Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.1995
Peer-review started: January 18, 2024
First decision: January 26, 2024
Revised: February 2, 2024
Accepted: March 13, 2024
Article in press: March 13, 2024
Published online: May 15, 2024
Processing time: 111 Days and 23.8 Hours
Limited knowledge exists regarding the casual associations linking blood metabolites and the risk of developing colorectal cancer.
To investigate causal associations between blood metabolites and colon cancer.
The study utilized a two-sample Mendelian randomization (MR) analysis to investigate the causal impact of 486 blood metabolites on colorectal cancer. The primary method of analysis used was the inverse variance weighted model. To further validate the results several sensitivity analyses were performed, including Cochran's Q test, MR-Egger intercept test, and MR robust adjusted profile score. These additional analyses were conducted to ensure the reliability and robustness of the findings.
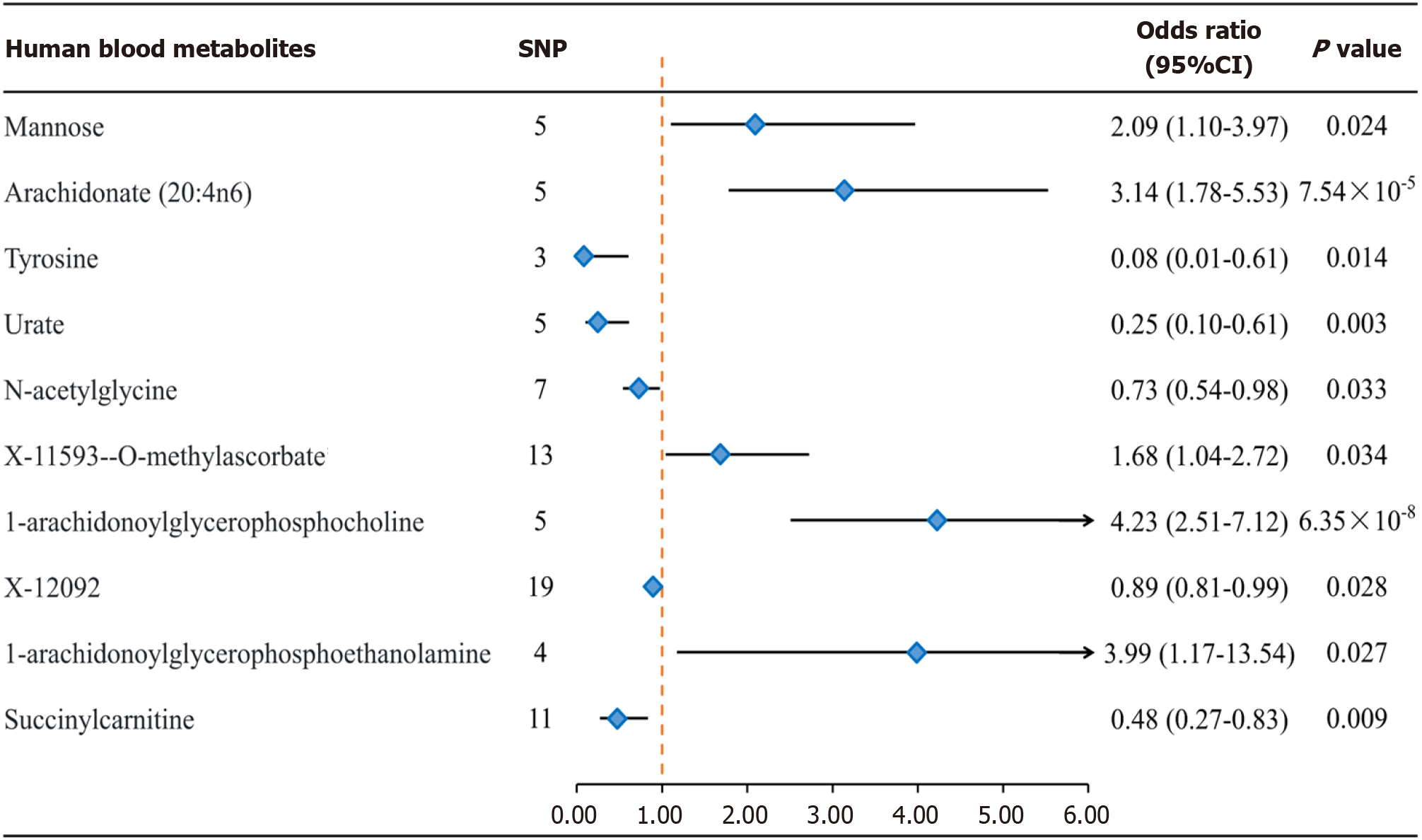
After rigorous selection for genetic variation, 486 blood metabolites were included in the MR analysis. We found Mannose [odds ratio (OR) = 2.09 (1.10-3.97), P = 0.024], N-acetylglycine [OR = 3.14 (1.78-5.53), P = 7.54 × 10-8], X-11593-O-methy
This study showed a causal relationship between 10 blood metabolites and colorectal cancer, of which 5 blood metabolites were found to be causal for the development of colorectal cancer and were confirmed as risk factors. The other five blood metabolites are protective factors.
Core Tip: The study utilized a two-sample Mendelian randomization analysis to investigate the causal impact of 486 blood metabolites on colorectal cancer. The primary method of analysis used was the inverse variance weighted model. To further validate the results several sensitivity analyses were performed. Our findings showed a causal relationship between 10 blood metabolites and colorectal cancer, of which 5 blood metabolites were found to be causal for the development of colorectal cancer and were confirmed as risk factors.
- Citation: Hu KY, Cheng YQ, Shi ZL, Ren FP, Xiao GF. Casual associations between blood metabolites and colon cancer. World J Gastrointest Oncol 2024; 16(5): 1995-2005
- URL: https://www.wjgnet.com/1948-5204/full/v16/i5/1995.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i5.1995
Colorectal cancer is globally ranked as the third most prevalent type of cancer, and it is the second leading cause of cancer-related mortality. In 2018, there were approximately 18 million new cases reported, resulting in 860000 deaths[1]. Projections based on population data suggest that the annual burden of colorectal cancer will exceed 3 million new cases and 16 million deaths by 2040[1-3]. Disparities in colorectal cancer incidence between countries and insights obtained from international migration studies have indicated a potential correlation between diet, lifestyle factors, and the development of the disease[3].
Metabolites, serving as substrates and products of metabolism, are indispensable cellular components. They not only drive fundamental cellular activities but also play a critical role as functional intermediates in predicting or influencing the onset and progression of diseases[4-6]. Observational studies have identified notable differences in blood metabolites between colorectal cancer patients and healthy individuals, primarily involving inflammation-related pathways, amino acids, and lipid metabolism[5]. For instance, Zhao et al[7] conducted a study that utilized liquid chromatography mass spectrometry (LC-MS/MS) metabolomics to examine small metabolites in serum samples from colorectal cancer patients. The results indicated a significant increase in S-(3-methylbutyryl)-dihydrolipoamide-E and N-nonyl-glycine, while S-phenyl-d-cysteine demonstrated a substantial decrease in colorectal cancer patients compared to the control group. However, traditional observational studies are prone to confounding factors and reverse causality, which have resulted in ongoing debates concerning the causal relationship between colorectal cancer and blood metabolites.
Mendelian Randomization (MR) is a statistical method used to assess causality in diseases of interest by utilizing single nucleotide polymorphisms (SNPs) as instrumental variables (IVs) for associated risk factors[8,9]. This method estimates the causal relationship between an exposure and an outcome based on genetic variation[10]. Similar to randomized controlled trials[10,11], genetic variants are randomly assigned to offspring along with gametes before the development of disease. As a result, they are less susceptible to confounding factors and reverse causality. While MR studies have been conducted on blood metabolites in various diseases, no studies have been performed on colorectal cancer.
This study utilized the metabolite database from a highly comprehensive metabolite. study for exposure assessment A systematic two-sample MR analysis was employed, using the GECCO, CORECT, CCFR, and other European cohorts' genome-wide association studies (GWAS) data as the phenotypic data for colorectal cancer[12]. The study extensively examines the causal association between 486 blood metabolites and colorectal cancer, thereby providing insights into the etiology and pathogenesis of metabolic-related colorectal cancer. These findings have important implications for risk prediction and treatment approaches.
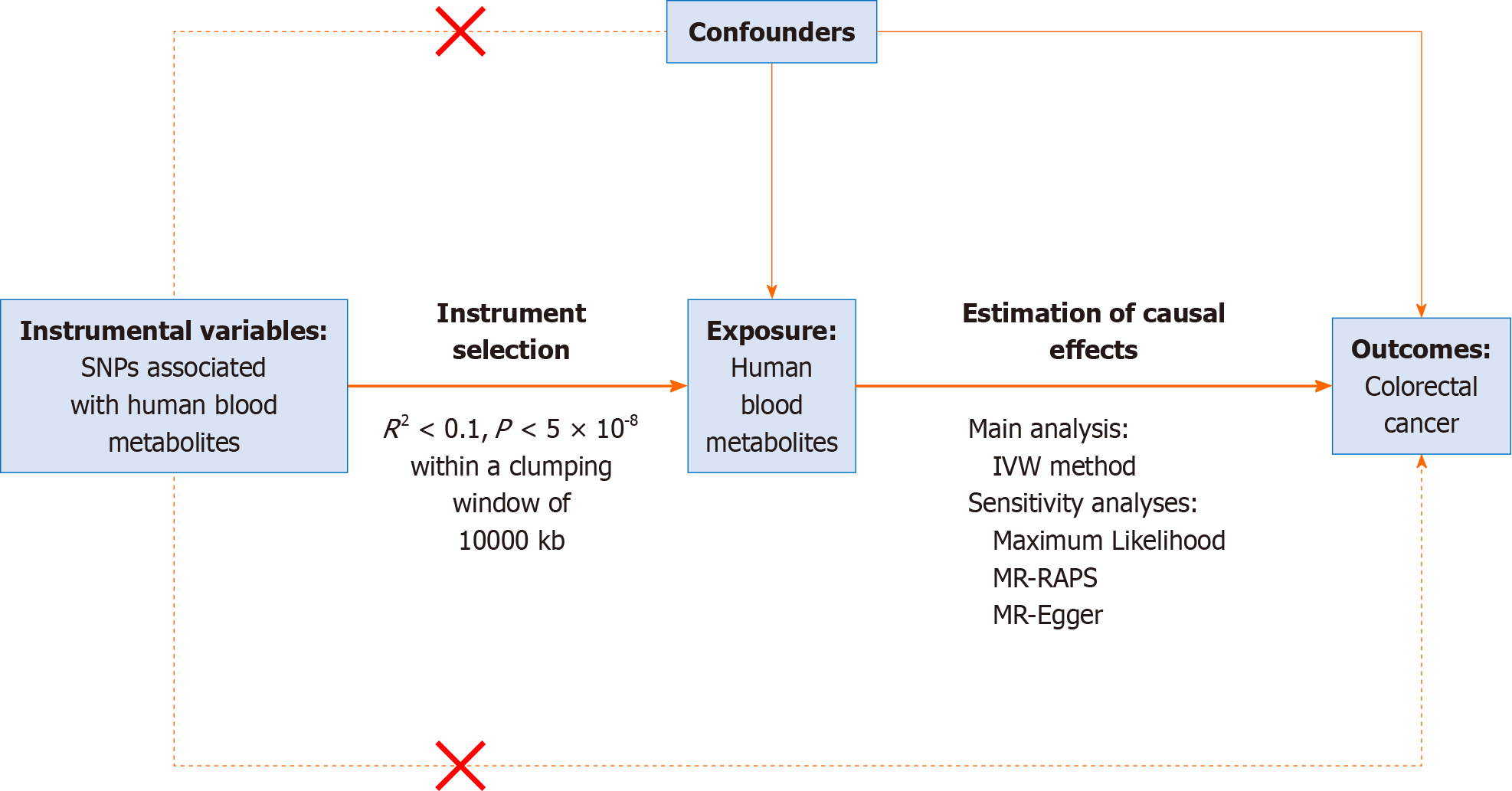
A two-sample MR analysis was conducted to assess the causal impact of human blood metabolites on the risk of colorectal cancer, using summary statistics obtained from GWAS. Three assumptions needed to be met by the chosen: IVs (1) They must exhibit a strong association with the exposure of interest, i.e., human blood metabolites; (2) The IVs must be independent of unobserved confounders; and (3) The IVs must have a relationship with the outcome, i.e., colorectal cancer, solely through their influence on the exposure of interest, rather than via confounding factors. In this MR study, human blood metabolites were considered as the exposure, while colorectal cancer was treated as the outcome. The entire process is depicted in the flow chart shown in Figure 1.
This study utilizes MR to investigate the causal associations between blood metabolites in humans (exposure) and colorectal cancer (outcome). The study assumes that the IVs are specifically related to metabolites, and do not have any connection to confounding factors. Additionally, it assumes that the IVs are not associated with the risk of developing colorectal cancer with respect to both metabolites and confounding factors. Ethical approval was obtained from the FinnGen steering committee for all selected GWASs in the FinnGen Consortium, and individuals provided written informed consent.
The metabolite database used in this study was obtained from a comprehensive metabolite study conducted by Tan et al[13] The study utilized GWAS data of human blood metabolites, which was obtained from the metabolomics GWAS server (https://metabolomics.helmholtz-muenchen.de/gwas/) The study cohort consisted of 7824 individuals of European descent, and a total of 2.1 million SNPs were tested for 486 different metabolites. Among these metabolites, 177 were classified as unknown. In addition, 309 metabolites were classified based on their chemistry using the Kyoto Encyclopedia of Genes and Genomes database. These metabolites were further assigned to eight broad metabolomes, which include amino acids, peptides, lipids, cofactors and vitamins, carbohydrates, energy, nucleotides, and xenobiotics (Tables 1 and 2).
| Ref. | Study cohort | Cohort description | Case/control | GWAS sample size | Population | Blood metabolites | |
| Exposure | |||||||
| Shin et al[33], 2014 | Twins United Kingdom | An adult twin British registry cohort study | - | 7824 | German | European | 453 |
| KORA | Population-based cohort studies | British | |||||
| Outcome | |||||||
| Huyghe et al [12], 2019 | GECCO, CORECT, CCFR, etc. | Cohort studies; Case-control studies | 19948/12124 | 32072 | European | - | |
| Metabolites | Maximum likelihood | MR-RAPS | MR-egger intercept | |||
| OR (95%CI) | P value | OR (95%CI) | P value | OR (95%CI) | P value | |
| Mannose | 2.10 (1.10-4.02) | 0.024 | 2.10 (1.10-4.03) | 0.025 | 1.02 (0.97-1.08) | 0.500 |
| Arachidonate (20:4n6) | 3.19 (1.79-5.68) | 8.48 × 10-5 | 3.19 (1.79-5.69) | 8.60 × 10-5 | 0.99 (0.90-1.10) | 0.868 |
| Tyrosine | 0.08 (0.01-0.63) | 0.016 | 0.08 (0.01-0.67) | 0.019 | 0.93 (0.74-1.15) | 0.491 |
| Urate | 0.25 (0.10-0.62) | 0.003 | 0.25 (0.10-0.62) | 0.003 | 1.01 (0.97-1.05) | 0.586 |
| N-acetylglycine | 0.73 (0.54-0.98) | 0.034 | 0.73 (0.54-0.98) | 0.036 | 0.97 (0.94-1.00) | 0.095 |
| X-11593-O-methylascorbate | 1.72 (1.06-2.80) | 0.028 | 1.69 (1.04-2.76) | 0.034 | 1.00 (0.97-1.02) | 0.871 |
| 1-arachidonoylglycerophosphocholine | 4.26 (2.49-7.27) | 1.13 × 10-7 | 4.26 (2.48-7.31) | 1.47 × 10-7 | 0.98 (0.95-1.02) | 0.365 |
| X-12092 | 0.89 (0.81-0.99) | 0.027 | 0.89 (0.81-0.99) | 0.028 | 1.00 (0.98-1.02) | 0.952 |
| 1-arachidonoylglycerophosphoethanolamine | 4.13 (1.91-8.91) | 3.04 × 10-4 | 4.13 (1.92-8.86) | 2.76 × 10-4 | 0.98 (0.88-1.10) | 0.756 |
| Succinylcarnitine | 0.48 (0.27-0.84) | 0.011 | 0.47 (0.27-0.84) | 0.010 | 1.02 (0.99-1.04) | 0.181 |
Summary statistics of colorectal cancer phenotypes were obtained from a GWAS conducted[12,14] The study utilized cohorts from various populations, including GECCO, CORECT, CCFR, and other European populations. The GWAS data comprised a total of 19948 cases and 12124 controls, yielding a final dataset of 32072 samples. The research data sources are publicly available in the IEU GWAS database (https://gwas.mrcieu.ac.uk), and further details can be found in Table 1[12].
In this MR study, we adjusted the association threshold to P < 1.0 × 10-5, in accordance with the findings of Cai et al[15]. For the investigation of metabolites, we identified eleven SNPs that exhibited no linkage disequilibrium with other SNPs
We conducted a MR analysis to assess the causal association between blood metabolites and colorectal cancer. For the main analysis, we employed the standard inverse variance weighted (IVW) method, which combines the Wald ratios of individual SNPs with outcomes to obtain pooled estimates of causality. This method considers potential overdispersion and is widely recognized and utilized in the field of MR analysis.
In addition, several other MR analyses have been used as complementary approaches to the IVW method. These include MR-Egger regression and the weighted median method, which aim to improve the robustness of estimates in a wider range of scenarios. MR-Egger regression is particularly useful for assessing and testing unbalanced pleiotropy and substantial heterogeneity, although it requires a larger sample size compared to IVW to achieve the same of level precision in assessing underexposure variation. On the other hand, the weighted median method can provide consistent effect estimates as long as at least half of the weighted variance is valid, even in the presence of horizontal pleiotropy.
Sensitivity analysis plays a crucial role in MR studies as it helps to identify potential genetic polymorphisms and heterogeneity in MR estimates. In order to achieve this, we conducted additional analyses using maximum likelihood, MR-RAPS, and MR-Egger intercept tests with the aim of detecting the presence of pleiotropy and evaluating the robustness of our findings. To assess the reliability of the causal estimation assumed by the IVW method, we employed MR-Egger[16], maximum likelihood[17], and robust adjusted Profile score (MR-RAPS)[18]. Moreover, to examine heterogeneity among SNPs, we utilized Cochran Q values obtained from the IVW and MR-Egger models. Consistent with common practice, we set a significance threshold of P < 0.05 to indicate the presence of significant heterogeneity. Lastly, to evaluate potential horizontal pleiotropy, we utilized MR-Egger regression, where an intercept close to 0 and a P-value greater than 0.05 suggest the absence of pleiotropy in the SNPs.
The identification of eligible candidate metabolites related to colorectal cancer was based on the following criteria: (1) A significant agreement among the three MR Methods in terms of direction and magnitude (all P < 0.05); (2) the absence of heterogeneity; and (3) the absence of pleiotropy at any level. All analyses were performed using the R package TwoSampleMR (version 0.5.6) in R (version 4.0.0).
After a comprehensive quality control process, IVW identified a total of 104 IVs associated with colorectal cancer (Supplementary Tables 1 and 2). The number of SNPs ranged from 5 to 19 for each metabolite, with all F values > 10 considered for SNPs. After applying the Bonferroni correction, the IVW analysis revealed evidence of association between colorectal cancer and only 10 metabolites (Figures 2 and 3), and these results remained robust even after supplementary and sensitivity analyses. Among the known metabolites, 5 showed a positive correlation with colorectal cancer, while 5 showed a negative correlation. Specifically, genetically determined high levels of Mannose [odds ratio (OR) = 2.09 (1.10-3.97), P = 0.024], N-acetylglycine [OR = 0.73 (0.54-0.98), P = 0.033], X-11593-O-methylascorbate [OR = 1.68 (1.04-2.72), P = 0.034], 1-arachidonoylglycerophosphocholine [OR = 4.23 (2.51-7.12), P = 6.35 × 10-8], and 1-arachidonoylglycerophosphoethanolamine 4 [OR = 0.48 (0.27-0.83), P = 0.027] were associated with the occurrence and development of colorectal cancer. Additionally, the IVW method identified genetically determined high levels of Arachidonate (20:4n6) [OR = 3.14 (1.78-5.53), P = 7.54 × 10-5], Tyrosine [OR = 0.08 (0.01-0.61), P = 0.015] did not have a causal relationship with colorectal cancer. Urate [OR = 0.25 (0.10-0.61), P = 0.003], X-12092 [OR = 0.89 (0.81-0.99), P = 0.028], and Succinylcarnitine [OR = 0.48 (0.27-0.83), P = 0.009] also did not show a causal relationship with colorectal cancer. Notably, the strongest positive correlation was observed between 1-arachidonoylglycerophosphocholine [OR (95%CI): 4.23 (2.51-7.12)] and 1-arachidonoylglycerophosphoethanolamine [OR (95%CI): 3.99 (1.17-13.54)], while the strongest negative correlation was observed for Tyrosine [OR (95%CI): 0.08 (0.01-0.61); Figures 2 and 3].
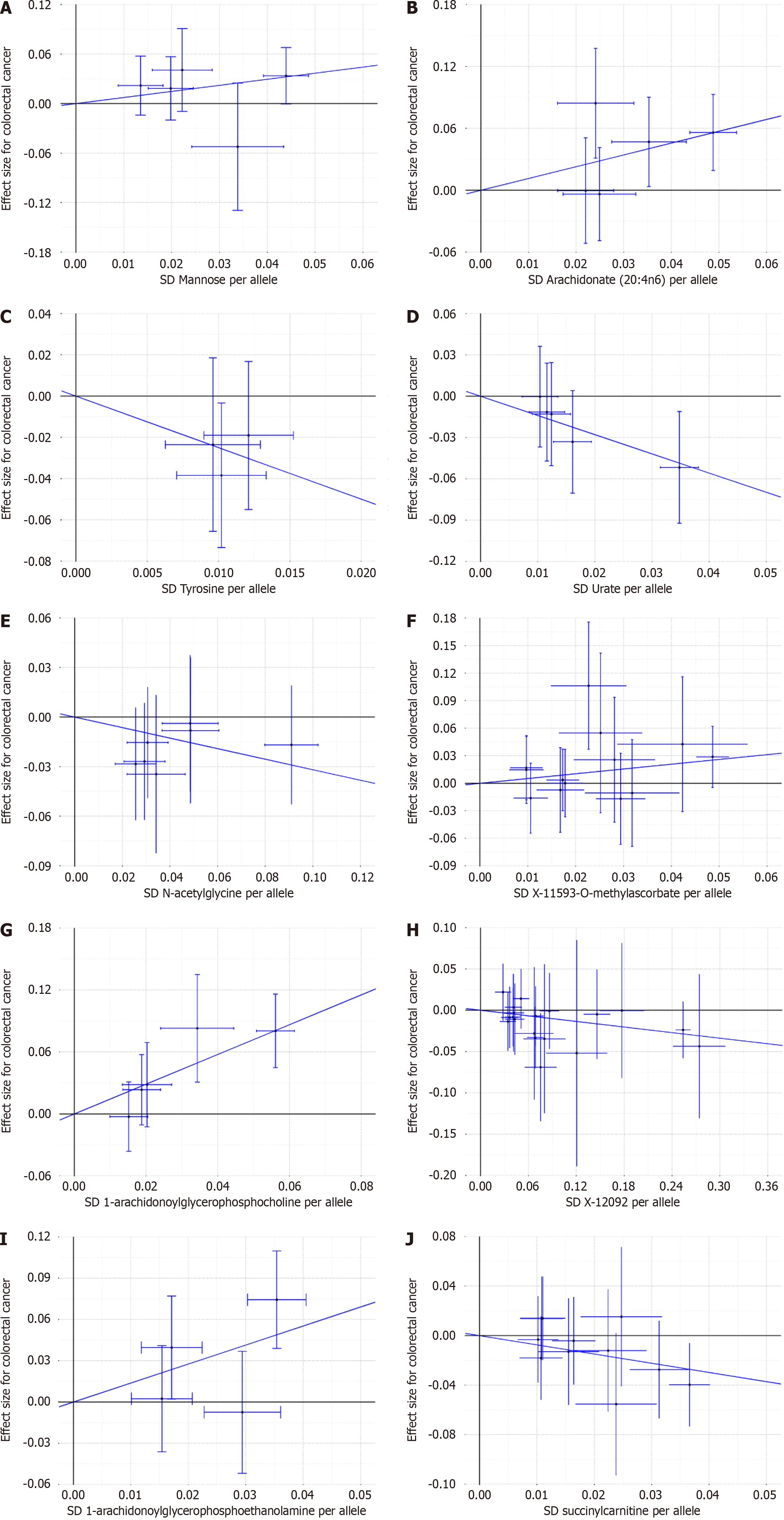
To assess the robustness of the findings, we conducted several sensitivity analyses, including Maximum Likelihood, MR-RAPS, and MR-Egger Intercept. Our examination did not detect any heterogeneity in the IVs pertaining to blood metabolites that exhibited significant associations with colorectal cancer, as indicated by Cochran's IVW Q test (P > 0.05). Furthermore, the MR-Egger regression intercept analysis did not provide substantial evidence of directional pleiotropy. It is noteworthy that the direction of effect remained consistent across all three methods, aligning with the IVW method. Furthermore, the IVW radial MR Results demonstrated that the corrected findings were in agreement with the pre-corrected results (Supplementary Tables 3-12 and Table 2).
The robustness of causality is substantiated by the Maximum likelihood and MR Estimates produced by MR-Egger, which consistently demonstrate the same direction and magnitude. The P values derived from Cochran Q test indicate the absence of heterogeneity. Moreover, the MR-Egger intercept suggests the absence of horizontal pleiotropy (Table 2). Likewise, the MR-RAPS analysis also fails to reveal any significant level of pleiotropy.
This study represents the first investigation utilizing MR analysis to explore the causal link between human blood metabolites and colorectal cancer. The findings demonstrate that specific genes, namely Mannose, Arachidonate (20:4 n6), X-11593-O-methylascorbate, 1-arachidonoylglycerophosphocholine, and 1-arachidonoylglycerophosphoethanolamine, are associated with an increased risk of colorectal cancer, suggesting their potential role in promoting its development. Conversely, genetically determined levels of Tyrosine, Urate, N-acetylglycine, X-12092, and Succinylcarnitine exhibit an inverse relationship with colorectal cancer risk, indicating a potential protective effect. Furthermore, these results were robust and verified through multiple analytical approaches.
These findings are consistent with previous studies in the field of tumor metabolism. For example, lipid metabolites such as arachidonate, X-11593-O-methylascorbate, and 1-arachidonoylglycerophosphocholine have been proposed to play a crucial role in tumor development[19]. These metabolites may regulate inflammatory pathways and enhance the body's immune response to tumor progression[13]. This was demonstrated in a metabonomics study involving a cancer survival cohort of 1812 Finnish men, which identified 49 metabolites associated with cancer survival. These metabolites include phosphatidylcholine, glutamate, arachidonic acid (20:4n6), and glutamylamino acids such as gamma-glutamylglutamic acid, gamma-glutamylglycine, and gamma-glutamylleucine. Higher levels of these lipid molecules were found to be associated with increased cancer-specific mortality[20-22]. The dysregulation of inflammatory pathways in cancer may be attributed to elevated levels of specific lipid molecules. Moreover, the reprogramming of lipid metabolism has been identified as a novel marker for malignant tumors[23]. The expression profile of lipid metabolism-related genes in colorectal cancer has also been linked to a high tumor mutation burden[24]. Furthermore, mannose has been identified as a marker for the malignant progression of early colorectal cancer. Alterations in sugar molecules have been implicated in the occurrence and progression of various types of cancer, including colorectal cancer[25].
The available physiological evidence on the relationship between X-11593-O-methylascorbate and colorectal cancer in humans is currently inadequate. Nonetheless, an animal experiment revealed that two new synthetic derivatives of ascorbic acid, namely 3-O-ethyl ascorbic acid and 3-O-dodecylmethylascorbate, were observed to enhance cancer pro
Urate, which acts as an antioxidant, has the potential to reduce the risk of cancer by reducing oxidative stress. This finding is consistent with previous studies exploring the role of antioxidants in preventing cancer. These findings provide new insights into the metabolic mechanisms involved in colon cancer and may have implications for future therapeutic strategies[27-29].
Multiple cancer-related cellular pathways have been identified, with protein phosphorylation and dephosphorylation, particularly on tyrosine residues, being prominent regulatory mechanisms. The intricate equilibrium between these processes is tightly controlled by protein tyrosine kinase (PTK) and protein tyrosine phosphatase (PTP)[30,31]. An abnormal activity of oncogenic PTK has been observed in a significant proportion of human cancers. PTPs, on the other hand, have long been considered as tumor suppressors due to their ability to counterbalance the effects of pho
Our study provides valuable insights into the metabolic mechanisms underlying colon cancer, which have important implications for future prevention and treatment strategies. To fully understand the relationship between these meta
This study has several strengths. Firstly, unlike previous MR investigations that focused on single or conventional exposure factors, this groundbreaking study integrated metabolomics with genomics to analyze the causal association between 486 human blood metabolites and colorectal cancer. Secondly, multiple techniques were employed to ensure the validity of the study findings.
However, there are several limitations that should be acknowledged in this study. Firstly, the study relied on summary statistics rather than individual data, which precluded subgroup analysis. In future MR studies, the use of individual-level data is crucial to obtain a more comprehensive perspective. Secondly, it is important to note that MR estimates are not adjusted for multiple testing. Nonetheless, we addressed this concern by conducting repeated analyses using an independent GWAS dataset, which significantly enhanced the validity of the findings. Lastly, it is crucial to consider that all participants in this study were of European ancestry. Therefore, caution should be exercised when generalizing the conclusions to individuals from other ethnic backgrounds. Future studies should aim to incorporate diverse populations to ensure the generalizability of the findings while recognizing the importance of individualized treatment approaches.
In conclusion, this study underscores the importance of genetic factors in establishing a causal relationship between metabolites and colorectal cancer. This finding provides a new perspective on the etiology of colorectal cancer and offers potential preventive strategies through the integration of metabolomics and genomics. Nevertheless, due to the limited number of studies directly linking these metabolites to colorectal cancer, additional experimental and clinical investigations are necessary to validate these findings and elucidate the underlying mechanisms.
Our study presents the first systematic assessment of the causal relationship between plasma metabolites and colorectal cancer. To achieve this, we utilized SNPs identified through GWAS as IVs in a MR study design. By employing this approach, we have successfully identified a number of significant blood metabolites that are associated with colorectal cancer. These findings lay the foundation for a more comprehensive understanding of the etiology of colorectal cancer, providing insights into the interplay between colorectal cancer and plasma metabolites in the disease's development.
Limited knowledge exists regarding the casual associations linking blood metabolites and the risk of developing colorectal cancer.
Colorectal cancer development is associated with the presence of five specific blood metabolites. These metabolites have been identified as causal agents and have been validated as risk factors for the disease.
To investigate causal associations between blood metabolites and colon cancer.
The study utilized a two-sample Mendelian randomization (MR) analysis to investigate the causal impact of 486 blood metabolites on colorectal cancer. The primary method of analysis used was the inverse variance weighted (IVW) model. To further validate the results several sensitivity analyses were performed, including Cochran's Q test, MR-Egger intercept test, and Mendelian randomization robust adjusted profile score (MR-RAPS). These additional analyses were conducted to ensure the reliability and robustness of the findings.
After rigorous selection for genetic variation, 486 blood metabolites were included in the MR analysis. We found Mannose [odds ratio (OR) = 2.09 (1.10-3.97), P = 0.024], N-acetylglycine [OR = 3.14 (1.78-5.53), P =7.54 × 10-8], X-11593-O-methylascorbate [OR = 1.68 (1.04-2.72), P = 0.034], 1-arachidonoylglycerophosphocholine [OR 4.23 (2.51-7.12), P = 6.35 × 10-8] and 1- arachidonoylglycerophosphoethanolamine 4 [OR = 3.99 (1.17-13.54), P = 0.027] were positively causally associated with colorectal cancer, and we also found a negative causal relationship between Tyrosine [OR = 0.08 (0.01-0.63), P = 0.014], Urate [OR =0.25 (0.10-0.62), P = 0.003], N-acetylglycine [0.73 (0.54-0.98), P = 0.033], X-12092 [OR =0.89 (0.81-0.99), P = 0.028], Succinylcarnitine [OR =0.48 (0.27-0.84), P = 0.09] with colorectal cancer. A series of sensitivity analyses were performed to confirm the rigidity of the results.
This study showed a causal relationship between 10 blood metabolites and colorectal cancer, of which 5 blood metabolites were found to be causal for the development of colorectal cancer and were confirmed as risk factors. The other five blood metabolites are protective factors.
A significant inverse relationship has been observed between the remaining five blood metabolites and the development of colorectal cancer, establishing them as protective.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Rasa HK, Turkey S-Editor: Li L L-Editor: A P-Editor: Zheng XM
| 1. | Nfonsam V, Wusterbarth E, Gong A, Vij P. Early-Onset Colorectal Cancer. Surg Oncol Clin N Am. 2022;31:143-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 2. | Hampel H, Kalady MF, Pearlman R, Stanich PP. Hereditary Colorectal Cancer. Hematol Oncol Clin North Am. 2022;36:429-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 3. | Sinha R. Colorectal cancer. Clin Radiol. 2021;76:870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 4. | Fan J, de Lannoy IA. Pharmacokinetics. Biochem Pharmacol. 2014;87:93-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 203] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 5. | Jackson AJ, Robbie G, Marroum P. Metabolites and bioequivalence: past and present. Clin Pharmacokinet. 2004;43:655-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 6. | Oh KK, Choi I, Gupta H, Raja G, Sharma SP, Won SM, Jeong JJ, Lee SB, Cha MG, Kwon GH, Jeong MK, Min BH, Hyun JY, Eom JA, Park HJ, Yoon SJ, Choi MR, Kim DJ, Suk KT. New insight into gut microbiota-derived metabolites to enhance liver regeneration via network pharmacology study. Artif Cells Nanomed Biotechnol. 2023;51:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Reference Citation Analysis (2)] |
| 7. | Zhao YS, Chen F, Li L. Are circulating metabolites important in pharmacokinetic drug-drug interactions? A retroanalysis of clinical data. Curr Drug Metab. 2014;15:767-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 8. | Birney E. Mendelian Randomization. Cold Spring Harb Perspect Med. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 356] [Article Influence: 118.7] [Reference Citation Analysis (0)] |
| 9. | Bowden J, Holmes MV. Meta-analysis and Mendelian randomization: A review. Res Synth Methods. 2019;10:486-496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 461] [Cited by in RCA: 1007] [Article Influence: 167.8] [Reference Citation Analysis (0)] |
| 10. | Burgess S, Thompson SG; CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40:755-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2753] [Cited by in RCA: 2341] [Article Influence: 167.2] [Reference Citation Analysis (0)] |
| 11. | Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377-389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1644] [Cited by in RCA: 2905] [Article Influence: 363.1] [Reference Citation Analysis (0)] |
| 12. | Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, Conti DV, Qu C, Jeon J, Edlund CK, Greenside P, Wainberg M, Schumacher FR, Smith JD, Levine DM, Nelson SC, Sinnott-Armstrong NA, Albanes D, Alonso MH, Anderson K, Arnau-Collell C, Arndt V, Bamia C, Banbury BL, Baron JA, Berndt SI, Bézieau S, Bishop DT, Boehm J, Boeing H, Brenner H, Brezina S, Buch S, Buchanan DD, Burnett-Hartman A, Butterbach K, Caan BJ, Campbell PT, Carlson CS, Castellví-Bel S, Chan AT, Chang-Claude J, Chanock SJ, Chirlaque MD, Cho SH, Connolly CM, Cross AJ, Cuk K, Curtis KR, de la Chapelle A, Doheny KF, Duggan D, Easton DF, Elias SG, Elliott F, English DR, Feskens EJM, Figueiredo JC, Fischer R, FitzGerald LM, Forman D, Gala M, Gallinger S, Gauderman WJ, Giles GG, Gillanders E, Gong J, Goodman PJ, Grady WM, Grove JS, Gsur A, Gunter MJ, Haile RW, Hampe J, Hampel H, Harlid S, Hayes RB, Hofer P, Hoffmeister M, Hopper JL, Hsu WL, Huang WY, Hudson TJ, Hunter DJ, Ibañez-Sanz G, Idos GE, Ingersoll R, Jackson RD, Jacobs EJ, Jenkins MA, Joshi AD, Joshu CE, Keku TO, Key TJ, Kim HR, Kobayashi E, Kolonel LN, Kooperberg C, Kühn T, Küry S, Kweon SS, Larsson SC, Laurie CA, Le Marchand L, Leal SM, Lee SC, Lejbkowicz F, Lemire M, Li CI, Li L, Lieb W, Lin Y, Lindblom A, Lindor NM, Ling H, Louie TL, Männistö S, Markowitz SD, Martín V, Masala G, McNeil CE, Melas M, Milne RL, Moreno L, Murphy N, Myte R, Naccarati A, Newcomb PA, Offit K, Ogino S, Onland-Moret NC, Pardini B, Parfrey PS, Pearlman R, Perduca V, Pharoah PDP, Pinchev M, Platz EA, Prentice RL, Pugh E, Raskin L, Rennert G, Rennert HS, Riboli E, Rodríguez-Barranco M, Romm J, Sakoda LC, Schafmayer C, Schoen RE, Seminara D, Shah M, Shelford T, Shin MH, Shulman K, Sieri S, Slattery ML, Southey MC, Stadler ZK, Stegmaier C, Su YR, Tangen CM, Thibodeau SN, Thomas DC, Thomas SS, Toland AE, Trichopoulou A, Ulrich CM, Van Den Berg DJ, van Duijnhoven FJB, Van Guelpen B, van Kranen H, Vijai J, Visvanathan K, Vodicka P, Vodickova L, Vymetalkova V, Weigl K, Weinstein SJ, White E, Win AK, Wolf CR, Wolk A, Woods MO, Wu AH, Zaidi SH, Zanke BW, Zhang Q, Zheng W, Scacheri PC, Potter JD, Bassik MC, Kundaje A, Casey G, Moreno V, Abecasis GR, Nickerson DA, Gruber SB, Hsu L, Peters U. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet. 2019;51:76-87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 384] [Cited by in RCA: 384] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 13. | Tan P, Dong X, Mai K, Xu W, Ai Q. Vegetable oil induced inflammatory response by altering TLR-NF-κB signalling, macrophages infiltration and polarization in adipose tissue of large yellow croaker (Larimichthys crocea). Fish Shellfish Immunol. 2016;59:398-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 14. | Grasso CS, Giannakis M, Wells DK, Hamada T, Mu XJ, Quist M, Nowak JA, Nishihara R, Qian ZR, Inamura K, Morikawa T, Nosho K, Abril-Rodriguez G, Connolly C, Escuin-Ordinas H, Geybels MS, Grady WM, Hsu L, Hu-Lieskovan S, Huyghe JR, Kim YJ, Krystofinski P, Leiserson MDM, Montoya DJ, Nadel BB, Pellegrini M, Pritchard CC, Puig-Saus C, Quist EH, Raphael BJ, Salipante SJ, Shin DS, Shinbrot E, Shirts B, Shukla S, Stanford JL, Sun W, Tsoi J, Upfill-Brown A, Wheeler DA, Wu CJ, Yu M, Zaidi SH, Zaretsky JM, Gabriel SB, Lander ES, Garraway LA, Hudson TJ, Fuchs CS, Ribas A, Ogino S, Peters U. Genetic Mechanisms of Immune Evasion in Colorectal Cancer. Cancer Discov. 2018;8:730-749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 380] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 15. | Cai H, Cai B, Zhang H, Sun W, Wang Y, Zhou S, Ye Z, Zhang Z, Liang J. Major depression and small vessel stroke: a Mendelian randomization analysis. J Neurol. 2019;266:2859-2866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 16. | Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2275] [Cited by in RCA: 6282] [Article Influence: 628.2] [Reference Citation Analysis (0)] |
| 17. | Xue H, Shen X, Pan W. Constrained maximum likelihood-based Mendelian randomization robust to both correlated and uncorrelated pleiotropic effects. Am J Hum Genet. 2021;108:1251-1269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 222] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 18. | Zhou F, Li S, Xu H. Insomnia, sleep duration, and risk of anxiety: A two-sample Mendelian randomization study. J Psychiatr Res. 2022;155:219-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 19. | Yang P, Wang W, Chi S, Mai K, Song F, Wang L. Effects of dietary lysine on regulating GH-IGF system, intermediate metabolism and immune response in largemouth bass (Micropterus salmoides). Aquac Rep. 2020;17:100323. [DOI] [Full Text] |
| 20. | Krishnan ST, Winkler D, Creek D, Anderson D, Kirana C, Maddern GJ, Fenix K, Hauben E, Rudd D, Voelcker NH. Staging of colorectal cancer using lipid biomarkers and machine learning. Metabolomics. 2023;19:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 21. | Ocvirk S, O'Keefe SJD. Dietary fat, bile acid metabolism and colorectal cancer. Semin Cancer Biol. 2021;73:347-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 158] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 22. | Pakiet A, Kobiela J, Stepnowski P, Sledzinski T, Mika A. Changes in lipids composition and metabolism in colorectal cancer: a review. Lipids Health Dis. 2019;18:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 217] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 23. | Kaźmierczak-Siedlecka K, Marano L, Merola E, Roviello F, Połom K. Sodium butyrate in both prevention and supportive treatment of colorectal cancer. Front Cell Infect Microbiol. 2022;12:1023806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 42] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 24. | D'Angelo S, Motti ML, Meccariello R. ω-3 and ω-6 Polyunsaturated Fatty Acids, Obesity and Cancer. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 150] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 25. | Zhang Q, Deng T, Zhang H, Zuo D, Zhu Q, Bai M, Liu R, Ning T, Zhang L, Yu Z, Ba Y. Adipocyte-Derived Exosomal MTTP Suppresses Ferroptosis and Promotes Chemoresistance in Colorectal Cancer. Adv Sci (Weinh). 2022;9:e2203357. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 117] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 26. | Yang Y, Luo D, Shao Y, Shan Z, Liu Q, Weng J, He W, Zhang R, Li Q, Wang Z, Li X. circCAPRIN1 interacts with STAT2 to promote tumor progression and lipid synthesis via upregulating ACC1 expression in colorectal cancer. Cancer Commun (Lond). 2023;43:100-122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 38] [Reference Citation Analysis (0)] |
| 27. | Liu F, Huang Y, Li ZW, Liu XR, Liu XY, Lv Q, Shu XP, Li LS, Zhang W, Tong Y, Zeng MH, Peng D. Hyperuricemia remission after colorectal cancer surgery for colorectal cancer patients. Sci Rep. 2023;13:18867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 28. | Teng H, Wang Y, Sui X, Fan J, Li S, Lei X, Shi C, Sun W, Song M, Wang H, Dong D, Geng J, Zhang Y, Zhu X, Cai Y, Li Y, Li B, Min Q, Wang W, Zhan Q. Gut microbiota-mediated nucleotide synthesis attenuates the response to neoadjuvant chemoradiotherapy in rectal cancer. Cancer Cell. 2023;41:124-138.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 64] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 29. | Zhang X, Zhao H, Man J, Yin X, Zhang T, Yang X, Lu M. Investigating Causal Associations of Diet-Derived Circulating Antioxidants with the Risk of Digestive System Cancers: A Mendelian Randomization Study. Nutrients. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 30. | Kuo ML, Huang TS, Lin JK. Preferential requirement for protein tyrosine phosphatase activity in the 12-O-tetradecanoylphorbol-13-acetate-induced differentiation of human colon cancer cells. Biochem Pharmacol. 1995;50:1217-1222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 31. | Woźniak M, Gamian E, Łaczmańska I, Sąsiadek MM, Duś-Szachniewicz K, Ziółkowski P. Immunohistochemical and Western blot analysis of two protein tyrosine phosphatase receptors, R and Z1, in colorectal carcinoma, colon adenoma and normal colon tissues. Histol Histopathol. 2014;29:635-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 32. | Thanikachalam K, Khan G. Colorectal Cancer and Nutrition. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 479] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 33. | Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, Walter K, Menni C, Chen L, Vasquez L, Valdes AM, Hyde CL, Wang V, Ziemek D, Roberts P, Xi L, Grundberg E; Multiple Tissue Human Expression Resource (MuTHER) Consortium, Waldenberger M, Richards JB, Mohney RP, Milburn MV, John SL, Trimmer J, Theis FJ, Overington JP, Suhre K, Brosnan MJ, Gieger C, Kastenmüller G, Spector TD, Soranzo N. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543-550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1097] [Cited by in RCA: 1102] [Article Influence: 100.2] [Reference Citation Analysis (1)] |