Published online Jul 15, 2023. doi: 10.4251/wjgo.v15.i7.1200
Peer-review started: February 4, 2023
First decision: March 21, 2023
Revised: March 28, 2023
Accepted: May 6, 2023
Article in press: May 6, 2023
Published online: July 15, 2023
Processing time: 157 Days and 19.8 Hours
Worldwide, gastric cancer (GC) is a common lethal solid malignancy with a poor prognosis. Cuproptosis is a novel type of cell death mediated by protein lipoylation and may be related to GC prognosis.
To offer new insights to predict GC prognosis and provide multiple therapeutic targets related to cuproptosis-related genes (CRGs) for future therapy.
We collected data from several public data portals, systematically estimated the expression level and prognostic values of CRGs in GC samples, and investigated related mechanisms using public databases and bioinformatics.
Our results revealed that FDX1, LIAS, and MTF1 were differentially expressed in GC samples and exhibited important prognostic significance in The Cancer Genome Atlas (TCGA) cohort. We constructed a nomogram model for overall survival and disease-specific survival prediction and validated it via calibration plots. Mecha-nistically, immune cell infiltration and DNA methylation prominently affected the survival time of GC patients. Moreover, protein-protein interaction network, KEGG pathway and gene ontology enrichment analyses demonstrated that FDX1, LIAS, MTF1 and related proteins play key roles in the tricarboxylic acid cycle and cuproptosis. Gene Expression Omnibus database validation showed that the expression levels of FDX1, LIAS, and MTF1 were consistent with those in the TCGA cohort. Top 10 perturbagens has been filtered by Connectivity Map.
In conclusion, FDX1, LIAS, and MTF1 could serve as potential prognostic biomarkers for GC patients and provide novel targets for immunotarget therapy.
Core Tip: In this study, the molecular biological mechanisms of cuproptosis-related genes (CRGs) were explored in gastric cancer, and clinical prognostic models for gastric cancer treatment were constructed by interactively analysing the links among CRGs and clinical information using bioinformatics. We constructed a significant prognostic nomogram model for gastric cancer and found that FDX1, LIAS, and MTF1 could serve as potential prognostic biomarkers for gastric cancer patients and provide novel targets for immunotarget therapy.
- Citation: Yan JN, Guo LH, Zhu DP, Ye GL, Shao YF, Zhou HX. Clinical significance and potential application of cuproptosis-related genes in gastric cancer. World J Gastrointest Oncol 2023; 15(7): 1200-1214
- URL: https://www.wjgnet.com/1948-5204/full/v15/i7/1200.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i7.1200
Currently, gastric cancer (GC) is a common malignant tumour with a high incidence and mortality rate worldwide, imposing a substantial economic burden on society[1]. The detailed pathogenesis of GC is currently unclear, and more than 35% of patients are initially diagnosed with distant metastasis and poor prognosis[2]. Although novel treatments, such as chemotherapy, surgery, radiotherapy and combination therapy, are constantly being updated, the prognosis of GC patients remains suboptimal[3]. Hence, it is urgent to understand the molecular mechanisms of GC and establish an effective prognostic model for clinical application.
Copper is an important cofactor for essential enzymes, and dysregulation of copper homeostasis can trigger cytotoxicity. Recent research points out that copper ionophores induce a distinct form of regulated cell death mediated by protein lipoylation of the tricarboxylic acid (TCA) cycle[4]. This special process is also called cuproptosis. Moreover, lipoylated proteins are tightly associated with a variety of human tumours, and cells with high levels of lipoylated proteins are sensitive to cuproptosis, which suggests that cuproptosis is strongly correlated with the biological behaviour of malignant tumour cells[4]. Additionally, it has been confirmed that abnormalities in intermediates in the TCA cycle are related to mitochondrial functions and GC morbidity[5]. All of this evidence suggests that cuproptosis influences the development and distal survival time of GC patients.
In our study, we systematically analysed the molecular alterations in cuproptosis-related genes (CRGs) and constructed a novel prognostic nomogram model in GC using bioinformatics technology. Our findings offer new insights into predicting GC prognosis and provide multiple therapeutic targets for future therapy.
We chose several open-source databases to retrieve the expression profiles, clinical information and survival data of GC and normal tissues, such as The Cancer Genome Atlas (TCGA) database (https://genome-cancer.ucsc.edu/) and the Genotype-Tissue Expression (GTEx) project. A total of 414 GC samples, 36 adjunct nontumor samples and 174 normal tissues were analysed in this study. All data were available in public open-access databases, and additional approval from the local ethics committee was not needed.
After a literature search, we selected 19 genes (ATP7A, ATP7B, CDKN2A, DBT, DLAT, DLD, DLST, FDX1, GCSH, GLS, LIAS, LIPT1, LIPT2, MTF1, NFE2L2, NLRP3, PDHA1, PDHB, SLC31A1) that function closely with cuproptosis[4]. We first compared the differentially expressed CRGs in GC from the TCGA cohort and in normal tissues in the GTEx cohort using the R statistical computing environment (3.6.3; R Foundation for Statistical Computing). P < 0.05 was considered statistically significant. We logged into the cBioPortal website (https://www.cbioportal.org/) and surveyed the mutation information for differentially expressed CRGs in GC[6].
Cox proportional hazards regression was performed to filter the prognosis-related genes, and P < 0.2 was considered statistically significant in the multivariate Cox proportional hazards regression model.
We first calculated the risk score for each sample using regression coefficients to identify the prognostic signature of CRGs for overall survival (OS) and disease-specific survival (DSS). The patients were further divided into high-risk and low-risk groups according to the median risk score. Subsequently, we analysed the survival data for each prognosis-related CRG in the high-risk and low-risk groups using the Kaplan-Meier method via the R package survival v 3.2-10.
Moreover, we established an OS and DSS nomogram model based on these prognosis-related CRGs. The concordance index (C-index) was used to obtain the discrimination of the nomogram, and calibration plots were generated to display the association between the predicted and observed risk results.
Methylation analysis of prognosis-related CRGs was performed via Methsurv (https://biit.cs.ut.ee/methsurv/), a web tool to perform multivariable survival analysis using DNA methylation data[7-9].
We determined the survival significance of prognosis-related CRGs and the immune infiltration levels of several immune cell types. Survival Genie is a web tool used to perform survival analysis of single-cell RNA-seq data and a variety of other molecular inputs for several cancer types[10]. We first applied Survival Genie to investigate correlations between prognosis-related CRGs and immune infiltration levels. Then, we detected the immune infiltration level of multifarious immune cells in the TCGA cohort using the R package “GSVA”[11]. TIMER, an online portal for systematic analysis of immune infiltrates across diverse cancer types (http://timer.cistrome.org), was used to validate the results[12-14]. Spearman’s correlation analysis was performed to determine the association between quantitative variables.
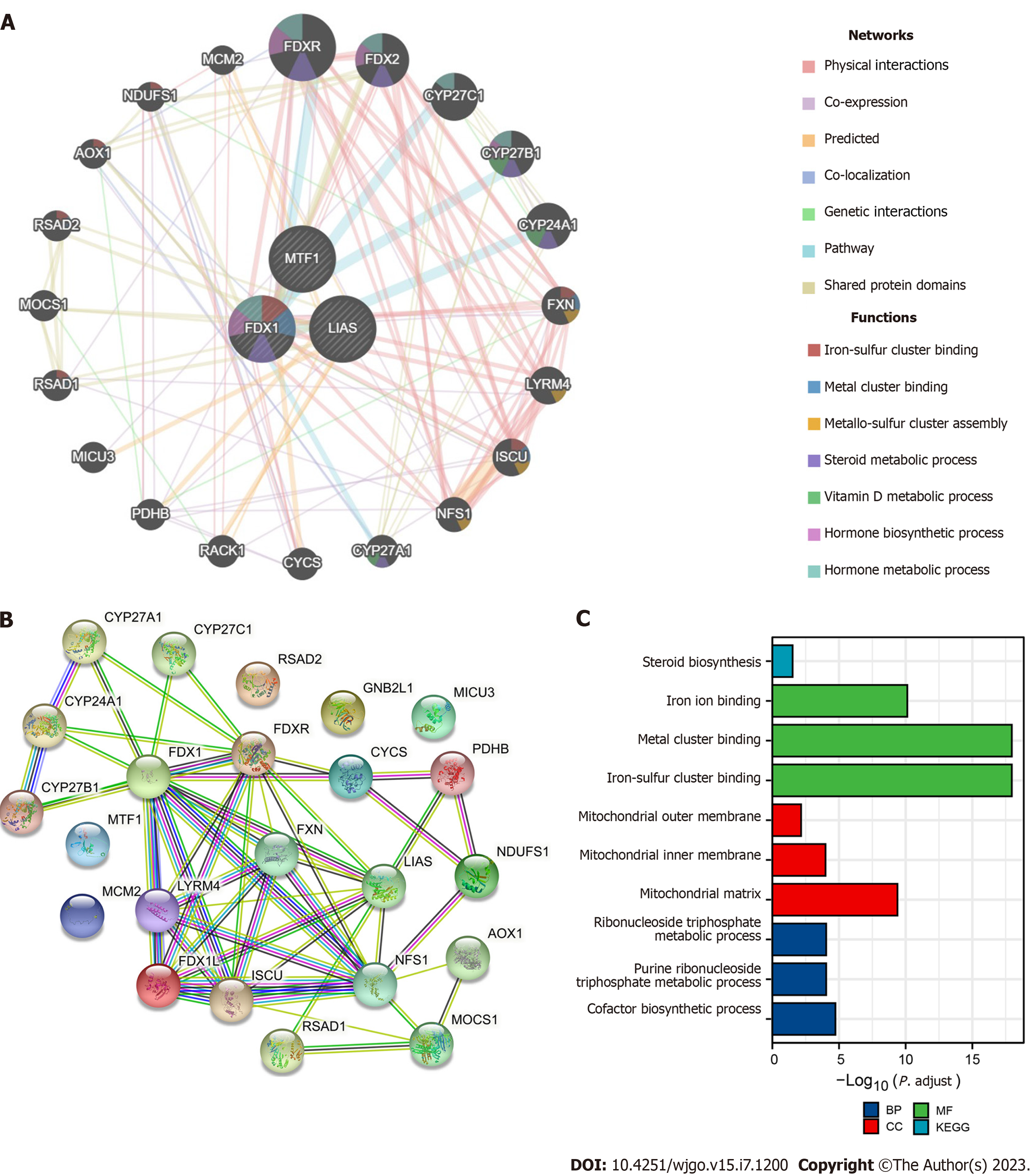
The GeneMANIA prediction server is a web interface for generating hypotheses about biological network integration for gene prioritization and predicting gene function[15]. We input the prognosis-related CRGs and output the nearest gene for each locus. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) website (https://string-db.org/) contains various protein-protein correlation data, which were used to build a prognosis-related CRG interacting protein-protein interaction (PPI) network. A confidence score > 0.7 was considered significant[16]. We input the genes preserved from GeneMANIA and output the networks. The nodes in the PPI network were further used to perform KEGG pathway enrichment analysis and gene ontology (GO) classification via the R packages “clusterProfiler” and “ggplot2”. A P value < 0.05, min enrichment > 3, and min overlap > 3 were considered significant[17]. Connectivity Map (https://clue.io/, CMap) is a systematic tool to discover functional connections among diseases and was utilized to find perturbagens to the expression of CRGs[18-20]. We selected the “Query” module and further filtered the top 10 perturbagens of “FDR_q_nlog 10” with an explicit “moa”.
The TNM plot is a web tool from the National Center for Biotechnology Information (www.tnmplot.com) used for comparison of gene expression in various tumours[21]. We chose the “compare Tumour and Normal” and “Gene chip data” modules for validation using Gene Expression Omnibus (GEO) samples. P < 0.05 was deemed statistically significant.
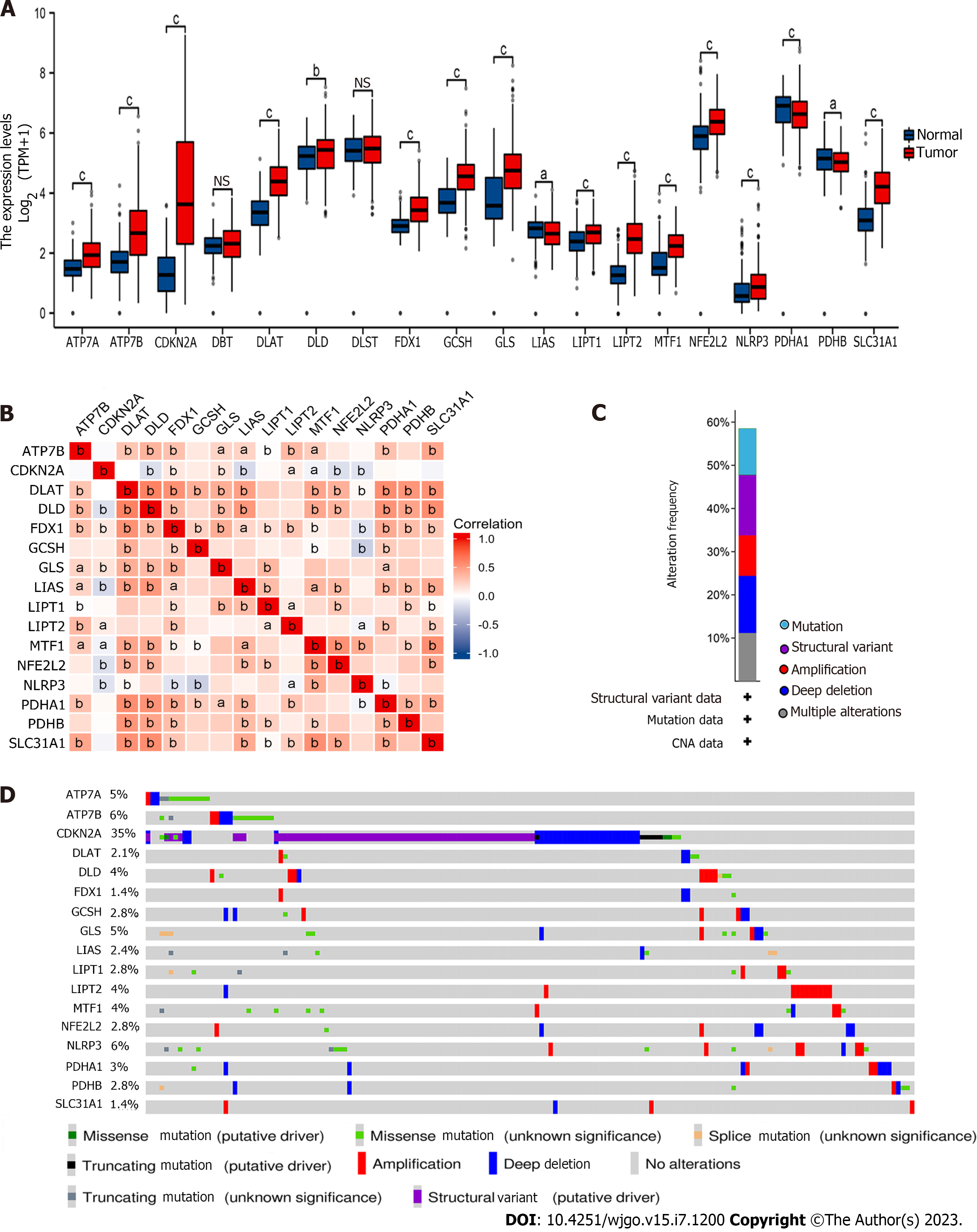
As previously mentioned, we contrasted the expression levels of CRGs in the GC cohort displayed in Figure 1A. We found that ATP7A, ATP7B, CDKN2A, DLAT, DLD, FDX1, GCSH, GLS, LIAS, LIPT1, LIPT2, MTF1, NFE2L2, NLRP3, PDHA1, PDHB, and SLC31A1 were differentially expressed in GC (P < 0.05). Then, we performed coexpression analysis of these CRGs and visualized them via a heatmap, which showed a high correlation (Figure 1B). For example, FDX1 was significantly positively associated with LIAS and negatively associated with MTF1.
Furthermore, we determined the gene mutation patterns of these CRGs in GC. The overall mutation landscape is shown in Figure 1C, and we list the particular patterns of each gene mutation in Figure 1D.
We further investigated the relationship between the expression of CRGs and prognosis in GC samples. We first constructed a multivariable Cox regression model to estimate the roles of CRGs in OS and DSS in the TCGA cohort. Our results showed that FDX1 (P = 0.059) and MTF1 (P = 0.088) were remarkably associated with OS in GC samples, as shown in Table 1. Similarly, FDX1 (P = 0.181), LIAS (P = 0.045), and MTF1 (P = 0.117) were remarkably associated with DSS in GC samples, as shown in Table 2. Hence, we selected FDX1, LIAS, and MTF1 as prognosis-related CRGs. The clinical information for FDX1, LIAS, and MTF1 in the TCGA cohort is shown in Supplementary Tables 1-3.
| Gene | Total, n | Univariate analysis | Multivariate analysis | ||
| Hazard ratio (95%CI) | P value | Hazard ratio (95%CI) | P value | ||
| ATP7A | 370 | 1.037 (0.725-1.483) | 0.842 | ||
| ATP7B | 370 | 0.922 (0.781-1.088) | 0.334 | ||
| CDKN2A | 370 | 0.985 (0.887-1.094) | 0.782 | ||
| DLAT | 370 | 0.785 (0.577-1.069) | 0.124 | ||
| DLD | 370 | 0.961 (0.678-1.363) | 0.825 | ||
| FDX1 | 370 | 0.737 (0.533-1.018) | 0.064 | 0.735 (0.534-1.011) | 0.059 |
| GCSH | 370 | 1.054 (0.769-1.446) | 0.744 | ||
| GLS | 370 | 1.052 (0.845-1.310) | 0.650 | ||
| LIAS | 370 | 0.730 (0.498-1.068) | 0.105 | ||
| LIPT1 | 370 | 1.168 (0.713-1.916) | 0.537 | ||
| LIPT2 | 370 | 1.014 (0.794-1.294) | 0.912 | ||
| MTF1 | 370 | 0.642 (0.410-1.006) | 0.053 | 0.661 (0.411-1.064) | 0.088 |
| NFE2L2 | 370 | 0.701 (0.477-1.031) | 0.071 | 0.809 (0.534-1.225) | 0.317 |
| NLRP3 | 370 | 1.279 (0.946-1.729) | 0.110 | ||
| PDHA1 | 370 | 0.873 (0.632-1.206) | 0.409 | ||
| PDHB | 370 | 1.051 (0.686-1.611) | 0.818 | ||
| SLC31A1 | 370 | 0.834 (0.653-1.065) | 0.146 | ||
| Gene | Total, n | Univariate analysis | Multivariate analysis | ||
| Hazard ratio (95%CI) | P value | Hazard ratio (95%CI) | P value | ||
| ATP7A | 349 | 1.032 (0.656-1.625) | 0.891 | ||
| ATP7B | 349 | 0.957 (0.776-1.180) | 0.680 | ||
| CDKN2A | 349 | 1.019 (0.894-1.162) | 0.774 | ||
| DLAT | 349 | 0.701 (0.471-1.043) | 0.080 | 1.185 (0.708-1.982) | 0.518 |
| DLD | 349 | 0.657 (0.415-1.039) | 0.072 | 0.926 (0.529-1.622) | 0.788 |
| FDX1 | 349 | 0.668 (0.441-1.013) | 0.057 | 0.722 (0.448-1.164) | 0.181 |
| GCSH | 349 | 1.280 (0.863-1.900) | 0.220 | ||
| GLS | 349 | 1.041 (0.788-1.376) | 0.778 | ||
| LIAS | 349 | 0.509 (0.310-0.836) | 0.008 | 0.578 (0.338-0.989) | 0.045 |
| LIPT1 | 349 | 1.117 (0.594-2.101) | 0.731 | ||
| LIPT2 | 349 | 1.014 (0.745-1.381) | 0.928 | ||
| MTF1 | 349 | 0.581 (0.329-1.023) | 0.060 | 0.604 (0.321-1.135) | 0.117 |
| NFE2L2 | 349 | 0.584 (0.360-0.947) | 0.029 | 0.709 (0.414-1.215) | 0.211 |
| NLRP3 | 349 | 1.082 (0.716-1.634) | 0.710 | ||
| PDHA1 | 349 | 0.676 (0.441-1.036) | 0.072 | 0.780 (0.469-1.297) | 0.338 |
| PDHB | 349 | 0.809 (0.465-1.408) | 0.454 | ||
| SLC31A1 | 349 | 0.768 (0.564-1.046) | 0.094 | 1.029 (0.717-1.476) | 0.878 |
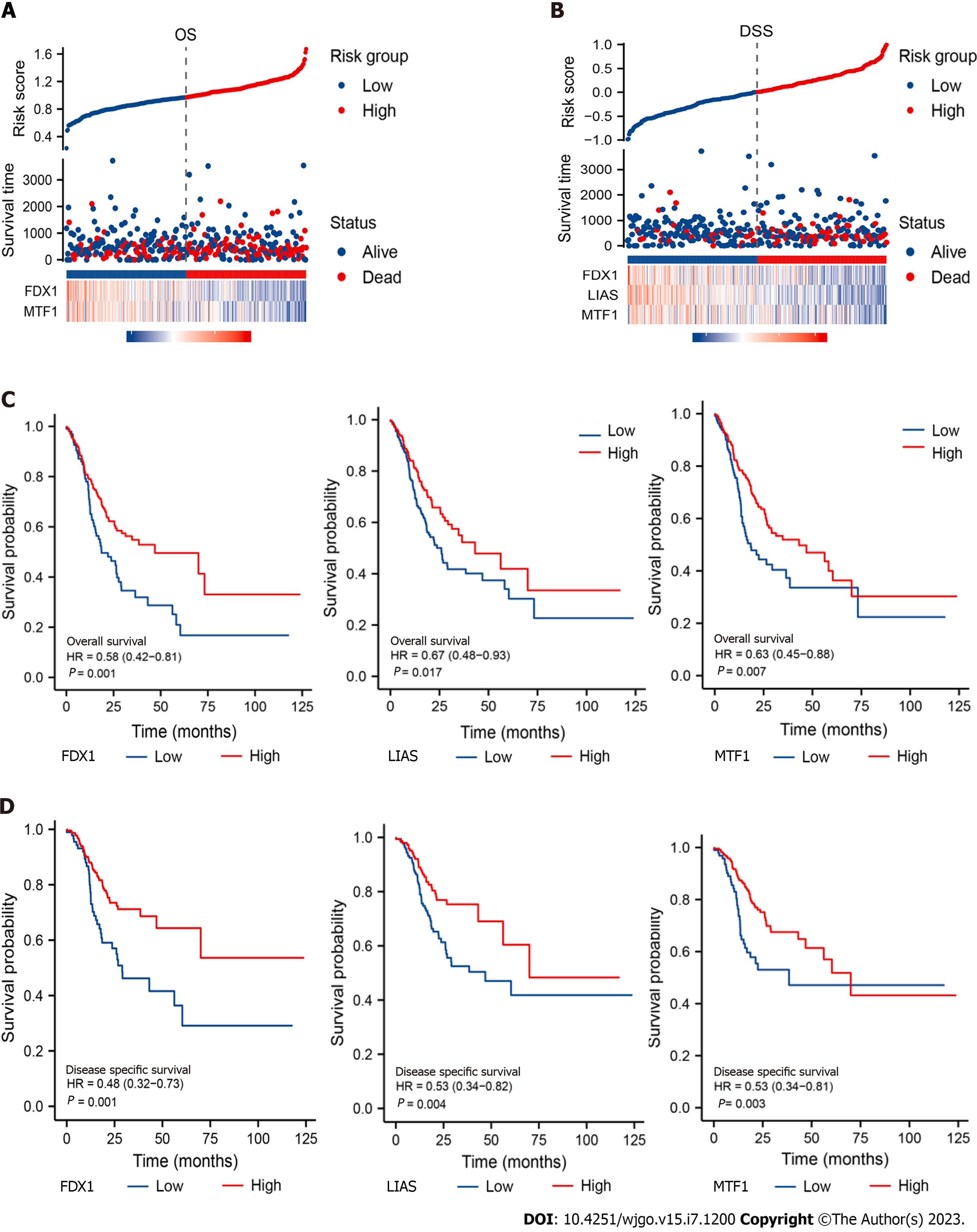
According to the outcomes of the Cox regression model, we used regression coefficients to build the OS/DSS risk score model. Risk score OS = -0.308 × FDX1 - 0.413 × MTF1 + 2.812. Risk score DSS = -0.373 × FDX1 - 0.601 × LIAS - 0.413 × MTF1 + 3.534. We separated the samples into high- and low-risk groups in terms of the risk score displayed in Figure 2A and B. Then, we built a survival curve via the Kaplan-Meier method to evaluate the prognostic value for each CRG. Our results suggested that all of these CRGs were prominently associated with OS and DSS in GC (Figure 2C and D), which was in keeping with the previous results.
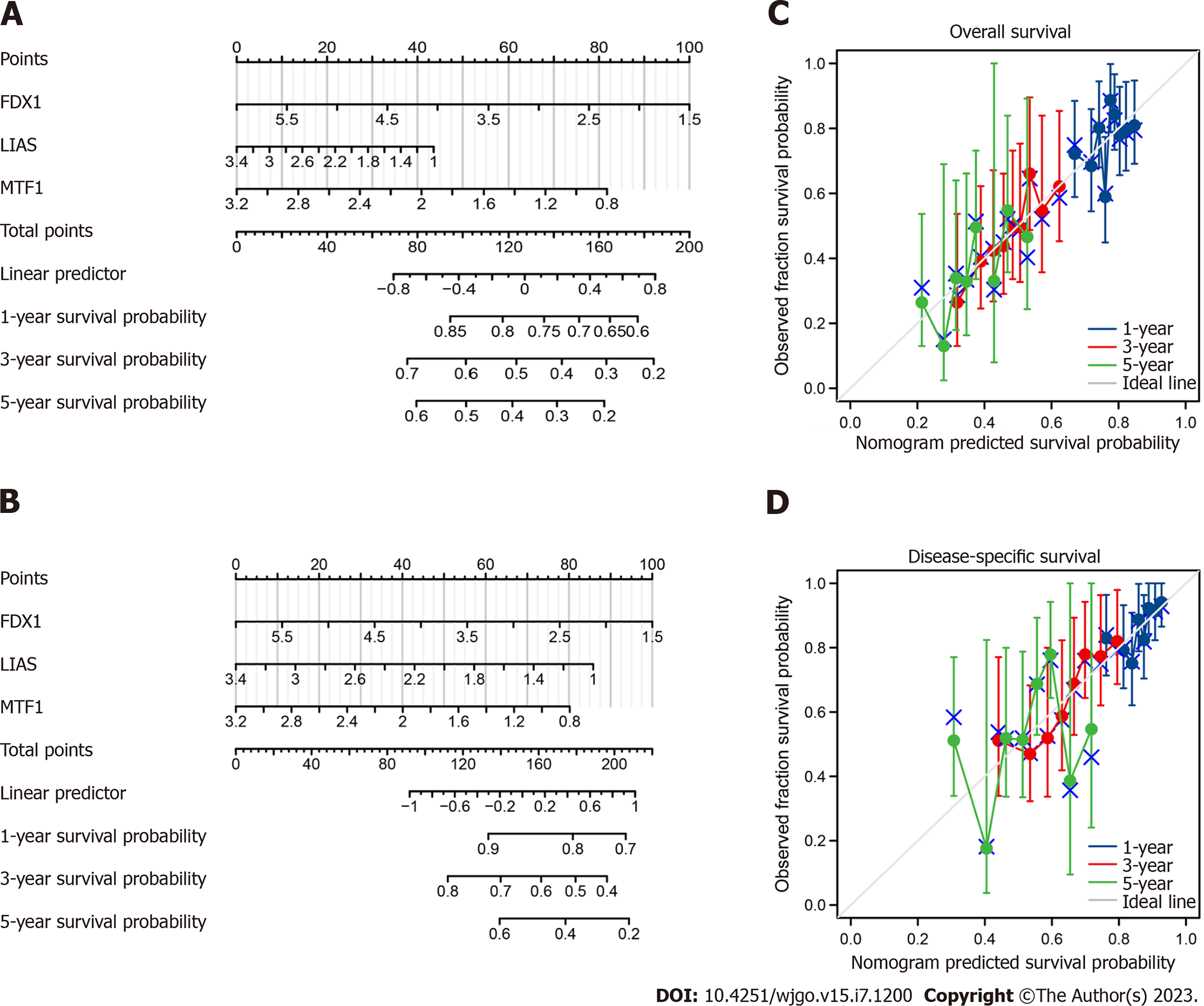
To better guide clinical application, we generated nomograms from the prognosis-related CRGs and the observed OS and DSS at 1, 3 and 5 years of survival (Figure 3A and B). The C-index was calculated to be 0.673 for OS and 0.623 for DSS. The nomogram calibration curves demonstrated ideal agreement between prediction and observation at 1, 3 and 5 years (Figure 3C and D), indicating that our nomogram models are worthy of a multicentre, prospective clinical study.
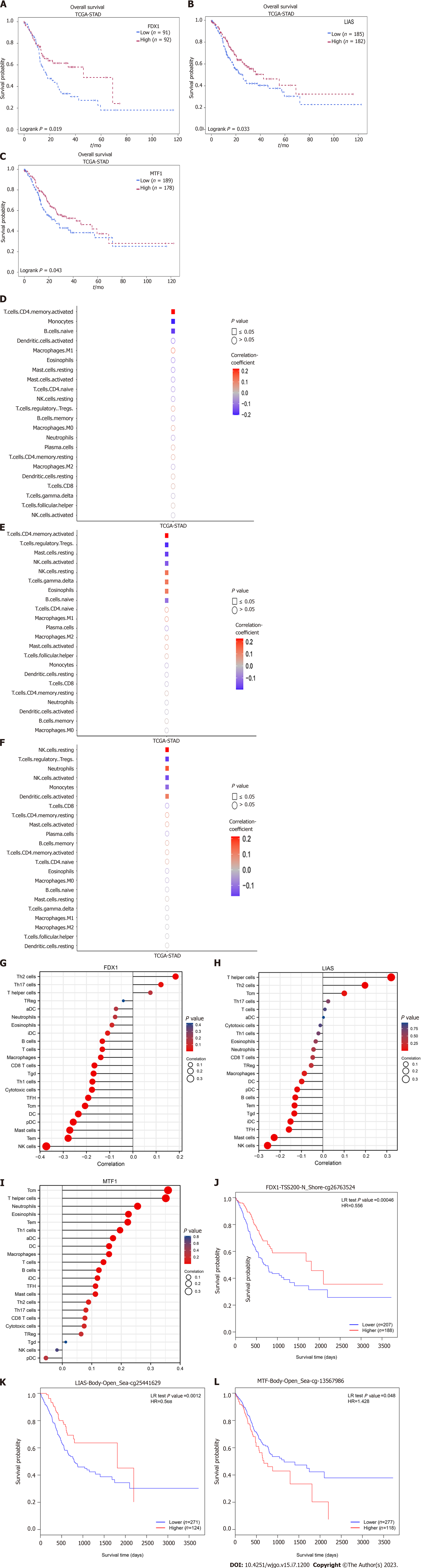
The dynamic relationship between malignant tumours and immune cells in the microenvironment plays important roles in cancer development[22]. We evaluated the correlations between FDX1, LIAS, MTF1 and distal survival probability from single-cell RNA-seq (scRNA-seq) data using Survival Genie. We found that FDX1, LIAS, and MTF1 were remarkably related to survival time, as shown in Figure 4A-C. Then, we investigated the immune cell infiltration level using scRNA-seq data, and our results showed that the expression of FDX1 was correlated with CD4 T+ memory cells, monocytes, and naive B cells, as shown in Figure 4D. LIAS was associated with CD4 T+ memory cells, Tregs, mast cells, NK cells, gamma delta T cells, eosinophils, and naive B cells, as shown in Figure 4E. MTF1 was significantly related to NK cells, Tregs, neutrophils, monocytes, and activated dendritic cells, as shown in Figure 4E. On this basis, we detected the immune cell infiltration level in GC tissues and visualized the results as lollipop plots in Figure 4F-I. The length of the bars in the lollipop plots is relative to the correlation levels, and the colour of the cycles is relative to the P value. Subsequently, we used TIMER to validate our results and found that the expression of FDX1, LIAS, and MTF1 and immune infiltration of macrophages were prominently correlated with the OS time of GC patients, which was consistent with our results (Supplementary Figure 1). Meanwhile, higher levels of methylation in MTF1 and lower levels of methylation in FDX1, LIAS were associated with poor prognosis in GC patients (Figure 4J-L). All of the evidence suggests that the prognosis-related CRGs can regulate immune cell infiltration and the tumour microenvironment to influence the survival times of GC patients.
To explore the biofunction of prognosis-related CRGs, we input FDX1, LIAS, and MTF1 into GeneMANIA to test their interactions and gathered 23 genes in the network (Figure 5A). Then, we inputted these genes into STRING to investigate the functions of their coding proteins, which were visualized as a PPI network (Figure 5B). Moreover, we performed KEGG pathway enrichment analysis and gene ontology classification to understand the related signalling pathways and biological functions in the PPI network. The results in Figure 5C show that FDX1, LIAS, and MTF1 play key roles in prognosis and immune cell infiltration by mediating iron ion binding and mitochondrial metabolism, which are closely associated with the TCA cycle and necroptosis. Furthermore, we performed CMap to explore the top 10 perturbagens to the expression of genes in the PPI network. We compared the expression levels of the genes in the PPI network using the TCGA cohort shown in Supplementary Figure 2 and identified upregulated genes in CMap. Our results revealed that fluconazole, KD-025, and clofarabine may be potential perturbagens of prognostic CRGs (Table 3).
| Perturbagen | Moa | Raw_cs | FDR_q_nlog 10 |
| Fluconazole | Sterol demethylase inhibitor | 0.79 | 1.03 |
| KD-025 | Rho associated kinase inhibitor | 0.77 | 0.95 |
| Clofarabine | Ribonucleoside reductase inhibitor | 0.76 | 0.89 |
| Tramadol | Opioid receptor agonist, Norepinephrine reuptake inhibitor, Serotonin reuptake inhibitor | 0.76 | 0.89 |
| Doxorubicin | Topoisomerase inhibitor | 0.75 | 0.88 |
| AXD-5438 | CDK inhibitor | 0.73 | 0.80 |
| BRD-K67174965 | Mucolytic | 0.73 | 0.79 |
| Faropenem | Lactamase inhibitor | 0.72 | 0.76 |
| Clocortolone-pivalate | Steroid | 0.72 | 0.68 |
| Ganglioside | Src activator | 0.71 | 0.46 |
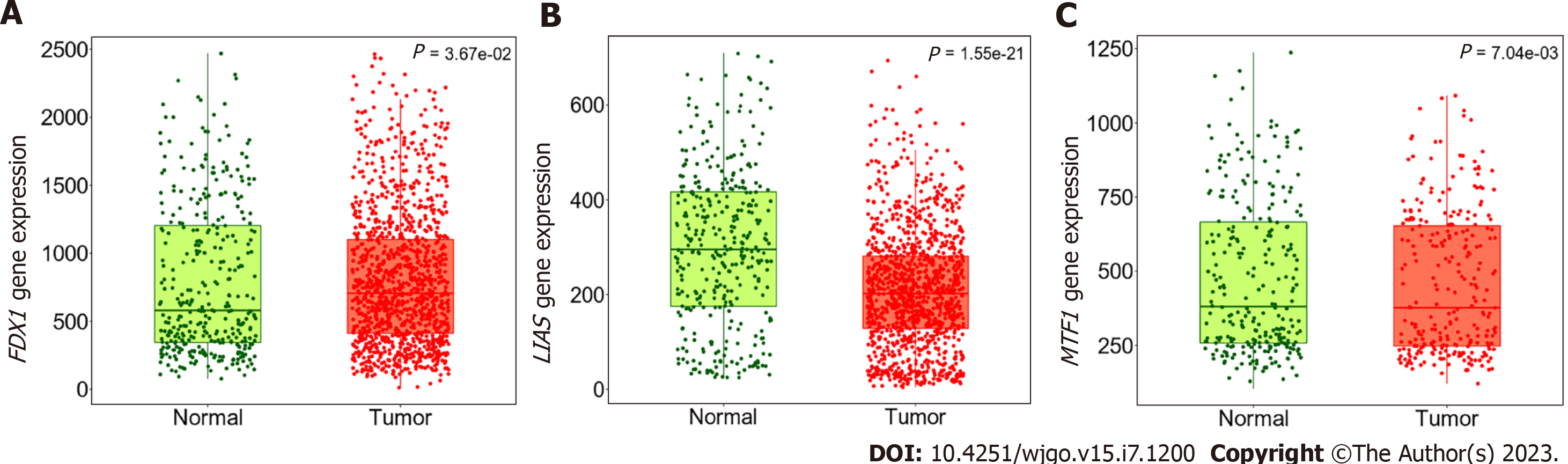
To identify promising prognosis-related CRGs, we validated the expression level using the GEO database for preliminary verification. In the GEO dataset, FDX1 was remarkably higher in GC patients (P = 3.67 × 10-2), and MTF1 was significantly overexpressed in the GC group (P = 7.04 × 10-3). LIAS was prominently downregulated in GC samples (P < 0.001), which was in line with the TCGA cohort data and revealed the role of LIAS as a tumour suppressor gene and the role of FDX1 and MTF1 as cancer promotors (Figure 6).
Despite aggressive multimodal therapy, GC is still a devastating disease with a very poor prognosis[23]. The pathogenesis of GC is complicated, and the in-depth mechanisms and molecular signalling pathways remain to be elucidated. Luckily, the development of bioinformatics can help to open different perspectives on analysing clinical samples from multiple dimensions and improve the efficiency and accuracy of studies focusing on several genes and cancer[24]. Cuproptosis is an unusual mechanism of cell death that is helpful in explaining the pathological mechanisms related to copper overload disease and suggests a new method of treating cancer with copper toxicity[4]. To the best of our knowledge, no previous studies have estimated the relationship between CRGs and the progression of GC. Hence, our study focused on the prognostic signature and explored the biofunction and oncological mechanism of CRGs in GC via bioinformatics.
There are distinct advantages in our research. We first filtered the differentially expressed CRGs in the TCGA cohort and defined their prognostic significance via multivariable Cox regression and Kaplan-Meier methods. Then, we constructed and validated a nomogram model for clinical application. Moreover, we explored the mechanisms of how prognosis-related CRGs influence distal prognosis at the DNA methylation level and immune cell infiltration level. Finally, we discovered the functions of FDX1, LIAS, and MTF1 and validated their differential expression via the GEO database.
The prognostic models constructed in our study consist of three CRGs (FDX1, LIAS, and MTF1). FDX1 has been confirmed to encode a reductase that decreases Cu2+ to its more toxic form, Cu1+. LIAS encodes lipoyl synthase, a critical component of the lipoic acid pathway. Deletion of FDX1 and LIAS can confer resistance to copper-induced cell death[4]. Existing studies have revealed that FDX1 plays a key role in steroidogenesis and mediates ageing and tumour suppression via the FDXR-p73 axis[25]. Furthermore, downregulated expression of FDX1 is correlated with more advanced tumour-node-metastasis stages and poor prognosis in clear cell renal cell carcinoma[26]. Burr et al[27] noted that LIAS was an important regulator controlling the stability of HIFα and that disruption of LIAS decreased the activity of HIFα, which may further facilitate tumour formation[27]. Higher LIAS expression was also considered a prognostic biomarker indicating better distant metastasis-free survival time in breast cancer[28]. MTF1 is a key transcription factor in charge of intracellular zinc efflux associated with the TCA cycle, is overexpressed in glioma and regulates malignant biological behaviours by modulating the TAF15/LINC00665/MTF1 (YY2)/GTSE1 axis[29]. Similarly, it has been demonstrated that elevated MTF1 is important for hepatocellular carcinoma tumour growth and migration and is regulated by the METTL3-METTL14-WTAP axis[30]. However, there are few studies on these genes in GC. Our study identified differentially expressed CRGs in GC and assessed their prognostic value and their biofunctions. Additionally, our prognostic model focusing on CRG expression displayed a fantastic performance in survival prediction, which warrants larger sample sizes and longitudinal research.
We further explored the potential mechanisms associated with prognosis in GC. Infiltration of immune cells within the tumour is typically related to distal prognosis and response to immunotherapy[31]. We delineated 22 unique clusters of immune cells in GC via scRNA-seq and examination of tissue samples. Our results showed that FDX1, LIAS, and MTF1 in scRNA-seq samples affected multiple types of immune cells, such as CD4 T+ memory cells, monocytes, naive B cells, NK cells, and Tregs. Similarly, in GC tissues, these genes impacted Th2 cells, T helper cells, DCs, iDCs, pDCs, B cells, T cells, Tgd cells, and NK cells and thus are important prognostic factors and could be promising targets for conventional immunosuppressant therapy or combination immunosuppression. Likewise, analysis of the levels of DNA methylation also suggested the prognostic significance of FDX1, LIAS and MTF1. The existing results indicate intrinsic connections between DNA methylation and prognosis, which are worthy of further validation.
Moreover, we performed functional analysis of FDX1, LIAS, and MTF1 using GeneMANIA, STRING, KEGG pathway enrichment analysis and GO classification. Functional analysis showed that the proteins associated with FDX1, LIAS, and MTF1 are involved in the TCA cycle, cuproptosis and several signalling pathways. FDX1, LIAS, MTF1 and related genes can modulate the progression of iron ion binding and mitochondrial metabolism to influence the survival time and immune cell infiltration. In addition, it is important to explore biological targets to develop novel drugs, and perturbagens are indispensable mediators in these efforts to discover biological connections[32]. We found 16 upre
Finally, we validated the differential expression of FDX1, LIAS, and MTF1 in the GEO database to make our results more robust. Interestingly, the expression levels of FDX1, LIAS, and MTF1 in the GEO database were in line with those in the TCGA cohort, which further supports the merits of application and warrants attention in future research.
In conclusion, our study systematically analysed the prognostic significance and interactive landscapes of CRGs in GC samples using bioinformatics. The prognostic risk score based on the expression signature of FDX1, LIAS, and MTF1 had important implications in the prediction of OS and DSS in GC patients, and these CRGs were associated with infiltration of various immune cell types, providing novel insights into therapeutic strategies for GC patients.
Gastric cancer (GC) is one of the most common digestive system cancers with high mortality rates worldwide.
Cuproptosis is strongly correlated with the biological behaviour of malignant tumour cells and no previous studies have estimated the relationship between cuproptosis related genes (CRGs) and the progression of GC.
Our study aims to offer new insights to predict GC prognosis and provide multiple therapeutic targets for future therapy about CRGs.
We collected data from several public data portals and systematically estimated the expression level and prognostic values of CRGs in GC samples and related mechanisms using public databases and bioinformatics.
We found that FDX1, LIAS, and MTF1 were differentially expressed in GC samples and exhibited important prognostic significance. We constructed a nomogram model for overall survival and disease-specific survival prediction and validated it via calibration plots. Mechanistically, immune cell infiltration and DNA methylation prominently affected the survival time of GC patients. Moreover, protein-protein interaction network, KEGG pathway and gene ontology enrichment analyses demonstrated that FDX1, LIAS, MTF1 and related proteins played key roles in the tricarboxylic acid cycle and cuprotosis. Top 10 perturbagens were filtered as well.
Our findings suggested that FDX1, LIAS, and MTF1 had important implications for the prediction of OS and DSS in GC patients, which were associated with various immune cell infiltrations, providing novel insights into therapeutic strategies for GC patients.
Considerable effort needs to be expended in exploring the therapeutic strategies via CRGs in GC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Casella C, Italy; Wang CY, Taiwan S-Editor: Li L L-Editor: Filipodia P-Editor: Ji MX
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64672] [Article Influence: 16168.0] [Reference Citation Analysis (176)] |
| 2. | Ding XQ, Wang ZY, Xia D, Wang RX, Pan XR, Tong JH. Proteomic Profiling of Serum Exosomes From Patients With Metastatic Gastric Cancer. Front Oncol. 2020;10:1113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 3. | Wang P, Zhang W, Wang L, Liang W, Cai A, Gao Y, Chen L. RCC2 Interacts with Small GTPase RalA and Regulates Cell Proliferation and Motility in Gastric Cancer. Onco Targets Ther. 2020;13:3093-3103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 4. | Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, Rossen J, Joesch-Cohen L, Humeidi R, Spangler RD, Eaton JK, Frenkel E, Kocak M, Corsello SM, Lutsenko S, Kanarek N, Santagata S, Golub TR. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 2493] [Article Influence: 831.0] [Reference Citation Analysis (1)] |
| 5. | Hu JD, Tang HQ, Zhang Q, Fan J, Hong J, Gu JZ, Chen JL. Prediction of gastric cancer metastasis through urinary metabolomic investigation using GC/MS. World J Gastroenterol. 2011;17:727-734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 44] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 6. | Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9144] [Cited by in RCA: 12812] [Article Influence: 985.5] [Reference Citation Analysis (0)] |
| 7. | Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10:277-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 388] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 8. | Anuraga G, Wang WJ, Phan NN, An Ton NT, Ta HDK, Berenice Prayugo F, Minh Xuan DT, Ku SC, Wu YF, Andriani V, Athoillah M, Lee KH, Wang CY. Potential Prognostic Biomarkers of NIMA (Never in Mitosis, Gene A)-Related Kinase (NEK) Family Members in Breast Cancer. J Pers Med. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 9. | Xing C, Wang Z, Zhu Y, Zhang C, Liu M, Hu X, Chen W, Du Y. Integrate analysis of the promote function of Cell division cycle-associated protein family to pancreatic adenocarcinoma. Int J Med Sci. 2021;18:672-684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 10. | Dwivedi B, Mumme H, Satpathy S, Bhasin SS, Bhasin M. Survival Genie, a web platform for survival analysis across pediatric and adult cancers. Sci Rep. 2022;12:3069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 47] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 11. | Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7222] [Cited by in RCA: 9287] [Article Influence: 773.9] [Reference Citation Analysis (0)] |
| 12. | Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509-W514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2451] [Cited by in RCA: 3367] [Article Influence: 673.4] [Reference Citation Analysis (0)] |
| 13. | Kao TJ, Wu CC, Phan NN, Liu YH, Ta HDK, Anuraga G, Wu YF, Lee KH, Chuang JY, Wang CY. Prognoses and genomic analyses of proteasome 26S subunit, ATPase (PSMC) family genes in clinical breast cancer. Aging (Albany NY). 2021;13:17970. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 83] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 14. | Laham AJ, El-Awady R, Lebrun JJ, Ayad MS. A Bioinformatics Evaluation of the Role of Dual-Specificity Tyrosine-Regulated Kinases in Colorectal Cancer. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 15. | Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214-W220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2336] [Cited by in RCA: 3263] [Article Influence: 217.5] [Reference Citation Analysis (0)] |
| 16. | Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605-D612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1211] [Cited by in RCA: 4828] [Article Influence: 1207.0] [Reference Citation Analysis (0)] |
| 17. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11591] [Cited by in RCA: 22245] [Article Influence: 1711.2] [Reference Citation Analysis (0)] |
| 18. | Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929-1935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3474] [Cited by in RCA: 3733] [Article Influence: 196.5] [Reference Citation Analysis (0)] |
| 19. | Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, Lahr DL, Hirschman JE, Liu Z, Donahue M, Julian B, Khan M, Wadden D, Smith IC, Lam D, Liberzon A, Toder C, Bagul M, Orzechowski M, Enache OM, Piccioni F, Johnson SA, Lyons NJ, Berger AH, Shamji AF, Brooks AN, Vrcic A, Flynn C, Rosains J, Takeda DY, Hu R, Davison D, Lamb J, Ardlie K, Hogstrom L, Greenside P, Gray NS, Clemons PA, Silver S, Wu X, Zhao WN, Read-Button W, Haggarty SJ, Ronco LV, Boehm JS, Schreiber SL, Doench JG, Bittker JA, Root DE, Wong B, Golub TR. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell. 2017;171:1437-1452.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1427] [Cited by in RCA: 2131] [Article Influence: 266.4] [Reference Citation Analysis (0)] |
| 20. | Wang CY, Chiao CC, Phan NN, Li CY, Sun ZD, Jiang JZ, Hung JH, Chen YL, Yen MC, Weng TY, Chen WC, Hsu HP, Lai MD. Gene signatures and potential therapeutic targets of amino acid metabolism in estrogen receptor-positive breast cancer. Am J Cancer Res. 2020;10:95-113. [PubMed] |
| 21. | Bartha Á, Győrffy B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 630] [Article Influence: 157.5] [Reference Citation Analysis (0)] |
| 22. | Acs B, Ahmed FS, Gupta S, Wong PF, Gartrell RD, Sarin Pradhan J, Rizk EM, Gould Rothberg B, Saenger YM, Rimm DL. An open source automated tumor infiltrating lymphocyte algorithm for prognosis in melanoma. Nat Commun. 2019;10:5440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 23. | Zhang K, Zhang L, Mi Y, Tang Y, Ren F, Liu B, Zhang Y, Zheng P. A ceRNA network and a potential regulatory axis in gastric cancer with different degrees of immune cell infiltration. Cancer Sci. 2020;111:4041-4050. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 24. | Zhang Y, Ma S, Wang M, Shi W, Hu Y. Comprehensive Analysis of Prognostic Markers for Acute Myeloid Leukemia Based on Four Metabolic Genes. Front Oncol. 2020;10:578933. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 25. | Zhang J, Kong X, Zhang Y, Sun W, Wang J, Chen M, Chen X. FDXR regulates TP73 tumor suppressor via IRP2 to modulate aging and tumor suppression. J Pathol. 2020;251:284-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Wang T, Liu Y, Li Q, Luo Y, Liu D, Li B. Cuproptosis-related gene FDX1 expression correlates with the prognosis and tumor immune microenvironment in clear cell renal cell carcinoma. Front Immunol. 2022;13:999823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 43] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 27. | Burr SP, Costa AS, Grice GL, Timms RT, Lobb IT, Freisinger P, Dodd RB, Dougan G, Lehner PJ, Frezza C, Nathan JA. Mitochondrial Protein Lipoylation and the 2-Oxoglutarate Dehydrogenase Complex Controls HIF1α Stability in Aerobic Conditions. Cell Metab. 2016;24:740-752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 28. | Cai Y, He Q, Liu W, Liang Q, Peng B, Li J, Zhang W, Kang F, Hong Q, Yan Y, Peng J, Xu Z, Bai N. Comprehensive analysis of the potential cuproptosis-related biomarker LIAS that regulates prognosis and immunotherapy of pan-cancers. Front Oncol. 2022;12:952129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 49] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 29. | Ruan X, Zheng J, Liu X, Liu Y, Liu L, Ma J, He Q, Yang C, Wang D, Cai H, Li Z, Liu J, Xue Y. lncRNA LINC00665 Stabilized by TAF15 Impeded the Malignant Biological Behaviors of Glioma Cells via STAU1-Mediated mRNA Degradation. Mol Ther Nucleic Acids. 2020;20:823-840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 30. | Yang Y, Qian Cai Q, Sheng Fu L, Wei Dong Y, Fan F, Zhong Wu X. Reduced N6-Methyladenosine Mediated by METTL3 Acetylation Promotes MTF1 Expression and Hepatocellular Carcinoma Cell Growth. Chem Biodivers. 2022;19:e202200333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 31. | Wu L, Shi W, Long J, Guo X, Michailidou K, Beesley J, Bolla MK, Shu XO, Lu Y, Cai Q, Al-Ejeh F, Rozali E, Wang Q, Dennis J, Li B, Zeng C, Feng H, Gusev A, Barfield RT, Andrulis IL, Anton-Culver H, Arndt V, Aronson KJ, Auer PL, Barrdahl M, Baynes C, Beckmann MW, Benitez J, Bermisheva M, Blomqvist C, Bogdanova NV, Bojesen SE, Brauch H, Brenner H, Brinton L, Broberg P, Brucker SY, Burwinkel B, Caldés T, Canzian F, Carter BD, Castelao JE, Chang-Claude J, Chen X, Cheng TD, Christiansen H, Clarke CL; NBCS Collaborators, Collée M, Cornelissen S, Couch FJ, Cox D, Cox A, Cross SS, Cunningham JM, Czene K, Daly MB, Devilee P, Doheny KF, Dörk T, Dos-Santos-Silva I, Dumont M, Dwek M, Eccles DM, Eilber U, Eliassen AH, Engel C, Eriksson M, Fachal L, Fasching PA, Figueroa J, Flesch-Janys D, Fletcher O, Flyger H, Fritschi L, Gabrielson M, Gago-Dominguez M, Gapstur SM, García-Closas M, Gaudet MM, Ghoussaini M, Giles GG, Goldberg MS, Goldgar DE, González-Neira A, Guénel P, Hahnen E, Haiman CA, Håkansson N, Hall P, Hallberg E, Hamann U, Harrington P, Hein A, Hicks B, Hillemanns P, Hollestelle A, Hoover RN, Hopper JL, Huang G, Humphreys K, Hunter DJ, Jakubowska A, Janni W, John EM, Johnson N, Jones K, Jones ME, Jung A, Kaaks R, Kerin MJ, Khusnutdinova E, Kosma VM, Kristensen VN, Lambrechts D, Le Marchand L, Li J, Lindström S, Lissowska J, Lo WY, Loibl S, Lubinski J, Luccarini C, Lux MP, MacInnis RJ, Maishman T, Kostovska IM, Mannermaa A, Manson JE, Margolin S, Mavroudis D, Meijers-Heijboer H, Meindl A, Menon U, Meyer J, Mulligan AM, Neuhausen SL, Nevanlinna H, Neven P, Nielsen SF, Nordestgaard BG, Olopade OI, Olson JE, Olsson H, Peterlongo P, Peto J, Plaseska-Karanfilska D, Prentice R, Presneau N, Pylkäs K, Rack B, Radice P, Rahman N, Rennert G, Rennert HS, Rhenius V, Romero A, Romm J, Rudolph A, Saloustros E, Sandler DP, Sawyer EJ, Schmidt MK, Schmutzler RK, Schneeweiss A, Scott RJ, Scott CG, Seal S, Shah M, Shrubsole MJ, Smeets A, Southey MC, Spinelli JJ, Stone J, Surowy H, Swerdlow AJ, Tamimi RM, Tapper W, Taylor JA, Terry MB, Tessier DC, Thomas A, Thöne K, Tollenaar RAEM, Torres D, Truong T, Untch M, Vachon C, Van Den Berg D, Vincent D, Waisfisz Q, Weinberg CR, Wendt C, Whittemore AS, Wildiers H, Willett WC, Winqvist R, Wolk A, Xia L, Yang XR, Ziogas A, Ziv E; kConFab/AOCS Investigators, Dunning AM, Pharoah PDP, Simard J, Milne RL, Edwards SL, Kraft P, Easton DF, Chenevix-Trench G, Zheng W. A transcriptome-wide association study of 229,000 women identifies new candidate susceptibility genes for breast cancer. Nat Genet. 2018;50:968-978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 166] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 32. | Li P, Bai C, Zhan L, Zhang H, Zhang Y, Zhang W, Wang Y, Zhao J. Specific gene module pair-based target identification and drug discovery. Front Pharmacol. 2022;13:1089217. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |