Published online May 15, 2025. doi: 10.4251/wjgo.v17.i5.103667
Revised: January 8, 2025
Accepted: February 28, 2025
Published online: May 15, 2025
Processing time: 169 Days and 22.2 Hours
Colorectal cancer (CRC) is the third-most prevalent cancer and the cancer with the second-highest mortality rate worldwide, representing a high public health bur
To use bibliometric approaches to analyze and visualize the current research state and development trend of DL in CRC as well as to anticipate future research directions and hotspots.
Datasets were retrieved from the Web of Science Core Collection for the period January 2011 to December 2023. Scimago Graphica (1.0.45), VOSviewer (1.6.20) and CiteSpace (6.3.1) were used to analyze and visualize the nation, institution, journal, author, reference and keyword indicators. Origin (2022) was utilized for plotting, and Excel (2021) was used to construct the tables.
A total of 1275 publications in 538 journals from 74 countries and 2267 institutions were collected. The number of annual publications has increased over time. China (371, 29.1%), the United States (265, 20.8%) and Japan (155, 12.2%) contributed significantly to the number of articles published, accounting for 62.1% of the total publications. The United States had the strongest ties to other nations. Sun Yat-sen University in China had the highest number of publications (32). The journal with the most publications was Scientific Reports (34, Q2), whereas Gastrointestinal Endoscopy had the most co-citations (1053, Q1). Kather JN, was the author with the most articles (12) and co-citations (287). The most frequently cited reference was Deep Residual Learning for Image Recognition. Keywords were divided into six clusters, with “colorectal cancer” (12.34) having the highest outbreak intensity.
This study highlights the current status and most active directions of the use of DL in CRC. This approach has important applications in the identification, diagnosis, localization, classification and prognosis of the disease and will remain a central focus in the future.
Core Tip: This bibliometric analysis evaluated the application of deep learning in colorectal cancer and identifies valuable future directions for studying the diagnosis, treatment and prognosis of colorectal cancer. It is recommended to optimize deep learning models, such as convolutional neural networks and transformers, strengthen multicenter collaboration, and focus on emerging hotspots, such as microsatellite instability and autoencoder-based models.
- Citation: Qi LY, Li BW, Chen JQ, Bian HP, Xue JN, Zhao HX. Research status and trends of deep learning in colorectal cancer (2011-2023): Bibliometric analysis and visualization. World J Gastrointest Oncol 2025; 17(5): 103667
- URL: https://www.wjgnet.com/1948-5204/full/v17/i5/103667.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i5.103667
Colorectal cancer (CRC) is a group of diseases that pose significant public health and socioeconomic challenges[1]. According to GLOBOCAN 2022, CRC incidence and mortality rank third and second among malignancies, respectively, with more than 1.92 million new cases and 900000 new deaths annually, accounting for almost 10% of total cancer incidence and deaths[2,3].
Artificial intelligence (AI) is a broadly applicable technology capable of leveraging data-driven algorithms, emulating computers, and even outperforming human intelligence to some extent[4,5]. Machine learning (ML) is a subfield of AI, and deep learning (DL) is a branch of ML that can identify patterns within a limited dataset, enabling the extraction of more complex features[4,6]. Over the years, AI algorithms have progressed from conventional ML to advanced DL techniques, and DL is particularly adept at recognizing complex features in medical images, such as radiological and pathology images[7]. Convolutional neural networks (CNNs) are among the most widely used DL models in medical imaging and are capable of automatically extracting representative characteristics from medical images[8]. A newly emerging category of neural network transformers is gaining popularity in medical research, with enhanced performance, resilience and data efficiency[9].
Bibliometrics is a comprehensive research strategy that employs mathematical and statistical tools to analyze research literature quantitatively and qualitatively[10,11]. It is extremely useful for examining the current state of diseases and forecasting future research hotspots[12]. In studies related to CRC, DL has been frequently applied in image diagnosis, image histology, image segmentation, patient risk stratification, histopathology, prognostic biomarker prediction and colon polyp detection in recent years[5,13-18]. Currently, there is no bibliometric analysis of DL in CRC. As a result, in this study, a combination of visualization and bibliometric analysis was used to summarize the research literature on DL as applied to CRC to identify the countries of origin, institutions of origin, authors, publishing journals, references and keywords of such research. By analyzing interrelationships and influences, we explore research hotspots and future development trends in this field to propose new ideas for the diagnosis and treatment of CRC.
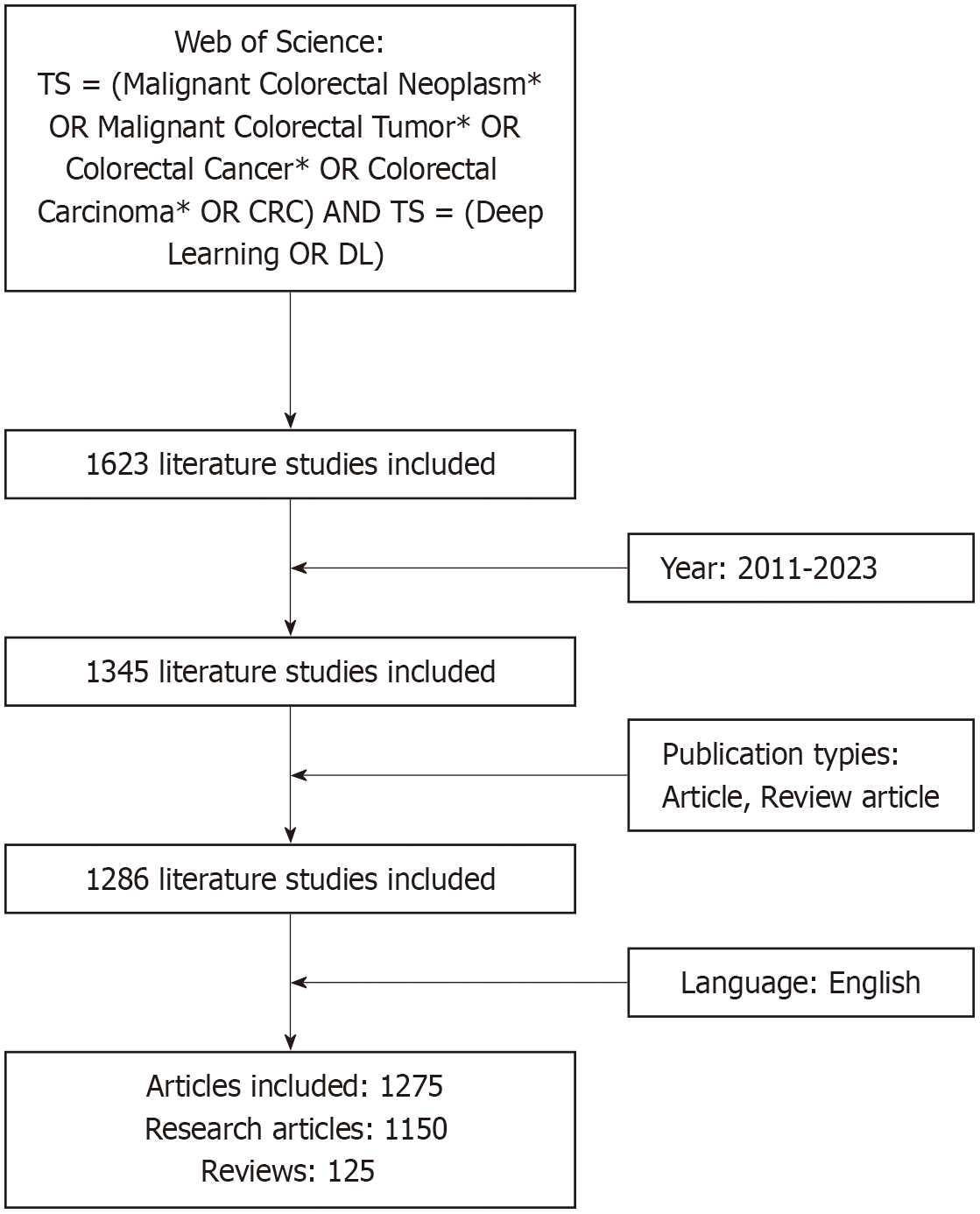
On October 20, 2024, relevant literature was retrieved from the Web of Science Core Collection database with the search strategy TS = (Malignant Colorectal Neoplasm* OR Malignant Colorectal Tumor* OR Colorectal Cancer* OR Colorectal Carcinoma* OR CRC) AND TS = (Deep Learning OR DL). The search time limit was January 1, 2011 to December 31, 2023, the language limit was English, the literature type limit was article, review article, and the literature screening flowchart was as shown in Figure 1.
This study conducted bibliometric and visualization studies via Scimago Graphica (1.0.45), CiteSpace (6.3.1), and VOSviewer (1.6.20). Scimago Graphic is data visualization software that enables researchers to rapidly and directly interpret and present data. It can generate a range of chart types, such as maps and networks. CiteSpace can display the structure, regularity, and distribution of scientific information via data visualization. VOSviewer creates bibliometric networks and analyzes research hotspots and trends on the basis of the terms that appear the most frequently in network visualization maps. Scimago Graphica (1.0.45) was used to analyze national indicators, whereas CiteSpace (6.3.1) was utilized to analyze indicators such as institutions, references and keywords. VOSviewer (1.6.20) was used to analyze the journal, author and reference indicators.
In the visualization maps and timeline diagrams created by these software programs, the size of the node indicates the frequency of occurrence. The connecting lines between nodes indicate collaboration or co-occurrence relationships. The thicker the connecting line is, the closer the relationship. Different colors in the network image represent different clusters, whereby the reference and keyword citation burst rates aided in identifying the most recent research trends for the subject. CiteSpace’s mediator centrality was an important parameter, whereby a value greater than 0.1 indicates that a node was a relatively more significant one (identified by a purple-red circle in the visualization graph), which was a major measure of such a node’s bridging role in the overall network structure[19].
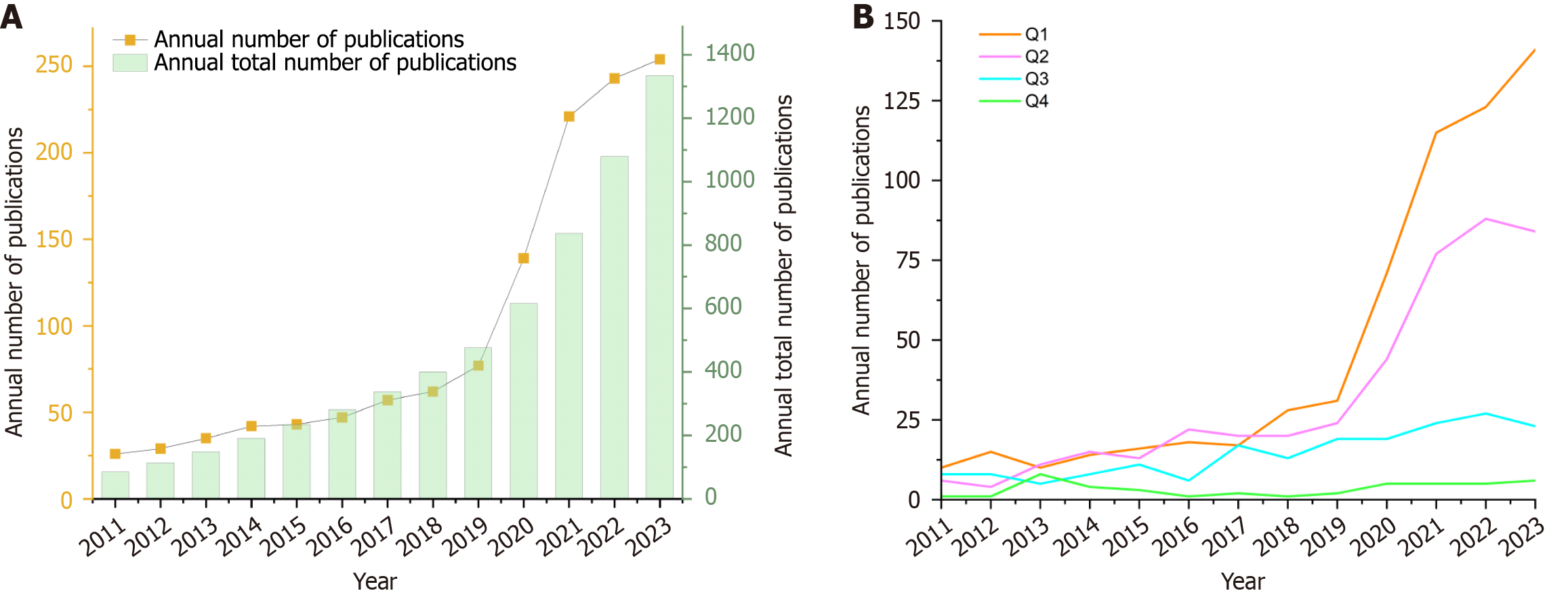
For the period January 2011 to December 2023, a total of 1345 papers were obtained via Web of Science, of which 1275 were ultimately included in the analysis. The trend of the number of publications in Figure 2A shows that the number of publications was low and expanded slowly from 2011 to 2019, increased from 2019 to 2021, and continued to grow steadily from 2021 to 2023. After 2019, the number of publications increased to more than 100 every year, and the number of publications in 2023 was approximately ten times greater than that in 2011. The trend in total annual publications indicated a large increase in the number of articles published after 2019. Figure 2B classifies the included studies into Journal Citation Reports quartiles and reveals an overall pattern of growth in publications in Q1, Q2, and Q3, with the number of articles published in Q1 in particular increasing rapidly after 2019, reaching 141 in 2023.
The search and screening produced a total of 1275 publications from 74 countries. Table 1 shows that among the top 10 countries in terms of the number of publications, the country with the highest number of publications was China (371), followed by the United States (265), and Japan (155), and that the top five countries had more than 100 publications each.
| Rank | Country | Documents |
| 1 | China | 371 |
| 2 | United States | 265 |
| 3 | Japan | 155 |
| 4 | South Korea | 121 |
| 5 | United Kingdom | 101 |
| 6 | Germany | 96 |
| 7 | Italy | 71 |
| 8 | Spain | 52 |
| 9 | India | 49 |
| 10 | Netherlands | 45 |
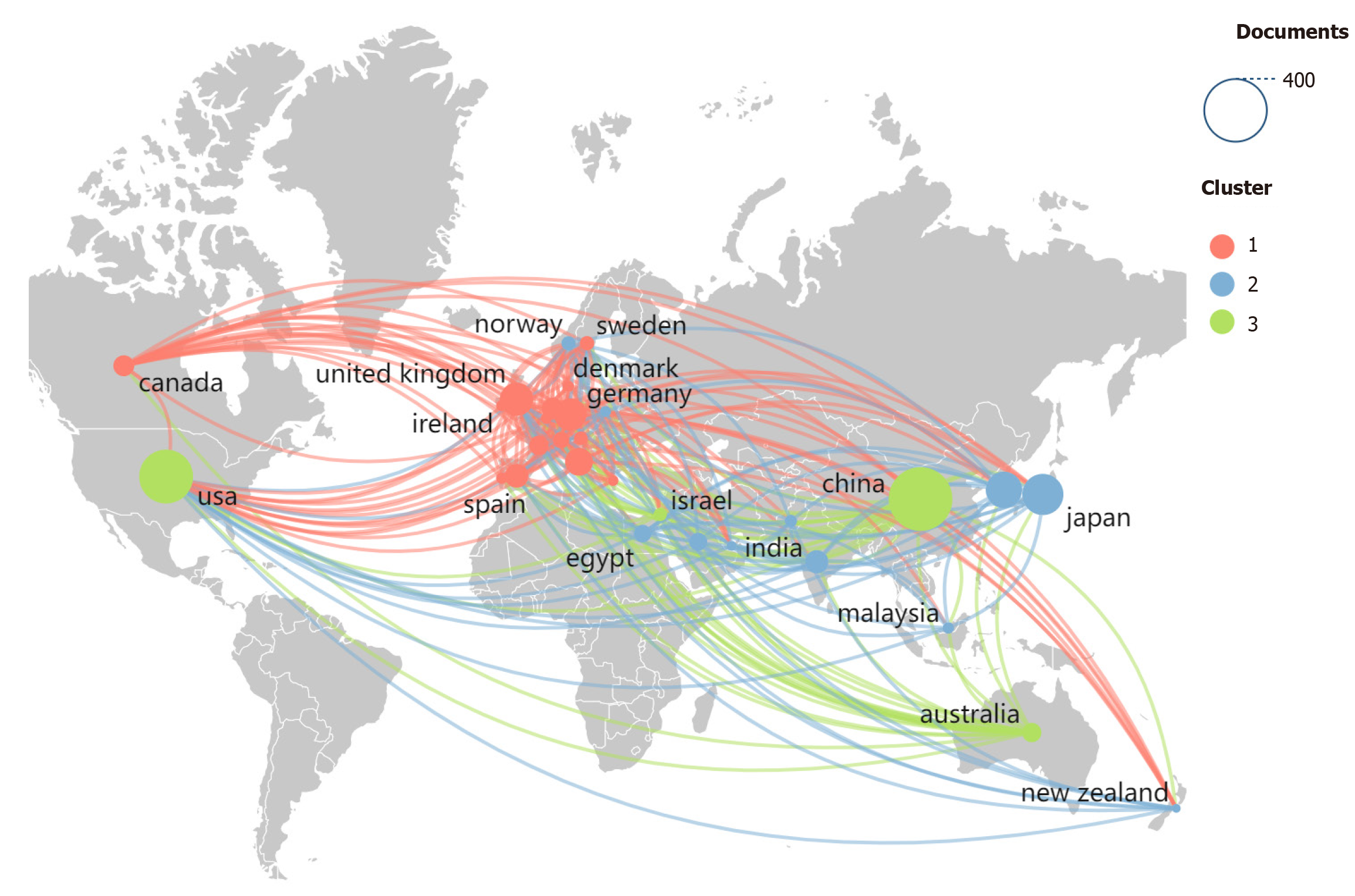
The network of countries publishing on DL in CRC research is shown in Figure 3. Here, we used VOSviewer (1.6.20) to export the data of the top 30 countries in terms of the number of publications to Excel and then imported the data into Scimago Graphica (1.0.45). “Label” was changed to “Country”, and “cluster” was changed to “String”. After entering the View interface, we dragged the weight <documents> tag into “Size”, clustered the tag according to “Color”, labeled the tag either “Label”, “Tooltip” or “Unit”, and clicked “Map” in “Layout” to complete the drawing of the country cooperation map. In this map, the most closely related countries were the United States, the United Kingdom, and Germany. The United States has developed collaborative networks or ties with dozens of countries on the subject of interest, whereas China has a major advantage in terms of the number of articles published, but its collaboration with other countries is limited.
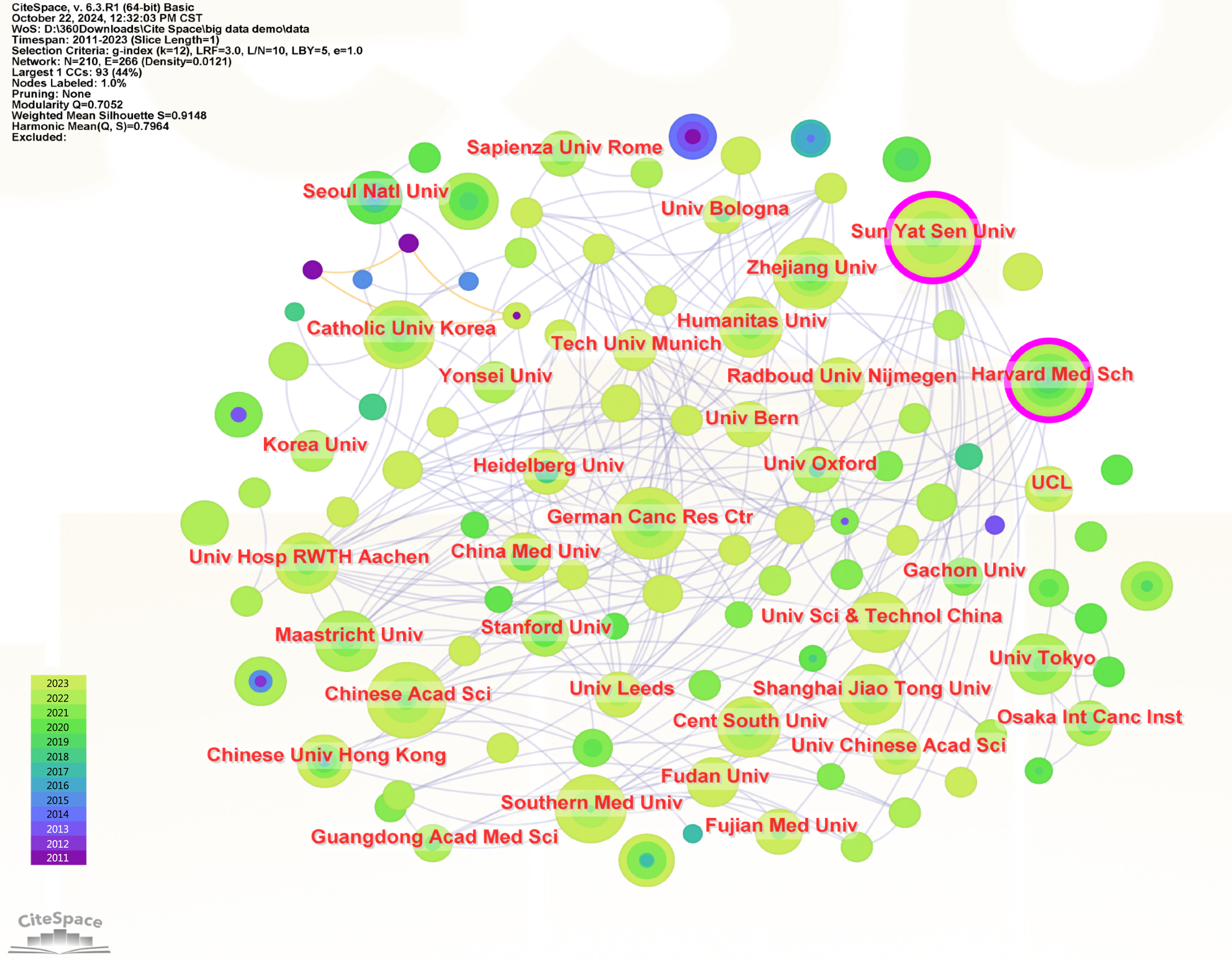
The screening yielded 1275 papers from 2267 institutions, with the top ten institutions in terms of the number of publications listed in Table 2. Sun Yat-sen University had the most publications (32), followed by the Chinese Academy of Sciences, Harvard Medical School and Seoul National University (20 each).
| Rank | Institution | Country | Count | Centrality | Proportion (%) |
| 1 | Sun Yat-sen University | China | 32 | 0.1 | 2.5 |
| 2 | Chinese Academy of Sciences | China | 20 | 0.06 | 1.5 |
| 3 | Harvard Medical School | America | 20 | 0.12 | 1.5 |
| 4 | Seoul National University | Korea | 20 | 0.01 | 1.5 |
| 5 | Zhejiang University | China | 19 | 0.03 | 1.4 |
| 6 | Catholic University of Korea | Korea | 18 | 0.08 | 1.4 |
| 7 | Southern Medical University | China | 18 | 0.01 | 1.4 |
| 8 | German Cancer Research Center | Germany | 17 | 0.06 | 1.3 |
| 9 | National Cancer Centre | Japan | 16 | 0.00 | 1.2 |
| 10 | Rheinisch-Westfälische Technische Hochschule Aachen | Germany | 16 | 0.02 | 1.2 |
Interinstitutional collaboration is represented by the visualization map in Figure 4. Larger nodes indicate a greater number of publications, more connecting lines indicate closer collaboration, and purple circles outside the nodes indicate greater centrality. Nodes with high centrality play important roles as bridges in the literature network, with Harvard Medical School (0.12) and Sun Yat-sen University (0.10) having the highest centrality. As a result, the large number of papers published by Chinese universities and the close cooperation network of United States institutions have contributed significantly to the development of this area of study.
Table 3 shows that during the timeframe covered by this analysis, 538 journals published articles on the use of DL in CRC. The top ten journals with the most published articles were Scientific Reports (34), IEEE Access (31), and Frontiers in Oncology (29), whereas Medical Image Analysis had the highest impact factor (10.7), followed by Computers in Biology and Medicine (7.0).
| Rank | Journals | Counts | Citations | IF | JCR | Co-cited journals | Co-citations | IF | JCR |
| 1 | Scientific Reports | 34 | 1212 | 3.8 | Q1 | Gastrointestinal Endoscopy | 1053 | 6.7 | Q1 |
| 2 | IEEE Access | 31 | 643 | 3.4 | Q2 | Scientific Reports | 969 | 3.8 | Q1 |
| 3 | Frontiers in Oncology | 29 | 254 | 3.5 | Q2 | Proceedings of the IEEE | 946 | 23.2 | Q1 |
| 4 | Cancers | 28 | 453 | 4.5 | Q1 | Gastroenterology | 923 | 25.7 | Q1 |
| 5 | Diagnostics | 27 | 267 | 3.0 | Q1 | Lecture Notes in Computer Science | 886 | / | / |
| 6 | World Journal of Gastroenterology | 22 | 454 | 4.3 | Q1 | Journal of Clinical Oncology | 762 | 42.1 | Q1 |
| 7 | Medical Image Analysis | 16 | 927 | 10.7 | Q1 | New England Journal of Medicine | 689 | 96.2 | Q1 |
| 8 | Applied Sciences-Basel | 15 | 187 | 2.5 | Q1 | IEEE Transactions on Medical Imaging | 671 | 8.9 | Q1 |
| 9 | Computers in Biology and Medicine | 15 | 516 | 7.0 | Q1 | Gut | 633 | 23.0 | Q1 |
| 10 | International Journal of Colorectal Disease | 13 | 418 | 2.5 | Q1 | Endoscopy | 621 | 11.5 | Q1 |
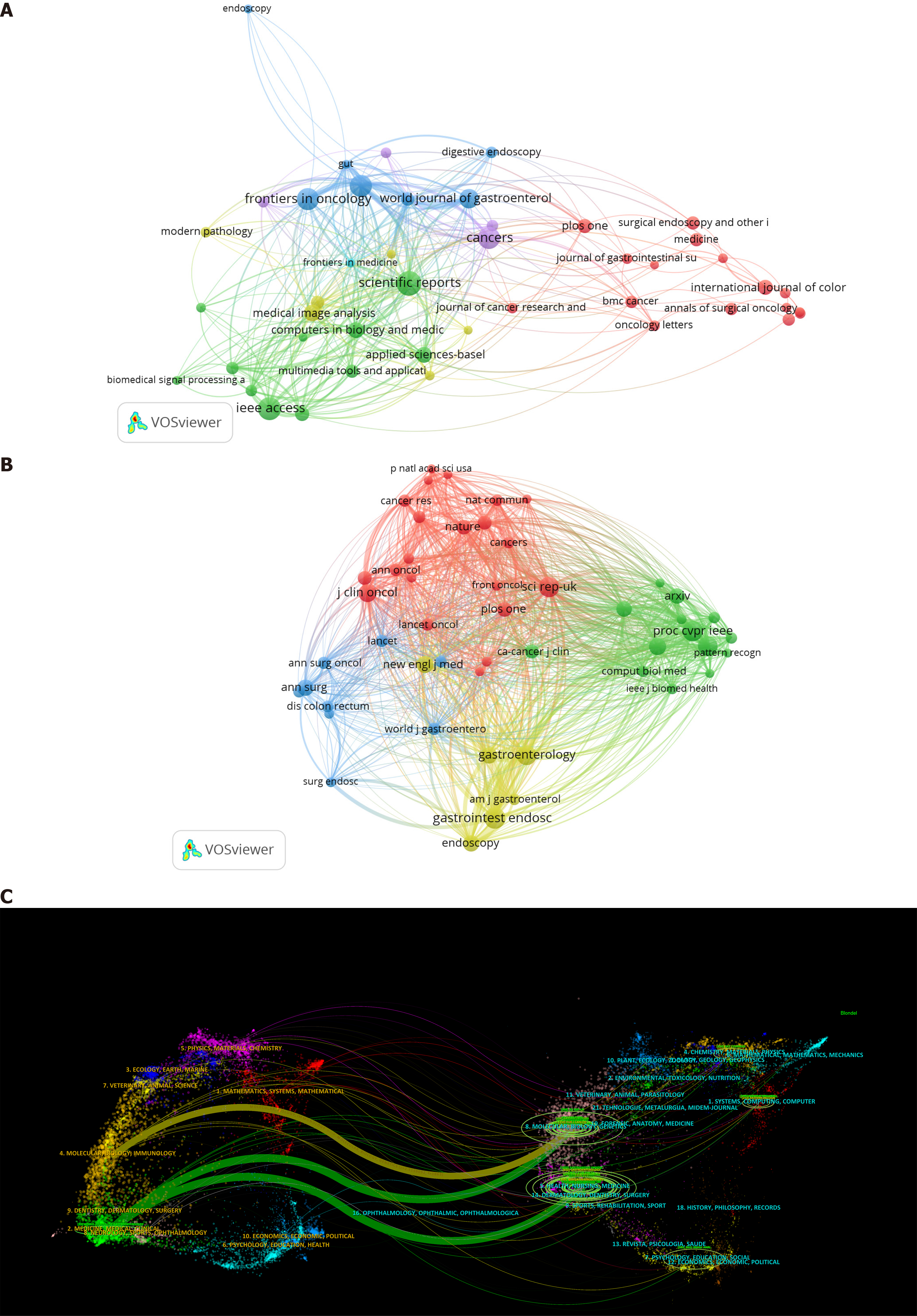
There were 7739 co-cited journals, with Gastrointestinal Endoscopy (1053), Scientific Reports (969) and Proceedings of the IEEE (946) having the most citations among the top ten and the New England Journal of Medicine (96.2) and Journal of Clinical Oncology (42.1) having the highest impact factors. The quantity of citations revealed the extent to which publications were used by scholars and researchers in the area. Nine journals were cited in Q1, indicating that these journals had a significant impact on the field. Table 4 displays the top ten journal publishers, with Elsevier, Springer Nature, and MDPI publishing the most papers, each with more than 100.
| Rank | Journal publishers | Counts |
| 1 | Elsevier | 239 |
| 2 | Springer Nature | 239 |
| 3 | MDPI | 135 |
| 4 | Wiley Inter Science | 99 |
| 5 | IEEE | 68 |
| 6 | Frontiers Media S.A. | 51 |
| 7 | Nature Portfolio | 51 |
| 8 | LWW | 37 |
| 9 | Baishideng Publishing Group Inc | 29 |
| 10 | Taylor & Francis | 24 |
Figure 5 presents the analysis of journals and co-cited journals. The top effect is more pronounced in Figure 5A, where the distribution of issuing journals in the area is unequal, with the majority of the papers appearing in a relatively small number of journals. The journals in Figure 5B were classified into four types on the basis of their co-citation frequency. Here, papers in the same category may have comparable research orientations and internal logics. The double map overlay of Figure 5C shows that the citing journals (left side) were mainly distributed in the fields of molecular biology, immunology, clinical care, medical care, and medicine. The cited journals (right side) were mainly concentrated in the fields of molecular biology, genetics, health, nursing, and medicine.
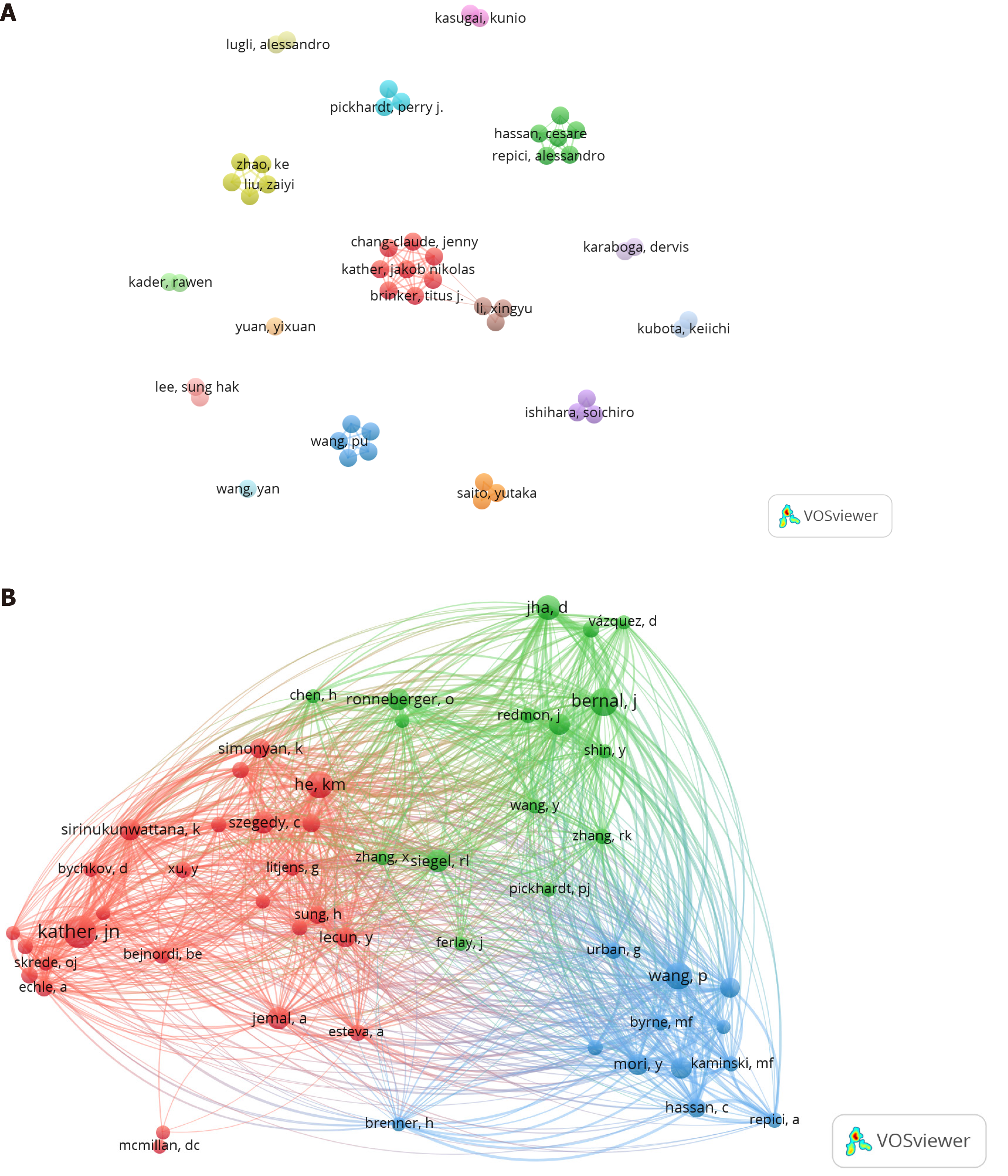
The analysis included 8101 authors. Table 5 shows that among the top ten authors in terms of the number of publications, Kather JN, Lee SH, Liu Z, and Pickhardt PJ, made the most contributions to the area. These researchers hail from Germany, South Korea, China, and the United States. Kather JN, from Germany, had the greatest citation frequency (287 citations) and was the leading researcher in the use of DL in CRC. Figure 6A depicts 16 author collaboration networks in the study area, with central collaborative research between the groups of Kather JN and Li X. Figure 6B shows the correlation between co-cited authors and that Kather JN, Bernal J, and He KM, had the highest citation frequencies.
| Rank | Author | Country | Documents | Co-cited author | Frequency |
| 1 | Kather JN | Germany | 12 | Kather JN | 287 |
| 2 | Lee SH | Korea | 9 | Bernal J | 198 |
| 3 | Liu Z | China | 9 | He KM | 196 |
| 4 | Pickhardt PJ | America | 9 | Wang P | 193 |
| 5 | Brenner H | Germany | 8 | Jha D | 162 |
| 6 | Brinker TJ | Germany | 8 | Jemal A | 134 |
| 7 | Hoffmeister M | Germany | 8 | Ronneberger O | 130 |
| 8 | Aaito Y | Japan | 8 | Siegel RL | 130 |
| 9 | Zhao K | China | 8 | Szegedy C | 130 |
| 10 | Ishihara S | Japan | 7 | Mori Y | 123 |
The included publications cited a total of 40035 references. Table 6 shows the top ten most-cited references. The most-cited reference was Kaiming He (149), who focused on the application of DL in image recognition, followed by Olaf Ronneberger, and Ahmedin Jemal DVM, who had 124 and 117 citations, respectively. These ten references can be divided into three categories: Cancer statistics, DL image recognition, and DL for clinical prediction in gastrointestinal illnesses.
| Rank | Title | Journal | First author | Year | Citations |
| 1 | Deep residual learning for image recognition | 2016 IEEE Conference on Computer Vision and Pattern Recognition | Kaiming He | 2016 | 149 |
| 2 | U-Net: Convolutional networks for biomedical image segmentation | Medical Image Computing and Computer | Olaf Ronneberger | 2015 | 124 |
| 3 | Global cancer statistics | CA: A Cancer Journal for Clinicians | Ahmedin Jemal DVM | 2011 | 117 |
| 4 | Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study | PLoS Medicine | Jakob Nikolas Kather | 2019 | 87 |
| 5 | Very deep convolutional networks for large-scale image recognition | Computer Science | Karen Simonyan | 2015 | 87 |
| 6 | WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs saliency maps from physicians | Computerized Medical Imaging and Graphics | Jorge Bernal | 2015 | 83 |
| 7 | ImageNet classification with deep convolutional neural networks | Research-Article | Alex Krizhevsky | 2017 | 81 |
| 8 | Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer | Brief Communication | Jakob Nikolas Kather | 2019 | 79 |
| 9 | Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy | Gastroenterology | Gregor Urban | 2018 | 75 |
| 10 | Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries | CA: A Cancer Journal for Clinicians | Hyuna Sung | 2021 | 72 |
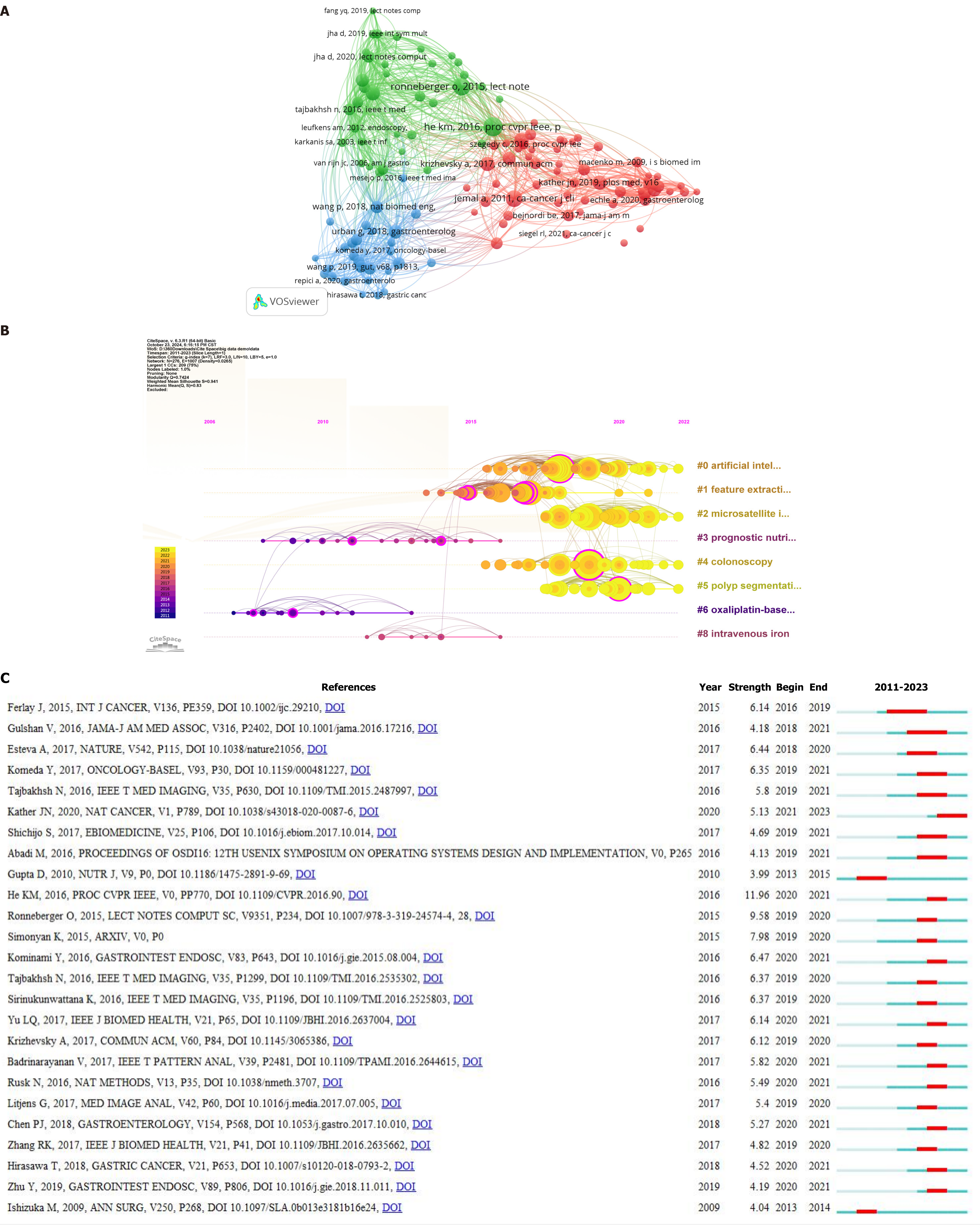
There were 122 references that were cited more than 20 times. References cited more than 20 times were imported into VOSviewer for visual analysis (Figure 7A) and divided into three main clusters: A red cluster (58 articles), which focused on DL in cancer, a green cluster (36 articles), which focused on DL and computers, and a blue cluster (28 articles), which focused on DL and colonoscopy.
A timeline chart (Figure 7B) can be used to show how the references are categorized and distributed over time. The number and types of references cited increased dramatically after 2015 compared with before 2015. The eight clusters revealed that DL was the primary study approach in the publications, with “artificial intelligence” (cluster 0), feature extraction (cluster 1), microsatellites (cluster 2), colonoscopy (cluster 4), and polyp segmentation (cluster 5) being associated. The articles’ outcomes were oriented toward the treatment and prognosis of CRC and were related to oxaliplatin (cluster 6), intravenous iron (cluster 8), and prognostic nutrition (cluster 3). The number of references in these clusters was low, and the citation time was mostly concentrated before 2015. After 2015, there was a transition to CRC diagnostic approaches based on AI, DL and other technologies.
Figure 7C depicts the top 25 papers with the highest number of citations. This figure helps one understand which papers focused on a specific subject over a period of time. The figure shows that the first citation explosion occurred in 2009, with citation intensities ranging from 3.99 to 11.96. The most-cited paper was Deep Residual Learning for Image Recognition by He KM, which presented a residual learning framework to optimize DL approaches.
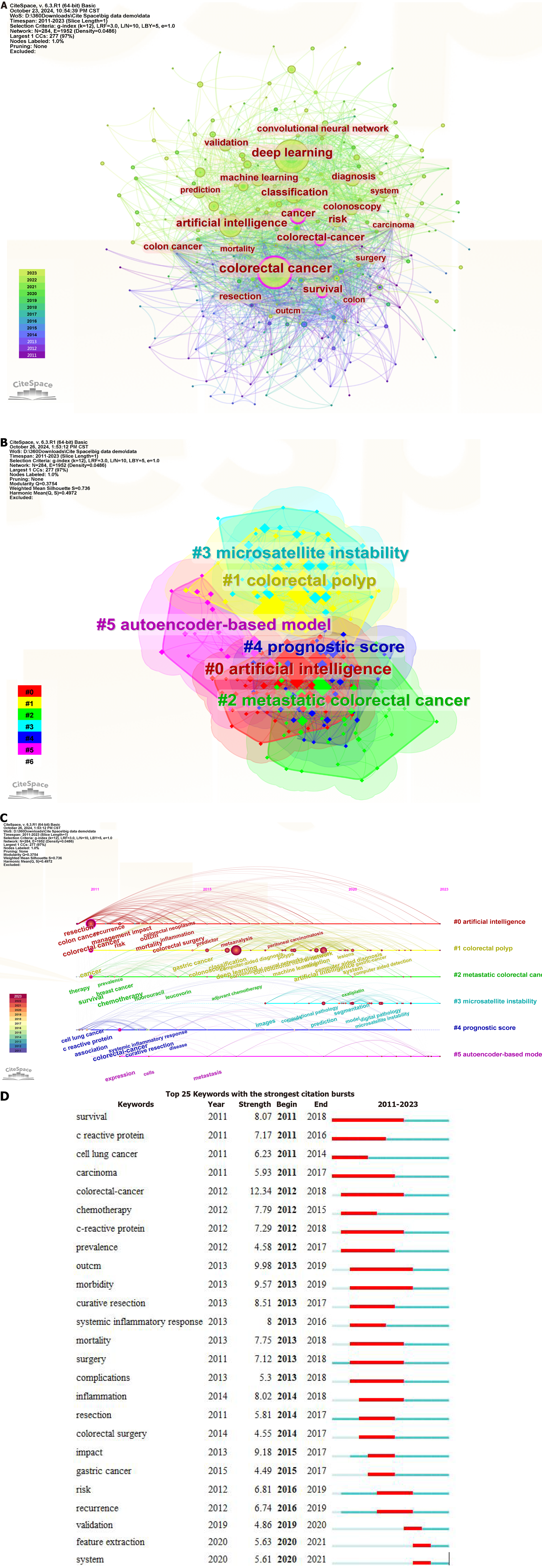
VOSviewer was used to analyze 4869 keywords in 1275 articles, with 81 keywords appearing more than 20 times. Table 7 shows the top ten keywords in terms of frequency of occurrence. In addition, CiteSpace was used to create a keyword co-occurrence chart (Figure 8A). The keywords with the highest frequency of occurrence were “colorectal cancer”, “deep learning”, “artificial intelligence”, “classification” and “cancer”, whereas the keywords with the highest centrality were “colorectal cancer” (0.16), “colon” (0.13), “cancer” (0.12), “survival” (0.11), and “colorectal cancer” (0.10).
| Rank | Keywords | Counts |
| 1 | Deep learning | 354 |
| 2 | Colorectal-cancer | 297 |
| 3 | Colorectal cancer | 293 |
| 4 | Cancer | 172 |
| 5 | Classification | 147 |
| 6 | Artificial intelligence | 144 |
| 7 | Colonoscopy | 136 |
| 8 | Survival | 121 |
| 9 | Risk | 106 |
| 10 | Diagnosis | 98 |
Cluster analysis (Figure 8B) was performed on the terms identified via keyword co-occurrence analysis to better understand the most popular areas of DL application to CRC. Six clusters were categorized: “#0 artificial intelligence”, “#1 colorectal polyps”, “#2 metastatic colorectal cancer”, “#3 microsatellite instability”, “#4 prognostic score” and “#5 autoencoder-based model”. Here, “#0”, “#3”, and “#5” were related to DL, whereas “#1”, “#2”, and “#4” were related to CRC. Figure 8C presents a keyword timeline chart, which reveals that there was less research about “#3 microsatellite instability” from 2011 to 2023 (in fact, only beginning in 2017), whereas there were more studies about “#0 artificial intelligence” and “#1 colorectal polyps”. Figure 8D displays the top 25 keywords with the highest citation explosion rates. The initial citation explosion occurred in 2011, with keyword intensities ranging from 4.49 to 12.34. The keywords with the highest burst intensity (12.34) and the longest length (2011-2018) were “colorectal-cancer” and “survival”, respectively, while the most recent keywords were “validation” (2019-2020), “feature extraction” (2020-2021) and “system” (2020-2021).
With the rapid development of research in numerous domains of medicine, it has become increasingly important for researchers to understand recent achievements in their respective professions and in cross-fertilized areas. Compared with meta-analysis and systematic evaluation methodologies, bibliometric analysis offers a more objective and straightforward visual approach to validating and analyzing the current literature[20].
Number of publications: A search of the Web of Science database revealed that the number of papers on the application of DL in CRC research has been increasing annually. The small number of publications and slow growth between 2011 and 2019 could be attributed to the fact that DL research in the field was still in its early stages. The significant increase in the number of publications from 2019 to 2021, particularly in Q1 and Q2, indicated that DL was undergoing rapid and high-quality progress in the area during this time, with 2019 representing the turning point. The number of publications in Q1 has been increasing and reached 141 in 2023, indicating that high-quality papers will continue to be produced in the future. On the one hand, the occurrence of DL and CNNs in AI domains has resulted in breakthroughs in the processing of complicated data, such as medical imagery[21]. On the other hand, the integration of CRC-related examination modalities, such as imaging, pathology, and colorectoscopy, with DL has aided in the detection and treatment of this disease[6]. Although the number of publications in 2024 is not mentioned, the use of DL in CRC research will remain a future hotspot on the basis of current research trends.
National cooperation: The top three nations in terms of the number of publications on the topic of interest were China, the United States, and Japan; however, Southern Europe, New Zealand, and Eastern Europe had the highest global incidence rates of CRC in 2022[22]. The disparity between the incidence rate and the ranking of the number of publications could be attributed to a variety of factors, including countries’ dietary patterns, population size, and scientific research support. The United States established cooperation networks or ties with dozens of countries in this subject area, whereas China was relatively weak, suggesting that Chinese scholars in this field can strengthen cooperation and exchange with other countries in their subsequent research.
Institutional distribution: China hosted four of the top ten institutions in terms of publishing volume, followed by Korea and Germany, with two each, and the United States and Japan, with one each. The large number of Chinese research institutions in this distribution may be because China has had the highest number of new CRC cases in recent years, which has piqued the interest of researchers[23]. Harvard University in the United States was more central to research in the field of DL applied to CRC, serving as an intermediary, and might be an important guide with respect to new ideas and research topics in this field. However, there were fewer research institutions in the United States, which might encourage the government there to increase investment support for this field.
Journal-related hotspots: Medical Image Analysis had the highest impact factor and the second-highest number of citations, indicating that it has been publishing high-quality, high-impact research on our subject. Scientific Reports had the most publications and citations, which suggests that scholars will continue to choose it as a venue for future publications in the field. Among the co-cited journals, Gastrointestinal Endoscopy had the most citations, and the New England Journal of Medicine had the highest impact factor, which should encourage researchers to pay more attention to these highly cited journals with high impact factors when citing the literature, thereby promoting field development. Additionally, researchers can consider journal publishers such as Elsevier, Springer, MDPI, Wiley, and IEEE as options. The distribution of journals publishing papers in Figure 5A was unbalanced, which suggests that certain journals could broaden the scope of their publications to include DL or CRC in the future.
Author-related hotspots: In terms of authors, Kather JN, had the most publications. Kather is from Germany and works at Memorial Sloan Kettering Cancer Center; his research has focused on the use of DL in cancer treatment[9,24]. Furthermore, he was the author with the most citations for his publications, indicating his great influence in the field of DL applied to CRC, a fact that could encourage other researchers to strengthen their grasp of the field by reading his publications. The author collaboration network in Figure 6A shows that research in this field has been scattered, with fewer connections between research groups. Research collaboration between groups should be strengthened in the future.
Reference hotspots: The top ten references cited reflects current trends in DL regarding the diagnosis and treatment of CRC, with the majority of these studies adopting DL for image identification and gastrointestinal disease prediction. Among them, “Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study” used CNN tissue slice image identification to predict colorectal rectal cancer survival[25]. Another study, entitled “Deep Residual Learning for Image Recognition”, focused on DL and investigated a residual learning framework to simplify DL neural networks[26]. The reference timeline graph clearly demonstrates the progression of the field over time. This graph enables academics to better grasp the research activity in the field and its importance in different eras. After 2015, the focus of research shifted to AI, feature extraction and colorectoscopy, with a greater emphasis on the early diagnosis and treatment of CRC, replacing the pre-2015 focus on the treatment of CRC[27,28]. This shift should not only improve quality of life for patients but also save major public health expenses and resources. “Deep Residual Learning for Image Recognition” had the highest citation explosion rate, implying that scholars can better find key material and track academic trends in this discipline than previously.
Keywords hotspots: The keywords that appeared more frequently in the keyword co-occurrence chart were “colorectal cancer”, “deep learning”, “artificial intelligence”, “classification” and “cancer”, demonstrating that DL and classification were hotspots in CRC research. According to the term clustering analysis, the largest clusters were “#0 artificial intelligence”, “#1 colorectal polyps” and “#2 metastatic colorectal cancer”. A family history of colorectal polyps is a risk factor for CRC, and individuals with early-stage CRC sometimes exhibit no conventional clinical symptoms or only vague indicators, resulting in a poor incidence of early diagnosis[29,30]. Dysregulation of metabolic reprogramming promotes tumor cell proliferation, metastasis, and resistance to therapy, and the majority of carbon units are generated by nucleotide biosynthesis via the serine-glycine-one-carbon pathway and the pentose phosphate pathway, facilitating the timely replication of DNA, which promotes cancer cell proliferation and resistance to chemotherapy, thus increasing the risk of metastasis and recurrence[31]. As a result, the application of AI, such as DL, to imaging, pathology and colorectoscopy devices to improve the sensitivity of CRC screening and diagnosis to enhance patient prognosis is a popular topic among researchers, which is consistent with our study. The timeline chart analysis demonstrates that AI in CRC is increasingly transitioning to “#3 microsatellite instability” and “#5 autoencoder-based model”, which can provide better informed and more accurate guidance for early clinical diagnosis and treatment decisions. The duration of the keyword burst was longer before 2019 and shorter after 2019, indicating that DL in the CRC field entered a rapid development phase after 2019, possibly due to detection equipment updates and rapid AI development.
DL is important in the diagnosis, classification, staging and prognosis of CRC patients. Wagner et al[9] revealed that transformer-based DL outperformed other methods in predicting CRC biomarker status, which can increase diagnostic speed and accuracy. Modern deep neural networks can perform tasks beyond standard visual interpretation, advancing the discipline’s ability to contribute to precision oncology and potentially simplifying diagnosis in the future[32]. Alalwani et al[33] discovered that DL models have been used to classify colorectal tumors utilizing a number of data types, most notably histopathological and endoscopic imagery, with the majority of researchers employing CNNs as their classification model. Lu et al[34] developed a DL system for CRC patients in which contrast-enhanced computed tomography imaging was used to predict the disease stage before surgery. A retrospective, multicenter investigation[17] revealed that the predictive efficacy of DL-based risk scores was independent of identified clinical risk markers and could robustly predict clinical outcomes in CRC patients. Computed tomography-based DL models can help radiologists in early screening, identification detection, localization of lesions, tumor staging, and prognosis associated with CRC[13,34-37]; pathohistology-based DL models can help pathologists in diagnostic identification, lymph node metastasis, gene mutation, tumor classification, and prognosis associated with CRC[25,38-41]; and colorectoscopy-based DL models can help gastroenterologists detect colorectal polyps and the depth of submucosal infiltration in CRC[42,43]. Continued bibliographic analysis on the topic will provide valuable insights into current trends and emerging areas of interest in the field.
This study has several significant drawbacks. First, we included only relevant literature from the Web of Science Core Collection, not from other databases. Second, our investigation period stopped at the end of December 2023, and therefore studies from 2024 were not included, possibly causing us to miss the most recent findings. These issues could be sources of bias in the study.
This paper highlights the current state of research on and popular applications of DL in CRC worldwide. In recent years, the number of publications in this sector has continuously increased. To date, China and the United States have made considerable contributions in this subject area, and they are expected to remain leaders in the future. The use of DL in CRC diagnosis and prognosis is highly important, particularly for DL models based on CNNs and transformers. To further expand the potential of DL applications in research on CRC, institutions and authors must strengthen international and multicenter exchange and cooperation.
The authors would like to thank the editors and the anonymous reviewers for their valuable comments and suggestions regarding how to improve this paper.
| 1. | Scanu AM, De Miglio MR. Therapeutic Landscapes in Colorectal Carcinoma. Medicina (Kaunas). 2023;59:821. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 2. | Li HM, Liu Y, Hao MD, Liang XQ, Yuan DJ, Huang WB, Li WJ, Ding L. Research status and hotspots of tight junctions and colorectal cancer: A bibliometric and visualization analysis. World J Gastrointest Oncol. 2024;16:3705-3715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 3. | Zhang X, Yang L, Liu S, Cao LL, Wang N, Li HC, Ji JF. [Interpretation on the report of global cancer statistics 2022]. Zhonghua Zhong Liu Za Zhi. 2024;46:710-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 4. | Acs B, Rantalainen M, Hartman J. Artificial intelligence as the next step towards precision pathology. J Intern Med. 2020;288:62-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 224] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 5. | Rompianesi G, Pegoraro F, Ceresa CD, Montalti R, Troisi RI. Artificial intelligence in the diagnosis and management of colorectal cancer liver metastases. World J Gastroenterol. 2022;28:108-122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 71] [Cited by in RCA: 72] [Article Influence: 24.0] [Reference Citation Analysis (3)] |
| 6. | Bousis D, Verras GI, Bouchagier K, Antzoulas A, Panagiotopoulos I, Katinioti A, Kehagias D, Kaplanis C, Kotis K, Anagnostopoulos CN, Mulita F. The role of deep learning in diagnosing colorectal cancer. Prz Gastroenterol. 2023;18:266-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 7. | Wu Y, Li Y, Xiong X, Liu X, Lin B, Xu B. Recent advances of pathomics in colorectal cancer diagnosis and prognosis. Front Oncol. 2023;13:1094869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 8. | Wong PK, Chan IN, Yan HM, Gao S, Wong CH, Yan T, Yao L, Hu Y, Wang ZR, Yu HH. Deep learning based radiomics for gastrointestinal cancer diagnosis and treatment: A minireview. World J Gastroenterol. 2022;28:6363-6379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (1)] |
| 9. | Wagner SJ, Reisenbüchler D, West NP, Niehues JM, Zhu J, Foersch S, Veldhuizen GP, Quirke P, Grabsch HI, van den Brandt PA, Hutchins GGA, Richman SD, Yuan T, Langer R, Jenniskens JCA, Offermans K, Mueller W, Gray R, Gruber SB, Greenson JK, Rennert G, Bonner JD, Schmolze D, Jonnagaddala J, Hawkins NJ, Ward RL, Morton D, Seymour M, Magill L, Nowak M, Hay J, Koelzer VH, Church DN; TransSCOT consortium, Matek C, Geppert C, Peng C, Zhi C, Ouyang X, James JA, Loughrey MB, Salto-Tellez M, Brenner H, Hoffmeister M, Truhn D, Schnabel JA, Boxberg M, Peng T, Kather JN. Transformer-based biomarker prediction from colorectal cancer histology: A large-scale multicentric study. Cancer Cell. 2023;41:1650-1661.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 96] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 10. | Huang P, Feng Z, Shu X, Wu A, Wang Z, Hu T, Cao Y, Tu Y, Li Z. A bibliometric and visual analysis of publications on artificial intelligence in colorectal cancer (2002-2022). Front Oncol. 2023;13:1077539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 11. | Meng G, Xu H, Yang S, Chen F, Wang W, Hu F, Zheng G, Guo Y. Bibliometric analysis of worldwide research trends on breast cancer about inflammation. Front Oncol. 2023;13:1166690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 12. | Cai M, Ni Z, Yuan Z, Yu J, Zhang D, Yao R, Zhou L, Yu C. Past and present: a bibliometric study on polycystic ovary syndrome. J Ovarian Res. 2023;16:42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 13. | Yao L, Li S, Tao Q, Mao Y, Dong J, Lu C, Han C, Qiu B, Huang Y, Huang X, Liang Y, Lin H, Guo Y, Liang Y, Chen Y, Lin J, Chen E, Jia Y, Chen Z, Zheng B, Ling T, Liu S, Tong T, Cao W, Zhang R, Chen X, Liu Z. Deep learning for colorectal cancer detection in contrast-enhanced CT without bowel preparation: a retrospective, multicentre study. EBioMedicine. 2024;104:105183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 14. | Bedrikovetski S, Dudi-Venkata NN, Kroon HM, Seow W, Vather R, Carneiro G, Moore JW, Sammour T. Artificial intelligence for pre-operative lymph node staging in colorectal cancer: a systematic review and meta-analysis. BMC Cancer. 2021;21:1058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 95] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 15. | Li X, Jonnagaddala J, Cen M, Zhang H, Xu S. Colorectal Cancer Survival Prediction Using Deep Distribution Based Multiple-Instance Learning. Entropy (Basel). 2022;24:1669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Wang R, Dai W, Gong J, Huang M, Hu T, Li H, Lin K, Tan C, Hu H, Tong T, Cai G. Development of a novel combined nomogram model integrating deep learning-pathomics, radiomics and immunoscore to predict postoperative outcome of colorectal cancer lung metastasis patients. J Hematol Oncol. 2022;15:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 147] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 17. | Jiang X, Hoffmeister M, Brenner H, Muti HS, Yuan T, Foersch S, West NP, Brobeil A, Jonnagaddala J, Hawkins N, Ward RL, Brinker TJ, Saldanha OL, Ke J, Müller W, Grabsch HI, Quirke P, Truhn D, Kather JN. End-to-end prognostication in colorectal cancer by deep learning: a retrospective, multicentre study. Lancet Digit Health. 2024;6:e33-e43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 33] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 18. | Repici A, Badalamenti M, Maselli R, Correale L, Radaelli F, Rondonotti E, Ferrara E, Spadaccini M, Alkandari A, Fugazza A, Anderloni A, Galtieri PA, Pellegatta G, Carrara S, Di Leo M, Craviotto V, Lamonaca L, Lorenzetti R, Andrealli A, Antonelli G, Wallace M, Sharma P, Rosch T, Hassan C. Efficacy of Real-Time Computer-Aided Detection of Colorectal Neoplasia in a Randomized Trial. Gastroenterology. 2020;159:512-520.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 393] [Article Influence: 78.6] [Reference Citation Analysis (0)] |
| 19. | Xiaojian Y, Zhanbo Q, Jian C, Zefeng W, Jian L, Jin L, Yuefen P, Shuwen H. Deep learning application in prediction of cancer molecular alterations based on pathological images: a bibliographic analysis via CiteSpace. J Cancer Res Clin Oncol. 2024;150:467. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 20. | Wang X, Cao X, Dai Z, Dai Z. Bibliometric analysis and visualisation of research hotspots and frontiers on omics in osteosarcoma. J Cancer Res Clin Oncol. 2024;150:393. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 21. | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36149] [Cited by in RCA: 20060] [Article Influence: 2006.0] [Reference Citation Analysis (0)] |
| 22. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 8189] [Article Influence: 8189.0] [Reference Citation Analysis (2)] |
| 23. | Yang Y, Han Z, Li X, Huang A, Shi J, Gu J. Epidemiology and risk factors of colorectal cancer in China. Chin J Cancer Res. 2020;32:729-741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 24. | Shmatko A, Ghaffari Laleh N, Gerstung M, Kather JN. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat Cancer. 2022;3:1026-1038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 205] [Article Influence: 68.3] [Reference Citation Analysis (0)] |
| 25. | Kather JN, Krisam J, Charoentong P, Luedde T, Herpel E, Weis CA, Gaiser T, Marx A, Valous NA, Ferber D, Jansen L, Reyes-Aldasoro CC, Zörnig I, Jäger D, Brenner H, Chang-Claude J, Hoffmeister M, Halama N. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019;16:e1002730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 555] [Cited by in RCA: 492] [Article Influence: 82.0] [Reference Citation Analysis (0)] |
| 26. | He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27-30; Las Vegas, NV, United States. United States: IEEE, 2016. |
| 27. | Ai D, Wang Y, Li X, Pan H. Colorectal Cancer Prediction Based on Weighted Gene Co-Expression Network Analysis and Variational Auto-Encoder. Biomolecules. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 28. | Wang Y, He X, Nie H, Zhou J, Cao P, Ou C. Application of artificial intelligence to the diagnosis and therapy of colorectal cancer. Am J Cancer Res. 2020;10:3575-3598. [PubMed] |
| 29. | Duan B, Zhao Y, Bai J, Wang J, Duan X, Luo X, Zhang R, Pu Y, Kou M, Lei J, Yang S. Colorectal Cancer: An Overview. In: Morgado-Diaz JA, editor. Gastrointestinal Cancers [Internet]. Brisbane (AU): Exon Publications; 2022. [PubMed] |
| 30. | Montminy EM, Jang A, Conner M, Karlitz JJ. Screening for Colorectal Cancer. Med Clin North Am. 2020;104:1023-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 31. | Zou S, Qin B, Yang Z, Wang W, Zhang J, Zhang Y, Meng M, Feng J, Xie Y, Fang L, Xiao L, Zhang P, Meng X, Choi HH, Wen W, Pan Q, Ghesquière B, Lan P, Lee MH, Fang L. CSN6 Mediates Nucleotide Metabolism to Promote Tumor Development and Chemoresistance in Colorectal Cancer. Cancer Res. 2023;83:414-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 32. | Ruusuvuori P, Valkonen M, Latonen L. Deep learning transforms colorectal cancer biomarker prediction from histopathology images. Cancer Cell. 2023;41:1543-1545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 33. | Alalwani R, Lucas A, Alzubaidi M, Shah HA, Alam T, Shah Z, Househ M. Deep Learning in Colorectal Cancer Classification: A Scoping Review. Stud Health Technol Inform. 2023;305:616-619. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 34. | Lu N, Guan X, Zhu J, Li Y, Zhang J. A Contrast-Enhanced CT-Based Deep Learning System for Preoperative Prediction of Colorectal Cancer Staging and RAS Mutation. Cancers (Basel). 2023;15:4497. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 35. | Wesp P, Grosu S, Graser A, Maurus S, Schulz C, Knösel T, Fabritius MP, Schachtner B, Yeh BM, Cyran CC, Ricke J, Kazmierczak PM, Ingrisch M. Deep learning in CT colonography: differentiating premalignant from benign colorectal polyps. Eur Radiol. 2022;32:4749-4759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 36. | Sahoo PK, Gupta P, Lai YC, Chiang SF, You JF, Onthoni DD, Chern YJ. Localization of Colorectal Cancer Lesions in Contrast-Computed Tomography Images via a Deep Learning Approach. Bioengineering (Basel). 2023;10:972. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 37. | Li CH, Cai D, Zhong ME, Lv MY, Huang ZP, Zhu Q, Hu C, Qi H, Wu X, Gao F. Multi-Size Deep Learning Based Preoperative Computed Tomography Signature for Prognosis Prediction of Colorectal Cancer. Front Genet. 2022;13:880093. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 38. | Yu G, Sun K, Xu C, Shi XH, Wu C, Xie T, Meng RQ, Meng XH, Wang KS, Xiao HM, Deng HW. Accurate recognition of colorectal cancer with semi-supervised deep learning on pathological images. Nat Commun. 2021;12:6311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 69] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 39. | Chuang WY, Chen CC, Yu WH, Yeh CJ, Chang SH, Ueng SH, Wang TH, Hsueh C, Kuo CF, Yeh CY. Identification of nodal micrometastasis in colorectal cancer using deep learning on annotation-free whole-slide images. Mod Pathol. 2021;34:1901-1911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 40. | Liu Y, Huang K, Yang Y, Wu Y, Gao W. Prediction of Tumor Mutation Load in Colorectal Cancer Histopathological Images Based on Deep Learning. Front Oncol. 2022;12:906888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 41. | Haj-Hassan H, Chaddad A, Harkouss Y, Desrosiers C, Toews M, Tanougast C. Classifications of Multispectral Colorectal Cancer Tissues Using Convolution Neural Network. J Pathol Inform. 2017;8:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 42. | Sánchez-Peralta LF, Bote-Curiel L, Picón A, Sánchez-Margallo FM, Pagador JB. Deep learning to find colorectal polyps in colonoscopy: A systematic literature review. Artif Intell Med. 2020;108:101923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 43. | Minami S, Saso K, Miyoshi N, Fujino S, Kato S, Sekido Y, Hata T, Ogino T, Takahashi H, Uemura M, Yamamoto H, Doki Y, Eguchi H. Diagnosis of Depth of Submucosal Invasion in Colorectal Cancer with AI Using Deep Learning. Cancers (Basel). 2022;14:5361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |