: July 28, 2015
Accepted: September 10, 2015
Article in press: September 16, 2015
Published online: October 15, 2015
Processing time: 171 Days and 0.3 Hours
Pancreatic cancer is a highly lethal cancer type, for which there are few viable therapeutic options. But, with the advance of sequencing technologies for global genomic analysis, the landscape of genomic alterations in pancreatic cancer is becoming increasingly well understood. In this review, we summarize current knowledge of genomic alterations in 12 core signaling pathways or cellular processes in pancreatic ductal adenocarcinoma, which is the most common type of malignancy in the pancreas, including four commonly mutated genes and many other genes that are mutated at low frequencies. We also describe the potential implications of these genomic alterations for development of novel therapeutic approaches in the context of personalized medicine.
Core tip: With the advance of sequencing technologies for global genomic analysis, the landscape of genomic alterations in pancreatic cancer is becoming increasingly well understood. In this review, we summarize the latest knowledge of genomic alterations in pancreatic ductal adenocarcinoma including commonly mutated genes and many other genes that are mutated at low frequencies. We also describe the potential implications of these genomic alterations for development of novel therapeutic approaches in the context of personalized medicine.
- Citation: Takai E, Yachida S. Genomic alterations in pancreatic cancer and their relevance to therapy. World J Gastrointest Oncol 2015; 7(10): 250-258
- URL: https://www.wjgnet.com/1948-5204/full/v7/i10/250.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v7.i10.250
Pancreatic cancer was the seventh leading cause of death in the world in 2012, and is responsible for about 331000 deaths per year[1]. The 5-year survival of pancreatic cancer patients is approximately 5%, and this figure has remained constant in recent decades. Because of the absence of effective methods for early detection and the aggressive nature of this disease, the majority of patients present with locally advanced or metastatic cancer which is not eligible for surgical resection. Chemotherapeutic options for treatment of advanced pancreatic cancer are still limited, and gemcitabine has been the standard chemotherapeutic drug for patients with advanced disease for many years, even though this drug alone provides only a modest survival advantage[2-4]. Since the approval of gemcitabine in United States, many randomized clinical trials have been performed to evaluate combinations of gemcitabine with other drugs, such as 5-fluorouracil (5-FU), cisplatin, oxaliplatin and irinotecan[5], but few of them show a significant survival advantage compared with gemcitabine alone. The combination of gemcitabine with the epidermal growth factor receptor (EGFR) inhibitor, erlotinib, does confer a survival advantage over gemcitabine monotherapy, but the overall survival of patients with advanced disease was extended by only 10 d on average[6]. The combination of gemcitabine with nab-paclitaxel (albumin-bound paclitaxel) was recently shown to be superior to gemcitabine alone, probably because of depletion of tumor stroma, which leads to improved delivery of gemcitabine to tumor cells[7]. Other than gemcitabine-based chemotherapies, 5-FU-based chemotherapeutic regimens have also been evaluated. FOLFIRINOX (folinic acid, fluorouracil, irinotecan and oxaliplatin) improved the median overall survival from 6.8 to 11.1 mo compared with gemcitabine, although significant toxicities associated with this regimen limit its utility in a wide range of patients[8]. It seems that a deeper understanding of the molecular biology of pancreatic cancer is needed to develop novel therapeutic approaches.
In recent years, advances in sequencing technologies have enabled us to perform genome-wide analysis to establish the genetic alterations underlying pancreatic carcinogenesis and progression. In this review, we summarize current knowledge of genomic alterations in pancreatic ductal adenocarcinoma (PDAC), which is the most common type of malignancy in the pancreas, and we discuss their implications for development of novel therapeutic strategies.
Jones et al[9] have shown that PDAC harbors an average of 63 genome alterations, of which the majority are point mutations. Four key genes are frequently altered in PDAC: KRAS, CDKN2A, TP53 and SMAD4. The most common gene alteration is in KRAS (v-ki-ras2 Kirsten rat sarcoma viral oncogene homolog), where mutations occur in codons 12, 13 and 61[9,10]. More than 90% of PDAC contains KRAS mutation, and such mutations are also present in about 45% of low-grade pancreatic intraepithelial neoplasia (PanIN) lesions[11,12]. KRAS encodes a GTPase that activates various downstream signaling pathways, including the mitogen-activated protein kinase (MAPK) cascades[13]. Mutations in KRAS result in constitutive activation. Ras proteins are involved in a variety of cellular functions, including proliferation, differentiation and survival[14,15]. P16, cyclin-dependent kinase inhibitor 2A gene (CDKN2A) is also inactivated in up to 90% of PDAC, due to intragenic mutation in association with allelic loss, homozygous deletion, or hypermethylation of the gene promoter[16-18]. CDKN2A encodes a cyclin-dependent kinase inhibitor that controls G1-S transition in the cell cycle. Mutations in CDKN2A are thought to be subsequent to those of KRAS, because of the higher prevalence of KRAS mutations in early-stage precursor lesions and the fact that most PanIN lesions containing CDKN2A inactivation also harbor KRAS mutation[19]. TP53 is one of the most frequently mutated genes in many types of cancer[20-22], and is inactivated in about 75% of PDAC, mainly due to point mutations or small deletions[21,22]. p53 is a transcription factor that determines cell fate by inducing expression of a variety of genes related to cell cycle arrest and apoptosis, and plays an important role as a master regulator of cellular stress responses. SMAD4 (DPC4, SMAD family member 4 gene) is inactivated in up to 55% of PDAC by homozygous deletion or intragenic mutation in association with allelic loss[23]. SMAD4 encodes a transcription factor that mediates signaling of the transforming growth factor-β (TGF-β) superfamily. TP53 and SMAD4 genes are mutated in late-stage precursor lesions, typically in high-grade PanIN[24,25].
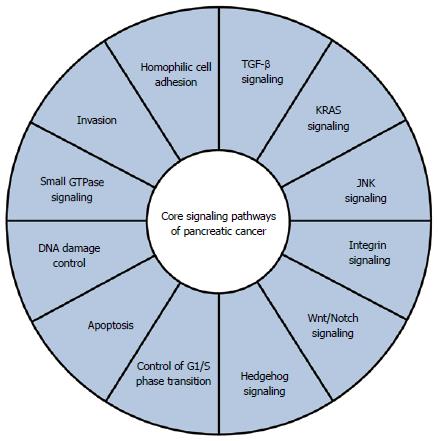
In addition to these four frequently altered genes, various other genes are mutated at relatively low frequencies in pancreatic cancer. Jones et al[9] reported alterations in genes related to chromatin remodeling (ARID1A, MLL3). Furthermore, they proposed that core signaling pathways exist in pancreatic cancer (Figure 1), and noted that the pathway components altered in individual tumors may vary widely[9]. Whole-exome sequencing analysis of 99 pancreatic cancers found many significantly mutated genes, including genes related to chromatin remodeling (EPC1, ARID2) and DNA damage repair (ATM)[26]. In addition to the core signaling pathways mentioned above[9], they identified significant alterations in genes related to the axon guidance pathway, including ROBO1/2 and SLIT2[26]. More recently, whole-genome analysis of 100 PDACs provided a comprehensive picture of the genomic alterations in this disease[27]. In addition to genes known to be important in PDAC (TP53, SMAD4, CDKN2A, ARID1A and ROBO2), chromosomal rearrangements affecting KDM6A and PREX2 were identified. KDM6A is related to chromatin remodeling, and is mutated in renal cell carcinoma and medulloblastoma[28,29]. The RAC1 guanine nucleotide exchange factor, PREX2, is mutated in melanoma[30]. Copy number analysis also uncovered a number of amplifications in genomic regions including KRAS and GATA6[27], in accordance with a previous report[31]. Most importantly, they demonstrated that a small fraction of patients (1%-2%) harbor focal amplifications in druggable genes, including ERBB2, MET, FGFR1, CDK6, PIK3CA and PIK3R3[27].
Some germline mutations are known to be associated with familial clusters of pancreatic cancer. For example, inactivation of BRCA2, which encodes a protein involved in DNA damage repair, is related to familial pancreatic cancer. Indeed, BRCA2 mutation is associated with a 3.5- to 10-fold increased risk of pancreatic cancer, as well as increased risk of breast cancer and ovarian cancer[32,33]. Germline mutations in the Fanconi anemia genes, such as FANCC, FANCG and PALB2 (also known as FANCN), are also implicated in familial pancreatic cancer[34-37]. In addition, germline mutation of ATM has recently been identified in subsets of familial pancreatic cancer[38].
The development of powerful sequencing technologies has led to a detailed knowledge of the human cancer genome, and it has become evident that some types of cancer can be effectively treated by targeted therapies based on their specific gene alterations. Here we discuss potential approaches for gene alteration-based treatment of pancreatic cancer.
The most prevalent oncogenic alteration, in KRAS, seems an obvious target for cancer therapy, because mutant KRAS protein has been experimentally demonstrated to play a pivotal role in maintenance of PDAC[39,40]. Activating mutations at KRAS codons 12, 13 and occasionally 61 are currently the most common gene alterations in pancreatic cancer. A therapeutic effect of blocking G12D mutant KRAS has been demonstrated by using siRNA and a novel siRNA delivery system, both in vitro and in vivo[41]. Although great efforts have been made to develop small-molecular inhibitors of mutant KRAS, no clinically effective antagonist has yet been identified[42]. Instead, some indirect approaches, such as targeting post-transcriptional processes, have been tried. Farnesylation of KRAS allows the protein to associate with the membrane and interact with Ras activating proteins, including Ras-GEFs. Farnesyltransferase is the key enzyme involved in addition of a 15-carbon isoprenoid chain to KRAS protein. However, despite in vitro and xenograft studies[43], farnesyltransferase inhibitors, such as tipifarnib, have proven unsuccessful in combination with gemcitabine[44,45]. This can be attributed to the existence of an alternative post-transcriptional mechanism, geranyl-geranylation, that compensates for inhibition of farnesyltransferase[46]. A dual inhibitor of farnesyltransferase and geranylgeranyltransferase (L-778,123) was tested in a Phase I clinical trial in combination with radiotherapy for locally advanced PDAC, and showed acceptable toxicity[47]. Some groups have recently investigated strategies targeting localization of KRAS to the membrane. Deltarasin is a small molecule that binds to the farnesyl-binding pocket of the delta subunit of phosphodiesterase (PDEδ) and inhibits translocation of KRAS to the membrane by blocking the interaction between PDEδ and farnesylated KRAS[48,49]. On the other hand, Salirasib blocks KRAS activation by dislodging the farnesylated protein from the membrane[50]. The results of preclinical and clinical trials suggest that salirasib may be effective[51].
Targeting downstream effectors of KRAS may be an alternative approach to block the KRAS signaling pathway. The MEK/MAPK and PI3K/Akt/mTOR pathways are the principal downstream pathways of KRAS. But, although several MEK inhibitors, such as CI-1040 and PD0325901, have been investigated in clinical trials, they failed to deliver meaningful therapeutic benefit[52,53]. In addition, trametinib, another MEK1/2 inhibitor, was recently tested in combination with gemcitabine for patients with metastatic pancreatic cancer, but failed to improve the clinical outcome[54]. Activation of the PI3K/Akt/mTOR pathway also plays an important role in maintenance of pancreatic cancer[55-57]. An inhibitor of PI3K, LY294002, was reported to induce apoptosis in vitro and to inhibit tumor growth in vivo[58]. In addition, everolimus, a mammalian target of rapamycin (mTOR) inhibitor, has been reported to inhibit tumor growth in vivo[59]. However, everolimus had minimal activity in patients with gemcitabine-resistant PDAC in a phase II study[60,61]. It was recently found that tumors with activated KRAS and mutant TP53 did not respond to mTOR inhibition, whereas tumors with KRAS activation and PTEN loss are responsive to mTOR inhibition[62].
Since the MEK/MAPK and PI3K/Akt/mTOR pathways are both downstream of KRAS, it is possible that inhibition of one pathway induces compensatory activation of the other pathway. Therefore, inhibition of both pathways may have a synergistic effect in treatment of pancreatic cancer[63,64]; thus, simultaneous blockade of MEK/MAPK and PI3K/Akt/mTOR seems to warrant further investigation as a candidate therapy for pancreatic cancer.
In addition to KRAS, CDKN2A, TP53 and SMAD4 are also commonly altered in pancreatic cancer. However, therapeutic approaches targeting these proteins are considered to be difficult for various reasons, including cellular location and multifunctionality. Although a number of therapeutic strategies targeting these genes have been examined for various types of cancer, none has yet been implemented for treatment of pancreatic cancer.
Focusing on signaling pathways in pancreatic cancer may be a better strategy than targeting particular gene alterations for treatment of pancreatic cancer. The core signaling pathways of pancreatic cancer[9] include several druggable pathways. For example, the Wnt/Notch pathway is important, and inhibition of the Notch pathway by inhibiting γ-secretase has been suggested as a potential treatment strategy[65]. The combination of γ-secretase inhibitor MRK003 with gemcitabine has been shown to provide a survival benefit in vivo[66]. It has also been reported that pancreatic cancer cells that harbor inactivating mutations of RNF43 are sensitive to LGK974, a Wnt pathway inhibitor currently in a phase 1 clinical trial[67]. Inhibition of the Hedgehog pathway with a natural hedgehog antagonist, cyclopamine, decreases growth of various types of tumor, including PDAC[68,69]. Clinical use of cyclopamine, however, is problematic because of its side effects and suboptimal pharmacokinetics. A novel, orally bioavailable, small-molecular Hedgehog inhibitor, IPI-269609, has been shown to inhibit tumor initiation and metastasis of pancreatic cancer[70]. Interestingly, blockade of the Hedgehog pathway has also been proposed as a means to target the tumor stroma and improve delivery of gemcitabine in vivo[71]. Small-molecular inhibitor Saridegib (IPI-926) was tested in combination with gemcitabine in patients with pancreatic cancer. However, the Phase I/IIb trial was stopped because patients receiving the combination had higher rates of progressive disease and lower overall survival in 2012[72].
Although the frequencies are low, mutations of several familial pancreatic cancer-related genes are associated with drug sensitivity. Inactivation of BRCA2 is found in about 7% of western PDAC patients[32,73]. BRCA2 plays a crucial role in homologous recombination-based DNA damage repair processes[74]. Poly ADP-ribose polymerase (PARP) is an important enzyme in the DNA repair mechanism mediated by BRCA2, and PARP inhibitors induce extreme genome instability and death of BRCA-mutated cancer cells[75]. As well as PARP inhibitors, DNA-crosslinking agents such as mitomycin C, cisplatin and carboplatin are also effective for treatment of BRCA-inactivated pancreatic cancer[76]. As PALB2 encodes a protein that interacts with BRCA2, PALB2 mutations are expected to disrupt BRCA2-mediated repair of DNA double strand breaks. PALB2 mutations in PDAC patients confer sensitivity to DNA-damaging agents[77]. Tumors with mutations in ATM, another familial pancreatic cancer-related gene, might also be sensitive to PARP inhibitors[78].
Overall, pancreatic cancer is characterized by substantial genomic heterogeneity with numerous infrequently mutated genes[9,26,27]. Although the common mutations in pancreatic cancer, KRAS, TP53, CDKN2A and SMAD4, are currently not druggable, stratified therapeutic strategies based on genomic alterations that occur at low frequency might be beneficial for treatment of pancreatic cancer. Recently, Jones et al[79] identified somatic alteration in potentially druggable genes in approximately 20% of PDAC patients. In Australia, the Individualized Molecular Pancreatic Cancer Therapy (IMPaCT) trial screens patients for actionable molecular phenotypes, with the aim of developing personalized therapies for pancreatic cancer[80]. IMPaCT is a randomized phase II clinical trial designed to assess standard therapy (gemcitabine) vs genotype-guided target therapies in patients with recurrent or metastatic pancreatic cancer. Initially, three subgroups with pre-defined actionable mutations, i.e., HER2-amplified (gemcitabine + trastuzumab), DNA damage response-defective (gemcitabine + PARP inhibitor) and anti-EGFR-responsive (gemcitabine + erlotinib), are being tested. This clinical trial was designed so that other arms could be added as novel subgroups or agents are identified. This approach could facilitate development of personalized therapies for pancreatic cancer.
Comprehensive genomic studies have provided extensive information on the pancreatic cancer genome, including its heterogeneity and core signaling pathways. These findings should be useful for the development of novel therapeutic strategies. For example, it might be helpful for early detection of pancreatic cancer to identify individuals with a genetic predisposition for the disease, including familial pancreatic cancer-related genes, so that periodic follow-up screening can be performed. Analysis of clonal evolution of pancreatic cancer indicates that it takes more than 10 years from occurrence of the initiating genomic alteration to formation of the parental clone[81]. Thus, there appears to be a substantial time window for early detection. Current sensitive sequencing technologies allow us to detect tumor DNA of various types of cancer in plasma (circulating tumor DNA, ctDNA)[82], and indeed, ctDNA has been detected in plasma from patients with early-stage breast and lung cancers[83,84]. Such an approach could also be applicable to patients with pancreatic cancer. More comprehensive genomic analysis may also be useful for identifying actionable mutations. Furthermore, ctDNA is thought to reflect the genetic heterogeneity of cancer, since it may contain tumor DNA derived from various regions, including metastases. Novel strategies based on genomic information seem likely to revolutionize pancreatic cancer therapy over the next few years, and may ultimately lead to fully personalized medicine.
P- Reviewer: Du YQ, Kleeff J S- Editor: Tian YL L- Editor: A E- Editor: Wu HL
| 1. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20493] [Article Influence: 2049.3] [Reference Citation Analysis (20)] |
| 2. | Verslype C, Van Cutsem E, Dicato M, Cascinu S, Cunningham D, Diaz-Rubio E, Glimelius B, Haller D, Haustermans K, Heinemann V. The management of pancreatic cancer. Current expert opinion and recommendations derived from the 8th World Congress on Gastrointestinal Cancer, Barcelona, 2006. Ann Oncol. 2007;18 Suppl 7:vii1-vii10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 3. | Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605-1617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2078] [Cited by in RCA: 2203] [Article Influence: 146.9] [Reference Citation Analysis (2)] |
| 4. | Burris HA, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, Cripps MC, Portenoy RK, Storniolo AM, Tarassoff P. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403-2413. [PubMed] |
| 5. | Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev Clin Oncol. 2010;7:163-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 555] [Cited by in RCA: 628] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 6. | Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, Au HJ, Murawa P, Walde D, Wolff RA. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2835] [Cited by in RCA: 2772] [Article Influence: 154.0] [Reference Citation Analysis (0)] |
| 7. | Von Hoff DD, Ramanathan RK, Borad MJ, Laheru DA, Smith LS, Wood TE, Korn RL, Desai N, Trieu V, Iglesias JL. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: a phase I/II trial. J Clin Oncol. 2011;29:4548-4554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 755] [Cited by in RCA: 870] [Article Influence: 62.1] [Reference Citation Analysis (2)] |
| 8. | Vaccaro V, Sperduti I, Milella M. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;365:768-779; author reply 769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 9. | Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801-1806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3216] [Cited by in RCA: 3023] [Article Influence: 177.8] [Reference Citation Analysis (0)] |
| 10. | Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1497] [Cited by in RCA: 1454] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 11. | Rozenblum E, Schutte M, Goggins M, Hahn SA, Panzer S, Zahurak M, Goodman SN, Sohn TA, Hruban RH, Yeo CJ. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 1997;57:1731-1734. [PubMed] |
| 12. | Löhr M, Klöppel G, Maisonneuve P, Lowenfels AB, Lüttges J. Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: a meta-analysis. Neoplasia. 2005;7:17-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 238] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 13. | Jancík S, Drábek J, Radzioch D, Hajdúch M. Clinical relevance of KRAS in human cancers. J Biomed Biotechnol. 2010;2010:150960. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 240] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 14. | Campbell SL, Khosravi-Far R, Rossman KL, Clark GJ, Der CJ. Increasing complexity of Ras signaling. Oncogene. 1998;17:1395-1413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 801] [Cited by in RCA: 790] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 15. | Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1287] [Cited by in RCA: 1376] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 16. | Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ, Kern SE. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 785] [Cited by in RCA: 754] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 17. | Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff I, Schmiegel W. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126-3130. [PubMed] |
| 18. | Wilentz RE, Geradts J, Maynard R, Offerhaus GJ, Kang M, Goggins M, Yeo CJ, Kern SE, Hruban RH. Inactivation of the p16 (INK4A) tumor-suppressor gene in pancreatic duct lesions: loss of intranuclear expression. Cancer Res. 1998;58:4740-4744. [PubMed] |
| 19. | Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140-2143. [PubMed] |
| 20. | Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K, Bigner SH, Davidson N, Baylin S, Devilee P. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342:705-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1896] [Cited by in RCA: 1959] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 21. | Barton CM, Staddon SL, Hughes CM, Hall PA, O’Sullivan C, Klöppel G, Theis B, Russell RC, Neoptolemos J, Williamson RC. Abnormalities of the p53 tumour suppressor gene in human pancreatic cancer. Br J Cancer. 1991;64:1076-1082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 268] [Cited by in RCA: 292] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 22. | Redston MS, Caldas C, Seymour AB, Hruban RH, da Costa L, Yeo CJ, Kern SE. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54:3025-3033. [PubMed] |
| 23. | Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1723] [Cited by in RCA: 1669] [Article Influence: 57.6] [Reference Citation Analysis (0)] |
| 24. | Wilentz RE, Iacobuzio-Donahue CA, Argani P, McCarthy DM, Parsons JL, Yeo CJ, Kern SE, Hruban RH. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002-2006. [PubMed] |
| 25. | Lüttges J, Galehdari H, Bröcker V, Schwarte-Waldhoff I, Henne-Bruns D, Klöppel G, Schmiegel W, Hahn SA. Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis. Am J Pathol. 2001;158:1677-1683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 168] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 26. | Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1690] [Cited by in RCA: 1639] [Article Influence: 126.1] [Reference Citation Analysis (0)] |
| 27. | Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2130] [Cited by in RCA: 1982] [Article Influence: 198.2] [Reference Citation Analysis (1)] |
| 28. | Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LF, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-on W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539-542. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1058] [Cited by in RCA: 1001] [Article Influence: 71.5] [Reference Citation Analysis (0)] |
| 29. | Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43-48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 600] [Cited by in RCA: 676] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 30. | Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 588] [Cited by in RCA: 577] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 31. | Zhong Y, Wang Z, Fu B, Pan F, Yachida S, Dhara M, Albesiano E, Li L, Naito Y, Vilardell F. GATA6 activates Wnt signaling in pancreatic cancer by negatively regulating the Wnt antagonist Dickkopf-1. PLoS One. 2011;6:e22129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 32. | Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, Yeo CJ, Jackson CE, Lynch HT, Hruban RH. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360-5364. [PubMed] |
| 33. | Klein AP. Genetic susceptibility to pancreatic cancer. Mol Carcinog. 2012;51:14-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 164] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 34. | van der Heijden MS, Yeo CJ, Hruban RH, Kern SE. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 2003;63:2585-2588. [PubMed] |
| 35. | Rogers CD, van der Heijden MS, Brune K, Yeo CJ, Hruban RH, Kern SE, Goggins M. The genetics of FANCC and FANCG in familial pancreatic cancer. Cancer Biol Ther. 2004;3:167-169. [PubMed] |
| 36. | Couch FJ, Johnson MR, Rabe K, Boardman L, McWilliams R, de Andrade M, Petersen G. Germ line Fanconi anemia complementation group C mutations and pancreatic cancer. Cancer Res. 2005;65:383-386. [PubMed] |
| 37. | Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, Lin JC, Palmisano E, Brune K, Jaffee EM. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 645] [Cited by in RCA: 598] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 38. | Roberts NJ, Jiao Y, Yu J, Kopelovich L, Petersen GM, Bondy ML, Gallinger S, Schwartz AG, Syngal S, Cote ML. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2:41-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 378] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 39. | Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243-247. [PubMed] |
| 40. | Fleming JB, Shen GL, Holloway SE, Davis M, Brekken RA. Molecular consequences of silencing mutant K-ras in pancreatic cancer cells: justification for K-ras-directed therapy. Mol Cancer Res. 2005;3:413-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 135] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 41. | Zorde Khvalevsky E, Gabai R, Rachmut IH, Horwitz E, Brunschwig Z, Orbach A, Shemi A, Golan T, Domb AJ, Yavin E. Mutant KRAS is a druggable target for pancreatic cancer. Proc Natl Acad Sci USA. 2013;110:20723-20728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 227] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 42. | Ledford H. Cancer: The Ras renaissance. Nature. 2015;520:278-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 43. | Omer CA, Kohl NE. CA1A2X-competitive inhibitors of farnesyltransferase as anti-cancer agents. Trends Pharmacol Sci. 1997;18:437-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 44. | Van Cutsem E, van de Velde H, Karasek P, Oettle H, Vervenne WL, Szawlowski A, Schoffski P, Post S, Verslype C, Neumann H. Phase III trial of gemcitabine plus tipifarnib compared with gemcitabine plus placebo in advanced pancreatic cancer. J Clin Oncol. 2004;22:1430-1438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 590] [Cited by in RCA: 579] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 45. | Appels NM, Beijnen JH, Schellens JH. Development of farnesyl transferase inhibitors: a review. Oncologist. 2005;10:565-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 218] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 46. | Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, Pai JK. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272:14459-14464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 666] [Cited by in RCA: 673] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 47. | Martin NE, Brunner TB, Kiel KD, DeLaney TF, Regine WF, Mohiuddin M, Rosato EF, Haller DG, Stevenson JP, Smith D. A phase I trial of the dual farnesyltransferase and geranylgeranyltransferase inhibitor L-778,123 and radiotherapy for locally advanced pancreatic cancer. Clin Cancer Res. 2004;10:5447-5454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 48. | Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PI. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature. 2013;497:638-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 460] [Cited by in RCA: 492] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 49. | Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, Ismail SA, Hedberg C, Hanzal-Bayer M, Venkitaraman AR. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol. 2012;14:148-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 272] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 50. | Weisz B, Giehl K, Gana-Weisz M, Egozi Y, Ben-Baruch G, Marciano D, Gierschik P, Kloog Y. A new functional Ras antagonist inhibits human pancreatic tumor growth in nude mice. Oncogene. 1999;18:2579-2588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 88] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 51. | Laheru D, Shah P, Rajeshkumar NV, McAllister F, Taylor G, Goldsweig H, Le DT, Donehower R, Jimeno A, Linden S. Integrated preclinical and clinical development of S-trans, trans-Farnesylthiosalicylic Acid (FTS, Salirasib) in pancreatic cancer. Invest New Drugs. 2012;30:2391-2399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 52. | Lorusso PM, Adjei AA, Varterasian M, Gadgeel S, Reid J, Mitchell DY, Hanson L, DeLuca P, Bruzek L, Piens J. Phase I and pharmacodynamic study of the oral MEK inhibitor CI-1040 in patients with advanced malignancies. J Clin Oncol. 2005;23:5281-5293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 304] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 53. | Haura EB, Ricart AD, Larson TG, Stella PJ, Bazhenova L, Miller VA, Cohen RB, Eisenberg PD, Selaru P, Wilner KD. A phase II study of PD-0325901, an oral MEK inhibitor, in previously treated patients with advanced non-small cell lung cancer. Clin Cancer Res. 2010;16:2450-2457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 189] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 54. | Infante JR, Somer BG, Park JO, Li CP, Scheulen ME, Kasubhai SM, Oh DY, Liu Y, Redhu S, Steplewski K. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer. 2014;50:2072-2081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 274] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 55. | Schlieman MG, Fahy BN, Ramsamooj R, Beckett L, Bold RJ. Incidence, mechanism and prognostic value of activated AKT in pancreas cancer. Br J Cancer. 2003;89:2110-2115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 206] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 56. | Agbunag C, Bar-Sagi D. Oncogenic K-ras drives cell cycle progression and phenotypic conversion of primary pancreatic duct epithelial cells. Cancer Res. 2004;64:5659-5663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 57. | Kong B, Wu W, Cheng T, Schlitter AM, Qian C, Bruns P, Jian Z, Jäger C, Regel I, Raulefs S. A subset of metastatic pancreatic ductal adenocarcinomas depends quantitatively on oncogenic Kras/Mek/Erk-induced hyperactive mTOR signalling. Gut. 2015;Jan 19; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 55] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 58. | Bondar VM, Sweeney-Gotsch B, Andreeff M, Mills GB, McConkey DJ. Inhibition of the phosphatidylinositol 3’-kinase-AKT pathway induces apoptosis in pancreatic carcinoma cells in vitro and in vivo. Mol Cancer Ther. 2002;1:989-997. [PubMed] |
| 59. | O’Reilly T, McSheehy PM, Wartmann M, Lassota P, Brandt R, Lane HA. Evaluation of the mTOR inhibitor, everolimus, in combination with cytotoxic antitumor agents using human tumor models in vitro and in vivo. Anticancer Drugs. 2011;22:58-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 60. | Javle MM, Shroff RT, Xiong H, Varadhachary GA, Fogelman D, Reddy SA, Davis D, Zhang Y, Wolff RA, Abbruzzese JL. Inhibition of the mammalian target of rapamycin (mTOR) in advanced pancreatic cancer: results of two phase II studies. BMC Cancer. 2010;10:368. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 141] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 61. | Wolpin BM, Hezel AF, Abrams T, Blaszkowsky LS, Meyerhardt JA, Chan JA, Enzinger PC, Allen B, Clark JW, Ryan DP. Oral mTOR inhibitor everolimus in patients with gemcitabine-refractory metastatic pancreatic cancer. J Clin Oncol. 2009;27:193-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 239] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 62. | Morran DC, Wu J, Jamieson NB, Mrowinska A, Kalna G, Karim SA, Au AY, Scarlett CJ, Chang DK, Pajak MZ. Targeting mTOR dependency in pancreatic cancer. Gut. 2014;63:1481-1489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 99] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 63. | Williams TM, Flecha AR, Keller P, Ram A, Karnak D, Galbán S, Galbán CJ, Ross BD, Lawrence TS, Rehemtulla A. Cotargeting MAPK and PI3K signaling with concurrent radiotherapy as a strategy for the treatment of pancreatic cancer. Mol Cancer Ther. 2012;11:1193-1202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 64. | Alagesan B, Contino G, Guimaraes AR, Corcoran RB, Deshpande V, Wojtkiewicz GR, Hezel AF, Wong KK, Loda M, Weissleder R. Combined MEK and PI3K inhibition in a mouse model of pancreatic cancer. Clin Cancer Res. 2015;21:396-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 109] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 65. | Hu H, Zhou L, Awadallah A, Xin W. Significance of Notch1-signaling pathway in human pancreatic development and carcinogenesis. Appl Immunohistochem Mol Morphol. 2013;21:242-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 66. | Cook N, Frese KK, Bapiro TE, Jacobetz MA, Gopinathan A, Miller JL, Rao SS, Demuth T, Howat WJ, Jodrell DI. Gamma secretase inhibition promotes hypoxic necrosis in mouse pancreatic ductal adenocarcinoma. J Exp Med. 2012;209:437-444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 67. | Jiang X, Hao HX, Growney JD, Woolfenden S, Bottiglio C, Ng N, Lu B, Hsieh MH, Bagdasarian L, Meyer R. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci USA. 2013;110:12649-12654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 338] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 68. | Feldmann G, Habbe N, Dhara S, Bisht S, Alvarez H, Fendrich V, Beaty R, Mullendore M, Karikari C, Bardeesy N. Hedgehog inhibition prolongs survival in a genetically engineered mouse model of pancreatic cancer. Gut. 2008;57:1420-1430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 139] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 69. | Kelleher FC. Hedgehog signaling and therapeutics in pancreatic cancer. Carcinogenesis. 2011;32:445-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 93] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 70. | Feldmann G, Fendrich V, McGovern K, Bedja D, Bisht S, Alvarez H, Koorstra JB, Habbe N, Karikari C, Mullendore M. An orally bioavailable small-molecule inhibitor of Hedgehog signaling inhibits tumor initiation and metastasis in pancreatic cancer. Mol Cancer Ther. 2008;7:2725-2735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 207] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 71. | Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457-1461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2581] [Cited by in RCA: 2534] [Article Influence: 158.4] [Reference Citation Analysis (0)] |
| 73. | Schutte M, da Costa LT, Hahn SA, Moskaluk C, Hoque AT, Rozenblum E, Weinstein CL, Bittner M, Meltzer PS, Trent JM. Identification by representational difference analysis of a homozygous deletion in pancreatic carcinoma that lies within the BRCA2 region. Proc Natl Acad Sci USA. 1995;92:5950-5954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 108] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 74. | Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 858] [Cited by in RCA: 887] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 75. | Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3070] [Cited by in RCA: 2854] [Article Influence: 178.4] [Reference Citation Analysis (0)] |
| 76. | van der Heijden MS, Brody JR, Gallmeier E, Cunningham SC, Dezentje DA, Shen D, Hruban RH, Kern SE. Functional defects in the fanconi anemia pathway in pancreatic cancer cells. Am J Pathol. 2004;165:651-657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 88] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 77. | Villarroel MC, Rajeshkumar NV, Garrido-Laguna I, De Jesus-Acosta A, Jones S, Maitra A, Hruban RH, Eshleman JR, Klein A, Laheru D. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol Cancer Ther. 2011;10:3-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 215] [Cited by in RCA: 198] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 78. | McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O’Connor MJ, Tutt AN, Zdzienicka MZ. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109-8115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1080] [Cited by in RCA: 1008] [Article Influence: 53.1] [Reference Citation Analysis (0)] |
| 79. | Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, Shukla M, Chesnick B, Kadan M, Papp E. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7:283ra53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 323] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 80. | Cowley MJ, Chang DK, Pajic M, Johns AL, Waddell N, Grimmond SM, Biankin AV. Understanding pancreatic cancer genomes. J Hepatobiliary Pancreat Sci. 2013;20:549-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 81. | Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114-1117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2041] [Cited by in RCA: 1937] [Article Influence: 129.1] [Reference Citation Analysis (0)] |
| 82. | Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2770] [Cited by in RCA: 3532] [Article Influence: 321.1] [Reference Citation Analysis (0)] |
| 83. | Beaver JA, Jelovac D, Balukrishna S, Cochran RL, Croessmann S, Zabransky DJ, Wong HY, Valda Toro P, Cidado J, Blair BG. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res. 2014;20:2643-2650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 313] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 84. | Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548-554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1341] [Cited by in RCA: 1641] [Article Influence: 149.2] [Reference Citation Analysis (0)] |