INTRODUCTION
Esophageal cancer (EC) is a malignant tumor that originates from the mucosal epithelium of the esophagus and is primarily classified into two main types: Esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC). According to the International Agency for Research on Cancer, there were 511000 new cases of EC and 445000 deaths worldwide in 2022[1]. Among these, China accounts for nearly half of both the new cases and deaths. Although the incidence and mortality rates of EC have been decreasing in recent years[2], it remains a significant malignant tumor that poses a serious threat to the health of the Chinese population.
EC typically presents with non-specific symptoms in its early stage. Patients often seek medical attention only when they experience progressive dysphagia or persistent retrosternal pain, at which point they are frequently in the middle to late stages of the disease. As a result, over 50% of patients are not eligible for curative surgical therapy. Despite the increase in the age-standardized 5-year survival rate for EC in China from 27.8% to 33.4% between 2008 and 2017 due to advancements in surgical techniques and the introduction of neoadjuvant therapy[3], the overall survival rate remains suboptimal. The prognosis of EC is highly dependent on clinical staging; early-stage patients can achieve a 5-year survival rate of up to 95%[4], while the median survival time for advanced-stage patients is only 6 to 8 months, with a 5-year survival rate of less than 5%[5]. Therefore, increasing the early detection rate is crucial for improving the survival outcomes of patients with EC.
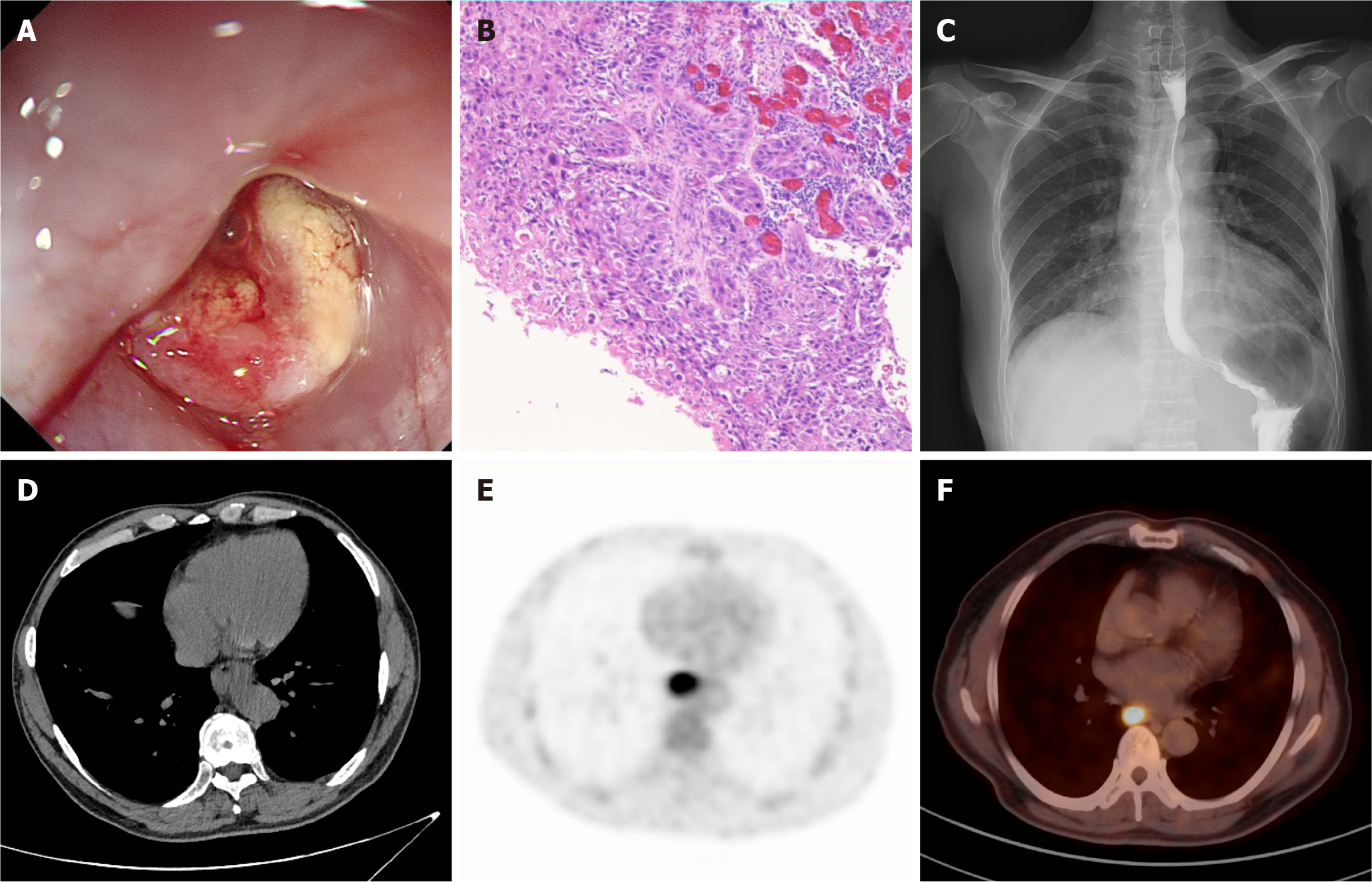
Due to the absence of specific biomarkers, the screening and diagnosis of EC primarily rely on imaging assessments, including endoscopy, computed tomography (CT), and other diagnostic modalities (Figure 1)[6]. Endoscopy combined with histopathological biopsy is currently the gold standard for diagnosing EC. However, due to the atypical morphological features of early EC and pre-cancerous lesions, the complexity of endoscopic procedures, and variations in physician expertise, diagnostic accuracy can vary significantly. Recently, the increasing utilization of chest CT has enhanced the potential for detecting early EC lesions. However, various factors, such as the insidious nature of EC lesions, low contrast, and indistinct borders, have been shown to significantly impact diagnostic accuracy. Some studies indicate that the accuracy of imaging radiologists in detecting EC on chest CT scans is less than 55%[7].
Figure 1 Common diagnostic modalities for esophageal cancer in current clinical practice.
A: Endoscopy; B: Pathological sections stained with hematoxylin-eosin; C: Esophageal barium swallow; D: Plain computed tomography; E: 18F-fluorodeoxyglucose positron emission tomography; F: 18F-fluorodeoxyglucose positron emission tomography-computed tomography.
In recent years, advancements in artificial intelligence (AI), particularly in deep learning (DL), have highlighted the potential of computer aided diagnosis (CAD) systems for automatic tumor detection and diagnosis[8,9]. CAD systems leverage DL and other technologies to train on large datasets of medical images [including endoscopy, CT, magnetic resonance imaging (MRI), etc.], enabling them to identify tumor characteristics, automatically locate potential tumor regions or abnormal findings, and provide reliable auxiliary diagnostic information. Currently, CAD systems have been extensively researched for their role in enhancing the automated detection and assessment of various tumors, including lung cancer[10], breast cancer[11,12], and pancreatic cancer[13]. The effective integration of DL methodologies to develop high-performance CAD systems that assist clinicians in achieving more accurate and efficient diagnoses has become a significant area of research within the field of medical imaging analysis.
Although DL has been widely applied in lesion detection across various diseases, research on the automatic detection of EC remains relatively limited. Current AI research related to EC primarily focuses on the analysis of esophageal endoscopy images and digital pathological images, employing techniques such as automatic feature extraction and supervised learning to facilitate automatic detection, diagnosis, and differential diagnosis. This includes the early detection of EC, differentiation between esophageal developmental abnormalities and EC, and the automatic evaluation of infiltration depth. While large-scale endoscopic screening has demonstrated promising results in enhancing the early detection of EC[14], the widespread adoption of routine endoscopy remains challenging due to the insidious nature of the initial symptoms of EC, which limits its broader application. Therefore, it is particularly important to explore the application prospects of DL techniques in imaging examinations, especially in CT images. This paper reviews the research status and progress of DL in the detection of EC, analyzing its diagnostic efficiency of different imaging modalities, including endoscopy, pathology, CT, etc. It also examines the challenges faced by DL in clinical practice and provides insights into future directions for its development in the field of EC diagnosis.
DL DETECTION MODELS
Convolutional neural networks (CNNs), as the core architecture of many DL models, have become a fundamental basis for current research in semantic segmentation and object detection tasks. CNN-based object detection methods are primarily categorized into two-stage detection and single-stage detection, both of which continue to evolve to optimize the balance between model accuracy and computational efficiency.
Two-stage detection network
The pioneering work of Girshick et al[15] introduced the region-based CNN (R-CNN), marking a significant breakthrough in the field of object detection. R-CNN generates candidate regions using a selective search algorithm and independently performs feature extraction and classification, but it suffers from computational redundancy. Fast R-CNN addressed this issue by introducing the region of interest pooling layer, which enables feature map sharing and accelerates feature extraction, partially resolving the speed problem. The core innovation of Faster R-CNN lies in the construction of the Region Proposal Network, which generates candidate regions end-to-end, eliminating the need for selective search[16]. Although Faster R-CNN increases inference time, it significantly improves detection accuracy. Consequently, Faster R-CNN has become a landmark in the field of object detection and is widely applied to two-stage detection tasks. Faster R-CNN has demonstrated significant clinical potential. The latest optimized iteration of faster R-CNN achieved an accuracy of 95.32%, precision of 94.63%, specificity of 94.84%, and sensitivity of 96.23% in detecting lung nodules in CT scans[17]. This algorithm has also shown exceptional performance across diverse medical imaging tasks, including cervical cancer cytology screening[18,19], dermoscopic image analysis for skin cancer diagnosis[20], and automated segmentation and classification of brain tumors[21]. In EC detection[22], Faster R-CNN has been preliminarily explored and demonstrated promising efficacy.
Single-stage detection network
You Only Look Once (YOLO) formulates the object detection task as a regression problem by dividing the image into grids and predicting bounding boxes along with class probabilities for each grid cell, thereby achieving end-to-end object detection. A primary advantage of YOLO lies in its computational efficiency and high accuracy, as it eliminates the need for candidate region generation, making it particularly suitable for real-time applications. The inaugural version, YOLOv1, was proposed by Redmon et al[23] in 2015. Subsequent iterations have incorporated innovations such as multi-scale prediction and cross-layer connections, with each iteration advancing state-of-the-art computer vision. However, research on YOLO-based applications in EC detection remains in its exploratory phase[24]. The single shot multibox detector (SSD) algorithm efficiently integrates multi-scale feature maps with prior boxes, demonstrating significantly improved performance in EAC endoscopic image recognition compared to traditional approaches[25]. However, the SSD model’s performance stability is heavily dependent on sample size, which restricts its application in scenarios with limited training data[26].
THE STATUS OF DL IN DIFFERENT MODALITY IMAGES
Digital pathological images
In the field of AI-assisted diagnosis of EC pathology, DL has made groundbreaking progress from bench to bedside. Research teams have pioneered a quantitative evaluation system for grading dysplasia in Barrett’s esophagus (BE), a precancerous lesion of EAC. This system is based on computational morphometry of epithelial nuclei, showing that nuclear texture heterogeneity is an independent predictor of disease progression (P = 0.004). Additionally, they have provided histomorphological evidence for computer-aided diagnosis. Subsequently, Faghani et al[27] developed a three-class DL model using the YOLO framework, enabling precise classification of non-dysplasia, low-grade dysplasia, and high-grade dysplasia in hematoxylin-eosin stained pathological images. This model achieved 81.3% sensitivity and 100% specificity for low-grade dysplasia, with specificities exceeding 90% across other categories, thereby reinforcing the potential of DL in EC histopathological grading.
However, traditional pathological image classification requires pathologist review of each slide, a laborious process that hinders the scalability of DL technologies. To address this issue, Bouzid et al[28] proposed a weakly supervised DL framework based on multiple instance learning, using a dataset built from hematoxylin-eosin stained sections paired with routine pathology reports to enable large-scale screening programs for BE. By deploying a semi-automated workflow, the framework achieved area under the receiver operating characteristic curve values of 91.4% and 87.3% on internal and external test sets, respectively, encompassing 1866 patients. This reduced pathologists' workload by 48% while preserving diagnostic accuracy, significantly enhancing the efficiency of BE screening. The integration of AI models into clinical pathology workflows is an active area of research[29], encompassing the development of visualization interfaces to accelerate diagnostic assessments. For instance, AI-driven rapid assisted visual search tools and attention heatmap overlays on pathological images can reduce review time by highlighting regions of interest. Future AI-assisted diagnostic tools must undergo multicenter clinical trials to validate their clinical efficacy and system robustness, a critical step toward advancing next-generation intelligent pathology systems.
Endoscopic images
In gastrointestinal endoscopic imaging, DL has been extensively used for colon polyp detection, histological grading of gastric and colonic polyps, early detection of gastrointestinal tumors and diagnosis of Helicobacter pylori infection[30]. These innovations culminated in the first Food and Drug Administration-approved AI-assisted endoscopy system, GI Genius™. However, no DL-based systems specifically targeting EC have received regulatory approval, primarily due to the high heterogeneity and diverse morphological presentations of early EC and precursor lesions. These presentations include erythematous patches, erosions, plaques, mucosal roughness, localized thickening, and disorganized mucosal vasculature, which often mimic benign conditions such as esophagitis and gastric heterotopia, hampering research progress.
In recent years, CAD systems for early tumor detection in BE have received considerable research attention. However, most studies[31-33] are predominantly single-center, small-sample, and retrospective, and have inadequately accounted for confounding by benign lesions, limiting the generalizability of their findings. De Groof et al[34] developed a CAD system using 494364 annotated endoscopic images from five independent datasets to pre-train a hybrid ResNet-UNet model, achieving an overall specificity, sensitivity, and accuracy of 88%, 90%, and 89%, respectively, in differentiating neoplasia from non-dysplastic BE. The BONS-AI consortium further enhanced the robustness of the system by integrating retrospective and prospective data from 15 international endoscopy centers, demonstrating improved generalizability for clinical use[35]. Using advanced pre-training, this CAD system autonomously detects and classifies BE in ex vivo volumetric laser endomicroscopy images/videos. Neoplasia is highlighted by green bounding boxes, with the system achieving 90% sensitivity and 80% specificity for images, and 80% sensitivity and 82% specificity for videos. Endoscopists assisted by CAD showed significantly improved sensitivity for neoplasia detection (image: 74% to 88%, video: 67% to 79%). However, the real-time in vivo performance of volumetric laser endomicroscopy requires further validation to optimize clinical feasibility.
Current research in AI-assisted lesion detection focuses primarily on improving image segmentation and classification accuracy, with limited attention paid to optimizing detection speed and developing real-time algorithms. Horie et al[36] used the SSD framework to analyze 1118 images in 27 seconds, achieving 98% accuracy in differentiating between superficial and advanced EC. The model successfully detected all seven sub-centimeter (< 10 mm) tumors, demonstrating exceptional microlesion detection capability. Tang et al[37] developed a real-time deep CNN that reduced the processing time of a single white light image to 15 milliseconds through parameter optimization.
However, previous lesion detection models have often excluded benign esophageal lesions during training, resulting in dataset selection bias and an increased risk of false positives. Wang et al[24] addressed this gap by integrating benign lesions such as reflux esophagitis, fungal esophagitis and gastric heterotopia into their YOLOv5-based model. The system generates lesion suspicion heatmaps with color intensity positively correlating with malignancy probability, demonstrating ≥ 93% diagnostic accuracy and specificity across white light image, narrow band imaging and Lugol’s chromoendoscopy, comparable to expert endoscopist performance. Single frame analysis took only 0.17 seconds, highlighting the real-time clinical utility. However, this single-center retrospective study focused only on ESCC, excluding cases of EAC and BE, and relied exclusively on static images, without addressing poor quality data, mucosal artefacts or endoscopic motion effects. These limitations prevent clinical generalizability.
Yuan et al[38] conducted the first global randomized controlled trial to evaluate real-time AI support for the diagnosis of early ESCC in a real-world clinical setting, enrolling 11715 patients in 12 Chinese medical centers. The AI system, based on the YOLOv5 algorithm, was seamlessly integrated into standard endoscopy devices and annotated suspicious lesions with malignancy probabilities on the endoscopic screen via a single-screen interface without disrupting clinical workflow. The AI-assisted group showed a significantly lower miss rate than the conventional group (RR = 0.25, P = 0.079). In addition, Li et al[39] developed the ENDOANGEL-ELD system, which enables real-time localization of high-risk esophageal lesions during narrow band imaging using bounding boxes and attention heatmaps, achieving 89.7% sensitivity, 98.5% specificity and 98.2% accuracy. Nevertheless, a comprehensive evaluation of the clinical effectiveness and cost-effectiveness of AI systems remains imperative to confirm their ability to improve diagnostic outcomes in the early detection of EC.
CT images
Contrast-enhanced CT (CECT), a routine imaging modality for the diagnosis of EC, provides clear visualization of irregular esophageal wall thickening, focal or diffuse contrast enhancement, enlarged lymph nodes and periesophageal tissue invasion. Takeuchi et al[40] pioneered an AI diagnostic system using a fine-tuned VGG16 architecture (a deep CNN) trained on 1500 CECT and non-contrast CT images, including 457 EC cases and 1000 normal controls. The system achieved an accuracy of 84.2%, an F1 score of 0.742, a sensitivity of 71.7%, and a specificity of 90.0%. Gradient-weighted class activation mapping was implemented to generate lesion-localizing heatmaps to improve diagnostic interpretability. A major limitation is the 2-dimensional (2D) image-based modelling approach, whereas clinical practice relies on 3D spatial analysis, highlighting the need for future development of 3D convolutional networks. Chen et al[22] developed an EC detection system based on a modified Faster R-CNN framework incorporating Online Hard Example Mining. Evaluated on 1520 CECT images of the gastrointestinal tract, the system achieved an F1 score of 95.71%, a mean average precision of 92.15% and a detection speed of 5.3 seconds per image. The modified Faster R-CNN framework demonstrated superior performance to both the traditional Faster R-CNN and Inception-v2 models by effectively capturing multi-scale feature information through convolutional and parallel architecture designs. The integration of Online Hard Example Mining significantly improved detection accuracy. Yasaka et al[41] evaluated the efficacy of a DL model in assisting radiologists with different levels of expertise (one attending radiologist and three radiology residents) to detect EC on CECT images. The DL model significantly improved the area under the curve (AUC) across readers (pre-intervention: 0.96/0.93/0.96/0.93, post-intervention: 0.97/0.95/0.99/0.96), with statistically significant improvements observed for less experienced residents. Additionally, the DL model enhanced diagnostic confidence and performance among junior radiologists, leading to significant improvements in diagnostic efficiency.
Early studies demonstrate that esophageal wall thickening on non-contrast CT represents a significant imaging biomarker of EC, enabling radiologists to recommend confirmatory endoscopic evaluation based on this finding. Sui et al[7] developed an improved VB-Net segmentation network incorporating multi-scale feature information to achieve precise esophageal wall delineation on CT images. This model automatically quantifies esophageal wall thickness and localizes lesions, demonstrating 88.8% sensitivity and 90.9% specificity. Clinical validation also showed that AI-assisted radiologists (three practitioners) achieved a mean increase in sensitivity from 27.5% to 77.5% and an increase in accuracy from 53.6% to 75.8%, significantly reducing missed diagnosis rates. Lin et al[42] employed a dual-center retrospective dataset with the nnU-Net model for esophageal segmentation, followed by decision tree classification based on extracted radiomic features, achieving an AUC of 0.890. Using DL algorithms, the AUC for physician diagnosis improved from 0.855/0.820/0.930 to 0.910/0.955/0.965 (P < 0.01). However, current models have limitations. These include: (1) Overdependence on esophageal wall thickness, which fails to capture other morphological alterations (e.g., texture features), thereby limiting detection of small lesions; and (2) Insufficient discriminatory power to differentiate malignant lesions from benign esophageal pathologies such as esophagitis and leiomyomas.
Low-dose CT (LDCT) has been widely adopted in cancer screening due to its reduced radiation exposure. However, the inherent limitations of LDCT, including lower spatial resolution and elevated noise levels, introduce additional challenges for EC detection. The subtle anatomical features of the esophageal wall are often poorly visualized. Imaging artifacts from feeding tubes or stents may further obscure lesion boundaries, complicating precise margin delineation in EC. Traditional machine learning approaches (e.g., region-based segmentation and handcrafted feature extraction) rely heavily on pronounced gray-level intensity variations, rendering them inadequate to address the unique diagnostic complexities of EC. In contrast, DL techniques, particularly CNNs, demonstrate superior capability through hierarchical feature learning to capture complex tumor morphology and peritumoral tissue patterns. Even in scenarios with poorly defined lesion boundaries, CNNs maintain clinically acceptable localization and identification accuracy, highlighting their potential for clinical translation. However, research on automated EC detection using DL in LDCT remains exploratory.
To improve the diagnosis of EC on LDCT, the following approaches can be pursued: (1) Multimodal integration of CECT, MRI and positron emission tomography (PET) allows cross-modal feature fusion, using anatomical detail of CT, soft tissue characterization of MRI and metabolic profiling of PET to compensate for the inherent limitations of LDCT; (2) Advanced image processing techniques (e.g., noise suppression and contrast enhancement algorithms) can optimize LDCT image quality for improved DL inputs; and (3) The development of LDCT-specific neural networks incorporating multimodal data, clinical parameters and histopathological correlations could improve the detection of early malignancies. Systematic implementation of these approaches may accelerate the clinical translation of AI-based diagnostic systems.
PET/CT and barium swallow study
18F-fluorodeoxyglucose PET/CT is recommended for initial clinical staging of EC due to its superior capability in detecting distant metastases and enabling comprehensive assessment of tumor burden[43,44]. The integration of multimodal imaging techniques (e.g., PET/CT and PET/MRI) with DL frameworks has significant potential to advance EC diagnosis. However, current research on DL applications in PET/CT-based EC analysis remains limited, with existing studies mainly focusing on radiomics-driven staging prediction, treatment response monitoring and prognostic stratification[45,46]. In particular, automated lesion detection in PET/CT images represents a critical unmet need in this field.
Barium swallow imaging demonstrates high sensitivity for EC diagnosis by identifying imaging features including mucosal disruption, ulceration, filling defect and luminal narrowing[47], which are critical for detection. While Yang et al[48] pioneered CAD systems for this modality, early implementations based on traditional machine learning (e.g., support vector machines and k-nearest neighbors) required labor-intensive manual region of interest annotation and limited radiomic feature extraction. Zhang et al[49] introduced an automated DL system trained on five independent datasets, which achieved 90.3% accuracy, 92.5% sensitivity, and 88.7% specificity through probability-weighted lesion localization, while significantly reducing radiologist interpretation time and improving workflow efficiency. However, no subsequent studies have reported DL applications for EC detection in barium swallow imaging, highlighting a critical research gap in automated diagnostic solutions for this imaging modality.
CHALLENGES AND FUTURE PERSPECTIVES
Despite the demonstrated potential of DL in automated EC detection, current research efforts remain predominantly confined to experimental validation and proof-of-concept prototyping. Few implementations have achieved clinical adoption, with real-world diagnostic performance consistently underperforming experimental benchmarks. This section systematically analyzes the critical translational challenges and proposes strategic solutions to bridge the innovation-to-clinical translation gap.
Building high-quality datasets
A major challenge in training DL models is the lack of large, diverse patient populations and high-quality, annotated datasets. Current research relies predominantly on retrospective, single-center data, which often suffer from selection bias, fragmentation, and inconsistencies across sites. In addition, the quality of medical imaging data is affected by variations in acquisition protocols, imaging equipment and clinical settings. The lack of publicly available datasets for EC detection hinders the reproducibility and validation of models across healthcare institutions. These challenges significantly affect the generalizability and clinical applicability of models.
Federated learning, a decentralized approach to training, provides a viable solution by enabling multi-institutional collaboration while maintaining data privacy, thereby overcoming barriers to data sharing[50,51]. This approach not only improves model performance, but also allows for continuous updates and incremental learning, improving model adaptability to evolving clinical practice. The emergence of generative AI opens up new avenues for efficient data use, with research showing that generative adversarial networks can augment datasets and synthesize images, potentially reducing reliance on training data by more than 40% without compromising diagnostic accuracy[52]. In addition, innovative unsupervised and self-supervised learning methods facilitate the effective use of diverse imaging datasets, significantly reducing the annotation burden associated with clinical AI development. Health data, which are essential for medical AI, is increasingly seen as a valuable asset by healthcare organizations[51]. Since 2024, a number of global healthcare organizations, such as the Mayo Clinic, the National Health Service, Stanford Health Care and Xuanwu Hospital, have initiated the creation of specialized disease datasets and facilitated data sharing through trading mechanisms. The future development of medical AI will depend on the establishment of robust data production and distribution cycles, supported by standardized health data ecosystems and advanced technologies.
Promoting multimodal feature fusion
Health data are inherently multimodal, encompassing radiomic, pathological, and genomic information that is essential for managing EC. Current DL models primarily concentrate on single-modality analyses, such as CT or histopathology, which, while effective for specific tasks, fall short in integrating multimodal imaging techniques, including PET-CT or PET-MRI, alongside high-dimensional omics data. Early fusion methods relied on basic feature concatenation[53], which failed to fully exploit the correlations and complementary insights available between different modalities. Recent advancements have shifted towards transformer architectures that enable modality-agnostic learning[54], employing adaptive attention mechanisms and facilitating knowledge transfer across modalities through co-learning strategies. These frameworks promote adaptable input-output configurations and enhance model robustness via shared representation learning. This underscores the critical need for interdisciplinary collaboration to fully realize the clinical applications of these innovations.
Balancing model transparency and accuracy
A primary concern regarding DL models is the methodology by which they derive their conclusions. This inherent lack of interpretability poses challenges for their acceptance and practical use in clinical environments. The opacity of these models complicates the establishment of clinical trust and adherence to regulatory standards. While existing interpretability techniques, such as gradient-weighted class activation mapping, provide some insights, they often highlight correlations rather than causative factors, making it difficult to understand the rationale behind the model's decisions. This highlights the growing need for frameworks that enhance explainability, such as local interpretable model-agnostic explanations and Shapley Additive Explanations, which are designed to clarify localized decision-making processes. Furthermore, regulatory frameworks like the Food and Drug Administration’s AI/ML Software as Medical Device guidelines (2024) and the European Union’s AI Act (2024) mandate continuous monitoring and independent validation for high-risk medical AI systems after deployment. New protocols like TRIPOD-ML suggest the creation of dynamic validation ecosystems that encompass data drift detection, real-time performance assessment, and iterative model refinement to maintain clinical safety. Achieving a balance between interpretability and accuracy, alongside ensuring adaptive compliance, presents a significant challenge in the clinical application of AI technologies.
Establishing a standardized evaluation framework
AI models often show impressive results in retrospective studies; however, their practical application in clinical settings is hindered by significant validation challenges. These challenges arise from the variability in medical imaging, differing clinical workflows, and operator-dependent factors. Additionally, two unresolved methodological issues exacerbate this translational difficulty: The lack of agreement on optimal DL architectures and the absence of standardized evaluation protocols, which complicates comparisons across studies. A clear example of this standardization issue can be seen in the assessment of esophageal edge irregularities, where variations in inclusion criteria can lead to a 15% fluctuation in Dice score results[55]. Moreover, traditional performance metrics such as accuracy and AUC-receiver operating characteristic are often insufficient in clinical contexts, particularly in early cancer detection, where the stakes of false negatives (which may delay necessary interventions) are much higher than those of false positives (which could lead to unnecessary confirmatory tests). Recent methodological advancements have proposed the use of decision curve analysis to assess clinical net benefit, thus aligning model evaluation more closely with patient-centered considerations, including the differential weighting of misclassification risks and the implications for treatment pathways.
Enhancing cross-domain collaboration
The practical application of DL models in clinical settings is hindered by strict legal and privacy regulations, leading to higher operational costs. Privacy-preserving technologies like federated learning allow for model training across institutions without requiring the sharing of raw data[50]. Additionally, methods such as homomorphic encryption and differential privacy enhance secure data management. The complexities of health data ownership underscore the need for innovative governance structures. For example, Genomics England’s dynamic consent model empowers patients to control their data, whereas The Cancer Genome Atlas treats de-identified datasets as public resources for research. Achieving a balance between data privacy, regulatory compliance, and scientific progress demands collaborative efforts to establish adaptable legal frameworks and standardized technical protocols that foster the development of medical AI while safeguarding patient rights.
Optimizing AI-clinical workflow integration
Challenges in integrating DL models into clinical practice arise from ambiguous operational frameworks, which can lead to cognitive overload due to fragmented multi-platform workflows and complex parameter settings. Future implementations should focus on developing context-sensitive human-AI interfaces that utilize natural language processing and offer adaptive display options, such as visualizing lesions based on risk stratification. Furthermore, establishing closed-loop workflows that combine automated detection, quantitative analysis, and human validation could streamline the screening, confirmation, and reporting processes, thereby improving clinical adoption[56]. The high computational demands of DL models also pose challenges in low-resource healthcare settings, which often lack sophisticated computing infrastructure. This situation highlights the urgent need for effective interoperability between AI systems and hospital information networks to enhance decision support. Additionally, AI-driven early cancer detection requires substantial data storage and processing capabilities, raising concerns about cost and practicality in routine clinical applications. Successful clinical integration relies on securing regulatory approvals, ensuring compatibility with hospital information systems, and validating through prospective clinical trials. Addressing these challenges is essential for the effective implementation of AI technologies in healthcare environments.