Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.1965
Peer-review started: December 20, 2023
First decision: January 10, 2024
Revised: January 21, 2024
Accepted: February 20, 2024
Article in press: February 20, 2024
Published online: May 15, 2024
Processing time: 141 Days and 1.5 Hours
Yigong San (YGS) is a representative prescription for the treatment of digestive disorders, which has been used in clinic for more than 1000 years. However, the mechanism of its anti-gastric cancer and regulate immunity are still remains unclear.
To explore the mechanism of YGS anti-gastric cancer and immune regulation.
Firstly, collect the active ingredients and targets of YGS, and the differentially expressed genes of gastric cancer. Secondly, constructed a protein-protein inter
Firstly, obtained 55 common targets of gastric cancer and YGS. The Kyoto Encyclopedia of Genes and Genomes screened the microtubule-associated protein kinase signaling axis as the key pathway and IL6, EGFR, MMP2, MMP9 and TGFB1 as the hub genes. The 5 hub genes were involved in gastric carcinogenesis, staging, typing and prognosis, and their mutations promote gastric cancer progression. Finally, molecular docking results confirmed that the components of YGS can effectively bind to therapeutic targets.
YGS has the effect of anti-gastric cancer and immune regulation.
Core Tip: Gastric cancer seriously endangers human life and quality of life. Traditional Chinese medicine has rich experience in treating tumors, and Yigong San (YGS) is a representative prescription that can treat a variety of digestive system di
- Citation: Lu DD, Yuan L, Wang ZZ, Zhao JJ, Du YH, Ning N, Chen GQ, Huang SC, Yang Y, Zhang Z, Nan Y. To explore the mechanism of Yigong San anti-gastric cancer and immune regulation. World J Gastrointest Oncol 2024; 16(5): 1965-1994
- URL: https://www.wjgnet.com/1948-5204/full/v16/i5/1965.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i5.1965
Gastric cancer refers to malignant tumors originating from the epithelial origin of the stomach, and it ranks at the forefront of the global tumor disease spectrum with high morbidity, mortality and metastasis[1], which seriously affects human health and quality of life. The pathogenesis of gastric cancer is complex, and in addition to precancerous lesions such as Helicobacter pylori-induced chronic superficial gastritis, chronic atrophic gastritis, and heterogeneous hyperplasia[2], factors such as gene mutations, immune system dysregulation, and aberrant epigenetic regulation also influence the progression of the disease[3]. Current treatment plans based on surgical and radiotherapy vs chemotherapy approaches have resulted in unsatisfactory outcomes for patients, either due to missing the optimal time for surgery or resistance to radiotherapy and intolerance of side effects. Therefore, the development of new drugs with anti-cancer effects and less toxic side effects is of great importance to the clinic, and in recent years researchers have gradually focused on the anti-cancer efficacy of traditional Chinese medicine (TCM) compounds.
Yigong San (YGS) is a representative prescription initially invented by Qian Yi at the time of the Song Dynasty (960-1279) for treating pediatric digestive disorders, which was first recorded in the ancient book named Xiaoer Yaozheng Zhijue[4]. YGS consist of five Chinese herbs: Renshen (the root from Panax ginseng C. A. Mey., Araliaceae, RS), Baizhu (the root from Atractylodes macrocephala Koidz., Composites, BZ), Fuling (the sclerotium of Poriacocos (Schw.) Wolf., Polyporaceae, FL), Chenpi (the peel from Citrus reticulata Blanco, Rutaceae, CP), and Gancao (the root from Glycyrrhiza uralensis Fisch., Glycyrrhiza inflata Bat. or Glycyrrhiza glabra L., Leguminosae, GC). Additive and subtractive formulations based on YGS have been shown to be effective in digestive disorders; for example, the new drug compound glutamine capsule deve
Among the "14 phenotypes" of tumors proposed by Hanahan[9], proliferation and apoptosis are important phenotypes that regulate tumor growth and progression. The imbalance between the growth and death of tumor cells is an important factor in their progression and a driver of metastasis even after treatment. Therefore, how to induce cell death or inhibit proliferative capacity is a key point in tumor therapy. In addition, another phenotype, namely “immunity”, has gradually emerged in the onset, progression and treatment of tumors in recent years. As a kind of treatment with high specificity and low cytotoxicity, “tumor immunotherapy” can indirectly eliminate tumors by mobilizing the body’s immune activity and improving the capacity of the immune system. YGS is rich in active ingredients such as polysaccharides, saponins, and triterpenoids, which are inextricably linked to immunity. Studies have confirmed that ginseng polysaccharides of RS can enhance the efficacy of programmed cell death protein 1 (PD-1)/programmed cell death 1 ligand 1 (PD-L1) against non-small cell lung cancer (NSCLC)[10], and ginsenosides of RS can improve cyclophosphamide-induced intestinal immune disorders and barrier damage[11]; carboxymethyl polysaccharides in FL have been shown to activate the immune system to exert anti-tumor effects in a variety of tumor cells[12]; the active components of CP can effectively regulate immune-related cytokines, such as interferon gamma (IFN-γ), tumor necrosis factor alpha, interleukin (IL)1, IL2, etc., in order to maintain the dynamic balance between immune responses[13]; atractylenolide, one of the main active components of BZ in exerting its pharmacological effects, promotes the presentation of tumor antigens and enhances the cytotoxicity of T-cells in the tumor immune escaped to increase the tumor’s response to immunotherapy[14]; licochalcone A in GC can inhibit PD-L1 to exert anti-cancer effects[15]. However, there have been no reports of anti-cancer effects from YGS through modulated immunity. Based on studies of the compounds and active ingredients, we speculate that YGS may have the efficacy of modulating the immune function of the organism and delaying the development and pro
Due to the complexity of their constituents, the variety of targets, and the unknown pathways of action, traditional pharmacological methods cannot be used to study and explore them in an integrated and comprehensive manner. With the “Bioinformatics research strategy for complex diseases” proposed by Li et al[16] in 2003, and the concept of “network targets” proposed by Hopkins[17], as well as the launch of the first “Guidelines for evaluation methods in network pharmacology”[18] in 2021, which began to correlate the specific targets of active ingredients of medicines with the network of the biological process (BP) of diseases, the research on TCM compounding has been put into a fast track, which provides a wider opportunity for the research on the treatment of diseases by TCM compounding and is expected to unravel the mysterious veil of the pharmacological effects of compounding.
In this study, based on publicly available datasets, integrating network pharmacology and bioinformatics analysis techniques, we screened the targets and signaling pathways in which YGS may exert anti-cancer therapeutic effects, explored the clinical relevance, gene mutations, and immune infiltration of the targets, we found that YGS may exert anti-tumor effects by regulating processes such as the RAS/MEK/ ERK signaling pathway and enhancing immune responses. The whole flowchart of this study is shown in Figure 1: (1) Screening of hub genes for the treatment of gastric cancer by YGS; (2) analyzing the correlation between hub genes and clinic; (3) exploring the effects of mutation, methylation and immune infiltration of hub genes on gastric cancer; and (4) analyzing the relationship between hub genes and drug therapy.
Screening the active ingredients of YGS and predicting targets: TCM systems pharmacology database and analysis platform (TCMSP) website (https://old.tcmsp-e.com) collects a large amount of information about herbal medicines, such as active ingredients, molecular names, targets of action, drug-likeness (DL) and oral bioavailability (OB), etc., of which the DL and OB indexes are one of the most important parameters for evaluating pharmacokinetics. We first screened the active ingredients with the parameters DL ≥ 0.18 and OB ≥ 30%. Next, we plotted the intersection of active ingredients using the SRplot website (http://www.bioinformatics.com.cn) and constructed a network diagram of active ingredient-targets using Cytoscape 3.9.1 software.
Screening targets for gastric cancer: Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov) is a public da
Intersection targeted of drug and disease: Firstly, we plotted the number of DEGs and compound targets of gastric cancer in GraphPad Prism 8.0 software, and intersected the targets of YGS with the up- and down-regulated genes in GSE65801 using SRplot, and then analyzed the intersected targets using the Jvenn online site (http://jvenn.toulouse.inra.fr). The three intersecting targets are presented as heatmaps to analyze their expression differences, and the top 10 up- and down-regulated genes are shown after classification based on the magnitude of the Log FC values. Finally, the intersecting targets were imported into the BioLadder online website (https://www.bioladder.cn) for down-regulation analysis to evaluate the inter- and intra-group differences between the gastric cancer and the normal group.
Construct the protein-protein interaction network map: The STRING (Search Tool for Recurring Instances of Neigh
Analyzing the relevance of core targets: To further analyze the correlation between the core targets, we explored them the Gene Expression Profiling Interactive Analysis (GEPIA) 2.0 database (http://gepia2.cancer-pku.cn) and draw heat maps of within-group differences by the Chiplot (https://www.chiplot.online) website.
Gene Ontology analysis: The core targets of YGS for gastric cancer treatment were analyzed by Gene Ontology (GO), including BP, molecular function (MF) and cellular localization (CC) entries. We used the Database for Annotation, Visualization and Integrated Discovery (DAVID) website (https://david.ncifcrf.gov) to obtain the analyzed data, and set up the parameters as follows: input the target form as "Official gene symbol", and select the species as "Homo". "The results were plotted in Sangerbox (http://sangerbox.com).
Analysis of the Kyoto Encyclopedia of Genes and Genomes and Gene Set Enrichment Analysis: The core targets of YGS for gastric cancer treatment were analyzed using Kyoto Encyclopedia of Genes and Gene Set Enrichment Analysis (GSEA) pathway enrichment. We also used the DAVID website to obtain KEGG pathway information, present all pathway entries enriched in the form of a circle diagram, and take the top 18 results to draw a Sankey diagram to further display the results. KEGG Mapper (https://www.genome.jp/kegg) was utilized to explore the pathway information with the highest number of enriched targets. The gene names of the intersected targets were first standardized as UniProt IDs in UniProt (https://www.uniprot.org) and then transformed into KEGG ID format using KEGG Mapper. Up-regulated and down-regulated genes were replaced by red and blue, respectively, and the species was set as "hsa" for analysis. The top-ranked pathways were simplified and edited using Adobe Illustrator 2020 software to obtain potential signaling pathways and Hub genes for YGS therapy in gastric cancer. To avoid one-sided results due to screening conditions, we further analyzed all gastric cancer targets by GSEA analysis. We explored the "Pathway Enrichment" module in the CAMOIP website (https://www.camoip.net) and selected the "TCGA-Cohort" dataset. Core targets are entered separately to analyze their relevance to diseases and pathways.
Clinical relevance of the hub gene: The hub genes obtained by screening were explored for clinical relevance in order to analyze their relationship to gastric cancerogenesis, staging and prognosis. In the first step, samples derived from the TCGA + GTEx database were analyzed in Sangerbox using a nonparametric test to assess the correlation between mRNA expression levels of the hub gene and stomach cancer. Secondly, the GEPIA 2.0 website was used to analyze the correlation between the copy number level of the hub gene and stomach cancer. Next, protein expression box plots were obtained through the "CPTAC" module of the University of Alabama at Birmingham Cancer Data Analysis Portal (UALCAN) website to evaluate whether the hub genes were differentially expressed at the protein level. Next, the correlation between hub genes and gastric cancer subtypes was explored in the "Expression" module of the Gene Set Cancer Analysis (GSCA; http://bioinfo.life.hust.edu.cn) website. Finally, we used the GEPIA 2.0 website to analyze their relationship to the clinical stage of gastric cancer.
The expression and prognosis of hub genes: After the analysis of hub genes in terms of mRNA, copy number, protein, tumor subtype and clinical stage, further exploration of Hub genes in clinical gastric cancer samples followed. The Human Protein Atlas (HPA) database (https://www.proteinatlas.org) contains a large amount of mapping information of tumors and normal tissues. We used this database to obtain immunohistochemical (IHC) images of gastric cancer and normal gastric glandular tissue to analyze the expression of the hub gene in the tissues; IHC images were obtained to analyze their localization information in the tumor tissue. In order to observe whether the hub genes obtained from the screening were correlated with the survival prognosis of gastric cancer, we used the Kaplan Meier Plotter website (http://kmplot.com) to plot the survival curves and explore the prognostic survival of gastric cancer patients and the correlation between the hub genes.
Mutation of the hub genes: To explore whether mutations in hub genes correlate with gastric cancer, we performed specific analyses using the GSCA database, which contains mutation information for several tumor samples in TCGA. Among them, single nucleotide variations (SNVs) are mainly categorized into deleterious mutations and non-deleterious mutatioccns, and the former include seven kinds of Missense Mutation, Nonsense Mutation, Frame ShiftIns, SpliceSite, lFrame ShiftDel, InFrameDel, InFrameIns forms of mutation. We first analyze the loci and types of SNVs mutations in the hub genes, and present the frequency of deleterious mutations as a heatmap percentage. The effect of copy number variations (CNVs) was further explored, and the mutation data were processed by GISTIC 2.0 and presented as bubble plots. A typical tumor organization includes the driver mutation gene and the passages mutation gene, the former of which drives tumor cells to undergo selective advantageous growth during gastric cancer development and progression and plays an extremely important role in tumors. We explored the top 20 driver mutation genes in the CAMOIP database related to the hub gene, both proto-oncogenes and oncogenes.
Microsatellite instability: Microsatellites, also known as simple repeat sequences, are small, partially repeated nucleotide sequences in the genome. If the insertion or deletion of this repeating unit leads to the alteration of the gene sequence, we call it " Microsatellite instability (MSI)", which is an important molecular marker for tumors. We used the "Immunogenicity" module in CAMOIP to analyze the MSI of tumors, and selected the parameters "TCGA-Cohort" and "Stomach adenocarcinoma (STAD)”.
Methylation of the hub genes: The UALCAN website was used to explore the relationship between gene methylation and disease. We plotted methylation expression box plots in the "TCGA" module and obtained the correlation between methylation of hub genes and cytotoxic T lymphocytes through the Tumor Immune Dysfunction and Exclusion (TIDE) website (http://tide.dfci.harvard.edu), and analyzed the correlation between the hypermethylated and hypomethylated groups.
Repair systems of mutation: Although mutations in genes affect the process of tumorigenesis and progression, there are actually repair systems in the body that can help the recovery of damaged genes, such as mismatch repair (MMR) and homologous recombination repair (HRR). In MMR, it is mainly the MSH (mutS homolog)2, MSH3, MSH6, recombinant MLH1 (mutL homolog)1, recombinant MLH3 and PMS1 (PMS1 homolog 1) genes that play a role, while HRR is associated with 30 genes including ATM serine/threonine kinase (ATM), ATR serine/threonine kinase (ATR) and ATRX chromatin remodeler (ATRX). GEPIA 2.0 was used to calculate the correlation between the 35 genes and the hub genes, and the final results are presented as heatmaps.
Immune infiltration and immune checkpoints: Immunity is strongly correlated with tumorigenesis and progression. Among them, CIBERSORTx (https://cibersortx.stanford.edu) was used to estimate the abundance of 22 immune cell subtypes in mixed cell populations. We imported the gene chip data downloaded from GEO into the CIBERSORT online site and presented the results as boxplots. We then used the "Immune Infiltration Analysis" module of Sangerbox to plot the StromalScore, ImmuneScore, and EATIMATE score scatter plots of the hub gene and gastric cancer, respectively. The correlation between immune checkpoints and Hub genes is then analyzed using Sangerbox, and the results are presented as heatmaps.
Single-cell sequencing: The tumor immunology single cell center (TISCH; http://tisch.comp-genomics.org) database provides specific cell type annotations at the single cell level, which provides an easier way to explore the tumor microenvironment (TME), which contains information on 2045746 cells from 28 tumor types and 79 datasets in GEO and Array Express. We set the parameters in the TISCH website: select "STAD" for tumor, "Human" for species, and "no treatment" for treatment to annotate the single-cell maps in the final dataset.
Relevance between immune cells and gastric cancer: Based on the results of single-cell annotation mapping, we further analyzed the correlation of macrophages, monocytes, cluster of differentiation 8 (CD8) T cells and tumor-associated fibroblasts with Hub genes in gastric cancer using the TIMER 2.0 website (http://timer.cistrome.org).
Pharmacotherapy: Immunotherapy is becoming an important component of antitumor therapy. We first explored the relevance of hub genes to immunotherapy using the "Drug" module of the TISIDB website (http://cis.hku.hk/TISIDB). IFN-γ can synergize with drugs to enhance the killing effect on tumor cells, while IL6, EGFR, MMP2, MMP9 and TGFB1 induces apoptosis and inhibits the proliferation of tumor cells, both of which can have an impact on the treatment of tumors. Therefore, we use the CAMOIP website to analyze the correlation between these two and hub genes to provide a predictive rationale for immunotherapy in stomach cancer. Both the Genomics of Cancer Drug Sensitivity (GDSC) database and the Cancer Treatment Response Portal (CTRP) contain half maximal inhibitory concentration and corresponding mRNA gene expression information for a large number of molecules in cell lines, which can be used to predict the sensitivity of a wide range of antitumor drugs. We explore the sensitivity of the hub genes to anti-tumor drugs in GSCA and present the top 30 results as bubbles.
Molecular docking: Cytoscape software was used to construct a network graph of the hub genes and active ingredient. Molecular docking was performed to validate the screened hub genes and active ingredients. First, the structure data file format structure of the active ingredient was obtained using PubChem (https://pubchem.ncbi.nlm.nih.gov), which was processed by energy optimization and hydrogenation; then the 2D structure map of the hub gene was obtained from the protein data bank database (https://www.rcsb.org), which was similarly subjected to de-watering, de-liganding, hydrogenation, assignment of charge and adding atom type processing. AutoDock Tools and AutoDock Vina were used to create active pockets and calculate binding energies, and some docking results were visualized.
All analyses were performed using GraphPad Prism 8.0. Continuous data are expressed as the mean ± SD, and qualitative data was expressed as rate. Group comparison was analyzed using one-way ANOVA or t-test. P < 0.05 was considered statistically significant.
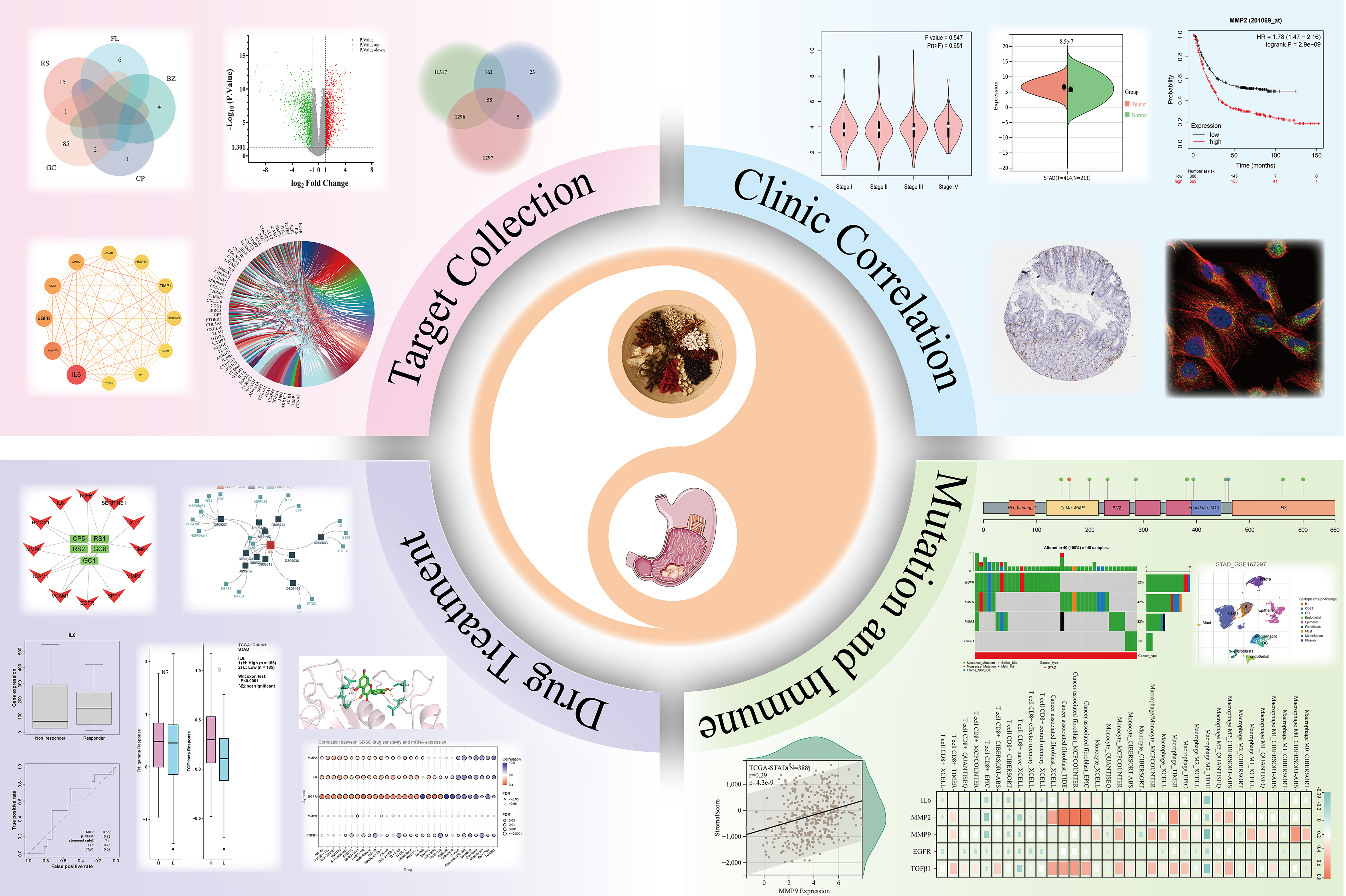
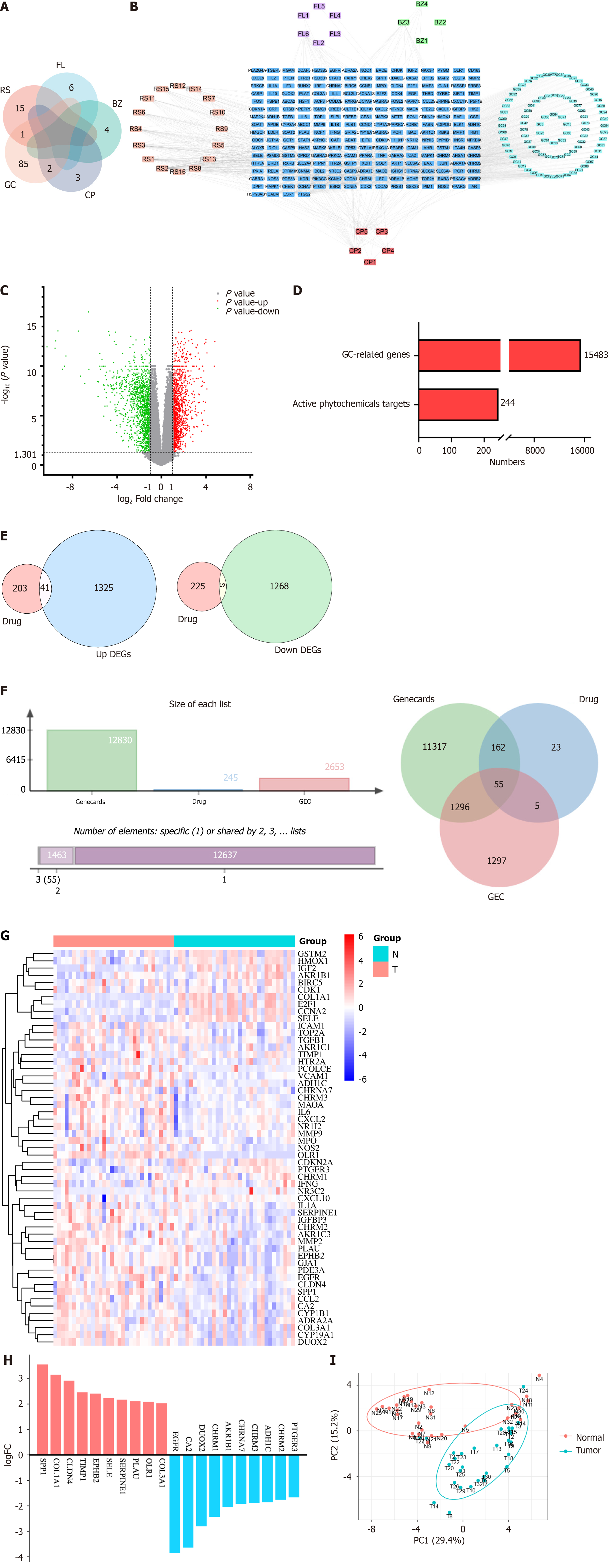
We obtained a total of 116 active ingredients for YGS from TCMSP, and the information on the active ingredients is presented in Supplementary Table 1. The number of active ingredients of each drug is shown in Figure 2A, among which there were a total of 16 active ingredients of RS, 6 active ingredients of FL, 4 active ingredients of BZ, 5 active ingredients of CP, and 88 active ingredients of GC. Cytoscape was used to construct the active ingredient-target network diagram of YGS, as shown in Figure 2B, the blue color in the middle represents the target, and the different colors in the outer circle represent the active ingredients of different drugs, and a total of 244 targets were obtained in which YGS exerted its pharmacological effects.
The dataset, numbered GSE65801, was selected for subsequent analysis in the GEO database and contains 64 samples, 32 each in the normal and gastric cancer groups. As shown in Figure 2C, the GSE65801 dataset contained a total of 22020 gastric cancer genes, among which there were a total of 2653 DEGs, with green color representing the down-regulated genes and red color representing the up-regulated genes, and meanwhile, we obtained 12,830 genes for gastric cancer from the Genecards database. After summarizing the total number of targets for drugs and gastric cancer, as shown in Figure 2D, there are 244 targets for the former and 15,483 targets for the latter.
As shown in Figure 2E, firstly, a total of 60 GEO and YGS crossover targets are obtained, of which 41 are up-regulated genes and 19 are down-regulated genes. Continuing to take the intersection of GEO, Genecards and YGS, as shown in Figure 2F and G), a total of 55 targets were obtained in Supplementary Table 2, of which about 2/3 showed an up-regulation trend in the gastric cancer group, and about 1/3 showed a down-regulation trend, which means that 138 active ingredients in YGS may exert their anti-gastric cancer pharmacological effects through these 55 targets. Log FC values for these 55 intersecting targets were ranked as a way to obtain the genes with the largest expression differences, as shown in Figure 2H, with SPP1, COL1A1, CLDN4D and EGFR, CA2, DUOX2 ranked higher. Intersecting targets were further subjected to PCoA downscaling to analyze how representative these 55 samples were, as shown in Figure 2I, where red and blue dots represent tumor and normal tissue, respectively, with only a small overlap between the two.
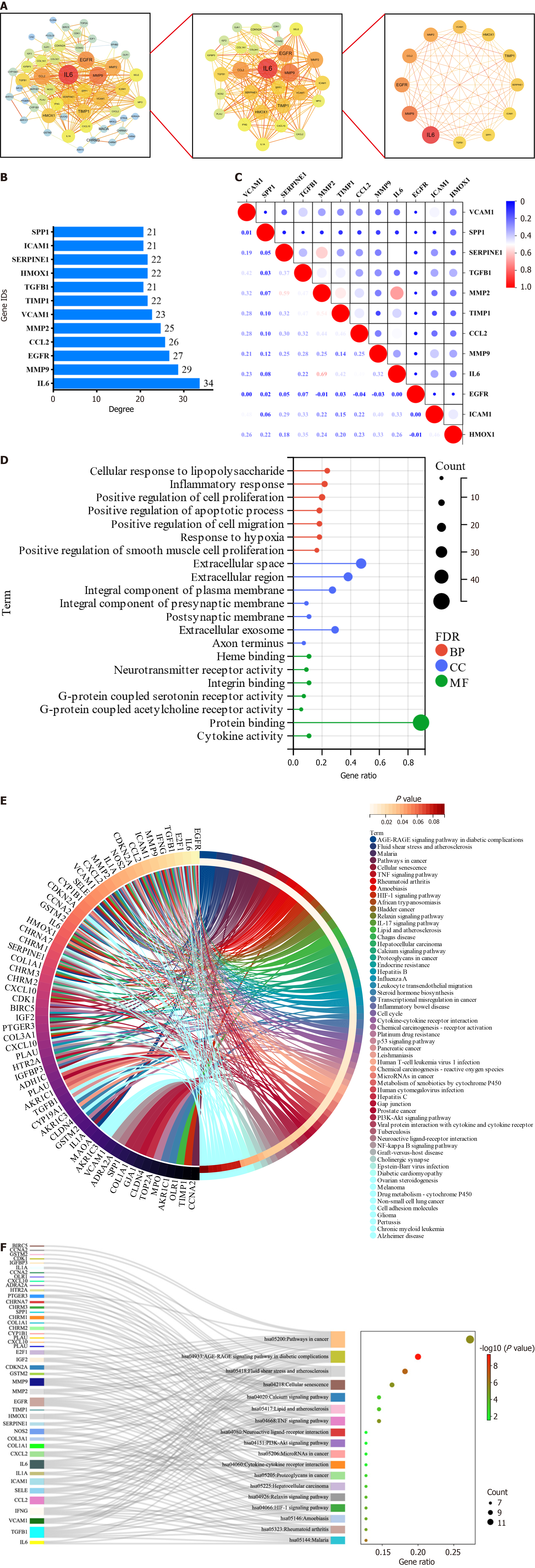
To further explore the key targets of YGS for gastric cancer treatment, we screened 55 targets twice using the Cytoscape software. As shown in Figure 3A, the screening was first performed with degree ≥ 8, and a total of 28 core targets were obtained; and continued with degree ≥ 21, and 12 targets that may be the most core targets for the treatment of gastric cancer by YGS were obtained, including IL6, MMP9, EGFR, CCL2, MMP2, VCAM1, TGFB1 and ICAM1. According to the values of degree used to judge the contribution of the targets, as shown in Figure 3B, IL6, MMP9, EGFR, MMP2, etc. are ranked high. Continuing the analysis of their correlation, Figure 3C shows that while most of the targets show positive correlation and synergy, IL6 and MMP2 show a clear trend of correlation. From these results, it is clear that these targets are strongly correlated with each other in the presence of strong interactions.
GO analysis was first performed on 55 targets to explore how they affect and regulate tumorigenesis and progression through their biological behaviors. In the GO analysis, we use red, blue, and green colors for the BP, CC, and MF biological entries, respectively, and indicate the number of targets enriched to that entry when the circle is large. As shown in Figure 3D, these 55 targets are enriched in 163 BP entries, which are mainly related to inflammation response, cell proliferation, apoptosis, migration and hypoxia response, among others; 38 CC entries suggest that the disjoint targets are mostly localized in the extracellular matrix, cell membranes, synapses, and exosomes, which deserves further exploration to see if this suggests that the targets obtained by screening are related to signaling between cells, etc.; 48 MF entries suggested that the intersected targets were mainly related to the regulation of the activity of neurotransmitter receptor, integrin binding, protein junctions, and cytokine activities, and other biological functions.
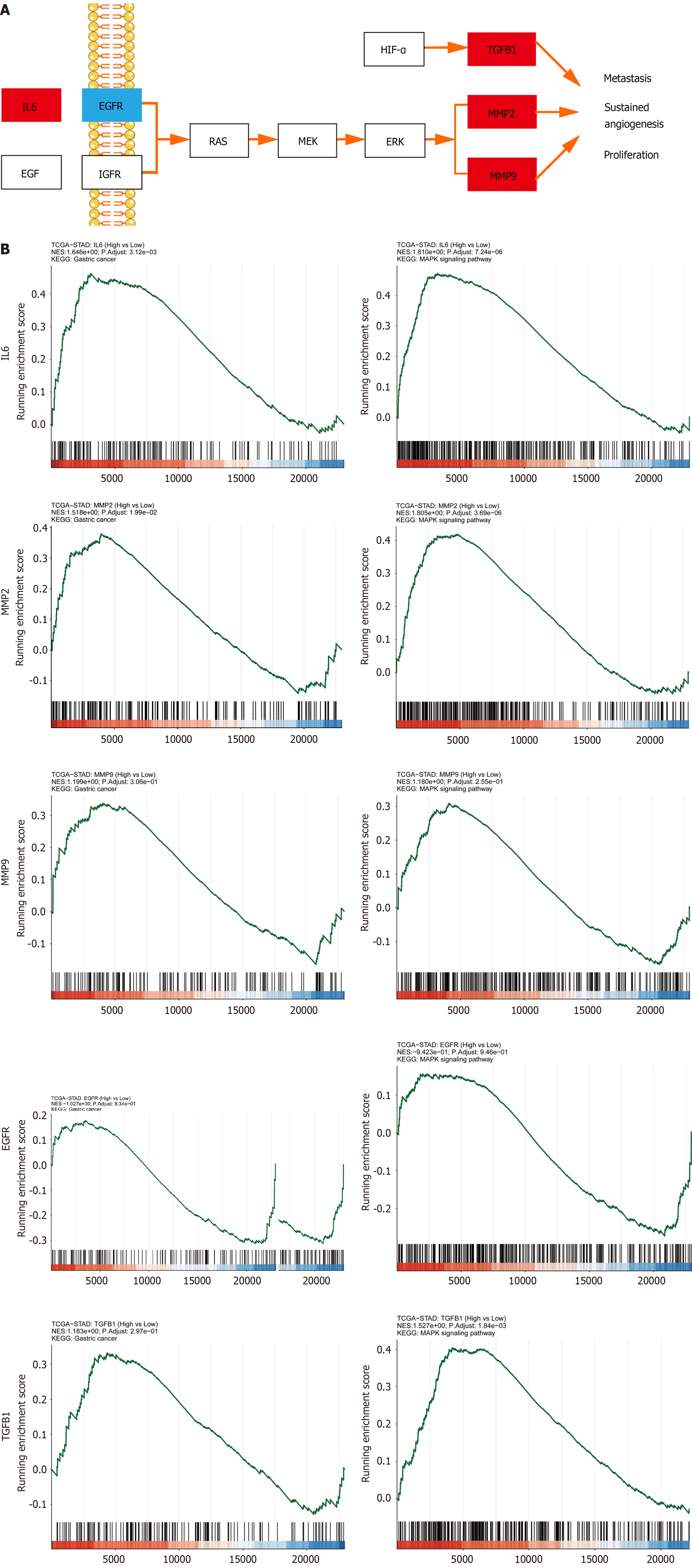
The KEGG analysis was continued to explore how YGS exerts anti-gastric cancer effects by modulating signaling pathways. As shown in Figure 3E, the 58 signaling pathways were 55 enriched pathways, and we further filtered the core pathways according to the number of targets enriched in the pathways, and as shown in Figure 3F, "pathways in cancers" was ranked first. After specific analysis of "pathways in cancer" using KEGG Mapper, we found that the MAPK signaling pathway was enriched with the highest number of intersecting targets, as shown in Figure 4A, IL6, TGFB1, MMP2 and MMP9 were up-regulated in this pathway, suggesting that this signaling pathway may be the key pathway for YGS to exert anti-gastric cancer, IL6, EGFR, MMP2, MMP9 and TGFB1 may be hub genes. The GSEA results shown in Figure 4B show that 5 hub genes are associated with gastric cancer and the MAPK signaling pathway, which is consistent with the results we obtained in the previous step. It has been suggested that YGS may exert its anti-gastric cancer pharmacological effects by modulating the BPs of tumor cells, such as proliferation, migration and apoptosis, through the MAPK signaling pathway.
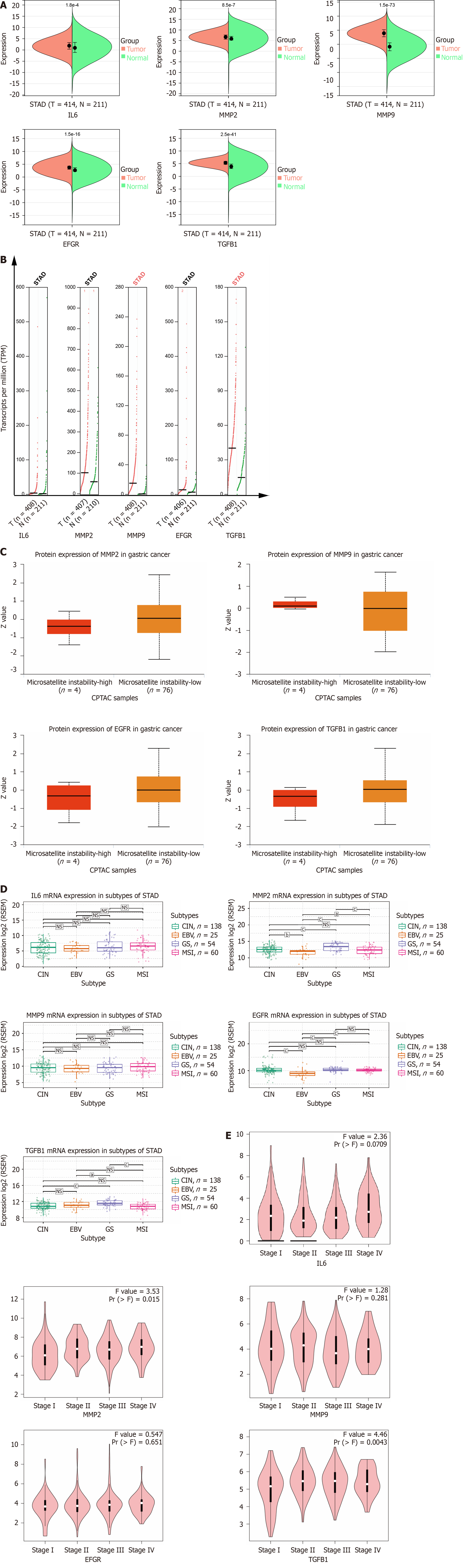
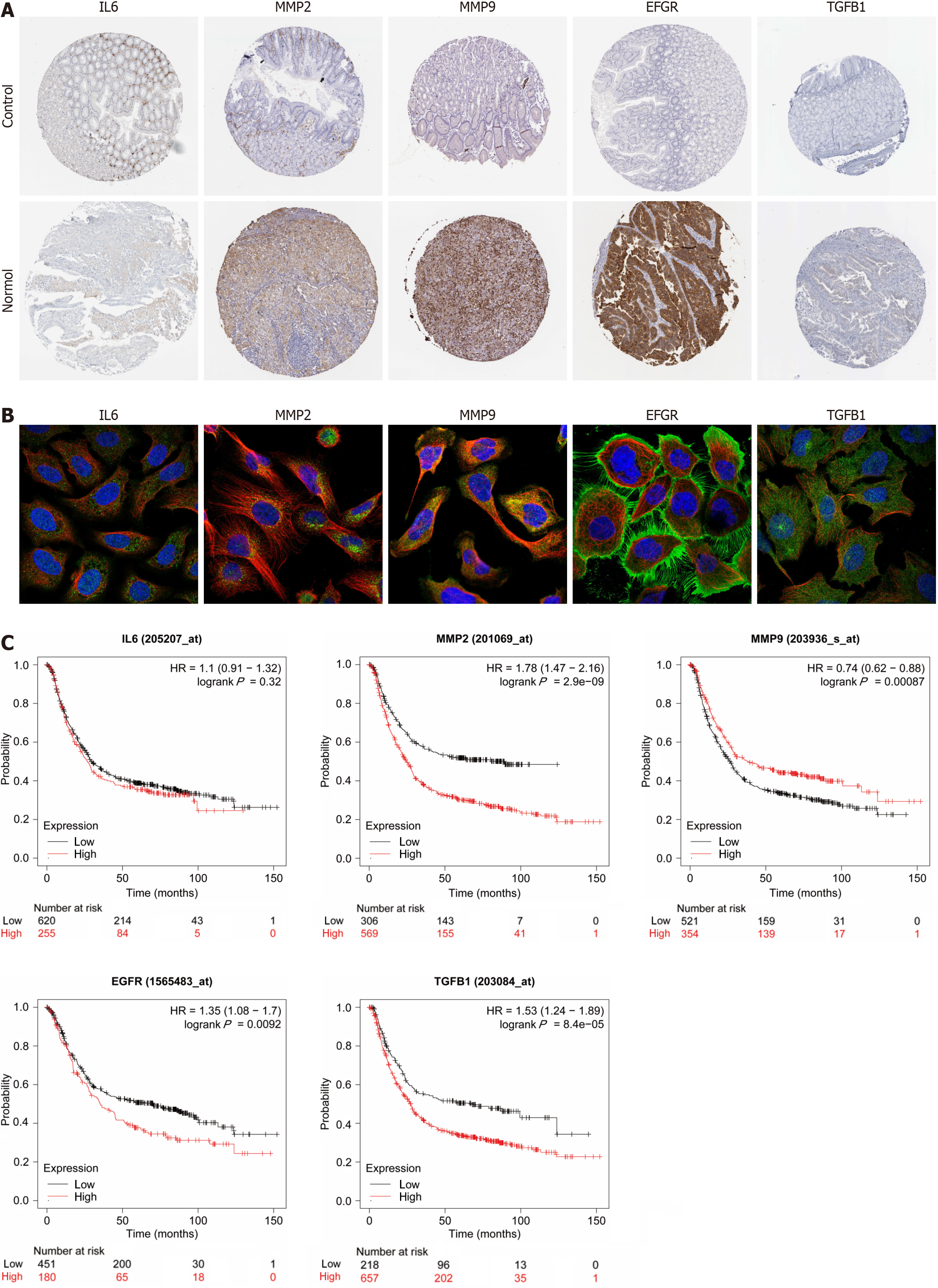
We analyzed the expression of hub genes at the transcriptional and translational levels to explore their association with gastric cancer. As shown in Figure 5A-C), all 5 hub genes show high expression in the gastric cancer group at the mRNA level. At the copy number level, MMP9 and TGFB1 are highly expressed in the stomach cancer group. While at the protein level, we observed low expression of MMP2, EGFR and TGFB1 in the gastric cancer microsatellite stabilizer group, in contrast to high expression of MMP9. Gastric cancer can be classified into four molecular subtypes, including EBV-positive (EBV), microsatellite-stable (MSI), genome-stable (GS), and chromosomally unstable (CIN). Different molecular subtypes were strongly associated with patient prognostic survival, with the EBV group having the best prognosis, followed by MSI and CIN, and GS having the worst prognosis. As shown in Figure 5D, the expression of MMP2 and TGFB1 increases in the GS subtype. Continuing the analysis of the hub genes with respect to the clinical stage of gastric cancer, Figure 5E shows that MMP2 and TGFB1 expression increases in intermediate and advanced gastric cancer. The expression of a gene may not be consistent at both the transcriptional and translational levels, and we proceed to analyze the protein expression of the hub gene, for which the HPA database is mainly used in this step. As shown in Figure 6A (patient and antibody information are in Supplementary Table 3), the plasmonic yellow part of the immunohistochemistry map is the region of increased protein expression, and the expression of all 5 hub genes is significantly increased in the gastric cancer group, which is consistent with the clinical correlation results we obtained. In order to understand the localization of hub genes in human tumor tissues for subsequent studies, we obtained their localization images in different tumor tissues, as shown in Figure 6B, blue fluorescence represents cell nuclei, red represents microtubule tissues, and green is the localization section of our hub genes, in which IL6 and MMP9 were mainly expressed in vesicles, MMP9 and TGFB1 were increased in cytoplasm, and EGFR was significantly expressed mainly in cell membranes and cellular junctions.
For the prognostic significance of hub genes, as shown in Figure 6C, in addition to IL6, MMP2, MMP9, EGFR and TGFB1 were correlated with the overall survival (OS) of patients with gastric cancer, among which the expression of MMP2, EGFR and TGFB1 was negatively correlated with the prognosis of the patients, which means that the higher the level of gene expression, the shorter the survival of patients with gastric cancer; on the contrary, the expression of MMP9 is positively correlated with the OS of patients.
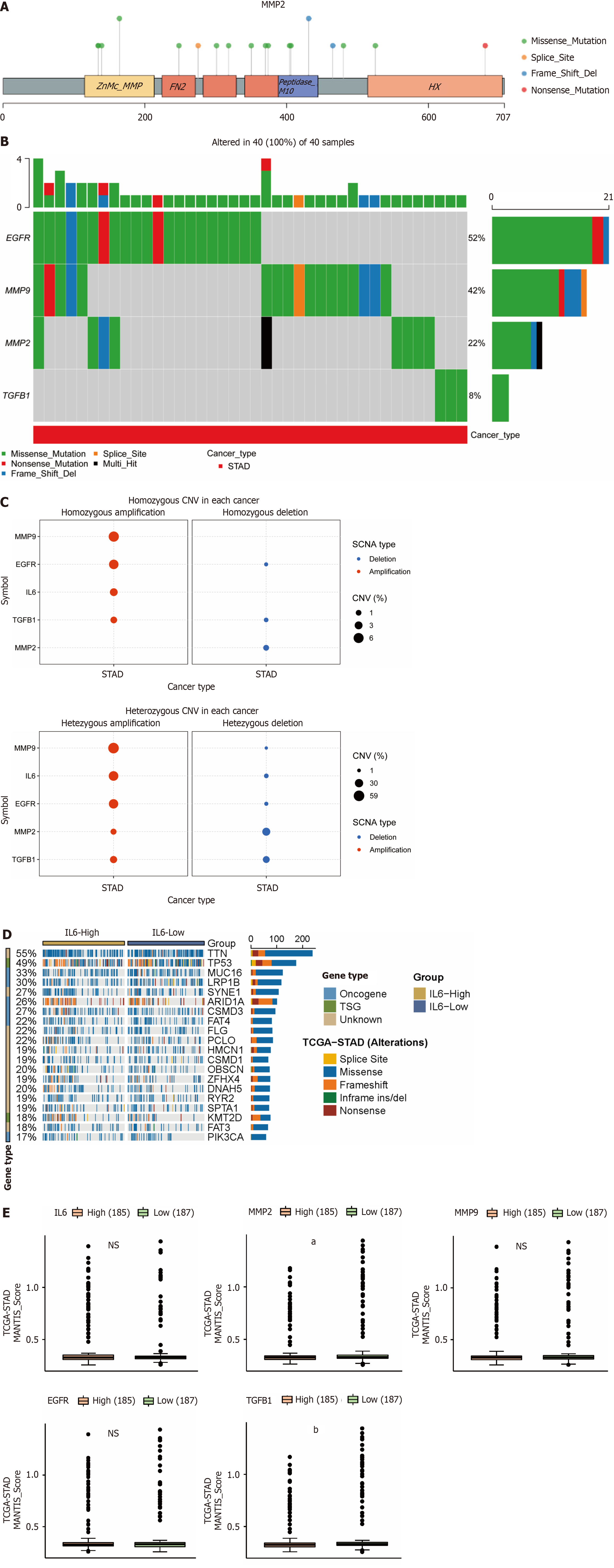
Tumorigenesis is characterized by genomic alterations, so we analyzed whether the 5 hub genes were altered at the genomic level by SNV and CNV mutations. We found that the hub genes were predominantly of the "missense-mutation" mutation type, as shown in Figure 7A for SNV mutation sites and types (MMP9, EGFR, and TGFB1 are in Supplementary Figure 1). We further analyzed the frequency of deleterious mutations in the hub genes, and as shown in Figure 7B, EGFR mutation frequency was the highest at 52%, followed by MMP9 at 42%, and notably, TGFB1 had the lowest mutation frequency at only 8%. CNVs include heterozygous and pure mutations, where pure mutations induce a more severe disease outcome. As shown in Figure 7C, where red and blue colors represent amplification and deletion, respectively, and the size of the bubble is proportional to the mutation frequency, we find that the 5 hub genes are predominantly mutated by amplification, with the highest proportion of MMP9 mutations. As shown in Figure 7D (MMP2, MMP9, EGFR, and TGFB1 are in Supplementary Figure 2), we observed that high IL6 expression promoted the expression of "driver mutation" such as LRP1B, ARID1A, CSMD3 and FAT4 proto-oncogenes. In addition, we also analyzed the correlation between MSI and hub gene in gastric cancer, as its alteration is also often seen in tumors and affects clinical outcomes and patient prognosis[19-21]. As shown in Figure 7E, MMP2 and TGFB1 are negatively correlated with MSI, that is, the higher the expression of the hub gene, the less MSI occurs.
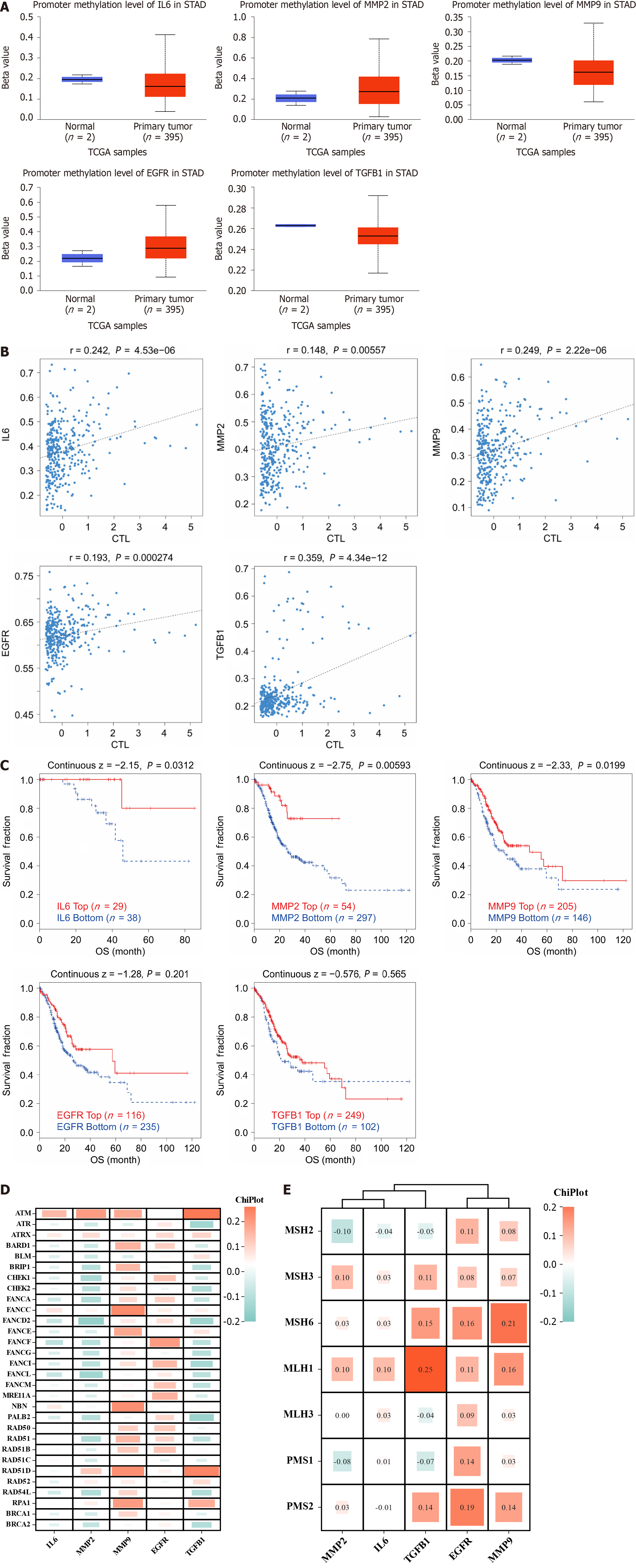
Since methylation occurs in the DNA of some tumors, altering chromatin structure, DNA conformation, DNA stability, and regulating gene transcription and expression processes, we analyzed the correlation between 5 hub genes and methylation. As shown in Figure 8A, IL6, MMP9 and TGFB1 showed humiliating levels in the gastric cancer group, while MMP2 and EGFR showed opposite results; furthermore, as shown in Figure 8B and C), the hub gene shows a positive correlation with both T lymphocyte toxicity and survival risk after methylation. Mutations in genes tend to be inextricably linked to the development and progression of gastric cancer, and genomic stability can in turn depend on different repair mechanisms, including MMR and HRR of DNA, so we analyzed the correlation between hub gene mutations and repair systems. As shown in Figure 8D and E), we found that MMP9 and EGFR showed significant positive correlation with the HRR repair system, while IL6, MMP2 and TGFB1 showed a negative correlation with the HRR; at the same time, repair-related genes showed mostly positive correlations with the 5 hub genes in the MMR repair system, with the strongest significance found in TGFB1, EGFR and MMP9.
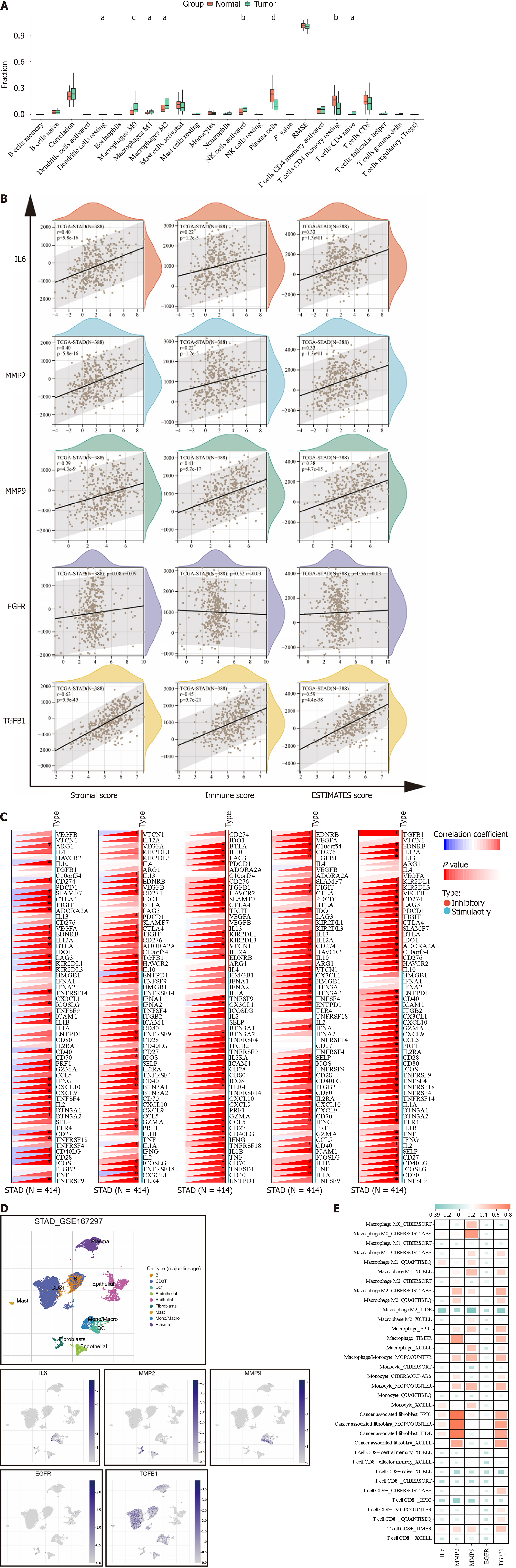
To evaluate the relevance of targets in gastric cancer to immune activity, we analyzed their correlation with 22 immune cells using CIBERSORT. As shown in Figure 9A, the tumor group showed high expression in macrophages and in NK cells, in contrast to low expression in plasma cells and cluster of differentiation 4 (CD4) T cells. The TME includes many cell types that play the dual roles of tumor promotion and tumor inhibition. Relevant scores are performed to obtain results in different dimensions, so that we can have a better picture of the TME. Among them, Stromal Score evaluates the fraction of stromal components in gastric cancer tissue, Immune Score represents the fraction of immune cells in gastric cancer tissue, and ESTIMATE Score evaluates tumor purity. As shown in Figure 9B, in addition to EGFR, IL6, MMP2, MMP9 and TGFB1 were positively associated with all three scores of gastric cancer. As shown in Figure 9C, these hub genes were also positively correlated with immune checkpoint molecules, suggesting that these genes may be involved in the immune point effect of gastric cancer. We used single-cell sequencing to annotate hub genes in cellular subpopulations, as shown in Figure 9D. In the GSE167297 dataset, IL6 was significantly expressed mainly in macrophages, monocytes, and endothelial cells; MMP2 is localized in the fibroblast; MMP9 was expressed in macrophages and dendritic cells (DCs), and TGFB1 was widely expressed in a wide range of cells, which is in agreement with our results that hub genes were mostly associated with macrophages and NK cells in the CIBERSORT. Five analysis algorithms, including CIBERSORT, CIBERSORT-ABS, QUANTISEQ, TIDE and XCELL methods, were further used to analyze the correlation of hub genes in macrophages, monocytes, tumor-associated fibroblasts and CD8+ T cells. As shown in Figure 9E, all 5 hub genes were positively correlated with macrophages and tumor fibroblasts, and conversely, they were negatively correlated with CD8+ T cells.
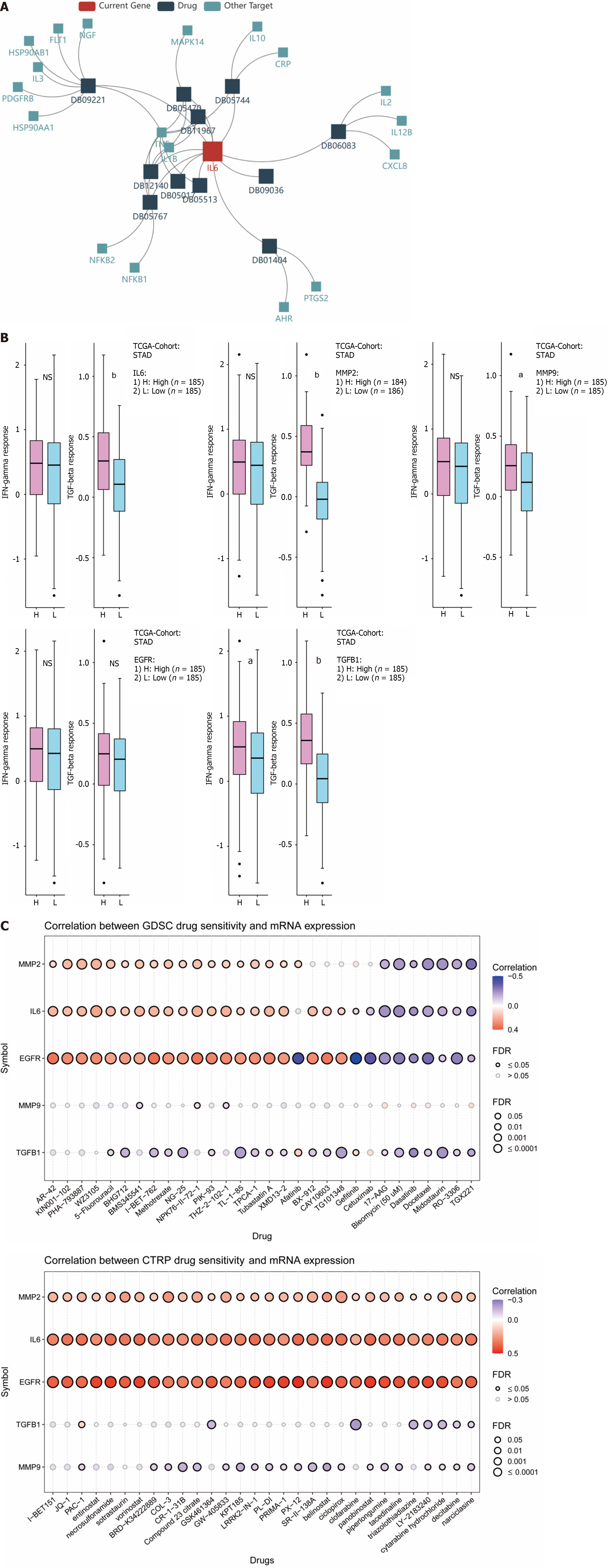
Immunotherapy refers to the treatment of tumors based on the principle of immunity by activating immune cells in the body and enhancing the anti-tumor immune response. TISIDB was first used to explore the relationship between hub genes and their targeted drugs. As shown in Figure 10A, the IL6-targeted drugs include DB01404, ginseng, which is also one of the important components of compound YGS. IFN-γ is an important activator of macrophages and an inducer of MHC II expression, and TGF-β can play a role as a cancer inhibitor in early-stage tumors. As shown in Figure 10B, TGFB1 was positively correlated with INF-γ expression; while all 5 hub genes were negatively correlated with TGF-β. Considering the poor clinical outcomes of gastric cancer patients receiving conventional chemotherapy, we tried to identify potential drugs with anti-hub genes. GDSC and CTRP were used to screen for sensitivity to anti-tumor drugs. As shown in Figure 10C, IL6, MMP2, MMP9, EGFR and TGFB1 show positive correlations with most of the drugs. The top 30 ranked in GDSC and CTRP None of the top 30 antitumor drugs in GDSC and CTRP contained the active ingredient of YGS, which also provides the basis for our next step of using YGS for the experimental validation of anti-gastric cancer and for the new adjuvant or surrogate treatment in the clinic.
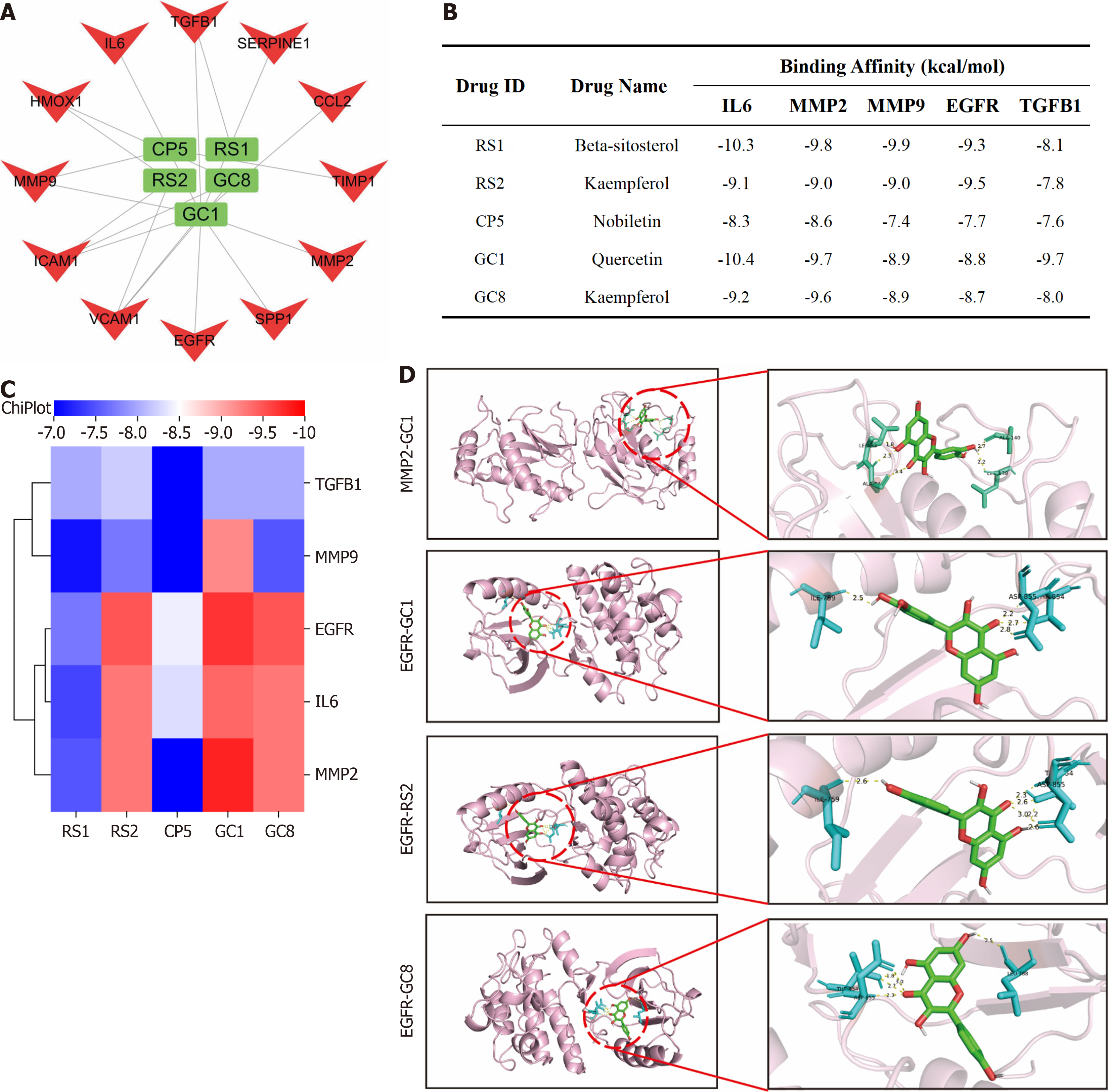
To further screen the key active ingredients and specific targets of action required for YGS to exert its therapeutic effect on gastric cancer, as shown in Figure 11A, CP5, RS1, RS2, GC1 and GC8 are likely to be the key active ingredients required for YGS to exert its anti-cancer effect. Molecular docking assays can be used to predict the binding ability of the active ingredient of a complex to a stomach cancer target, mimicking the absorption and metabolism of the drug in the human body. As shown in Figure 11B and C), the binding energies of CP5, RS1, RS2, GC1 and GC8 with the 5 hub genes were all < -7 kcal/mol, among which the binding energies of EGFR with GC1, RS2, GC8 and MMP2 and GC1 were the lowest, which were -9.5, -9.7, -9.5, and -9.8 kcal/mol, respectively and visualized this result as shown in Figure 11D.
The onset of gastric cancer is insidious, and most patients are diagnosed later than the appearance of clinical symptoms, so the scope of surgical resection is greatly reduced[22]. Although there are others ways for treatment, such as ra
Firstly, we obtained more than 100 active ingredients of YGS through TCMSP database, among which Nobiletin (CP5), Beta-sitosterol (RS1), Kaempferolf (RS2/GC8), Quercetin (GC1) might be the key to the pharmacological effects of the compound. After taking the intersection of gastric cancer and compound targets and screening them, we focused on 5 hub genes, namely IL6, MMP2, MMP9, EGFR and TGFB1, which we hypothesize are the primary targets of YGS for their anti-tumor effects. To further observe the relevance of these 5 hub genes to gastric cancer, we explored them at the level of mRNA, transcription, translation, and epigenetic regulation using publicly available databases. To our surprise, their expression in gastric cancer is indeed differential and correlated with the stage and prognosis of survival in gastric cancer patient. In terms of survival time of gastric cancer patients, MMP2, EGFR and TGFB1 are inversely proportional to survival time, that is, when the expression of these three genes is high, the survival time of patients is shorter, which may provide help for clinical judgment of the prognosis of patients. We continue to do functional and pathway enrichment of the intersected targets, and excitingly, we found that they are closely related to the BPs of apoptosis, proliferation and metastasis of tumor cells, and may exert anti-tumor effects through the MAPK signaling pathway. It is well known that the pathogenesis of tumors is very complex, in addition to the influence of environmental factors with which we are familiar, the abnormalities of the cell's metabolism and regulation also enter into the occurrence and progression of the disease, and this viewpoint is supported in our study, such as genes occurring in SNVs, CNVs mutations, mutations in the driver gene, the derangement of the regulation of methylation, and the alteration of the TME are all involved in the course of gastric cancer. Finally, anti-tumor drugs were screened, drawing our attention to the fact that RS has been shown to exert anti-gastric cancer pharmacology through immunomodulation, and that the remaining TCM have not been explored for this effect, providing an innovation in our research.
More and more studies have confirmed that tumors are not the exclusive domain of cancer cells alone, but a new environment including other surrounding cells and extracellular matrix, called " TME"[26], which has an important impact on tumor growth, invasion, metastasis, drug resistance, immune escape and other malignant phenotypes. TME has a bidirectional regulatory role: in the early stages, inflammatory cells, endothelial cells, macrophages, NK cells and DCs in the TME play a key role[27]. As the tumor progresses, the stromal cells can be "induced" to become various other cell types, and components of the tumor-suppressive microenvironment begin to be continuously suppressed, and the tumor-promoting microenvironment is continuously suppressed, leading to immune tolerance and tumor progression, which is one of the reasons for the chronic inflammatory nature of the tumor[28]. In our study, one of the screened hub genes, IL6, is an important pro-inflammatory cytokine that has been shown to be highly expressed in gastric cancer tissue.
IL6 is a typical oncogene, and its expression and dysregulation of downstream receptor signaling are common events in cancer, often suggestive of adverse clinical outcomes[29-32]. IL6 has an effect on various tumor growth and proliferation segments, such as inducing the transcriptional process of molecules such as the cell cycle G1/S-specific cytokine protein D1, proto-oncogenes MYC oncogene (MYC), and apoptosis regulator bcl x (BCL-XL), while also regulating the expression of proteins such as recombinant HIF1A, MMP2, and MMP9. In recent years, more and more studies have found that IL6 can participate in immune responses and promote tumor progression, such as inducing the production of pathogenic TH17 cells and MDSCs, inhibiting the activity of antigen-presenting DCs and anti-tumor cytotoxic CD8+ T cells and Treg cells, as well as switching tumor-associated macrophages from a tumor-killing m1-type phenotype to an immune-suppressing M2-type phenotype conversion, among others[33]. Tumor therapeutic modalities targeting IL6, including monoclonal antibodies that directly block cytokines or cytokine receptors, recombinant cytokine regimens, and small molecule therapies that interfere with the downstream pathways of cytokines, have been used progressively in the clinic. Through our analysis, we found that ginseng has a direct inhibitory effect on IL6, and some scholars have confirmed that ginseng has good immunomodulatory activity in humans, which also suggests that our compound, YGS, can exert anti-tumor activity through immunomodulatory pathways. It has been found that developing tumor cells can recruit bone marrow-derived cells to aid in their own differentiation toward a survival phenotype[34], and among the recruited cells are macrophages, which are one of the bone marrow-derived cells that promote EGFR expression as a means of increasing tumor cell invasive potential, angiogenesis, and other processes[35], which was confirmed by the high expression of EGFR in our study. In addition to this, macrophages induce TGFB1 production. TGFBI is a stromal cell protein that is mainly associated with the BPs of cell-matrix interactions and cell migration[36], and the deletion of this gene alters the microtubule stability[37] and is negatively correlated with the survival prognosis of patients. MMPs are a family of zinc-dependent endopeptidases[38] that mediate the degradation of a variety of molecules to promote cell adhesion and regulate cell and extracellular matrix interactions[39]. Ye et al[40] demonstrated a strong association between MMP2/MMP9 and immune profiles in mouse models of melanoma and lung cancer, and the use of an inhibitor of this molecule, SB-3CT, significantly enhanced the therapeutic efficacy of PD-1 or CTLA-4 blockade in the treatment of primary and metastatic tumors. Muniz-Bongers et al[41] similarly in melanoma confirmed this idea. Jenkins et al[42] suggested that immune checkpoint blockade in combination with other therapeutic strategies could improve the clinical outcome.
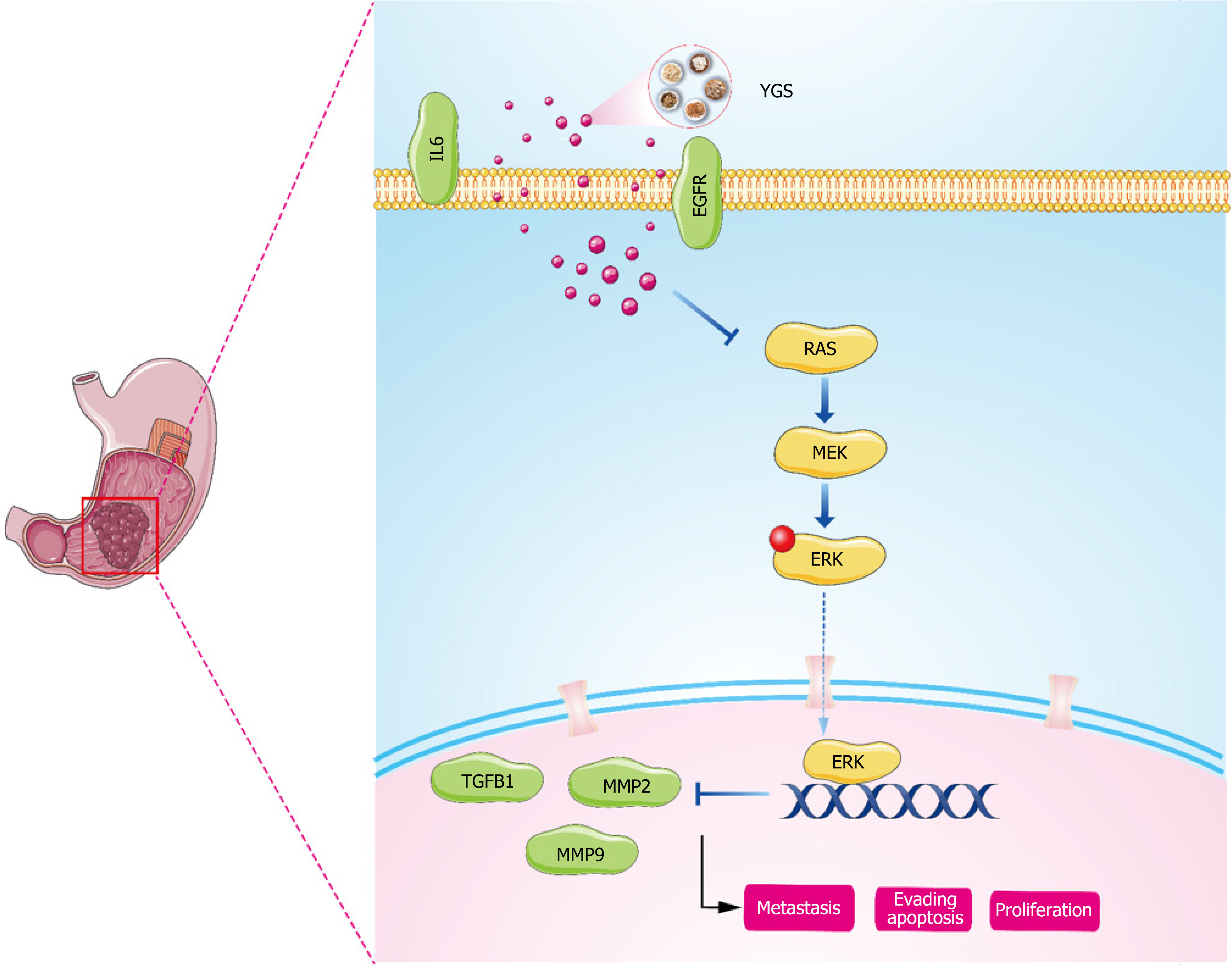
Our pathway enrichment analysis using biosignal analysis revealed that YGS was associated with the MAPK signaling pathway, i.e., MAPK signal transduction cascade axis as shown in Figure 12. Eukaryotes include a total of four: The ERK cascade, the p38MAPK cascade, the JNKs, and the ERK5 cascade, and the RAS/MEK/ERK signaling axis that we focused on in this study. The upstream molecule of this signaling axis, RAS, is stimulated by EGFR, TGFB1, IL6, etc. and rapidly binds to MEK to form a tetramer, and the cascade reaction phosphorylates downstream ERK, which then translocates to the nucleus and activates a large number of substrate molecules[43,44]. RAS, MER and ERK in this pathway are on
In summary, we have confirmed through network pharmacology and biosignature analysis that YGS exerts its anti-tumor effects through the MAPK signaling pathway, and participating in the modulation of immune regulatory mechanisms. However, our study still have some limitations. In the future, we will continue to validate our conjecture with in-vivo animal experiments using nude mouse tumorigenesis and in vitro cellular experiments; use gene silencing methods (lentiviral transfection technology) to verify the causal relationship between the upstream and downstream proteins of the MAPK signaling pathway; further detection of ERK regulation in gastric cancer was performed in combination with bi-luciferase and chromatin immunoprecipitation assays to explore the specific mechanisms of YGS anti-tumor activity.
Yigong San (YGS) is a representative prescription for the treatment of digestive disorders, which has been used in clinic for more than 1000 years. However, the mechanism of its anti-gastric cancer and regulate immunity are still remains unclear.
The treatment of gastric cancer includes drugs, chemotherapy, radiotherapy and surgery, but all of them have certain side effects.
To explore the mechanism of YGS anti-gastric cancer and immune regulation.
The main active ingredients and potential targets of YGS in the treatment of gastric cancer were screened by network pharmacology. Then bioinformatics technology was used to study the correlation between potential targets and the stage, prognosis and immune invasion of gastric cancer.
We obtained 55 common targets of gastric cancer and YGS. The IL6, EGFR, MMP2, MMP9 and TGFB1 as the hub genes. The 5 hub genes are involved in gastric carcinogenesis, staging, typing and prognosis, and their mutations promote gastric cancer progression.
YGS has the effect of anti-gastric cancer and immune regulation.
This study confirmed that YGS could be used as a new anti-gastric cancer drug.
We thank all the authors of this manuscript.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Baryshnikova NV, Russia S-Editor: Lin C L-Editor: A P-Editor: Zhao S
| 1. | Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654-2664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1282] [Cited by in RCA: 1468] [Article Influence: 163.1] [Reference Citation Analysis (0)] |
| 2. | National Health Commission of the People's Republic of China. National guidelines for diagnosis and treatment of gastric cancer 2022 in China (English version). Chin J Cancer Res. 2022;34:207-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 37] [Reference Citation Analysis (2)] |
| 3. | Lordick F, Carneiro F, Cascinu S, Fleitas T, Haustermans K, Piessen G, Vogel A, Smyth EC; ESMO Guidelines Committee. Electronic address: clinicalguidelines@esmo.org. Gastric cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2022;33:1005-1020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 689] [Article Influence: 229.7] [Reference Citation Analysis (0)] |
| 4. | Qian Yi. Xiaoer Yaozheng Zhijue. 1119. |
| 5. | Qu L, Tan W, Yang J, Lai L, Liu S, Wu J, Zou W. Combination Compositions Composed of l-Glutamine and Si-Jun-Zi-Tang Might Be a Preferable Choice for 5-Fluorouracil-Induced Intestinal Mucositis: An Exploration in a Mouse Model. Front Pharmacol. 2020;11:918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 6. | Zhang J, Wang X, Wang F, Tang X. Xiangsha Liujunzi Decoction improves gastrointestinal motility in functional dyspepsia with spleen deficiency syndrome by restoring mitochondrial quality control homeostasis. Phytomedicine. 2022;105:154374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 7. | Yue P, Zhong J, Huang J, Lan Z, Zhong S. The efficacy and safety of Xiangsha Liujunzi decoction in the treatment of chronic non-atrophic gastritis: A protocol for a systematic review and meta-analysis. Medicine (Baltimore). 2021;100:e24504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Rao K, Qin S, Yang Y, Zhan K, Wu H, Zheng H, Huang S. Shenling Baizhu Powder Alleviates TNBS-Induced Colitis in Rats by Improving Intestinal Epithelial Permeability and Inhibiting Inflammation Through the TLR5/MyD88/NF-κB Pathway. Front Pharmacol. 2022;13:883918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 664] [Cited by in RCA: 5033] [Article Influence: 1677.7] [Reference Citation Analysis (0)] |
| 10. | Huang J, Liu D, Wang Y, Liu L, Li J, Yuan J, Jiang Z, Hsiao WW, Liu H, Khan I, Xie Y, Wu J, Zhang Y, Fu Y, Liao J, Wang W, Lai H, Shi A, Cai J, Luo L, Li R, Yao X, Fan X, Wu Q, Liu Z, Yan P, Lu J, Yang M, Wang L, Cao Y, Wei H, Leung EL. Ginseng polysaccharides alter the gut microbiota and kynurenine/tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/programmed cell death ligand 1 (anti-PD-1/PD-L1) immunotherapy. Gut. 2022;71:734-745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 316] [Cited by in RCA: 312] [Article Influence: 104.0] [Reference Citation Analysis (0)] |
| 11. | Zhou R, He D, Xie J, Zhou Q, Zeng H, Li H, Huang L. The Synergistic Effects of Polysaccharides and Ginsenosides From American Ginseng (Panax quinquefolius L.) Ameliorating Cyclophosphamide-Induced Intestinal Immune Disorders and Gut Barrier Dysfunctions Based on Microbiome-Metabolomics Analysis. Front Immunol. 2021;12:665901. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 84] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 12. | Liu X, Wang X, Xu X, Zhang X. Purification, antitumor and anti-inflammation activities of an alkali-soluble and carboxymethyl polysaccharide CMP33 from Poria cocos. Int J Biol Macromol. 2019;127:39-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 13. | Zheng Y, Zeng X, Chen P, Chen T, Peng W, Su W. Integrating Pharmacology and Gut Microbiota Analysis to Explore the Mechanism of Citri Reticulatae Pericarpium Against Reserpine-Induced Spleen Deficiency in Rats. Front Pharmacol. 2020;11:586350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 14. | Xu H, Van der Jeught K, Zhou Z, Zhang L, Yu T, Sun Y, Li Y, Wan C, So KM, Liu D, Frieden M, Fang Y, Mosley AL, He X, Zhang X, Sandusky GE, Liu Y, Meroueh SO, Zhang C, Wijeratne AB, Huang C, Ji G, Lu X. Atractylenolide I enhances responsiveness to immune checkpoint blockade therapy by activating tumor antigen presentation. J Clin Invest. 2021;131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 100] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 15. | Yuan LW, Jiang XM, Xu YL, Huang MY, Chen YC, Yu WB, Su MX, Ye ZH, Chen X, Wang Y, Lu JJ. Licochalcone A inhibits interferon-gamma-induced programmed death-ligand 1 in lung cancer cells. Phytomedicine. 2021;80:153394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 16. | Li S, Wang YY, Ji L, Li YD. TCM and its case studies in the context of complex systems. Xitong Fanfzhen Xuebao. 2002;14: 1429-1431+1442. |
| 17. | Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2254] [Cited by in RCA: 2811] [Article Influence: 165.4] [Reference Citation Analysis (0)] |
| 18. | Guidelines for network pharmacological evaluation methods. Zhongguo Linchuangyuxinyao Zazhi. 2021;40:459. |
| 19. | Ben-David U, Amon A. Context is everything: aneuploidy in cancer. Nat Rev Genet. 2020;21:44-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 417] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 20. | Ettinger DS, Wood DE, Aggarwal C, Aisner DL, Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, D'Amico TA, Dilling TJ, Dobelbower M, Gettinger S, Govindan R, Gubens MA, Hennon M, Horn L, Lackner RP, Lanuti M, Leal TA, Lin J, Loo BW Jr, Martins RG, Otterson GA, Patel SP, Reckamp KL, Riely GJ, Schild SE, Shapiro TA, Stevenson J, Swanson SJ, Tauer KW, Yang SC, Gregory K; OCN, Hughes M. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 1.2020. J Natl Compr Canc Netw. 2019;17:1464-1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 527] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 21. | Picard E, Verschoor CP, Ma GW, Pawelec G. Relationships Between Immune Landscapes, Genetic Subtypes and Responses to Immunotherapy in Colorectal Cancer. Front Immunol. 2020;11:369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 348] [Article Influence: 69.6] [Reference Citation Analysis (0)] |
| 22. | Maconi G, Manes G, Porro GB. Role of symptoms in diagnosis and outcome of gastric cancer. World J Gastroenterol. 2008;14:1149-1155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 115] [Cited by in RCA: 147] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 23. | National Health Commission of the People's Republic of China. Notice of the Ministry of Health on Further regulating the management of raw materials for Health food. Mar 11, 2002. [cited 3 August 2022]. Available from: http://www.nhc.gov.cn/wjw/gfxwj/201304/e33435ce0d894051b15490aa3219cdc4.shtml. |
| 24. | Zhu J, Jiang XH, Qiang CX, Xu RF, Zhu YY. Effects of addition and reduction of Yigong SAN combined with Dongyuan acupuncture on gastrointestinal symptoms, gastrointestinal hormones and quality of life in patients with irritable bowel syndrome with spleen and stomach weakness. Sichuan Zhongyi. 2023;41:100-103. |
| 25. | He W, Chen MZ. Effects of Wuwei Yigong Powder on renal function and blood lipid in patients with chronic glomerulonephritis. Shaanxi Zhongyi. 2016;37:1142-1143. |
| 26. | Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2921] [Cited by in RCA: 3362] [Article Influence: 258.6] [Reference Citation Analysis (0)] |
| 27. | Albini A, Sporn MB. The tumour microenvironment as a target for chemoprevention. Nat Rev Cancer. 2007;7:139-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 580] [Cited by in RCA: 602] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 28. | Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339:286-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 849] [Cited by in RCA: 866] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 29. | Jones SA, Jenkins BJ. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat Rev Immunol. 2018;18:773-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 745] [Article Influence: 106.4] [Reference Citation Analysis (0)] |
| 30. | Li N, Grivennikov SI, Karin M. The unholy trinity: inflammation, cytokines, and STAT3 shape the cancer microenvironment. Cancer Cell. 2011;19:429-431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 200] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 31. | Taniguchi K, Karin M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin Immunol. 2014;26:54-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 533] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 32. | Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1338] [Cited by in RCA: 1661] [Article Influence: 151.0] [Reference Citation Analysis (0)] |
| 33. | Tsukamoto H, Fujieda K, Senju S, Ikeda T, Oshiumi H, Nishimura Y. Immune-suppressive effects of interleukin-6 on T-cell-mediated anti-tumor immunity. Cancer Sci. 2018;109:523-530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 120] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 34. | Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2228] [Cited by in RCA: 3065] [Article Influence: 255.4] [Reference Citation Analysis (0)] |
| 35. | Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2949] [Cited by in RCA: 2767] [Article Influence: 172.9] [Reference Citation Analysis (0)] |
| 36. | Mosher DF, Johansson MW, Gillis ME, Annis DS. Periostin and TGF-β-induced protein: Two peas in a pod? Crit Rev Biochem Mol Biol. 2015;50:427-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 37. | Steitz AM, Steffes A, Finkernagel F, Unger A, Sommerfeld L, Jansen JM, Wagner U, Graumann J, Müller R, Reinartz S. Tumor-associated macrophages promote ovarian cancer cell migration by secreting transforming growth factor beta induced (TGFBI) and tenascin C. Cell Death Dis. 2020;11:249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 38. | Verma RP, Hansch C. Matrix metalloproteinases (MMPs): chemical-biological functions and (Q)SARs. Bioorg Med Chem. 2007;15:2223-2268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 480] [Cited by in RCA: 555] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 39. | Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52-67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3948] [Cited by in RCA: 3800] [Article Influence: 253.3] [Reference Citation Analysis (0)] |
| 40. | Ye Y, Kuang X, Xie Z, Liang L, Zhang Z, Zhang Y, Ma F, Gao Q, Chang R, Lee HH, Zhao S, Su J, Li H, Peng J, Chen H, Yin M, Peng C, Yang N, Wang J, Liu J, Liu H, Han L, Chen X. Small-molecule MMP2/MMP9 inhibitor SB-3CT modulates tumor immune surveillance by regulating PD-L1. Genome Med. 2020;12:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 41. | Muniz-Bongers LR, McClain CB, Saxena M, Bongers G, Merad M, Bhardwaj N. MMP2 and TLRs modulate immune responses in the tumor microenvironment. JCI Insight. 2021;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 42. | Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, Bowden M, Deng J, Liu H, Miao D, He MX, Walker W, Zhang G, Tian T, Cheng C, Wei Z, Palakurthi S, Bittinger M, Vitzthum H, Kim JW, Merlino A, Quinn M, Venkataramani C, Kaplan JA, Portell A, Gokhale PC, Phillips B, Smart A, Rotem A, Jones RE, Keogh L, Anguiano M, Stapleton L, Jia Z, Barzily-Rokni M, Cañadas I, Thai TC, Hammond MR, Vlahos R, Wang ES, Zhang H, Li S, Hanna GJ, Huang W, Hoang MP, Piris A, Eliane JP, Stemmer-Rachamimov AO, Cameron L, Su MJ, Shah P, Izar B, Thakuria M, LeBoeuf NR, Rabinowits G, Gunda V, Parangi S, Cleary JM, Miller BC, Kitajima S, Thummalapalli R, Miao B, Barbie TU, Sivathanu V, Wong J, Richards WG, Bueno R, Yoon CH, Miret J, Herlyn M, Garraway LA, Van Allen EM, Freeman GJ, Kirschmeier PT, Lorch JH, Ott PA, Hodi FS, Flaherty KT, Kamm RD, Boland GM, Wong KK, Dornan D, Paweletz CP, Barbie DA. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov. 2018;8:196-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 416] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 43. | Macdonald SG, Crews CM, Wu L, Driller J, Clark R, Erikson RL, McCormick F. Reconstitution of the Raf-1-MEK-ERK signal transduction pathway in vitro. Mol Cell Biol. 1993;13:6615-6620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 66] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 44. | Marais R, Light Y, Paterson HF, Marshall CJ. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 1995;14:3136-3145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 430] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 45. | Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nat Rev Clin Oncol. 2018;15:709-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 255] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 46. | Moore AR, Rosenberg SC, McCormick F, Malek S. RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov. 2020;19:533-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 709] [Article Influence: 141.8] [Reference Citation Analysis (0)] |
| 47. | Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, Falchook GS, Price TJ, Sacher A, Denlinger CS, Bang YJ, Dy GK, Krauss JC, Kuboki Y, Kuo JC, Coveler AL, Park K, Kim TW, Barlesi F, Munster PN, Ramalingam SS, Burns TF, Meric-Bernstam F, Henary H, Ngang J, Ngarmchamnanrith G, Kim J, Houk BE, Canon J, Lipford JR, Friberg G, Lito P, Govindan R, Li BT. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med. 2020;383:1207-1217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1184] [Article Influence: 236.8] [Reference Citation Analysis (0)] |
| 48. | Allen JE, Krigsfeld G, Mayes PA, Patel L, Dicker DT, Patel AS, Dolloff NG, Messaris E, Scata KA, Wang W, Zhou JY, Wu GS, El-Deiry WS. Dual inactivation of Akt and ERK by TIC10 signals Foxo3a nuclear translocation, TRAIL gene induction, and potent antitumor effects. Sci Transl Med. 2013;5:171ra17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 283] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 49. | Sullivan RJ, Infante JR, Janku F, Wong DJL, Sosman JA, Keedy V, Patel MR, Shapiro GI, Mier JW, Tolcher AW, Wang-Gillam A, Sznol M, Flaherty K, Buchbinder E, Carvajal RD, Varghese AM, Lacouture ME, Ribas A, Patel SP, DeCrescenzo GA, Emery CM, Groover AL, Saha S, Varterasian M, Welsch DJ, Hyman DM, Li BT. First-in-Class ERK1/2 Inhibitor Ulixertinib (BVD-523) in Patients with MAPK Mutant Advanced Solid Tumors: Results of a Phase I Dose-Escalation and Expansion Study. Cancer Discov. 2018;8:184-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 302] [Article Influence: 37.8] [Reference Citation Analysis (0)] |