Published online Aug 15, 2025. doi: 10.4251/wjgo.v17.i8.106783
Revised: May 14, 2025
Accepted: July 7, 2025
Published online: August 15, 2025
Processing time: 117 Days and 21.2 Hours
Abnormal iron metabolism plays a critical role in paclitaxel (PTX) resistance in esophageal cancer cells. Qige San (QG) is a traditional Chinese herbal formula that is reported to improve short-term therapeutic effects of esophageal cancer.
To investigate the effects and regulatory mechanisms involved in QG-targeted PTX-resistant esophageal cancer cells.
Cell viability was assessed using the Cell Counting Kit-8 assay. Ferroptosis was evaluated by analyzing lipid reactive oxygen species accumulation and the Fe2+ concentration in PTX-resistant esophageal cancer cells. Expression of ferroptosis regulators was measured by western blot. Network pharmacology analysis was employed to identify potential targets of QG in PTX-resistant esophageal cancer cells.
Treatment with QG significantly suppressed the viability, proliferation, and mi
QG could repress the resistance of esophageal cancer cells to PTX via targeting the PI3K signaling pathway.
Core Tip: Abnormal iron metabolism plays a critical role in paclitaxel-resistance of esophageal cancer cells. Qige San (QG) is a traditional Chinese herbal formula that is reported to improve short-term therapeutic effects of esophageal cancer. We observed that QG could repress the resistance of esophageal cancer cells to paclitaxel via targeting the phosphoinositide 3-kinase/protein kinase B signaling pathway.
- Citation: Song J, Guo WY, Sun LB. Qige San regulates paclitaxel resistance in esophageal cancer by targeting ferroptosis. World J Gastrointest Oncol 2025; 17(8): 106783
- URL: https://www.wjgnet.com/1948-5204/full/v17/i8/106783.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i8.106783
Esophageal cancer is a common malignant tumor of the digestive tract that poses a serious threat to human health[1]. According to statistical reports, esophageal cancer currently ranks as the sixth-leading cause of cancer-related deaths worldwide with a 5-year overall survival rate of only 20%-40%[1]. China is a high-incidence area for esophageal cancer, and esophageal squamous cell carcinoma (ESCC) accounts for 95% of all esophageal cancer cases in the country[2,3]. Although advances in early diagnosis, radical surgery, and postoperative adjuvant therapies have improved patient outcomes in recent years, high mortality rates and distant organ metastasis continue to limit the effectiveness of clinical treatments[4-6]. Therefore, esophageal cancer treatment remains a significant challenge.
Chemotherapy can reduce the tumor size to some extent and induce cancer cell death. Unfortunately, long-term chemotherapy use may lead to drug resistance and tumor recurrence. The development of drug resistance not only severely limits the clinical application of chemotherapeutic agents but also enhances tumor metastasis[6]. More impor
Ferroptosis, an iron-dependent programmed cell death driven by lipid peroxidation, has emerged as a key mechanism influencing chemotherapy resistance in ESCC[7-9]. Recent studies revealed that overexpression of glutathione peroxidase 4 (GPX4) and SLC7A11, critical regulators of ferroptosis, correlated with paclitaxel (PTX) resistance in clinical ESCC specimens[10]. Conversely, iron chelation therapy partially restored chemosensitivity[11], suggesting that targeting ferroptosis could overcome drug resistance. Notably, GPX4 is upregulated in 68% of ESCC tumors compared with adjacent normal tissues, and its inhibition synergizes with cisplatin to suppress growth[12]. Similarly, SLC7A11-mediated cystine uptake sustains glutathione synthesis, protecting ESCC cells from ferroptotic death[13]. These findings position ferroptosis modulation as a promising strategy for resistant ESCC.
Qige San (QG) is a traditional Chinese herbal formula first recorded in the book “Yixue Xinwu.” Its primary functions are moistening dryness, relieving stagnation, resolving phlegm, and counteracting upward rebellion[14]. It is traditionally used to treat dysphagia and obstruction symptoms, such as difficulty swallowing, vomiting after eating, morning food intake followed by evening vomiting, gastric distension and pain, red tongue with little moisture, and dry stools. The main ingredients of QG include Radix Glehniae (Sha Shen, rich in tanshinones with demonstrated proferroptotic activity in lung and liver cancers), Salvia miltiorrhiza (Dan Shen), Poria (Fu Ling), Bulbus Fritillariae Cirrhosae (Chuan Bei Mu, core removed), Curcuma aromatica (Yu Jin), Amomum Shell (Sha Ren Ke), Lotus Pedicel (He Ye Di), and Bran (Chu Tou Kang)[15]. This formula was prioritized based on both clinical tradition and modern compound screening: Tanshinone IIA from Salvia miltiorrhiza directly binds to GPX4 to inhibit its activity; and emodin (present in Rheum officinale, a common QG additive) enhances iron accumulation via nuclear receptor coactivator 4 (NCOA4)-mediated ferritinophagy[16].
In cancer treatment QG has shown potential clinical applications. When used in combination with chemotherapy for esophageal cancer, it has been found to improve short-term therapeutic effects and enhance patient quality of life[16]. Compared with chemotherapy alone, the combination of QG and chemotherapy demonstrated better efficacy[17]. Addi
This study aimed to investigate the effects of QG on the induction of ferroptosis in PTX-resistant ESCC cells, validate the involvement of the PI3K/Akt pathway in QG-mediated chemosensitization, and identify the bioactive components of QG using network pharmacology.
The esophageal cancer cells TE-1 and EC109 were incubated in DMEM medium supplemented with 10% fetal bovine serum. Cells were maintained in a humidified incubator at 37 °C with 95% air and 5% CO2. PTX-resistant cell lines, TE-1/PTX and EC109/PTX, were established by our laboratory. Briefly, TE-1 and EC109 cells were initially exposed to 1 nM of PTX for 1 week. When cells returned to a normal growth rate after the period of recovery, the concentrations of PTX were gradually increased (5 nM, 10 nM, 20 nM, and 40 nM) until cells became resistant to 40 nM of PTX after approximately 10 months. The obtained PTX-resistant esophageal cancer cells were treated with QG (50 μmol/L), PI3K activator (740 Y-P, 20 μmol/L), and Ferrostatin-1 (Fer-1) (5 μmol/L) for 24 h.
The proliferation of PTX-resistant esophageal cancer cells was evaluated using the Cell Counting Kit-8 (CCK-8; Beyotime, China) according to the manufacturer’s instructions. Cells were seeded into 96-well plates at a density of 5000 cells/well and incubated overnight. After the specified treatments 10 μL of CCK-8 reagent was added to each well. The absorbance at 450 nm (OD450) was measured 24 h post-treatment using a microplate reader (Thermo, United States).
For the colony formation assay, cells (1 × 103) were first mixed with culture medium (1.5 mL) and placed onto a 6-well plate. After 3 weeks of incubation, colonies were stained with crystal violet (SolarBio, China). A microscope (Nikon, Japan) was used to visualize and count the colonies.
For the cell invasion assay 5 × 104 cells in serum-free media were placed in the top chamber (354480, BD Biosciences, San Jose, CA, United States), while complete media was added to the bottom chamber. After incubation for 24 h at 37 °C, the medium was removed, and the chambers were fixed with methanol for 30 min. Subsequently, the chambers were stained with crystal violet for another 30 min.
The intracellular iron and Fe2+ levels were quantified using an Iron Assay Kit (ab83366) following the manufacturer’s instructions. Malondialdehyde (MDA) is the end product of lipid peroxidation and was assessed using a lipid peroxi
Total protein was extracted from cells using RIPA lysis buffer containing protease and phosphatase inhibitors (Roche), and protein concentrations were determined using a bicinchoninic acid assay kit (Pierce). Equal amounts of protein (30 μg per lane) were resolved by 10% SDS-PAGE and transferred onto PVDF membranes (Millipore). Membranes were blocked with 5% skim milk in TBST for 1 h at room temperature and then incubated overnight at 4 °C with the following primary antibodies: Anti-GPX4 (Abcam, ab125066, 1:2000); anti-SLC7A11 (Abcam, ab175186, 1:2000); anti-acyl-CoA synthetase long chain family member 4 (ACSL4) (Proteintech, 22401-1-AP; 1:1000); anti-transferrin receptor 1 (TFR1) (Abcam, ab84036, 1:2000); anti-NCOA4 (Santa Cruz, sc-373739, 1:500); anti-phospho-Akt (Ser473) (Cell Signaling Technology, #4060, 1:1000); and anti-total Akt (Cell Signaling Technology, #4691, 1:1000). After washing three times with TBST, membranes were incubated with HRP-conjugated secondary antibodies (goat anti-rabbit, Sigma, A0545, 1:20000; goat anti-mouse, Sigma, A6154, 1:2000) for 1 h at room temperature. Protein bands were visualized using ECL detection rea
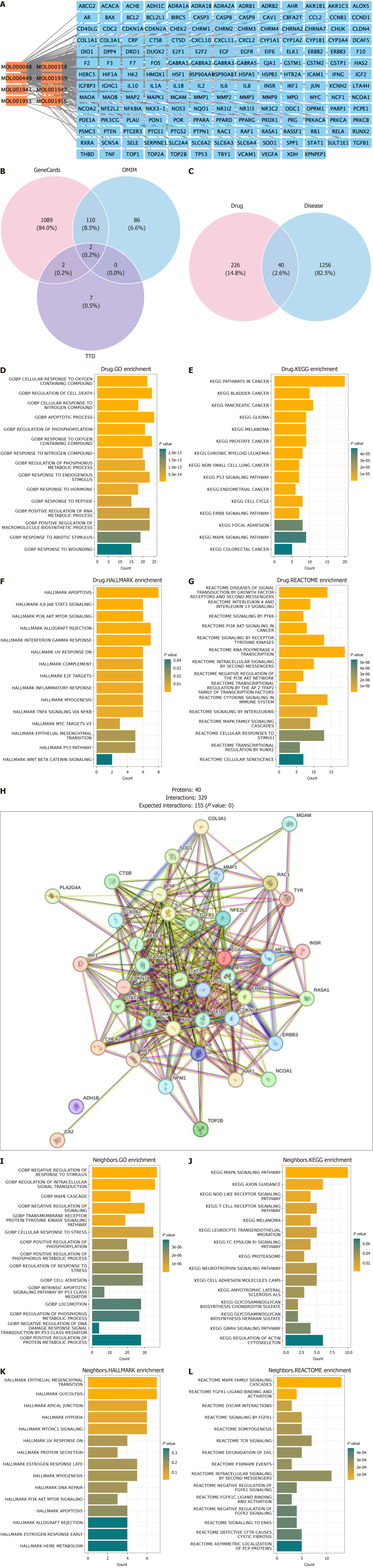
To identify the active ingredients of the drug, we used the Traditional Chinese Medicine Systems Pharmacology database, applying the filtering criteria of oral bioavailability ≥ 30% and drug-likeness ≥ 0.18. The UniProt database was then utilized to remove duplicates, standardize target information, and obtain gene abbreviations. Disease-related targets were retrieved from the GeneCards, Therapeutic Target Database, and Online Mendelian Inheritance in Man databases. The common targets shared between disease-related targets and traditional medicine-related targets were visualized using a Venn diagram. These common targets were imported into the STRING database with “Homo sapiens” as the protein species, while the other parameters remained default. A protein-protein interaction network for potential drug targets was constructed. This network was further imported into Cytoscape, where the degree value was used to filter key targets. Gene Ontology (biological process, cellular component, and molecular function) and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed using the R package clusterProfiler. A P value < 0.05 was considered statistically significant. The top enriched pathways were visualized in graphical form.
The data in this study are presented as mean ± standard error of the mean and analyzed using GraphPad Prism version 7.0 software. The differences between groups were assessed using the Student’s t-test for two groups and one-way analysis of variance for comparisons among multiple groups. A P value of < 0.05 was considered statistically significant. For western blot quantification normalized band intensities (phospho-Akt/total Akt ratio) were analyzed as continuous variables.
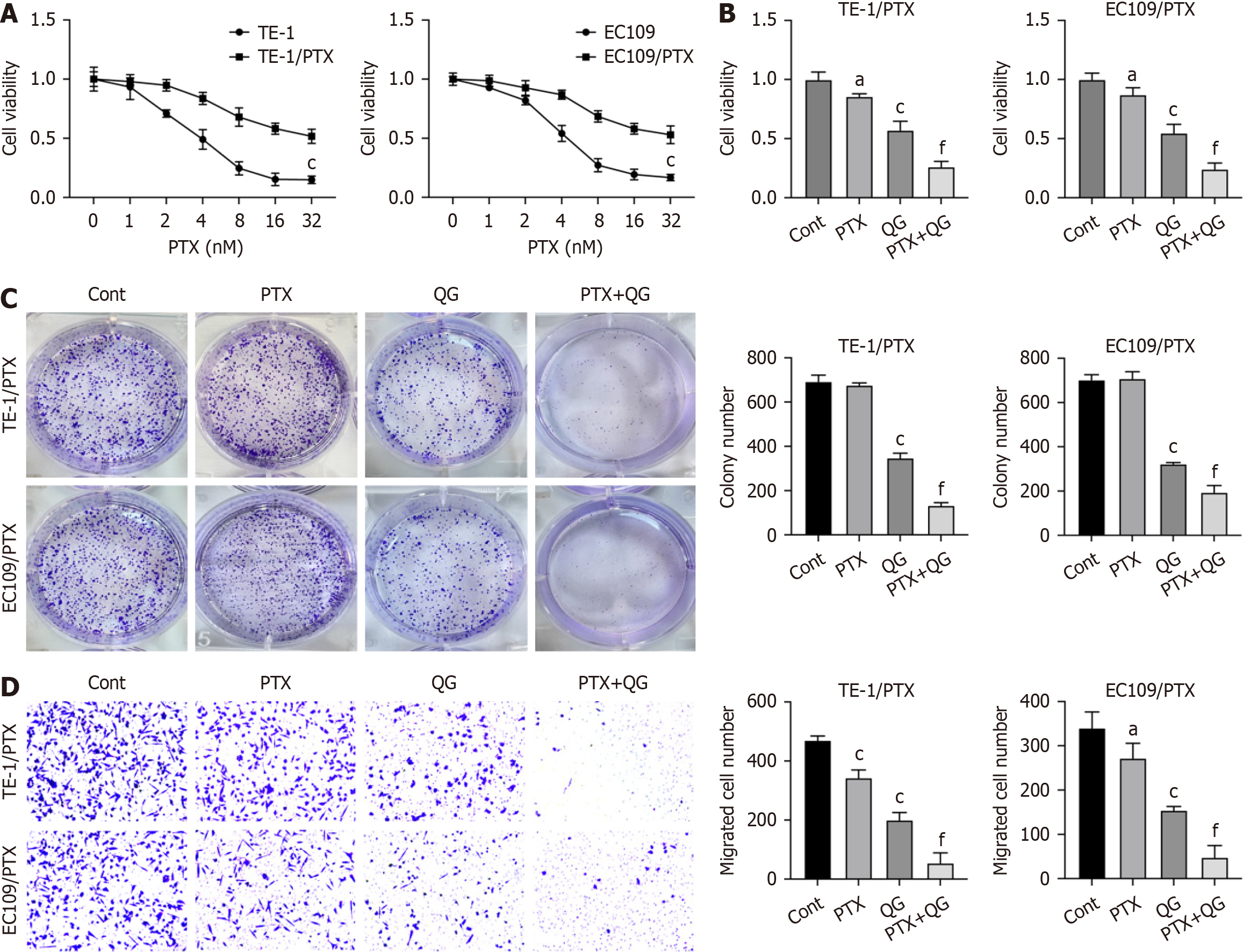
To investigate the effects of QG on PTX resistance of esophageal cancer, we established drug-resistant esophageal cancer cell lines (TE-1/PTX and EC109/PTX). CCK-8 results verified that TE-1/PTX and EC109/PTX showed notable resistance to PTX treatment compared with the parental TE-1 and EC109 cell lines (Figure 1A). Next, results from CCK-8 and colony formation demonstrated that QG treatment significantly enhanced the in vitro anti-growth effects of PTX against the PTX-resistant cells (Figure 1B and C). Moreover, QG treatment also suppressed the migration of esophageal cancer cells treated with PTX (Figure 1D).
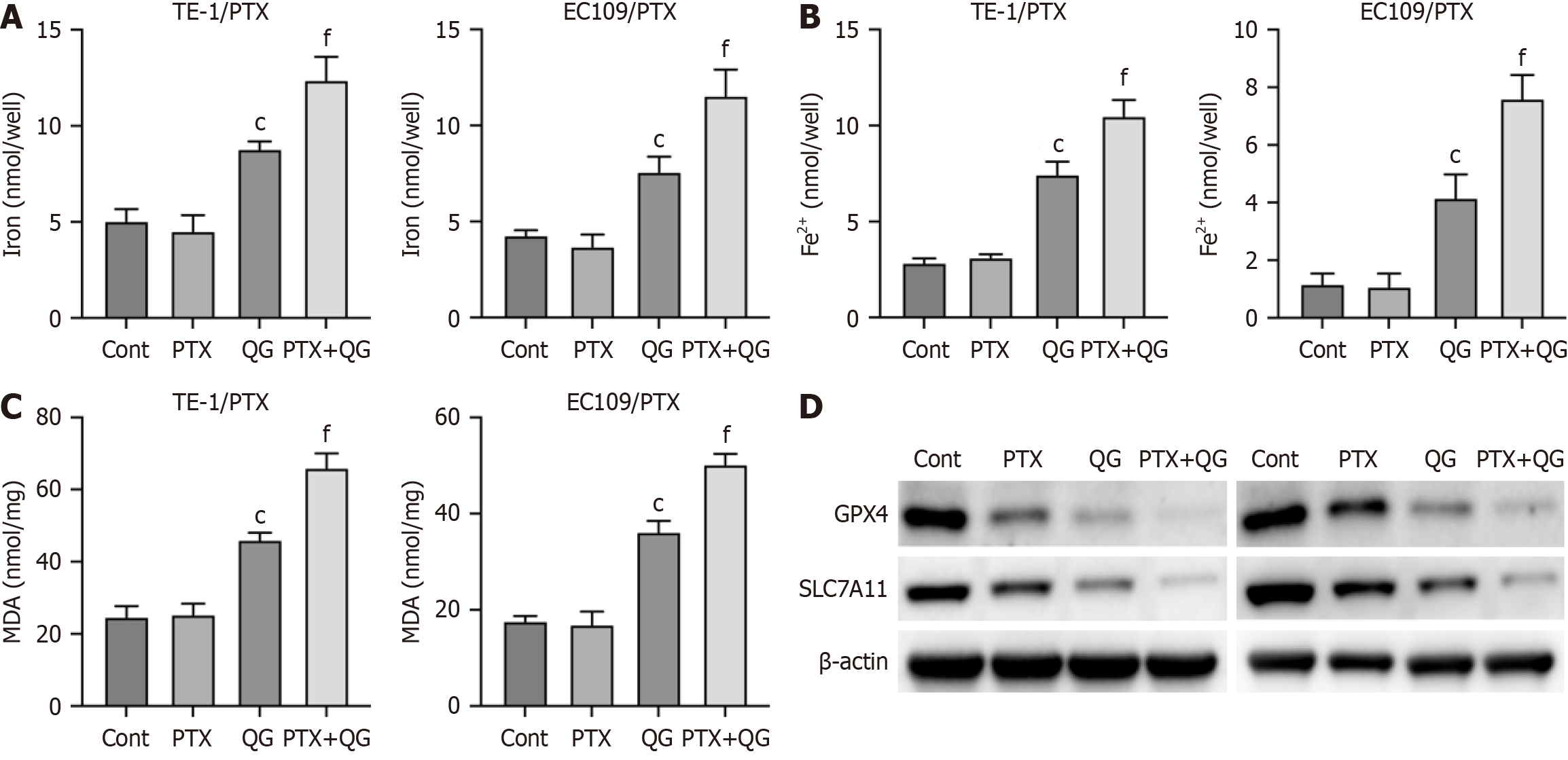
Next, we analyzed the effects of QG on ferroptosis of drug-resistant esophageal cancer cells. As shown in Figure 2A, treatment with PTX alone did not significantly alter the accumulation of total iron and Fe2+ (Figure 2A and B), production of MDA (Figure 2C), and the expression of ferroptosis regulators GPX4 and SLC7A11 (Figure 2D). QG alone enhanced the levels of total iron, Fe2+, and MDA and reduced the expression of GPX4 and SLC7A11, suggesting the induction of fer
Subsequently, we conducted a network pharmacology analysis to explore the potential regulatory mechanisms in
Functional analysis of these common genes revealed that they were predominantly involved in critical cellular processes and pathways, including cell death, apoptosis, the p53 pathway, and the PI3K/Akt pathway (Figure 3D-G). Moreover, we performed a protein-protein interaction network analysis of the overlapping genes (Figure 3H) and observed that these genes were primarily enriched in cellular processes such as response to stimuli, intracellular signal transduction, and stress response (Figure 3I). In addition, these genes were associated with several key signaling pathways, including the mitogen-activated protein kinase (MAPK) pathway, NOD-like receptor signaling, proteasome pathway, and PI3K/Akt pathway (Figure 3I-L). These findings suggest that the PI3K/Akt pathway is a potential regulatory signaling pathway involved in the treatment of esophageal cancer by QG.
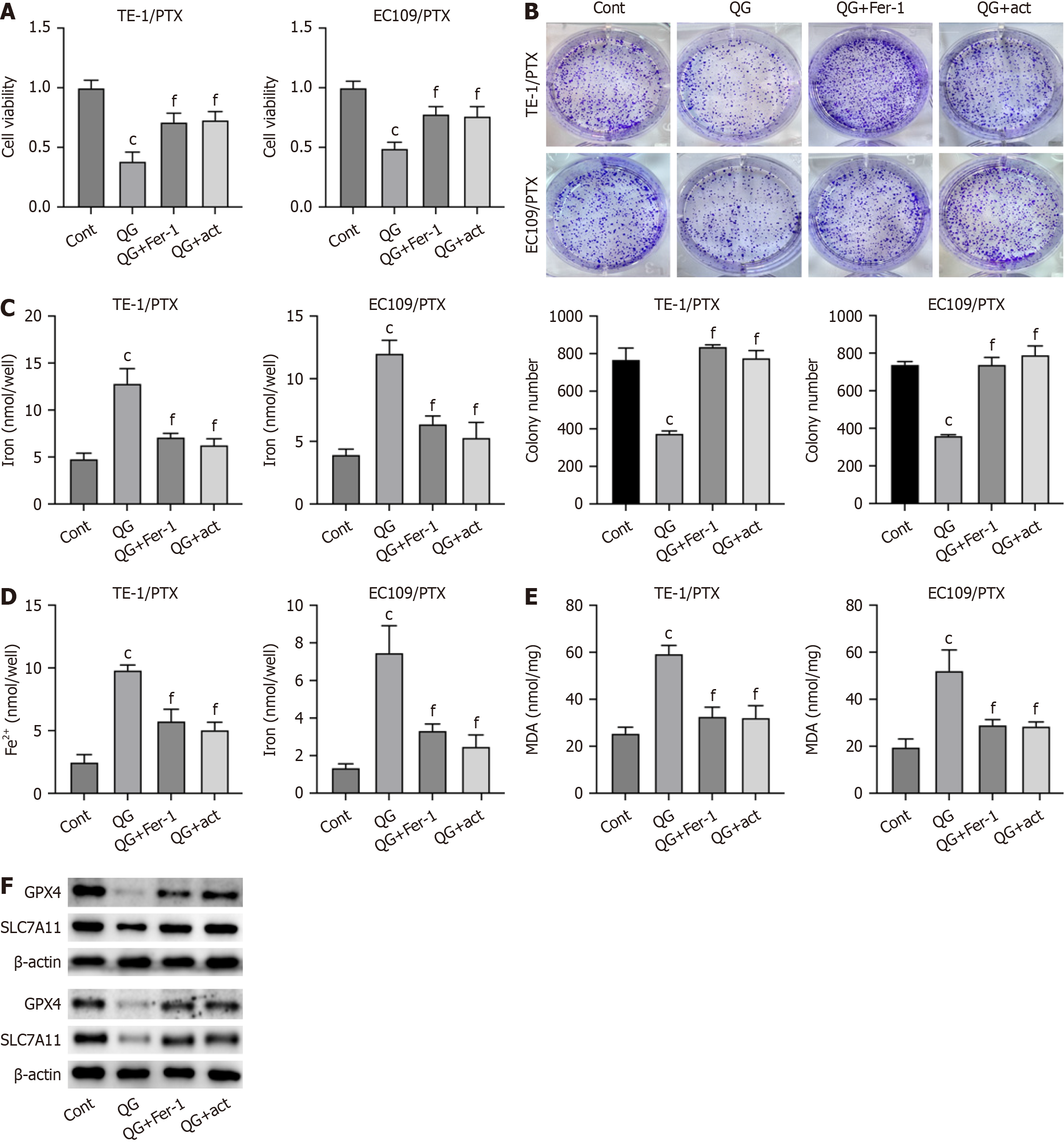
To verify that the PI3K/Akt signaling is involved in PTX-resistant cancer cell ferroptosis induced by QG treatment, we treated PTX-resistant esophageal cancer cells with QG alone or in combination with a PI3K activator or ferroptosis inhibitor (Fer-1). As shown in CCK-8 and colony formation results (Figure 4A and B), activation of PI3K and treatment with Fer-1 significantly recovered the viability and proliferation of PTX-resistant esophageal cancer cells. The PI3K activator and Fer-1 treatment also repressed the total iron and Fe2+ accumulation (Figure 4C and D) as well as the MDA level (Figure 4E) induced by QG treatment. Moreover, the reduced expression of GPX4 and SLC7A11 under QG treat
We demonstrated that QG treatment could induce ferroptosis and relieve drug resistance of esophageal cancer cells. The network pharmacology analysis on the potential targets of QG in esophageal cancer revealed PI3K/Akt signaling as the potential regulatory mechanism. Subsequent in vitro cell experiments confirmed that activation of the PI3K pathway partially impeded the anti-cancer effects of QG on drug resistant esophageal cancer. Notably, QG uniquely upregulated key ferroptosis drivers (ACSL4, TFR1, NCOA4) while suppressing Akt phosphorylation (Supplementary Figure 1), a dual effect not reported for classical ferroptosis inducers like erastin (system Xc- inhibitor) or RSL3 (GPX4 inhibitor). This multitarget action may explain the superior efficacy of QG in reversing PTX resistance compared with single-agent inducers.
Chemotherapy resistance is a significant challenge in cancer treatment and severely affects the efficacy of chemothe
The multi-component nature of QG raises questions about potential off-target effects. Our network pharmacology identified tanshinone IIA (from Salvia miltiorrhiza) and emodin (common in traditional Chinese medicine formulas) as top candidates responsible for the observed effects: Tanshinone IIA directly binds GPX4 to inhibit its activity[23], consistent with our findings of GPX4 downregulation. Emodin promotes NCOA4-mediated ferritinophagy[24], aligning with increased iron accumulation. While these components likely drive the primary anti-resistance effects, future studies should isolate individual compounds to delineate their specific contributions and minimize off-target interactions.
Cancer cells activate multiple signaling pathways to resist chemotherapy-induced damage, among which PI3K/Akt, MAPK, Wnt/β-catenin, and nuclear factor kappa B are particularly critical[25,26]. While network pharmacology pre
Our study demonstrated that QG overcame PTX resistance in esophageal cancer through a dual mechanism: First, by inducing ferroptosis as evidenced by the upregulation of ACSL4, TFR1, and NCOA4 alongside suppression of GPX4 and SLC7A11; and second, by inhibiting PI3K/Akt survival signaling, indicated by decreased p-Akt levels and the complete reversal of the effects of QG following PI3K pathway activation. The multi-component nature of QG, particularly the presence of compounds such as tanshinone IIA and emodin, enabled coordinated disruption of iron metabolism and lipid peroxidation, setting it apart from single-target ferroptosis inducers like erastin and RSL3. These findings suggest that QG is a promising multitarget therapeutic strategy for overcoming drug resistance in esophageal cancer although further research is needed to isolate its active constituents and optimize its clinical application.
| 1. | Kelly RJ. Emerging Multimodality Approaches to Treat Localized Esophageal Cancer. J Natl Compr Canc Netw. 2019;17:1009-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 112] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 2. | Demarest CT, Chang AC. The Landmark Series: Multimodal Therapy for Esophageal Cancer. Ann Surg Oncol. 2021;28:3375-3382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 3. | Huang FL, Yu SJ. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J Surg. 2018;41:210-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 539] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 4. | Borggreve AS, Kingma BF, Domrachev SA, Koshkin MA, Ruurda JP, van Hillegersberg R, Takeda FR, Goense L. Surgical treatment of esophageal cancer in the era of multimodality management. Ann N Y Acad Sci. 2018;1434:192-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 100] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 5. | Waters J, Sewell M, Molena D. Multimodal Treatment of Resectable Esophageal Cancer. Ann Thorac Surg. 2025;119:70-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 6. | Tong X, Jin M, Wang L, Zhang D, Yin Y, Shen Q. Prognostic biomarkers for immunotherapy in esophageal cancer. Front Immunol. 2024;15:1420399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 7. | Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22:266-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2184] [Cited by in RCA: 4202] [Article Influence: 1050.5] [Reference Citation Analysis (0)] |
| 8. | Mou Y, Wang J, Wu J, He D, Zhang C, Duan C, Li B. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol. 2019;12:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 392] [Cited by in RCA: 1238] [Article Influence: 206.3] [Reference Citation Analysis (0)] |
| 9. | Tong X, Tang R, Xiao M, Xu J, Wang W, Zhang B, Liu J, Yu X, Shi S. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J Hematol Oncol. 2022;15:174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 456] [Cited by in RCA: 473] [Article Influence: 157.7] [Reference Citation Analysis (0)] |
| 10. | Zhang C, Liu X, Jin S, Chen Y, Guo R. Ferroptosis in cancer therapy: a novel approach to reversing drug resistance. Mol Cancer. 2022;21:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 775] [Article Influence: 258.3] [Reference Citation Analysis (0)] |
| 11. | Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting Ferroptosis to Iron Out Cancer. Cancer Cell. 2019;35:830-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 900] [Cited by in RCA: 1620] [Article Influence: 270.0] [Reference Citation Analysis (0)] |
| 12. | Fan X, Fan YT, Zeng H, Dong XQ, Lu M, Zhang ZY. Role of ferroptosis in esophageal cancer and corresponding immunotherapy. World J Gastrointest Oncol. 2023;15:1105-1118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 5] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Wang Z, Wu S, Zhu C, Shen J. The role of ferroptosis in esophageal cancer. Cancer Cell Int. 2022;22:266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 14. | Zhou L, Wu YS, Yin SG, Wang HH, Chen YL. [Effect of Qige Powder on Angiogenesis Induced by Esophageal Cancer Cell Line Eca-9706]. Zhong Yao Cai. 2015;38:123-126. [PubMed] |
| 15. | Peng C, Li J, Ke X, Liu F, Huang KE. In silico and in vivo demonstration of the regulatory mechanism of Qi-Ge decoction in treating NAFLD. Ann Med. 2023;55:2200258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 16. | Wu C, Wang Z, Song X, Feng XS, Abnet CC, He J, Hu N, Zuo XB, Tan W, Zhan Q, Hu Z, He Z, Jia W, Zhou Y, Yu K, Shu XO, Yuan JM, Zheng W, Zhao XK, Gao SG, Yuan ZQ, Zhou FY, Fan ZM, Cui JL, Lin HL, Han XN, Li B, Chen X, Dawsey SM, Liao L, Lee MP, Ding T, Qiao YL, Liu Z, Liu Y, Yu D, Chang J, Wei L, Gao YT, Koh WP, Xiang YB, Tang ZZ, Fan JH, Han JJ, Zhou SL, Zhang P, Zhang DY, Yuan Y, Huang Y, Liu C, Zhai K, Qiao Y, Jin G, Guo C, Fu J, Miao X, Lu C, Yang H, Wang C, Wheeler WA, Gail M, Yeager M, Yuenger J, Guo ET, Li AL, Zhang W, Li XM, Sun LD, Ma BG, Li Y, Tang S, Peng XQ, Liu J, Hutchinson A, Jacobs K, Giffen C, Burdette L, Fraumeni JF Jr, Shen H, Ke Y, Zeng Y, Wu T, Kraft P, Chung CC, Tucker MA, Hou ZC, Liu YL, Hu YL, Liu Y, Wang L, Yuan G, Chen LS, Liu X, Ma T, Meng H, Sun L, Li XM, Li XM, Ku JW, Zhou YF, Yang LQ, Wang Z, Li Y, Qige Q, Yang WJ, Lei GY, Chen LQ, Li EM, Yuan L, Yue WB, Wang R, Wang LW, Fan XP, Zhu FH, Zhao WX, Mao YM, Zhang M, Xing GL, Li JL, Han M, Ren JL, Liu B, Ren SW, Kong QP, Li F, Sheyhidin I, Wei W, Zhang YR, Feng CW, Wang J, Yang YH, Hao HZ, Bao QD, Liu BC, Wu AQ, Xie D, Yang WC, Wang L, Zhao XH, Chen SQ, Hong JY, Zhang XJ, Freedman ND, Goldstein AM, Lin D, Taylor PR, Wang LD, Chanock SJ. Joint analysis of three genome-wide association studies of esophageal squamous cell carcinoma in Chinese populations. Nat Genet. 2014;46:1001-1006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 140] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 17. | Zhang ZQ, Du WJ, Liu LZ. [Observation on effects of qige tongye decoction combined with chemotherapy in treating esophageal carcinoma]. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2002;22:63-64. [PubMed] |
| 18. | Ippolito MR, Martis V, Martin S, Tijhuis AE, Hong C, Wardenaar R, Dumont M, Zerbib J, Spierings DCJ, Fachinetti D, Ben-David U, Foijer F, Santaguida S. Gene copy-number changes and chromosomal instability induced by aneuploidy confer resistance to chemotherapy. Dev Cell. 2021;56:2440-2454.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 124] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 19. | Delou JMA, Souza ASO, Souza LCM, Borges HL. Highlights in Resistance Mechanism Pathways for Combination Therapy. Cells. 2019;8:1013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 20. | Ouyang S, Li H, Lou L, Huang Q, Zhang Z, Mo J, Li M, Lu J, Zhu K, Chu Y, Ding W, Zhu J, Lin Z, Zhong L, Wang J, Yue P, Turkson J, Liu P, Wang Y, Zhang X. Inhibition of STAT3-ferroptosis negative regulatory axis suppresses tumor growth and alleviates chemoresistance in gastric cancer. Redox Biol. 2022;52:102317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 279] [Article Influence: 93.0] [Reference Citation Analysis (0)] |
| 21. | Wang Y, Wu X, Ren Z, Li Y, Zou W, Chen J, Wang H. Overcoming cancer chemotherapy resistance by the induction of ferroptosis. Drug Resist Updat. 2023;66:100916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 157] [Reference Citation Analysis (0)] |
| 22. | Zeng K, Li W, Wang Y, Zhang Z, Zhang L, Zhang W, Xing Y, Zhou C. Inhibition of CDK1 Overcomes Oxaliplatin Resistance by Regulating ACSL4-mediated Ferroptosis in Colorectal Cancer. Adv Sci (Weinh). 2023;10:e2301088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 60] [Reference Citation Analysis (0)] |
| 23. | Wang Y, Zheng L, Shang W, Yang Z, Li T, Liu F, Shao W, Lv L, Chai L, Qu L, Xu Q, Du J, Liang X, Zeng J, Jia J. Wnt/beta-catenin signaling confers ferroptosis resistance by targeting GPX4 in gastric cancer. Cell Death Differ. 2022;29:2190-2202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 250] [Article Influence: 83.3] [Reference Citation Analysis (0)] |
| 24. | He Y, Sun MM, Zhang GG, Yang J, Chen KS, Xu WW, Li B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct Target Ther. 2021;6:425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 862] [Article Influence: 215.5] [Reference Citation Analysis (0)] |
| 25. | Miricescu D, Totan A, Stanescu-Spinu II, Badoiu SC, Stefani C, Greabu M. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int J Mol Sci. 2020;22:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 477] [Article Influence: 95.4] [Reference Citation Analysis (0)] |
| 26. | Yu L, Wei J, Liu P. Attacking the PI3K/Akt/mTOR signaling pathway for targeted therapeutic treatment in human cancer. Semin Cancer Biol. 2022;85:69-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 362] [Article Influence: 90.5] [Reference Citation Analysis (0)] |
| 27. | Zhao Y, Fu Y, Zhang W, Zhao S, Li H. Evidence summary on management strategies for gastroesophageal reflux symptoms in patients following esophageal cancer surgery. Asia Pac J Oncol Nurs. 2025;12:100639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 28. | Song Q, Yu Z, Lu W, Zhuo Z, Chang L, Mei H, Cui Y, Zhang D. PD-1/PD-L1 inhibitors related adverse events: A bibliometric analysis from 2014 to 2024. Hum Vaccin Immunother. 2025;21:2424611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 29. | Mishra R, Patel H, Alanazi S, Kilroy MK, Garrett JT. PI3K Inhibitors in Cancer: Clinical Implications and Adverse Effects. Int J Mol Sci. 2021;22:3464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 183] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 30. | Jin ZY, Liu K, Wallar G, Zhou JY, Mu LN, Liu X, Li LM, He N, Wu M, Zhao JK, Zhang ZF. Environmental tobacco smoking (ETS) and esophageal cancer: A population-based case-control study in Jiangsu Province, China. Int J Cancer. 2025;156:1552-1562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 31. | Lee S, Rauch J, Kolch W. Targeting MAPK Signaling in Cancer: Mechanisms of Drug Resistance and Sensitivity. Int J Mol Sci. 2020;21:1102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 512] [Article Influence: 102.4] [Reference Citation Analysis (0)] |