Published online May 15, 2023. doi: 10.4251/wjgo.v15.i5.731
Peer-review started: December 7, 2022
First decision: February 23, 2023
Revised: March 7, 2023
Accepted: April 4, 2023
Article in press: April 4, 2023
Published online: May 15, 2023
Processing time: 156 Days and 4.2 Hours
Colorectal cancer (CRC), the third most common type of cancer worldwide, threaten human health and quality of life. With multidisciplinary, including surgery, chemotherapy and/or radiotherapy, patients with an early diagnosis of CRC can have a good prognosis. However, metastasis in CRC patients is the main risk factor causing cancer-related death. To elucidate the underlying molecular mechanisms of CRC metastasis is the difficult and research focus on the investigation of the CRC mechanism. On the other hand, the tumor microenvironment (TME) has been confirmed as having an essential role in the tumorigenesis and metastasis of malignancies, including CRCs. Among the different factors in the TME, exosomes as extracellular vesicles, function as bridges in the communication between cancer cells and different components of the TME to promote the progression and metastasis of CRC. MicroRNAs packaged in exosomes can be derived from different sources and transported into the TME to perform oncogenic or tumor-suppressor roles accordingly. This article focuses on CRC exosomes and illustrates their role in regulating the metastasis of CRC, especially through the packaging of miRNAs, to evoke exosomes as novel biomarkers for their impact on the metastasis of CRC progression.
Core Tip: Exosomes, the extracellular vesicles function as connectors in communication between cancer cells and different components of the tumor microenvironment (TME). The miRNAs packaged into exosomes were derived from different sources and transported into the TME, performing oncogenesis or tumor-suppressor roles.
- Citation: Wu Z, Fang ZX, Hou YY, Wu BX, Deng Y, Wu HT, Liu J. Exosomes in metastasis of colorectal cancers: Friends or foes? World J Gastrointest Oncol 2023; 15(5): 731-756
- URL: https://www.wjgnet.com/1948-5204/full/v15/i5/731.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i5.731
Colorectal cancer (CRC) is a common malignant tumor of the digestive system, which has a high incidence and mortality, accounting for 10% of all cancer incidences and 9.4% of deaths worldwide[1]. The major reason for the high mortality rate of patients with CRC is the high heterogeneity and metastasis. Tumor microenvironment (TME) provides the environment for the growing, developing and maturing processes of cancer cells, whose special structure and composition have a great influence on the growth and metastasis of malignancies, including CRC[2]. Therefore, the study of the CRC microenvironment enables us to have a deep understanding of the mechanism of tumorigenesis and metastasis of CRC and is of great significance for evoking novel therapeutic strategies for metastatic CRC.
Exosomes are extracellular vesicles (EVs) secreted by various cells, which are actively involved in biological growth and development, immune system response, anti-tumor activity, mediating tumor metastasis and other biochemical reactions in vivo and in vitro[3]. Exosomes originate from the endocytosis of cells and are released after a series of transport to form intralumenal vesicles (ILVs)[4]. It is confirmed that exosomes carry cargo, such as proteins and miRNAs, that promotes tumor initiation, metastasis, and therapeutic resistance of cancer cells through intercellular communication in TME[5]. In different types of exosome-loaded biomolecules, miRNA plays the main regulatory role in the expression of downstream genes. As one kind of non-coding single-stranded RNA molecule, miRNAs was proved to be involved in regulating the process of protein synthesis[6,7]. A large number of studies have shown that exosomal miRNAs are highly expressed in a variety of tumors, and since exosomal miRNAs can be isolated and detected from body fluids, the exosomal miRNAs may become novel markers for tumor diagnosis[8].
Exosomes, on the other hand, act as communication mediators, carrying contents that function not only between cancer cells, but also between cancer cells and stromal cells, which is one of the main mechanisms by which exosomes participates in tumor metastasis. In colon cancer cells, exosome miRNAs can play a regulatory role in the initiation and metastasis of colon cancer through different signaling pathways, such as WNT pathway[9] and transforming growth factor beta (TGF-β) pathway[10]. It also plays an essential role in regulating epithelial-mesenchymal transition (EMT) formation, extracellular matrix (ECM) remodeling and premetastatic niches (PMN) formation, which are vital for tumor metastasis. Therefore, this article focused on the investigation of miRNA in exosomes, comprehensively analyzed the function and mechanism of miRNA in CRC metastasis and its effects on ECM remodeling, EMT, angiogenesis and PMN formation during metastatic processes and described the application of miRNA in exosomes as the novel biomarkers for the diagnosis and treatment of metastatic CRC.
TME, a complex and constantly changing system as the "soil" of tumor cell growth and development, is mainly composed of ECM, stromal cells and immune cells, which can be divided into an immune microenvironment dominated by immune cells and the non-immune microenvironment dominated by stromal cells[5]. The former contains both innate and adaptive immune cells, such as macrophages/dendritic cells (DC) and T lymphocytes, mediating the immunosuppressive function. Among them, tumor-associated macrophages (TAMs) and regulatory T cells (Tregs) performed the main immunosuppressive role by helping the immune escape of tumor cells and promoting the malignant development of tumors. On the other hand, the non-immune microenvironment mainly including fibroblasts, stromal cells and endothelial cells, was also involved in the development of malignancies. Cancer-associated fibroblasts (CAFs) were found to release stromal cell-derived factors and pro-angiogenic factors to promote tumor cell growth and angiogenesis process, while vascular endothelial cells mainly mediate tumor angiogenesis, jointly contributing to tumorigenesis and metastasis. Due to its complexity and heterogeneity with a close impact on tumor cells, TME has been widely studied in the field of cancer therapy[11].
Since the role of TME in cancer has been reported before, the effect of T lymphocyte migration in TME was the focus of the research on tumorigenesis. It is discovered that innate immune responses not only indirectly control the production of T lymphocytes, but also directly shape TME through the production of cytokines. The following section will demonstrate the solely different roles of innate immune response cells in TME, including macrophages, DC, neutrophils, natural killer cells (NK) and bone marrow-derived suppressor cells (MDSC), as well as the non-immune microenvironment.
Immune cells: Among all innate immune cells, there is no doubted that macrophages derived from monocytes play an indispensable role, which is the first activated by pathogens and subsequently evoking the immune activation state[12]. During the tumorigenesis and development processes, TAMs are classified into classical inflammatory “M1” and alternative immunosuppressive “M2” activation modes[13].
M1 macrophages have pro-inflammatory, immune-stimulating and anti-tumor properties, which produce interleukin (IL)-1β, IL-6 and tumor necrosis factor α (TNF-α), participating in immune stress of the body[14]. Nevertheless, in the colitis model, the proinflammatory effect of M1 TAMs inducing an inflammatory response, is a risk factor for CRC, indicating that the effect of M1 TAMs on CRC will be judged by the specific environment[15]. M2 macrophages, highly infiltrated in most types of cancers, have immunosuppressive and tumor-promoting properties[16-18]. In colitis models, Wang et al[19] discovered that the density of M1 and M2 TAMs changed in the inflammation-carcinoma sequence, and the total number of TAMs gradually increased along with tumor metastasis[19]. Cultured with M1 TAMs conditioned medium, CRC cells were found to accelerate pro-apoptotic morphological changes, while those in the M2 TAMs medium promoted cell proliferation and increased the expression of anti-apoptotic markers[20]. TME with increased IL-4 cytokines, enhanced the immunosuppressive effect of M2 TAMs, promoting tumor growth and progression as well as the increase of M2 TAMs[21]. Vascular endothelial growth factor (VEGF) was also secreted by M2 TAMs to promote tumor angiogenesis through conjunction with other cells in TME[22,23].
On the flip side, M2 TAMs was also found to be involved in remodeling ECM and promoting EMT, to accelerate the invasion and metastasis of CRC[24,25]. Afik et al[26] conducted transcriptome and proteomic analyses in TAM of CRC and found that they are enriched in molecular features related to ECM remodeling[26], especially the expression of matrix metalloproteinase (MMP)[27]. TGF-β was also contributed to enhancing ECM remodeling and the EMT process, which is produced by M2 TAMs[28]. Herbeuval et al[29] demonstrated the production of IL-10 by CRC cells, which was induced by TAM-derived IL-6 and recruited transcription factor, signal transducers and activators of transduction3 (STAT3)[29], while M2 TAMs could promote CRC immune evasion by secreting immunosuppressive cytokines, IL-10 and TGF-β, and suppress the activities of T lymphocytes[30]. These findings indicate the promoting role of M2 TAMs in TME to accelerate CRC progression and invasion.
DCs, key players in the innate immune system, have high antigen presentation, through recognizing, capturing, and presenting the antigens to T cells in lymphoid organs. In cancer, DCs are specifically referred to as tumor-infiltrating DCs, which often exhibit immune stimulatory phenotypes in TME, through secreting inflammatory cytokines and prime effector T cells[31]. Orsini et al[32] confirmed that the antigen presentation ability of DCs was impaired in CRC patients, suggesting the immune stimulatory capacity of DCs can be inhibited by CRC cells to promote the development of cancers[32], while removed from the such environment, the ability to process antigens to T cells of DCs will be regained[33]. It is found that immunosuppressive factors, such as VEGF, IL-10, and TGF-β secreted by cancer cells were involved in inhibiting DCs maturation and antigen presentation[34,35]. In CRC, myeloid DCs are the most common subtypes, which are increased in frequency at the leading edge of tumor invasion and associated with lymph node invasion[36]. Hsu et al[37] found the high expression of C-X-C motif chemokine ligand 1 (CXCL1) in DCs obtained from CRC patients, enhancing the migration and stemness of cancer cells[37]. Additionally, the composition and function of DCs can be influenced by the unique TME of different types of cancer, even in different subtypes within the same malignancies[38,39].
Neutrophils, the primary responders in acute inflammation, are the first line of defense against pathogens, by producing neutrophil extracellular traps (NETs) and engulfing invading microorganisms[40]. It is reported that NETs dissolved ECM through MMP8/9 protease and improved tumor invasion and angiogenesis by releasing VEGF[41,42]. The EMT process was also induced by NETs for tumor cells to break through the vascular wall and enter the circulatory system, thus promoting the immune escape of tumor cells[40,41]. As part of the innate immune response, tumor-associated neutrophils (TANs) are similarly classified into the tumor-suppressive N1 and tumor-promoting N2 phenotypes. N1 TANs are the main type in the early stage of tumorigenesis, performing anti-tumor function through secreting type I interferon and activating IL-18 from NKs, while N2 TANs increase during the tumor development gradually, promoting tumor progression through increasing the level of reactive oxygen species and inhibiting the function of T and NK cells[43]. Interestingly, in CRC, the production of TGF-β in TME polarized TANs from N1 to N2 phenotype, contributing the immune evasion by activating TAN-secreted MMP-9 and inhibiting the proliferation of T cells[33,44].
In addition, other immune cells, such as MDSCs, NK and Tregs also have been reported to be involved in the occurrence, development and metastasis of CRC. The function of MDSCs in TME was confirmed as suppressing the immune by inhibiting T cells and innate immune regulation, as well as contributing to the formation of PMN, maintenance of tumor stemness and promotion of angiogenesis[45]. As non-specific innate immune cells, NKs performed cytotoxic effects through secreting killing mediators, such as perforin, NK cytotoxic factor, and TNF-α, thereby limiting the metastatic growth of tumor cells rather than the proliferation of primary tumor cells[46,47]. Enhancing NK cytotoxicity is speculated as a novel way to prevent cancer metastasis. Tregs, one of the important factors for main
Stromal cells: CAFs are one of the most abundant stromal components in solid tumors, which play an important role in tumorigenesis, angiogenesis, metastasis and invasion, and chemotherapy resistance of malignancies[50]. Similar to TAMs, CAFs promote the metastasis of cancer cells through remodeling ECM and promoting EMT[51]. By secreting collagen, fibronectin and MMP, as well as increasing VEGF levels, CAFs reorganize ECM components and form a directional migration trajectory available to tumor cells[52,53]. Additionally, expression of the collagen cross-linking enzyme lysine oxidase-like 2 (LOXL2) in CAFs is, to a certain extent, associated with a high recurrence rate, poor overall and disease-free survival in patients with CRC, since CAFs stimulate the EMT process through LOXL2 elevation[54]. Moreover, CAFs also promote immune evasion by restraining T cell function and promoting the polarization of TAMs, which was also inhibited by the high levels of TGF-β present at the edge of tumor invasion, derived from CAFs mainly[44,55]. Zhang et al[56] revealed that CXCL8 secreted by CAFs attracts monocytes to TME of CRC and promotes the polarization of M2 TAMs, further promoting immune suppression[56].
Mesenchymal stem cells (MSCs) are a kind of pluripotent stem cells with self-renewal and multidirectional differentiation abilities, participating in tissue generation and repair in a variety of tissues. In many types of tumors, the cancer-associated MSCs are reprogrammed by tumors and have significant effects on the structure and function of TME through enhancing EMT and angiogenesis processes. What is noteworthy is that MSCs are the only one capable to produce large amounts of exosomes[57]. Utilizing secreting exosomes, MSCs transfer genes carried by cancer cells to other tissues and promote the formation of PMN, thus affecting the proliferation and metastasis of cancer cells[58]. However, the function of MSC-derived exosomes in cancers is controversial and needs further investigation[59].
ECM: Along with immune and stromal cells, ECM is also the important structural and biochemical support in TME, composed of a variety of extracellular proteins and macromolecules, participating in and controlling cell growth, migration, metabolism and other activities. It is found that ECM is mainly composed of collagen, non-collagen, elastin, and proteoglycans[60]. During tumor progression, the structure and function of ECM can be remodeled by the cells in TME, including immune and stromal cells[61]. Regarding cancer metastasis, the remodeling enzymes MMP-2/9 against collagenase in CRC was increased to degrade type IV collagen, resulting in the loss of ECM support and enhancement of tumor cell viability and aggressiveness[62,63]. The fibrotic response is another reason for biomechanical changes, like tumor sclerosis, which is mainly caused by the TGF-β-mediated activation of CAFs[64]. Stiffness in CRC, usually in collagen-rich regions, is associated with metastasis and the EMT process[65,66].
As mentioned above, immune cells, stromal cells and ECM in TME all mediate the tumorigenesis, development and metastasis of types of cancer. During different processes, exosomes as bilayer vesicles containing complex RNA and proteins, have a non-negligible function in intercellular communication in the process of tumor metastasis. Uncovering the function and mechanism of exosomes will provide a new trend for anti-cancer research and benefit patients with CRC.
In recent years, exosomes, as a newly proposed concept, have attracted much interest in their role in tumor growth and metastasis. Numerous studies have shown that exosomes and their cargoes promote tumor cell genesis and metastasis through cell-to-cell communication in TMEs[5,67,68]. Under normal conditions, the individual cells and extracellular matrix of the body complement each other to form a healthy ecological niche[69,70]. When pre-metastasis niches are formed in the body, tumor stem cells begin to survive and proliferate, and induce other cells to participate in tumor formation and metastasis[71], so as to construct a TME suitable for tumor cell growth and propagation. It is confirmed that exosomes play an important role in this process. It can have large or small effects on different tumorigenic pathways in TME, including tumor dryness, angiogenesis, tumor metastasis and EMT formation[72]. In addition, other studies have shown that the trigger of tumor is not only caused by sufficient gene mutations, but more by functional changes of different interacting mediators in TME[73,74]. In view of the important role of exosomes in the occurrence, development and metastasis of cancer, we will elaborate on the origin, development, cargos and function of exosomes, as well as the role of cell-to-cell communication of exosomes in cancer.
Exosomes, first discovered in sheep reticulocytes, were simply considered as cellular vesicles for excreting cellular wastes. Soon, the new intercellular communication mode represented has made outstanding contributions to the monitoring, diagnosis and treatment of diseases. Particularly, tumor-derived exosomes (TDEs), are the main focus of the research in cancers. Exosomes and/or their vectors have been reported as biomarkers, therapeutic targets and even vectors for anti-cancer drugs[75]. Therefore, understanding the formation and transport of exosomes and their relationship with the TME is of great significance for studying the function of exosomes in cancer.
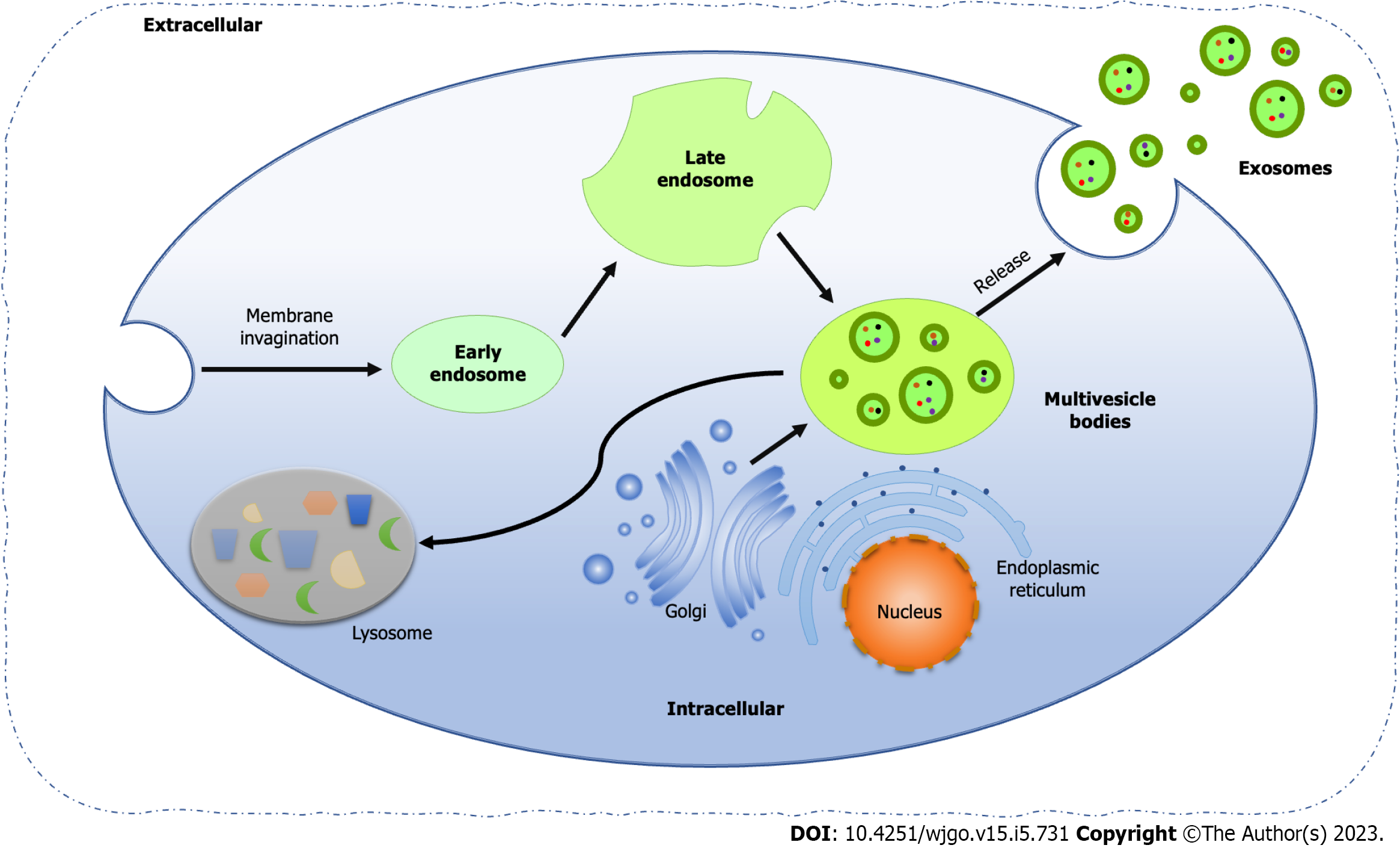
Exosomes are EVs encapsulated by lipid bilayers with a diameter of 40-160 nm, originating from the endocytic pathway of cells[75]. The biogenesis of exosomes includes four processes, that is membrane invagination, endosome formation, endosome maturation, and multivesicle bodies (MVBs) release (Figure 1). First, the cell membrane invaginates and generates small vesicles by endocytosis, which contain cell surface proteins and soluble proteins related to the extracellular environment. Next, the vesicles fuse to form early endosomes (EEs), which share their contents and membrane composition through clathrin and vesicle protein pathways, which is the main reason for the diversity and heterogeneity of exosomes. At the same time, the trans-Golgi network and endoplasmic reticulum also contribute to the formation of EEs. Along with the acidification of the contents and the entry of some "cargoes", such as cytoplasmic miRNAs, enzyme molecules and heat shock proteins (HSP), EEs gradually become mature late endosomes, also known as cellular MVBs containing ILVs. Finally, MVBs fuse with the plasma membrane and release ILVs to form exosomes, while the rest part fused with lysosomes or autophagosomes for degradation[76,77].
The endosomal sorting complex required for transport (ESCRT) is essential for the classical pathways to facilitate the formation of ILVs, composed of four complexes, ESCRT-0, -I, -II and -III[78]. Among them, ESCRT-0 and –I is responsible for the recognition of ubiquitinated proteins, while the combination of ESCRT-I/II initiates the activation of ESCRT-III, which combined with the ESCRT-I/II complex, cleaved the plasma membrane and releases buds into the endosomes to form ILVs[79].
Interestingly, ESCRT-independent exosome was first found in melanoma, involving CD63, one tetraspanin in the lysosome/endosome-associated organelle melanosomes[80]. Recently, Wei et al[81] reported an ESCRT-independent exosome pathway and demonstrated Ras-related protein Rab-31 (RAB31), a small GTP-binding protein related to vesicle-mediated transport, drives ILVs formation via the Flotillin domain of flotillin proteins and recruited GTPase-activating protein to prevent the fusion of MVEs with lysosomes and suppress MVEs degradation, thereby enabling the secretion of ILVs as exosomes[81].
Meanwhile, lipids, as the basic construction of exosome formation, located in the inner membrane of cellular MVBs with high density, such as lysobisphosphatidic acid (LBPA), leads to the composition of ILVs and then the exosomes[82]. Programmed cell death 6 interacting protein (PDCD6IP), functioning within the ESCRT pathway in the abscission stage of cytokinesis, interacted with LBPA, promoting the internal germination of the MVB membrane[83]. It is found that exosomes can alter the lipid composition of target cells by transferring molecules to them, especially cholesterol and sphingolipids, and subsequently affect the cellular homeostasis of targets[84]. Since the ceramide-rich fraction of endosomes is highly sensitive to inward plasma membrane germination, loss of sphingomyelin and subsequent converter of sphingomyelin to ceramide results in inhibition of ILVs formation[85].
Exosomes have different functions in different physiological and pathological processes, according to their size, content, origin, contents and influence on recipient cells, which is called exosome heterogeneity. In the process of MVBs formation, it restricts the uneven invaginations of the membrane, resulting in different total contents of liquid and solid contained in the vesicles formed by MVBs, which may be the cause of the size and content heterogeneity of exosomes[86]. Proteomic analysis of breast cancer cells and their exosomes identified epithelioid or mesenchymal origin cells according to the enrichment degree of different proteins and nucleic acids in exosomes, which reflects the specific sorting mechanism in the formation of exosomes[87]. Exosome heterogeneity gives them unique characteristics based on different types of cells or tissues of their origin, including absorption by specific cells and tendency to certain organs, which also provides the possibility for the location and migration of cancer cell metastasis[87].
Exosomes, involved in diverse processes as communicators between cells, are dependent on the presence of a great of biologically vital “cargos” in them, such as proteins, mRNAs, non-coding RNAs (ncRNAs) and various metabolic enzymes, all of which are bioactive substances to determine the type and function of exosomes[86]. The proteins in exosomes can be divided into the following four types, according to the discrepant structure and function, that is membrane transport and fusion-related proteins, tetraspanins, MVBs-related proteins and other proteins involved in cell adhesion and skeleton construction[88].
Membrane transport and fusion-related proteins include annexin, RAB and HSPs, which were involved in regulating plasma membrane fusion and release during exosome formation[89]. Among them, the regulating function of the RAB family is dependent on the surrounding environment and cell types. For example, Ostrowski et al[89] found that RAB27a and RAB27b control the exosome secretion pathway in different steps in cervical cancer[89], while in breast cancer, RAB7 was identified as the key regulator for exosome release in cancer cells[90]. However, in the central nervous system, the inhibition of RAB35 function leads to intracellular accumulation of endosomal vesicles and impairs exosome secretion in oligodendrocytes[91]. In the HSP family, HSP90 is a major intercellular chaperone protein, ensuring the normal folding and function of protein under normal conditions. However, in tumors, HSP90 plays the anti-apoptotic function by promoting abnormal protein folding, protein balance and proteolysis[92]. Lauwers et al[93] found that HSP90 is membrane-deformable to mediate the fusion of MVBs and plasma membranes and facilitate the exosome release[93], while exosomes lacking HSP90α, a key subtype of Hsp90, will lose important cell-to-cell communication from tumor cells to stromal cells to promote cell movement[94].
Tetraspanins are demonstrated to facilitate the entry of specific cargos into exosomes, including CD9, CD63, CD81, CD82, CD106, tetraspanin 8 (Tspan8), and intercellular adhesion molecules-1 (ICAM-1)[95]. PDCD6IP and TSG101 are the main MVBs-related proteins to regulate exosome formation in MVBs[96]. The analysis of exosomal protein composition displayed that a series of fusion and transfer proteins, as well as cytoskeletal proteins, such as actin, myosin, and tubulin, are non-specific and common in all exosomes[97]. Generally, the proteome of exosomes mirrors that of the protocell, but it is worth noting that the proteins in exosomes from cancer cells can selectively induce specific signals in the recipient cells, leading to the occurrence of carcinogenic changes[98,99].
Apart from proteins, ncRNAs referring to functional RNAs without encoding potential, also play an indispensable role in exosomes, including miRNAs, long ncRNAs (lncRNAs) and circular RNAs (circRNAs). Through binding to the 3' non-coding region of target mRNA, miRNA induced the inhibition of protein translation, involved in precise, fine and dynamic intercellular communication during human reproduction, pregnancy and embryonic development[6,7]. Molecular profiling indicated that miR-148a, let-7b, miR-148a, miR-375, and miR-99a associated with the expression of IL-10/13 in spermatogenic exosomes from multiple human donors are enriched, suggesting that exosomes may be involved in reproductive immunity through secreting miRNAs[100].
The analysis of tumor-related studies manifested that miRNA in exosomes is highly expressed during the development of lung cancer, prostate cancer and other cancers, therefore, it may be used as a potential biomarker or grading basis for cancer prognosis[101-103]. Moreover, miRNAs in exosomes can be isolated from body fluids and detected, which means that exosomal miRNAs have an advantage in becoming novel biomarkers for non-invasive utilization in vivo[8]. Puik et al[104] use miRNA profiling to identify miR-21, miR-26, miR-122 and miR-150 as potential blood biomarkers for the non-invasive diagnosis of cholangiocarcinoma[104]. In addition to being used as diagnostic markers, exosomes can also be used as predictive therapeutic markers. Sun et al[105] found that in the exosomes secreted by CSCs and corresponding mother cells, six miRNAs including miR-1246, miR-424-5p, miR628-5p, miR-1290, miR-675-3p and miR-590-3p were up-regulated, whereas five miRNAs such as miR-224-5p, let-7b-5p, miR-615-3p, miR-122-5p and miR-5787 were the opposite, which suggest that miRNAs may contribute to the early diagnosis of gastric cancers and are expected to be a potential biomarker for predicting patients with a high risk of gastric cancer[105]. Furthermore, exosomal miR-222-3p can be used as a predictive biomarker of gemcitabine sensitivity, while miR-208a can be used as a predictive biomarker of radiation response[106,107].
LncRNA is a kind of ncRNA with a transcription length of more than 200 nucleotides, which plays essential roles in a series of life activities, such as dose compensation effect, epigenetic regulation, cell cycle regulation and cell differentiation regulation[108]. In cancer cells, lncRNA has the function of "cell messenger", which can be selectively packaged into exosomes to regulate tumor growth, metastasis and angiogenesis[109]. For example, Conigliaro et al[110] discovered that exosomes secreted by CD90 cells and CSCs can be taken up by human umbilical vein endothelial cells (HUVECs) and deliver lncRNA H19 to the corresponding target cells through adhesion to CD90 cells and HUVECs, and subsequently, synthesis and release of VEGF to stimulate angiogenesis[110].
CircRNAs are another main type of ncRNA in exosomes, as endogenous RNAs in all eukaryotic cells. Different from traditional linear RNAs containing 5' and 3' ends, circRNA molecules presenting as a closed ring structure is not affected by RNA exonuclease, so their expression is more stable and not easily degraded. This strong stability may enable non-invasive detection in body fluids, and the absence of the 5’ to 3’ polar structure and poly-adenosine tail makes it inherently resistant to nucleic acid degrading enzymes targeting the 5’ and 3’ ends[111]. Recently, circRNAs rich in miRNA binding sites, are reported to serve as the miRNA sponge in cells, which dissolves the inhibitory effect of miRNA on its target genes and increases the expression level of target genes, making circRNA as a competitive inhibitor of miRNA to regulate the translation and function of the downstream protein[112]. Through regulating target genes and miRNA, circRNA plays an important role in the proliferation, invasion, metastasis and progression of tumor cells in a variety of cancer biological processes. For example, circ-IARS expression in exosomes is up-regulated in the plasma of patients with in situ metastatic lung cancer, which, however, down-regulate the levels of miR-122 and tight junction protein 1 (TJP1) significantly but up-regulate the levels of RhoA and RhoA-GTP in exosome, thereby increasing the expression and adhesion of F-actin, enhancing endothelial permeability and promoting tumor invasion and metastasis[113], suggesting that the expression level of circRNA is highly correlated with clinicopathology and may serve as biomarkers with diagnostic, prognostic and predictive properties[111].
The release of exosomes and uptake by recipient cells provides the basic mechanism of the intercellular communication function of exosomes, which is occurred in almost all types of cells in mammals[114]. After being released by cells, exosomes enter the blood, saliva, urine, cerebrospinal fluid, breast milk and other body fluids through autocrine and paracrine methods, after which reach other cells and tissues in the distance, producing a remote regulation effect. Interestingly, exosomes also influence the origin cell itself through the autocrine pathway based on specific receptors.
Within the exosome of chronic myeloid leukemia cells, cytokine TGF-β1 binds to its receptor and promotes tumor growth through an autocrine mechanism by activating anti-apoptotic pathways[115]. The exosomes were also involved in maintaining cellular homeostasis, through exosome secretion with harmful cytoplasmic DNA from cells[116]. The inhibition of exosomes will cause accumulated DNA in the cytoplasm, associated with increased reactive oxygen species-dependent DNA damage reaction, thus leading to cell cycle arrest or apoptosis. Therefore, cell secretion of such DNA-containing exosomes contributes to cell survival and homeostasis maintenance[116].
Through the paracrine pathway, exosomes mediate intercellular information transmission and microenvironment regulation, especially in the field of tumor therapy. Exosomes contain cargoes such as proteins, DNA, mRNA, ncRNA and metabolic enzymes described above, acting as external stimuli for recipient cells, triggering the uptake of exosomes and changing their biological phenotypes. The uptake of exosomes by recipient cells is not random but is accomplished through the recognition of exosome surface proteins that trigger interactions including endocytosis, receptor-ligand binding, and membrane fusion. Yang et al[117] found that breast cancer cells release and transfer exosomes containing programmed cell death ligand 1 (PD-L1) to other cancer cells with low or even no PD-L1 through the secretory pathway, to help cancer cells escape immune monitoring[117]. In CRC, Demory et al[118] reported that the transfer of mutant Kirsten rat sarcoma viral proto-oncogene (KRAS) to cancer cells with wild-type KRAS receptors via exosomes can promote the invasion of cancer cells[118].
In cancers, the intercellular communication of exosomes is not only limited between cancer cells but also occurred between cancer cells and stromal cells, which is one of the important mechanisms of distant metastasis of tumors. Shimoda et al[119] investigated the molecular mechanism of CAF-derived tumor progression and demonstrated the metalloproteinase ADAM10 as the important factor in CAF-derived exosomes to enhance the viability of cancer cells through activating Notch receptor and increasing aldehyde dehydrogenase expression[119]. Moreover, the absorption of CAF-derived exosomes by cancer cells caused the increase of glycolysis and glutamine-dependent reductive carboxylation, which promotes the growth of tumors under nutrients deficiency or nutrient stress, as the carry of amino acids, lipids and intermediates in exosomes[120].
Conversely, exosomes from cancer cells also act on stromal cells, imbuing them with the properties to be transformed into cancer cells and inducing the formation of pro-TME. Cho et al[121] demonstrated that exosomes from breast cancer cells triggered the transformation of MSCs in fat into tumor-associated myoblasts via the TGF-β-mediated signaling pathway[121]. Also, miR-9 in exosomes derived from triple-negative breast cancer can induce EMT in tumor cells through down-regulating E-cadherin in fibroblasts, and promote the transformation of fibroblasts into CAFs, thus stimulating tumor migration[122]. The hallmark, angiogenesis in cancer was also promoted by TDEs promoting endothelial cell proliferation and angiogenesis[123]. Nazarenko et al[123] found the cell surface Tspan8 as the contributor to exosome-induced endothelial cell activation, in which Tspan8-α4 integrin in exosomes facilitates the binding and absorption of exosomes to endothelial cells and promotes angiogenesis accordingly[123]. Even under hypoxia conditions, the stimulating effect of exosomes on angiogenesis was enhanced in cancers[124]. Hsu et al[125] found that hypoxic lung cancer-secreted exosomes with miR-23a not only cause the accumulation of hypoxia-inducing factor-1α (HIF-1α) in endothelial cells but also target TJP1 to increase vascular permeability and cancer migration possibility[125].
Interestingly, mRNA delivery by exosomes to recipient cells is a rare case, while it was increased in those with acute inflammation (peritonitis) or chronic inflammation (subcutaneous tumors)[126]. Engineered exosomes have been found to conduct certain functions in inducing innate and adaptive immune responses in cancers[127]. The mechanism mainly involves the antigen presentation, activation of the intracellular cGAS-STING signaling pathway, intercellular miRNA transfer, and immunoregulation of exosome surface presenting molecules, which may be related to different contents wrapped in exosomes and their effects on the recipient cells[114].
First, the antigen-presenting peptides were direct presented by antigen-presenting cells (APCs), such as DCs or Tregs to specific T cells and induce activation of them via carrying exosomes containing co-stimulatory signals[114]. A single intradermal injection of APC-derived exosomes significantly induced tumor eradication and growth delay in the mouse model[114]. Simultaneously, exosomes secreted by human DCs promote the production of interferon (IFN) production and thus enhance antigen presentation, regardless of the maturation of the exosomal origin cells[128].
Then, the activation of the intracellular cGAS-STING signaling pathway was induced by genomic DNA in exosomes to generate an anti-tumor response[129]. The production of IFN was enhanced after the contact between DCs and T cells with the uptake of exosomes through the activated cGAS-STING signaling pathway[130]. Although the uptake of exosomal DNA by recipient cells may change their signaling, such alternations may be beneficial in the context of cancer[116], as the inhibition of epidermal growth factor receptor (EGFR) in cancer cells may lead to an increase in DNA in their secreted exosomes, helping to induce cGAS-STING signaling in DCs to inhibit tumor growth[131]. In contrast, the uptake of tumor-derived exosomal DNA by circulating neutrophils enhances the production of tissue factors and IL-8, which may indirectly worsen cancer by promoting inflammatory responses[132].
Next, in the process of cell-to-cell communication, exosomes influence signaling pathways and gene expression in recipient cells through miRNA transfer to regulate the immune response. Immature DCs have a strong ability to phagocytose antigen, but their weak ability to present antigen makes the activation of DCs to specific T cells limited. In addition, miR-212-3p in TDEs promoted the immune escape of cancer cells by suppressing transcription factor RFXAP in DCs[133]. However, miR-222-3p promoted the polarization of TAMs to M2 phenotype and generated an immunosuppressive microenvironment through a down-regulating suppressor of cytokine signaling 3 (SOCS3)[134].
Finally, immunomodulatory molecules such as PD-L1 and Fas cell surface death receptor ligand (FasL) on TDEs accelerate the failure and apoptosis of T cells, thus regulating the immune response and promoting the progression of tumors[135,136]. On the contrary, mast cell-derived exosomes with CD86, lymphocyte function-associated antigen 1, and ICAM-1 on their surface, induce the proliferation of B and T immune cells and enhance anti-tumor activities[137].
Despite how the cargoes carried in exosomes act and affect recipient cells have been extensively studied, the mechanism of how exosomes selectively package those cargoes remains unclear. Through comparing miRNA content in CRCs exosomes of mutant and wild-type KRAS, Cha et al[138] found that the exosomes of wild-type KRAS cancer cells were enriched with miR-10b, while the others were enriched with miR-100[138], suggesting that exosomes selectively pack the cargo under an unknown condition for further investigation.
As described above, miRNA, a vital ncRNA, is responsible for negatively regulating the expression of up to 60% of the protein-coding genes, and play important role in the processes of malignancies[139]. Since Michael et al[140] described the association between miRNAs and CRC, the involvement of miRNAs in the occurrence of CRC evoked plenty of investigation to explore the molecular mechanism of miRNA regulating CRC[140,141]. As the main content of exosomes, miRNAs are also involved in certain control and regulatory functions on tumor proliferation, EMT and ECM remodeling, and the formation of PMNs.
It is accepted that EMT is an important biological process in which epithelial cells become cells with mesenchymal phenotypic characteristics and acquire the ability to migrate[142]. During the EMT process, epithelial cells lose cell polarity and the ability to connect with the basement membrane, so that the genome of cancer cells can be transferred between cells through exosomes, thus gaining aggressive abilities of migration and invasion, anti-apoptosis and ECM degradation[143]. First, epithelial cell-associated proteins located in the primary tumor of CRC are down-regulated, while mesenchymal adhesion proteins are up-regulated, and cancer cells, especially with mesenchymal characteristics, secrete abundant exosomes to invade local tissues[144]. Next, the locally proliferating cancer cells break through the basement membrane and propagate through the circulatory system to distant organs through specific signaling mechanisms involving exosomal miRNAs. Finally, these mesenchymal cells reprogram the microenvironment of distant metastases, inducing the formation of metastatic TMEs and angiogenesis[145,146].
EMT-related transcription factors are confirmed as the key regulators during this process. Snail family transcriptional repressor (Snail1/2), belonging to the Zinc finger transcription factor, can destroy the normal tight junction between cells, while zinc finger E-box binding homeobox (ZEB1/2) can inhibit the expression of adhesion protein in epithelial cells and promote the initiation of EMT, all of which can be regulated by miRNAs. In cancers, the inhibition of EMT by p53 to prevent metastasis, down-regulated Snail and ZEB1 via induction of miR-34, which also suppresses the expression of the stemness factors, BMI1, CD44, CD133 and c-MYC. Interestingly, Siemens et al[147] reported a double-negative feedback loop between miR-34 and Snail, that is Snail and ZEB1 conversely inhibit the expression of miR-34 through binding to E-box regions in miR-34 promoters[147]. MiR-200 family members are another group involved in the regulation of ZEB1 by forming a double-negative feedback loop, to reduce the migration and invasion of CRC cells[148]. Also, miR-429, the member of the miR-200 family, was found to reverse TGF-β-induced EMT by targeting one cut homeobox 2 (ONECUT2), thereby inhibiting cell migration and invasion, and its activity is significantly down-regulated in CRC[149]. The downregulation of other tumor suppressor factors, mainly miR-335, miR-132 and miR-192, is associated with the invasion and metastasis of CRC by increasing the expression of their ZEB2 target genes[150-152].
Additionally, twist family basic helix-loop-helix (bHLH) transcription factor (TWIST), containing the bHLH domain, inhibits mesenchymal cell protein expression to promote the EMT process[153], which can be suppressed by miR-145[154]. Prospero homeobox 1, another transcription factor, inhibit the expression of E-cadherin to promote the occurrence of EMT, which was achieved by binding to the promoter of pre-miR-9 and triggering its expression[155]. The FOX family of transcription factors, FOXQ1 and FOXM1, are also involved in the induction of EMT, and their expressions are negatively correlated with the low expression of miR-320 in CRC, which reduces the expression of E-cadherin[156].
TGF-β, the acceptable EMT-inducer, can activate the EMT process by regulating downstream factors, such as miR-187, which inhibit the expression of SMAD family member 4 (SMAD4), the maintainer of epithelial phenotype in CRC[10]. Furthermore, miR-20a overexpression can also facilitate EMT by inhibiting SMAD4 expression to promote the metastasis of CRC[157]. Interestingly, SMAD7 is the inhibitor of SMAD4, and the inhibition of SMAD7 can initiate TGF-β-induced EMT. A series of miRNAs, like miR-4775, miR-1269, and miR21 have been approved to promote the metastasis of CRC in a SMAD7/TGF-β-dependent manner[158-160].
Regarding the Wnt/β-catenin signaling pathway, the enhanced effect of miR-150 on EMT in CRC is generated by targeting the cAMP response element-binding protein signaling pathway[9]. Interestingly, Wnt-induced EMT is not only through the Wnt/β-catenin signaling pathway, but also partially activated through inhibiting negative transcription factors by miR-34a, miR-145 and miR-29b[161,162]. The loss of miR-145 function is negatively correlated with the EMT process and the downregulation of E-cadherin expression[163], while miR-29b inhibits β-catenin co-activators to block multiple β-catenin target genes and achieve the regulation of EMT in CRC[164].
After breaking through the basement membrane, CRC cells enter the circulation turning into circulating tumor cells (CTCs), responding to distant metastasis, referring to EMT-MET plasticity with promoting new invasion and metastasis, the important marker of metastasis when this characteristic exists in CTCs[165]. Increased activity of MMP or decrease the function of tissue inhibitors of metalloproteinases (TIMPs) promote CTCs detachment from the primary location, which can be regulated by miR-375 to suppress MMP2 level in CRC cells and correspondingly inhibit the proliferation, migration and invasion of CRC[166]. Cai et al[167] revealed that miR-194 promoted EMT-mediated metastasis in CRC through activating MMP2 function, while Xu et al[168] found that miR-20a performed a facilitated role during the EMT process through inhibiting TIMP2, resulting in increased activities of MMP2 and MMP9[167,168].
As distant metastasis of CRC is a major reason for clinical treatment failure and death in cancer patients, PMN is found to be a crucial factor of CRC metastasis, which is the formation of a microenvironment conducive to tumor metastasis at a specific site in the distant organ[169,170]. Tumor secretory factors, recruitment of inhibitory immune cells and inflammatory polarization of matrix components are key factors involved in the formation of PMN[171].
TDEs are the main component of tumor secretory factors, secreted by cancer cells at the primary site of the tumor and transmitted to distant sites through autocrine or paracrine to recruit immune cells[171]. The recruited immune cells, such as MDSCs, TAMs, and Tregs, induce the formation of the immunosuppressive microenvironment and subsequently secrete inflammatory cytokines to produce an inflammatory response and form an inflammatory microenvironment, which is conducive to the colonization and growth of CTCs. During this process, the increased tumor volume with a continuous proliferation of CRC cells leads to cells prone to hypoxia and nutrient deficiency, hence rapid angiogenesis can be found in the primary lesion. The pro-angiogenic factors secreted by the neovascularization will circulate with TDEs to the distant metastasis, promote angiogenesis at distant sites, and construct a perfect PMN to bear more tumor cells from distant metastasis[171]. So Liu et al[171] proposed immunosuppression, inflammatory response, angiogenesis and increased permeability, lymphangiogenesis, organicity and reprogramming as the six characteristics of PMN, which make PMN the best choice for the settlement and proliferation of metastatic cancer cells[171].
Immunosuppression, an important contributor to the formation and development of PMN, is also the major reason for the survival and development of tumors in vivo. Takano et al[172] found that plasma-derived exosome miR-203 induced the differentiation of monocytes in distal organs into M2 TAMs of immunosuppressant phenotype, while Zhao et al[173] reported that exosomal miR-934 also induced the differentiation of normal phenotype M1 TAMs into M2 TAMs, inducing the formation of immunosuppressive microenvironment[172,173]. SOCS3 was down-regulated by miR-222-3p in TDEs, which promoted STAT3-mediated M2 polarization of TAMs and contributed to the immunosuppressive microenvironment[134]. Wang et al[174] also demonstrated the enhancing role of exosomal miR-425-5p and miR-25-3p on M2 TAMs expression through the PI3K/AKT signaling pathway, to promote CRC metastasis to distant metastases[174].
TDEs in CRC were also reported to be involved in promoting T cell differentiation into Tregs, inhibiting normal immune cell function, and recruiting immunosuppressive cells into PMN[175]. Other immune cells, such as MDSCs, DCs, and NKs can also be transformed into immunosuppressive phenotypes in TME, together constituting the immunosuppressive microenvironment for tumor metastasis[176]. In addition to recruiting immunosuppressive cells to PMN, the immune escape of tumors ultimately needs to be realized by destroying the normal immunity of the body[177]. Inhibition of T cell function, disturbance of normal NK cell function and immature reversal of DCs are all able to lead to the destruction of the normal immune mechanism of the body[133]. Huang et al[178] demonstrated that lncRNA SNHG10 in TDEs participated in the TGF-β signaling pathway, inhibited the activity of NK cells, and damaged the normal anti-tumor immune function in CRC[178].
It should be noted that the activation of immune checkpoints is an effective pathway for the development of immunosuppression. PD-L1 derived from TDEs with highly similar function to the surface of tumor cells, can bind to its receptor on T cells to generate an immune examination response, effectively inhibiting the proliferation of T cells and inducing apoptosis, and destroying the anti-tumor function of positive T cells[136,179]. In addition, CSCs-derived exosomal miRNA-17-5p inhibits normal immune cell function and promotes immunosuppression by targeting speckle-type POZ protein and promoting the expression of PD-L1[180].
During the formation of PMN, ECM remodeling is one of the essential links. In situ tumor cells colonize and proliferate in distant metastases, secreting exosomes and producing inflammatory cytokines, causing hypoxia and inflammatory responses in normal cells of metastases. In such an environment, a large number of cytokines such as VEGF, macrophage migration inhibitory factor, TGF-β, and immunosuppressive cells are recruited to participate in the formation of PMN[181]. Similar to the ECM remodeling mechanism in the primary site, CAFs activated by TGF-β promote the fibrosis of ECM through the secretion of collagen and fibronectin, increase the hardness of ECM, and change its biomechanics[182]. It is found that CRC-derived exosomal miR-10b promotes CAFs formation and leads to ECM fibronectin through the PI3K/Akt/mTOR pathway[183]. Exosomal miR-1246 and miR-1290 advance the development of interstitial fibrosis by activating the expression of actin alpha 2 and pro-fibrotic factors[184], while exosomal miR-139-5p and miR-21-5p degrade ECM proteins by promoting the expression of MMP2 and MMP13, thus accelerating the formation of PMN[185].
Interestingly, inflammation is not only involved in inhibiting the process of cancer but also found to promote tumor occurrence and metastasis. Inflammatory M1 TAMs, as a pro-inflammatory, immunostimulating and anti-tumor factor producing IL-1β, IL-6 and TNF-α, contributed to the development of CRC in colitis[15]. Therefore, the inflammatory microenvironment caused by chronic inflammation significantly promotes the formation of PMN in distant organs during tumor growth and metastasis[186]. Pro-inflammatory cytokines such as IL-1β, IL-6 and TNF-α are important factors in the inflammatory microenvironment, which directly or indirectly stimulate tumor survival, proliferation and metastasis[187]. It is shown that low-density IL-1β, an important pro-inflammatory factor involved in innate immunity, could induce local inflammatory responses and lead to protective immune responses, while high concentration would result in inflammation-related cancer tissue damage[188].
Another chemokine, IL-6 stimulates the activation of T and B cells during the immune response to perform an anti-inflammatory role[189]. Pucci et al[190] found that CRC tumor cell-derived exosomal miRNAs increase IL-6 secretion, thereby promoting inflammatory responses[190]. High levels of IL-6 have been detected in serum detected in live tumors or biopsies from cancer patients, suggesting that the inflammatory effects of this cytokine may be related to the occurrence of cancer[191]. MiR-21 carried by exosomes promotes the release of pro-inflammatory IL-6 and IL-21 and induces them into circulation, thus inducing the formation of an inflammatory microenvironment[192].
Although originally TNF-α was reported as an anti-tumor cytokine, high-dose recombinant TNF-α has been verified to induce tumor necrosis and promote the progression of tumors in vivo[193]. In addition, owing to the special biological environment of CRC, intestinal bacteria also promote the formation of an inflammatory microenvironment through secreting exosomes. The induction of E.coli-derived exosome with miRNAs on the inflammatory microenvironment is achieved by increasing the expression of toll-like receptor (TLR) and promoting the secretion of pro-inflammatory cytokine IL-8[194]. The exosomal miR-149-3p derived from enterotoxin bacteria disrupts normal gene transcription and leads to DNA damage and oxidative stress, which promotes the formation of an inflammatory microenvironment[195]. Exosomal miRNA-21 and miRNA-29a promote CRC metastasis by acting on TLR7/TLR8 and inducing the formation of an inflammatory microenvironment in PMN[196].
Angiogenesis is another critical factor in PMN formation in CRC, which rapidly generates tumor cells providing oxygen, energy and nutrients for survival and metastasis in the case of hypoxia and nutrient deficiency. To form a suitable PMN for the metastasis of CRC, a variety of pro-angiogenic factors must reach the distal metastasis via exosomes through blood circulation and be expressed. VEGF, fibroblast growth factor, platelet-derived growth factor, basic fibroblast growth factor, TGF-β, TNF-α, and IL-8 are the main angiogenic stimulator carried by TDEs[197]. VEGF signaling pathway is the most promising target for angiogenesis and plays a key role in angiogenesis[198]. Equally important to these pro-angiogenic factors are cell- or plasma-derived exosomes from various human tumors identified as effective inducers of angiogenesis in vitro and in vivo, which have the function of inducing closely related to the miRNA carried in exosomes[199]. For instance, CRC-derived exosomal miR-21-5p improves the expression of VEGF and Cyclin D1, enhances vascular permeability and promotes angiogenesis[145]. Exosome miR-25-3p promotes the expression of VEGF receptor 2, and regulates tight junction protein Claudin-5, resulting in the production of PMN in the liver and other sites of CRC patients[200]. In addition, exosomal miRNA-92a-3p stimulates angiogenesis by increasing vascular endothelial cell division and participating in the regulation of the binding protein, Claudin-11[201]. Zhao et al[202] reported that CRC-derived exosome miR-1229 promoted metastasis of CRC by activating VEGF production and promoting angiogenesis[202]. Exosome-derived miRNA-183-5p accelerates the generation of neovascularization in CRC metastasis, whereas exosomes secreted by neovascularization in PMN promote the metastasis of tumor cells from the primary site to specific organs and tissues[143].
It is worth mentioning that exosomes have the role of inducing tumor drug resistance, which provides a new research direction to solve the drug resistance problem that has puzzled doctors and researchers for a long time. Exosomes secreted by drug-resistant cancer cells encapsulate chemotherapeutic drugs and transport them out of tumor cells[203], and the interaction of exosomes containing miRNA, mRNA and protein from cancer cells is also associated with tumor drug resistance[204]. In summary, the mechanism of exosomes inducing drug resistance mainly involves drug expulsion, activation of anti-apoptotic pathways, changes in signal transduction, and promotion of survival and proliferation of CSCs.
First of all, exosomes released by tumor cells help cells to expel cytotoxic drugs, related to the overexpression of P-glycoprotein[205]. Although no reports on CRC, it is demonstrated that exosomes directly or indirectly regulate drug efflux pumps and thus influence drug resistance by regulating P-glycoprotein expression in breast and ovarian cancers[203,206]. Second, acquired or intrinsic resistance to chemotherapy often prevents tumor cells from undergoing adequate levels of apoptosis, resulting in poor survival and treatment[207]. Inhibitors of the apoptotic pathway are used to sensitize tumor cells to chemotherapy. In the clinical treatment of CRC, cetuximab-resistant CRC cells RKO have been found to induce cetuximab resistance by down-regulating PTEN and increasing AKT phosphorylation, which is related to apoptosis escape[208]. Third, signaling pathways in drug-sensitive cells are altered by the uptake of drug-resistant cell-derived exosomes, including EGFR, Wnt/β-catenin, PI3K/AKT, PTEN, and mTORC signaling pathways that play important roles in tumor progression and drug resistance, whose abnormalities are associated with chemotherapy resistance[209,210]. Hu et al[211] reported that CRC cells secreted exosomes capable of inducing chemotherapy resistance, which caused drug resistance by promoting β-catenin stabilization and nuclear translocation and activating the Wnt/β-catenin pathway[211]. Furthermore, miR-30a, miR-222, or miR-100-5p carried by exosomes may induce drug resistance in drug-sensitive cells by regulating MAPK or mTOR pathways[212]. Lastly, exosomes induce drug resistance by promoting the growth and proliferation of CSCs[213]. Plenty of stromal cells, such as CAFs and MSCs, promote the growth of CSCs by secreting exosomes[214]. Exosomes derived from MSCs increase the proportion of CSCs by activating the Wnt signaling pathway and activating the 1/2 extracellular signal (ERK1/2), thus endowing CSCs with phenotypes, and inducing drug resistance in CRC[215].
Notably, CSC self-derived exosomes maintain stemness within TME by transporting their cargoes, thus enhancing resistance to different cancer therapies[216]. The cargos include Hedgehog, Wnt, β-catenin, and other CSC-specific mRNAs, as well as proteins needed by CSCs to maintain self-renewal and other stemness. TDEs have been reported to carry different types of integrins and related ligands that are involved in the formation of cancer cell colonization and PMN, while integrin is the key drug resistance factor in cancer therapy in maintaining the phenotype and behavior of stem cells[217].
CRC is a highly heterogeneous, highly metastatic and fatal cancer, and tumor cell metastasis is the main reason for the high mortality rate of this cancer. In the process of diagnosis and treatment of CRC, the lack of specific symptoms causes great difficulties in the early diagnosis of CRC due to its similarity to non-cancerous intestinal diseases. At present, the diagnosis of CRC depends on clinical evaluation and imaging diagnosis. However, routine diagnosis such as radiographic imaging or histopathological analysis fails to detect early systemic spread of CRC[218], and colon cancer markers such as carcinoembryonic antigen (CEA) and CA19-9 have low sensitivity and specificity[219]. In most clinical cases of CRC, surgery is the best treatment option, sometimes accompanied by chemoradiotherapy. However, due to limited diagnostic means, most patients are often diagnosed with advanced CRC and miss the optimal surgical opportunity. Therefore, the development of new and effective diagnostic biomarkers for CRC is essential for early detection and reduction of CRC mortality.
As EVs that play a key role in intercellular communication, exosomes contain proteins, miRNAs and other substances that are closely related to tumorigenesis, tumor cell survival, chemotherapy resistance and metastasis. Due to their non-invasive, high sensitivity and specificity, exosomes have advantages in being ideal biomarkers for early cancer screening and diagnosis at this stage[220]. In addition, some studies have shown that exosomal miRNAs can be used as drug carriers to transport drugs and participate in the immunotherapy of CRC[221]. Next, we describe the advantages of exosomes in CRC screening, diagnosis, treatment, and prognosis.
It is interesting and useful that exosomes can be detected by taking body fluids, such as blood, urine, saliva and cerebrospinal fluid for analysis, suggesting that exosomes could be an ideal non-invasive or less invasive biomarker for early cancer screening and diagnosis, with high specificity and sensitivity at an early stage[220]. Recently, transcriptomics research revealed that ncRNAs in exosomes are involved in different biological processes of CRC, and the high stability of exosome miRNAs in a variety of biological samples makes them an important candidate molecule for the discovery of new cancer biomarkers for CRC[222-224]. Wang et al[225] reported a group of six miRNAs including miR-21, let-7g, miR-31, miR-92a, miR-181b, and miR-203 as reliable biomarkers for CRC diagnosis, whose specificity and sensitivity exceed 40% compared to classical biomarkers, CEA and CA19-9[225], while the sensitivity of exosomes miR-1229, miR-223, miR-1224-5p and miR-150 are reach to 50%, whose expressions were significantly different between CRC patients and healthy individuals[226]. Increased serum levels of exosome miR-200 were significantly associated with CRC progression and liver metastasis[227]. Wang et al[158] confirmed that miR-125A-3p is highly expressed in the plasma of patients with early CRC but not in normal subjects, suggesting that miRNA in exosomes can be used as a biomarker for early CRC screening[158]. Moreover, compared with normal people, the expression of exosomal miR-92b is significantly decreased in CRC patients, indicating its higher accuracy in early CRC screening[228]. The expression of miR-23a and miR-1246 in exosomes was abundant in CRC patients. Decreased expression of exosome miRNA-23a and miRNA-1246 can be used as diagnostic markers for CRC in patients with primary resection[229]. In addition, circulating exosomal miR-17-5p and miR-92a-3p are associated with pathological staging and grading of CRC[230].
Apart from being biomarkers for CRC screening and diagnosis, exosomal miRNAs are also closely related to the prognosis of CRC and can be used as biomarkers for postoperative or therapeutic evaluation. Liu et al[231] found that low expression of plasma exosomal miR-4772-3p was closely associated with less lymph node metastasis, less tumor recurrence, and better prognosis in CRC patients[231]. Plasmid-derived exosome miRNA-193a is highly expressed in patients with middle and advanced CRC, suggesting that CRC patients have a longer survival time and a higher survival rate, since exosomal miR-193a could inhibit the mitosis and proliferation of tumor cells and induce cell apoptosis[232]. Peng et al[233] found that low expression of exosome miR-548-3c suggested poor prognosis, and its low expression in CRC liver metastases was positively correlated with angiogenesis and reduced overall survival rate[233].
Despite a large number of studies that have shown that exosomal miRNAs are potential biomarkers for a variety of cancers, their application in clinical biomarkers still faces many problems. Most current exosome miRNA studies have been limited to small patient cohorts or mice models, which means that miRNA levels in plasma exosomes vary widely in a single cohort and results are inconsistent across groups even when studying the same cancer type. Another common drawback is that the methods used to isolate exosomes from plasma are different from those used to extract miRNAs from exosomes. Studies lack common endogenous miRNA controls for quantifying exosome miRNAs. These problems affect the reliability of circulating exosome miRNAs as cancer biomarkers in clinical diagnosis or prognosis. Therefore, the techniques for isolating exosomes from body fluids and the methods for quantifying miRNAs or proteins also need to be further standardized.
In order to explore the potential of exosomes as novel biomarkers in clinical practice, the most important aspect is to optimize or standardize the measurement of exosomes. Nonetheless, to date, the isolation and purification of exosomes lacks a universally accepted gold standard. At present, the common method for exosome separation is ultra-centrifugation[234], which is controlled by different centrifugal forces and durations according to the density and size differences between exosomes and other components. However, the effectiveness of exosomes is limited due to many reasons such as excessive pressure, long time, high equipment requirements and the specificity of precipitation for separation during the centrifugation process[235]. Size exclusion chromatography is another common method for exosome separation[236], but owing to the high dilution degree of samples, this method cannot be used in applications requiring high concentration of exosomes. Additionally, quantitative reverse transcription polymerase chain reaction (qRT-PCR) is commonly used for the quantitative detection of exosome miRNAs[237], but this method is prone to produce false positive signals. Subsequently, researchers developed non-PCR miRNA quantitative spectroscopy based on proportional electrochemistry, local surface Plasma Resonance[238] and Surface-enhanced Raman spectroscopy[239,240]. Yet, its application has been hampered by expensive instruments and complex operation. Currently, it is attempted to detect exosome miRNAs using fluorescence method have achieved varying degrees of success, and this method has been attached great importance by researchers due to its inherent advantages of simple instruments, high sensitivity, and high throughput screening[241]. The only fly in the ointment is that the complexity of biological systems makes it necessary to develop fluorescent systems with anti-interference for exosome miRNAs as diagnostic biomarkers. It is worth noting that environmental fluctuations caused by such experimental conditions can be offset by ratio fluorescence measurements by calculating the emission intensity ratio of two different wavelengths. In general, the isolation and measurement methods of exosome miRNAs are under constant research and innovation, and the increasingly mature technological conditions make it possible for exosome miRNAs to be used as novel biomarkers for cancer.
In fact, the key obstacle to exosome research to date has not been the separation of impurities from exosome samples, but rather the lack of information on the ratio of actual exosomes to exovesicles in the "exosomes" collected by experimental techniques. Since exosomes overlap with these cellular microvesicles in structure and characteristics[242,243], the reliability of the analysis can be ensured as long as the composition of exosomes in the extracted samples can be accurately determined. From the perspective of exosome drug development, the current drug approval system in most countries only requires a high proportion and quantification of exosomes to meet the requirements for quality control and safety assessment of cell-derived compounds[242]. Therefore, how to effectively quantify the individual components of exosomes in collected samples will facilitate the utilization and clinical application of exosomes in the future.
At present, the treatment of CRC mainly includes surgery, adjuvant chemotherapy, radiotherapy and immunotherapy. Exosomes can be used as drug carriers to transport drugs or directly transport miRNA small molecules to participate in CRC chemotherapy, radiotherapy and immunotherapy[221]. Zaharie et al[244] demonstrated that exosome miR-375 promoted tumor cell apoptosis and inhibited CRC proliferation, invasion and metastasis by participating in the Bcl-2 signaling pathway[244]. Similarly, exosomal miR-140-3p inhibits CRC proliferation, growth, and liver metastasis by involving the Bcl-2 and Bcl-9 pathways[245]. Yan et al[246] found that CRC patients with high expression of exosome miR-548c-5p capable to inhibit the proliferation, invasion and metastasis of CRC cells by enhancing the expression of HIF-1α, had a better prognosis, predicting miR-548c-5p as an indicator for prognostic analysis[246]. In addition, Hu et al[247] demonstrated that exosome miR-214 inhibits CRC autophagy and promotes its sensitivity to radiotherapy[247].
Recently, due to the rapid development of therapeutic methods, targeted therapy has become an effective strategy for the treatment of CRC. Exosomes are natural nanoparticle biological carriers that have emerged as promising therapeutic tools for the delivery and transfer of drugs, miRNAs, small interfering RNAs (siRNA), short hairpin RNAs, and other compounds that remain stable in exosomes used to treat cancer and other diseases, based on their non-toxicity and non-immunogenicity[248]. As delivery carriers of natural drugs and functional RNA, exosomes have their natural advantages[249].
First, exosomes can be produced and absorbed, and are capable of stable delivery of therapeutic drugs, such as therapeutic miRNAs and proteins[250]. Currently, doxorubicin and paclitaxel have been used in targeted cancer therapy via exosomes with minimal immunogenicity and toxicity compared to liposome, metal and polymer nanomaterials[251-254]. Second, exosomes enhance endocytosis by targeting specific cells and tissues with specific proteins, thus promoting the transfer of their contents[255]. In animal tumor models, exosome-mediated chemotherapy is more effective than free agents. For example, the anti-mitotic chemotherapy drug paclitaxel can be ultrasound-loaded into exosomes and is 50 times more cytotoxic to drug-resistant cancer cells in vitro than free paclitaxel[256]. Also, it is found that exosomes coated with different chemotherapeutic drugs inhibit tumor growth when delivered to mice tumor tissues, but no equivalent side effects have been observed in free drugs[257]. Bioengineered exosomes have been used to deliver anticancer drugs and functional RNAs to cancer cells, including CSCs, in a cell-specific manner. Several strategies have been reported to improve the targeting specificity and tumor absorption of the exosome, for instance, transforming the exosome into lysosome-associated membrane protein 2b and tumor-targeting integrin to express target ligands[253]. Exosomes are surface modified through oligonucleotide binding, which could potentially alter not only cell function, but also transport between cells. Third, exosomes deliver therapeutic goods with better efficacy and fewer off-target effects than other biological carriers, such as liposomes, due to their small size, membrane-permeability, ease of crossing the blood-brain barrier, and faster penetration of tumor cells than liposomes[258]. Kim et al[256] found that macrophage-derived exosomes loaded with paclitaxel significantly increased cellular uptake of Lewis lung cancer cell line compared to paclitaxel-loaded liposomes[256,259].
Remarkably, exosome targeting of CSCs is a promising approach for the development of cancer therapy, as the growth of CSCs causes drug-sensitive cells to transform into drug-resistant cells, reducing the sensitivity of anti-cancer drug therapy. CSC signaling pathways such as Wnt, Notch, Hippo, Hedgehog, NF-κB and TGF-β are significant for maintaining a series of biological functions such as self-renewal, differentiation and tumorigenesis, which, therefore, is also the main way for exosome loaded inhibitors (miRNA or siRNA) to selectively target CSCs[213]. Previous studies have indicated that fibroblast-derived exosome with Wnt could induce dedifferentiation of tumor cells and thus increase chemotherapy resistance to CRC, suggesting that interference with the exosomal Wnt signaling pathway is helpful to improve chemotherapy sensitivity and treatment window[211]. Furthermore, specific producers of CSCs, such as CD44, CD24, CD133, and CD200 can also be used as exosome targets using bioengineering techniques[260]. Liu et al[261] manifested that exosomes designed to carry miR-21 inhibitors and chemotherapeutic agents enhance the killing effect on CRC tumor cells and inhibit CRC resistance[261]. RDEs carrying miR-3a improve immunosuppression and inhibit CRC proliferation and metastasis[262].
In addition to being a transport vehicle for targeted drugs, another promising clinical application area for exosomes is anti-cancer vaccination. Taking the DC exosome vaccine as an example, DC-derived exosomes express MHC-I and MHC-II molecules, which can effectively activate cytotoxic T cells and induce anti-tumor immunity. Currently, DC-derived exosome vaccines have been tested in phase I clinical trials[263,264]. The results showed that no grade 2 or higher toxicity was observed in these clinical trials, proving that exosome administration is safe. In one of the Phase I trials, exosomes were isolated from the ascites of colon cancer patients and injected into the patients as a vaccine. Ascites-derived exosomes were found to be safe, well tolerated, and capable of producing tumor-specific antitumor cytotoxic T cell responses after the granulocyte-macrophage colony-stimulating factor use in the immunotherapy of CRC[265]. However, the safety of TDE vaccines remains uncertain because TDEs carry a large number of oncogenes, mRNAs, and miRNAs that induce tumor progression and metastasis.
Though a large number of experimental models support the use of exosomes in cancer therapy, only a few clinical trials are in progress, thus the clinical use of exosomes in cancer and other diseases still needs to solve many challenges. First, how to effectively load exogenous therapeutic miRNAs or therapeutic agents into exosomes and enhance cell-specific delivery. Second, how to prevent autoimmune reactions when using non-autologous exosomes carrying MHC-I or II, and how to control the degree of cytotoxic T cell activation in vaccine use. Furthermore, how to prolong the half-life of bioengineered exosomes in vivo to avoid the rapid clearance of immune cells, liver or kidney, etc. Finally, it remains to be seen whether exosomes can overcome the digestive effects of the gut and be used as oral agents to treat cancer. Therefore, before the widespread use of exosomes in clinical trials, the quality standards of exosomes should be carefully established to improve their efficacy in vivo.
To sum up, exosomes have been successfully used as drug carriers in clinical therapy, and their safety and clinical application in targeted therapy and so on still need further exploration and research. In this review, the mechanism of exosomes in CRC metastasis was comprehensively described, including the formation and influencing factors of TME, the formation, function and role of exosomes in cancer, as well as the role of exosome miRNAs in the process of CRC metastasis. In TME, exosomes secreted by tumor-derived immune cells such as TAMs, DCs, MDSCs and NKs are critical for tumor growth and metastasis. In addition, stromal cells such as CAFs and MSCs as well as the ECM also play a significant role in tumor metastasis. Exosomes, as EVs carrying biological cargos, exert their function in primary tumors and metastases mainly of intercellular communication. In exosomes, DNA, mRNA, ncRNA and protein have different biological meanings and participate in the regulation of the body together. The role of exosome miRNA in early screening, diagnosis and prognosis of CRC, as well as in the treatment strategies for CRC along with chemotherapy, radiotherapy, immunotherapy and targeted therapy, provides a promising way for preventing and treating the metastasis in patients with CRC.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mu Y, China; Zhang L, China S-Editor: Fan JR L-Editor:A P-Editor: Zhang XD
| 1. | Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14:101174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 687] [Cited by in RCA: 1336] [Article Influence: 334.0] [Reference Citation Analysis (5)] |
| 2. | Chen H, Yao J, Bao R, Dong Y, Zhang T, Du Y, Wang G, Ni D, Xun Z, Niu X, Ye Y, Li HB. Cross-talk of four types of RNA modification writers defines tumor microenvironment and pharmacogenomic landscape in colorectal cancer. Mol Cancer. 2021;20:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 163] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 3. | Xu Z, Zeng S, Gong Z, Yan Y. Exosome-based immunotherapy: a promising approach for cancer treatment. Mol Cancer. 2020;19:160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 356] [Cited by in RCA: 335] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 4. | Kahroba H, Hejazi MS, Samadi N. Exosomes: from carcinogenesis and metastasis to diagnosis and treatment of gastric cancer. Cell Mol Life Sci. 2019;76:1747-1758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 5. | Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4060] [Cited by in RCA: 5752] [Article Influence: 522.9] [Reference Citation Analysis (0)] |
| 6. | Yang F, Ning Z, Ma L, Liu W, Shao C, Shu Y, Shen H. Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Mol Cancer. 2017;16:148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 234] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 7. | Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 578] [Cited by in RCA: 916] [Article Influence: 130.9] [Reference Citation Analysis (0)] |
| 8. | Nedaeinia R, Manian M, Jazayeri MH, Ranjbar M, Salehi R, Sharifi M, Mohaghegh F, Goli M, Jahednia SH, Avan A, Ghayour-Mobarhan M. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. 2017;24:48-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 164] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 9. | Guo YH, Wang LQ, Li B, Xu H, Yang JH, Zheng LS, Yu P, Zhou AD, Zhang Y, Xie SJ, Liang ZR, Zhang CM, Zhou H, Qu LH. Wnt/β-catenin pathway transactivates microRNA-150 that promotes EMT of colorectal cancer cells by suppressing CREB signaling. Oncotarget. 2016;7:42513-42526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 10. | Zhang F, Luo Y, Shao Z, Xu L, Liu X, Niu Y, Shi J, Sun X, Liu Y, Ding Y, Zhao L. MicroRNA-187, a downstream effector of TGFβ pathway, suppresses Smad-mediated epithelial-mesenchymal transition in colorectal cancer. Cancer Lett. 2016;373:203-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 11. | Wang M, Zhao J, Zhang L, Wei F, Lian Y, Wu Y, Gong Z, Zhang S, Zhou J, Cao K, Li X, Xiong W, Li G, Zeng Z, Guo C. Role of tumor microenvironment in tumorigenesis. J Cancer. 2017;8:761-773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1071] [Cited by in RCA: 954] [Article Influence: 119.3] [Reference Citation Analysis (0)] |
| 12. | Chen H, Shi R, Luo B, Yang X, Qiu L, Xiong J, Jiang M, Liu Y, Zhang Z, Wu Y. Macrophage peroxisome proliferator-activated receptor γ deficiency delays skin wound healing through impairing apoptotic cell clearance in mice. Cell Death Dis. 2015;6:e1597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 13. | Yang L, Zhang Y. Tumor-associated macrophages: from basic research to clinical application. J Hematol Oncol. 2017;10:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 673] [Cited by in RCA: 649] [Article Influence: 81.1] [Reference Citation Analysis (0)] |
| 14. | Grivennikov SI. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin Immunopathol. 2013;35:229-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 422] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 15. | Zhu W, Yu J, Nie Y, Shi X, Liu Y, Li F, Zhang XL. Disequilibrium of M1 and M2 macrophages correlates with the development of experimental inflammatory bowel diseases. Immunol Invest. 2014;43:638-652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 135] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 16. | Cardoso AP, Pinto ML, Pinto AT, Oliveira MI, Pinto MT, Gonçalves R, Relvas JB, Figueiredo C, Seruca R, Mantovani A, Mareel M, Barbosa MA, Oliveira MJ. Macrophages stimulate gastric and colorectal cancer invasion through EGFR Y(1086), c-Src, Erk1/2 and Akt phosphorylation and smallGTPase activity. Oncogene. 2014;33:2123-2133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 101] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 17. | Wang N, Liu W, Zheng Y, Wang S, Yang B, Li M, Song J, Zhang F, Zhang X, Wang Q, Wang Z. CXCL1 derived from tumor-associated macrophages promotes breast cancer metastasis via activating NF-κB/SOX4 signaling. Cell Death Dis. 2018;9:880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 229] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 18. | Zhao X, Qu J, Sun Y, Wang J, Liu X, Wang F, Zhang H, Wang W, Ma X, Gao X, Zhang S. Prognostic significance of tumor-associated macrophages in breast cancer: a meta-analysis of the literature. Oncotarget. 2017;8:30576-30586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 169] [Cited by in RCA: 251] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 19. | Wang W, Li X, Zheng D, Zhang D, Peng X, Zhang X, Ai F, Wang X, Ma J, Xiong W, Li G, Zhou Y, Shen S. Dynamic changes and functions of macrophages and M1/M2 subpopulations during ulcerative colitis-associated carcinogenesis in an AOM/DSS mouse model. Mol Med Rep. 2015;11:2397-2406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 20. | Lee YS, Song SJ, Hong HK, Oh BY, Lee WY, Cho YB. The FBW7-MCL-1 axis is key in M1 and M2 macrophage-related colon cancer cell progression: validating the immunotherapeutic value of targeting PI3Kγ. Exp Mol Med. 2020;52:815-831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 21. | Kim J, Bae JS. Tumor-Associated Macrophages and Neutrophils in Tumor Microenvironment. Mediators Inflamm. 2016;2016:6058147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 293] [Cited by in RCA: 546] [Article Influence: 60.7] [Reference Citation Analysis (0)] |
| 22. | Barbera-Guillem E, Nyhus JK, Wolford CC, Friece CR, Sampsel JW. Vascular endothelial growth factor secretion by tumor-infiltrating macrophages essentially supports tumor angiogenesis, and IgG immune complexes potentiate the process. Cancer Res. 2002;62:7042-7049. [PubMed] |
| 23. | Suarez-Lopez L, Sriram G, Kong YW, Morandell S, Merrick KA, Hernandez Y, Haigis KM, Yaffe MB. MK2 contributes to tumor progression by promoting M2 macrophage polarization and tumor angiogenesis. Proc Natl Acad Sci U S A. 2018;115:E4236-E4244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 24. | Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022-7029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 818] [Cited by in RCA: 855] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 25. | Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X, Liu Q, Dou R, Xiong B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 238] [Cited by in RCA: 568] [Article Influence: 94.7] [Reference Citation Analysis (1)] |
| 26. | Afik R, Zigmond E, Vugman M, Klepfish M, Shimshoni E, Pasmanik-Chor M, Shenoy A, Bassat E, Halpern Z, Geiger T, Sagi I, Varol C. Tumor macrophages are pivotal constructors of tumor collagenous matrix. J Exp Med. 2016;213:2315-2331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 263] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 27. | Illemann M, Bird N, Majeed A, Sehested M, Laerum OD, Lund LR, Danø K, Nielsen BS. MMP-9 is differentially expressed in primary human colorectal adenocarcinomas and their metastases. Mol Cancer Res. 2006;4:293-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 79] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 28. | Cai J, Xia L, Li J, Ni S, Song H, Wu X. Tumor-Associated Macrophages Derived TGF-β‒Induced Epithelial to Mesenchymal Transition in Colorectal Cancer Cells through Smad2,3-4/Snail Signaling Pathway. Cancer Res Treat. 2019;51:252-266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 29. | Herbeuval JP, Lelievre E, Lambert C, Dy M, Genin C. Recruitment of STAT3 for production of IL-10 by colon carcinoma cells induced by macrophage-derived IL-6. J Immunol. 2004;172:4630-4636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 95] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 30. | Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1523] [Cited by in RCA: 1660] [Article Influence: 87.4] [Reference Citation Analysis (2)] |
| 31. | Sumpter TL, Dangi A, Matta BM, Huang C, Stolz DB, Vodovotz Y, Thomson AW, Gandhi CR. Hepatic stellate cells undermine the allostimulatory function of liver myeloid dendritic cells via STAT3-dependent induction of IDO. J Immunol. 2012;189:3848-3858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 32. | Orsini G, Legitimo A, Failli A, Ferrari P, Nicolini A, Spisni R, Miccoli P, Consolini R. Defective generation and maturation of dendritic cells from monocytes in colorectal cancer patients during the course of disease. Int J Mol Sci. 2013;14:22022-22041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 33. | Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, Worthen GS, Albelda SM. Polarization of tumor-associated neutrophil phenotype by TGF-beta: "N1" versus "N2" TAN. Cancer Cell. 2009;16:183-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2035] [Cited by in RCA: 2526] [Article Influence: 157.9] [Reference Citation Analysis (0)] |
| 34. | Motta JM, Rumjanek VM. Sensitivity of Dendritic Cells to Microenvironment Signals. J Immunol Res. 2016;2016:4753607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 35. | Kobie JJ, Wu RS, Kurt RA, Lou S, Adelman MK, Whitesell LJ, Ramanathapuram LV, Arteaga CL, Akporiaye ET. Transforming growth factor beta inhibits the antigen-presenting functions and antitumor activity of dendritic cell vaccines. Cancer Res. 2003;63:1860-1864. [PubMed] |
| 36. | Pryczynicz A, Cepowicz D, Zaręba K, Gryko M, Hołody-Zaręba J, Kędra B, Kemona A, Guzińska-Ustymowicz K. Dysfunctions in the Mature Dendritic Cells Are Associated with the Presence of Metastases of Colorectal Cancer in the Surrounding Lymph Nodes. Gastroenterol Res Pract. 2016;2016:2405437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 37. | Hsu YL, Chen YJ, Chang WA, Jian SF, Fan HL, Wang JY, Kuo PL. Interaction between Tumor-Associated Dendritic Cells and Colon Cancer Cells Contributes to Tumor Progression via CXCL1. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 38. | Chaput N, Conforti R, Viaud S, Spatz A, Zitvogel L. The Janus face of dendritic cells in cancer. Oncogene. 2008;27:5920-5931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 39. | Michea P, Noël F, Zakine E, Czerwinska U, Sirven P, Abouzid O, Goudot C, Scholer-Dahirel A, Vincent-Salomon A, Reyal F, Amigorena S, Guillot-Delost M, Segura E, Soumelis V. Adjustment of dendritic cells to the breast-cancer microenvironment is subset specific. Nat Immunol. 2018;19:885-897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 166] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 40. | Uribe-Querol E, Rosales C. Neutrophils in Cancer: Two Sides of the Same Coin. J Immunol Res. 2015;2015:983698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 273] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 41. | Chen Q, Zhang L, Li X, Zhuo W. Neutrophil Extracellular Traps in Tumor Metastasis: Pathological Functions and Clinical Applications. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 42. | Schauer C, Janko C, Munoz LE, Zhao Y, Kienhöfer D, Frey B, Lell M, Manger B, Rech J, Naschberger E, Holmdahl R, Krenn V, Harrer T, Jeremic I, Bilyy R, Schett G, Hoffmann M, Herrmann M. Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines. Nat Med. 2014;20:511-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 614] [Cited by in RCA: 708] [Article Influence: 64.4] [Reference Citation Analysis (0)] |
| 43. | Zhou J, Nefedova Y, Lei A, Gabrilovich D. Neutrophils and PMN-MDSC: Their biological role and interaction with stromal cells. Semin Immunol. 2018;35:19-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 259] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 44. | Germann M, Zangger N, Sauvain MO, Sempoux C, Bowler AD, Wirapati P, Kandalaft LE, Delorenzi M, Tejpar S, Coukos G, Radtke F. Neutrophils suppress tumor-infiltrating T cells in colon cancer via matrix metalloproteinase-mediated activation of TGFβ. EMBO Mol Med. 2020;12:e10681. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 120] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 45. | Condamine T, Dominguez GA, Youn JI, Kossenkov AV, Mony S, Alicea-Torres K, Tcyganov E, Hashimoto A, Nefedova Y, Lin C, Partlova S, Garfall A, Vogl DT, Xu X, Knight SC, Malietzis G, Lee GH, Eruslanov E, Albelda SM, Wang X, Mehta JL, Bewtra M, Rustgi A, Hockstein N, Witt R, Masters G, Nam B, Smirnov D, Sepulveda MA, Gabrilovich DI. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci Immunol. 2016;1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 607] [Article Influence: 67.4] [Reference Citation Analysis (0)] |
| 46. | Glasner A, Levi A, Enk J, Isaacson B, Viukov S, Orlanski S, Scope A, Neuman T, Enk CD, Hanna JH, Sexl V, Jonjic S, Seliger B, Zitvogel L, Mandelboim O. NKp46 Receptor-Mediated Interferon-γ Production by Natural Killer Cells Increases Fibronectin 1 to Alter Tumor Architecture and Control Metastasis. Immunity. 2018;48:396-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 47. | Eyles J, Puaux AL, Wang X, Toh B, Prakash C, Hong M, Tan TG, Zheng L, Ong LC, Jin Y, Kato M, Prévost-Blondel A, Chow P, Yang H, Abastado JP. Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J Clin Invest. 2010;120:2030-2039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 387] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 48. | Olguín JE, Medina-Andrade I, Rodríguez T, Rodríguez-Sosa M, Terrazas LI. Relevance of Regulatory T Cells during Colorectal Cancer Development. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 49. | Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, Nagase H, Nishimura J, Yamamoto H, Takiguchi S, Tanoue T, Suda W, Morita H, Hattori M, Honda K, Mori M, Doki Y, Sakaguchi S. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 654] [Article Influence: 72.7] [Reference Citation Analysis (0)] |
| 50. | Garvey CM, Lau R, Sanchez A, Sun RX, Fong EJ, Doche ME, Chen O, Jusuf A, Lenz HJ, Larson B, Mumenthaler SM. Anti-EGFR Therapy Induces EGF Secretion by Cancer-Associated Fibroblasts to Confer Colorectal Cancer Chemoresistance. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 51. | Yu Y, Xiao CH, Tan LD, Wang QS, Li XQ, Feng YM. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-β signalling. Br J Cancer. 2014;110:724-732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 365] [Cited by in RCA: 518] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 52. | Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392-1400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1053] [Cited by in RCA: 1207] [Article Influence: 67.1] [Reference Citation Analysis (0)] |
| 53. | Erdogan B, Ao M, White LM, Means AL, Brewer BM, Yang L, Washington MK, Shi C, Franco OE, Weaver AM, Hayward SW, Li D, Webb DJ. Cancer-associated fibroblasts promote directional cancer cell migration by aligning fibronectin. J Cell Biol. 2017;216:3799-3816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 412] [Cited by in RCA: 401] [Article Influence: 50.1] [Reference Citation Analysis (0)] |
| 54. | Torres S, Garcia-Palmero I, Herrera M, Bartolomé RA, Peña C, Fernandez-Aceñero MJ, Padilla G, Peláez-García A, Lopez-Lucendo M, Rodriguez-Merlo R, García de Herreros A, Bonilla F, Casal JI. LOXL2 Is Highly Expressed in Cancer-Associated Fibroblasts and Associates to Poor Colon Cancer Survival. Clin Cancer Res. 2015;21:4892-4902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 55. | Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M, Ibiza S, Cañellas A, Hernando-Momblona X, Byrom D, Matarin JA, Calon A, Rivas EI, Nebreda AR, Riera A, Attolini CS, Batlle E. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554:538-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 917] [Cited by in RCA: 1325] [Article Influence: 189.3] [Reference Citation Analysis (0)] |
| 56. | Zhang R, Qi F, Zhao F, Li G, Shao S, Zhang X, Yuan L, Feng Y. Cancer-associated fibroblasts enhance tumor-associated macrophages enrichment and suppress NK cells function in colorectal cancer. Cell Death Dis. 2019;10:273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 246] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 57. | Gassmann P, Hemping-Bovenkerk A, Mees ST, Haier J. Metastatic tumor cell arrest in the liver-lumen occlusion and specific adhesion are not exclusive. Int J Colorectal Dis. 2009;24:851-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 58. | Matsumura H, Kondo T, Ogawa K, Tamura T, Fukunaga K, Murata S, Ohkohchi N. Kupffer cells decrease metastasis of colon cancer cells to the liver in the early stage. Int J Oncol. 2014;45:2303-2310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 59. | Tsilimigras DI, Brodt P, Clavien PA, Muschel RJ, D'Angelica MI, Endo I, Parks RW, Doyle M, de Santibañes E, Pawlik TM. Liver metastases. Nat Rev Dis Primers. 2021;7:27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 299] [Article Influence: 74.8] [Reference Citation Analysis (0)] |
| 60. | Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123:4195-4200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2308] [Cited by in RCA: 2780] [Article Influence: 198.6] [Reference Citation Analysis (0)] |
| 61. | Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol. 2010;22:697-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 587] [Cited by in RCA: 685] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 62. | Dong W, Li H, Zhang Y, Yang H, Guo M, Li L, Liu T. Matrix metalloproteinase 2 promotes cell growth and invasion in colorectal cancer. Acta Biochim Biophys Sin (Shanghai). 2011;43:840-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 63. | Zeng ZS, Cohen AM, Guillem JG. Loss of basement membrane type IV collagen is associated with increased expression of metalloproteinases 2 and 9 (MMP-2 and MMP-9) during human colorectal tumorigenesis. Carcinogenesis. 1999;20:749-755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 194] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 64. | Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2671] [Cited by in RCA: 3076] [Article Influence: 192.3] [Reference Citation Analysis (0)] |
| 65. | Brauchle E, Kasper J, Daum R, Schierbaum N, Falch C, Kirschniak A, Schäffer TE, Schenke-Layland K. Biomechanical and biomolecular characterization of extracellular matrix structures in human colon carcinomas. Matrix Biol. 2018;68-69:180-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 135] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 66. | Bauer J, Emon MAB, Staudacher JJ, Thomas AL, Zessner-Spitzenberg J, Mancinelli G, Krett N, Saif MT, Jung B. Author Correction: Increased stiffness of the tumor microenvironment in colon cancer stimulates cancer associated fibroblast-mediated prometastatic activin A signaling. Sci Rep. 2020;10:7606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 67. | Zhang HG, Grizzle WE. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol. 2014;184:28-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 297] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 68. | Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32:623-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 922] [Cited by in RCA: 899] [Article Influence: 74.9] [Reference Citation Analysis (0)] |
| 69. | Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 883] [Cited by in RCA: 1177] [Article Influence: 130.8] [Reference Citation Analysis (0)] |
| 70. | Riches A, Campbell E, Borger E, Powis S. Regulation of exosome release from mammary epithelial and breast cancer cells - a new regulatory pathway. Eur J Cancer. 2014;50:1025-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 193] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 71. | Miyoshi H, Ajima R, Luo CT, Yamaguchi TP, Stappenbeck TS. Wnt5a potentiates TGF-β signaling to promote colonic crypt regeneration after tissue injury. Science. 2012;338:108-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 378] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 72. | Deep G, Panigrahi GK. Hypoxia-Induced Signaling Promotes Prostate Cancer Progression: Exosomes Role as Messenger of Hypoxic Response in Tumor Microenvironment. Crit Rev Oncog. 2015;20:419-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 73. | DeCosse JJ, Gossens CL, Kuzma JF, Unsworth BR. Breast cancer: induction of differentiation by embryonic tissue. Science. 1973;181:1057-1058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 86] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 74. | Fujii H, Cunha GR, Norman JT. The induction of adenocarcinomatous differentiation in neoplastic bladder epithelium by an embryonic prostatic inductor. J Urol. 1982;128:858-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 37] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 75. | Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6920] [Cited by in RCA: 6514] [Article Influence: 1302.8] [Reference Citation Analysis (0)] |
| 76. | van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3060] [Cited by in RCA: 5598] [Article Influence: 799.7] [Reference Citation Analysis (0)] |
| 77. | McAndrews KM, Kalluri R. Mechanisms associated with biogenesis of exosomes in cancer. Mol Cancer. 2019;18:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 291] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 78. | Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol. 2013;5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 344] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 79. | Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172-177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 551] [Cited by in RCA: 507] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 80. | van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E, Raposo G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell. 2011;21:708-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 516] [Cited by in RCA: 711] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 81. | Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W, Zhang R, Wu Y, Gao S, Kang T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021;31:157-177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 293] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 82. | Bissig C, Lenoir M, Velluz MC, Kufareva I, Abagyan R, Overduin M, Gruenberg J. Viral infection controlled by a calcium-dependent lipid-binding module in ALIX. Dev Cell. 2013;25:364-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 83. | Laulagnier K, Motta C, Hamdi S, Roy S, Fauvelle F, Pageaux JF, Kobayashi T, Salles JP, Perret B, Bonnerot C, Record M. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem J. 2004;380:161-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 489] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 84. | Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241-D1244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 697] [Cited by in RCA: 837] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 85. | McGough IJ, Vincent JP. Exosomes in developmental signalling. Development. 2016;143:2482-2493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 135] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 86. | Bebelman MP, Smit MJ, Pegtel DM, Baglio SR. Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther. 2018;188:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 592] [Article Influence: 84.6] [Reference Citation Analysis (1)] |
| 87. | Wen SW, Lima LG, Lobb RJ, Norris EL, Hastie ML, Krumeich S, Möller A. Breast Cancer-Derived Exosomes Reflect the Cell-of-Origin Phenotype. Proteomics. 2019;19:e1800180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 88. | Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1483] [Cited by in RCA: 2618] [Article Influence: 436.3] [Reference Citation Analysis (0)] |
| 89. | Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19-30; sup pp 1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1525] [Cited by in RCA: 1962] [Article Influence: 122.6] [Reference Citation Analysis (0)] |
| 90. | Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1084] [Cited by in RCA: 1377] [Article Influence: 105.9] [Reference Citation Analysis (0)] |
| 91. | Hsu C, Morohashi Y, Yoshimura S, Manrique-Hoyos N, Jung S, Lauterbach MA, Bakhti M, Grønborg M, Möbius W, Rhee J, Barr FA, Simons M. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189:223-232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 545] [Cited by in RCA: 635] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 92. | Taha EA, Ono K, Eguchi T. Roles of Extracellular HSPs as Biomarkers in Immune Surveillance and Immune Evasion. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 136] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 93. | Lauwers E, Wang YC, Gallardo R, Van der Kant R, Michiels E, Swerts J, Baatsen P, Zaiter SS, McAlpine SR, Gounko NV, Rousseau F, Schymkowitz J, Verstreken P. Hsp90 Mediates Membrane Deformation and Exosome Release. Mol Cell. 2018;71:689-702.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 115] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 94. | Tang X, Chang C, Guo J, Lincoln V, Liang C, Chen M, Woodley DT, Li W. Tumour-Secreted Hsp90α on External Surface of Exosomes Mediates Tumour - Stromal Cell Communication via Autocrine and Paracrine Mechanisms. Sci Rep. 2019;9:15108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 95. | Mazurov D, Barbashova L, Filatov A. Tetraspanin protein CD9 interacts with metalloprotease CD10 and enhances its release via exosomes. FEBS J. 2013;280:1200-1213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 96. | Bobrie A, Colombo M, Krumeich S, Raposo G, Théry C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles. 2012;1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 427] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 97. | Poliakov A, Spilman M, Dokland T, Amling CL, Mobley JA. Structural heterogeneity and protein composition of exosome-like vesicles (prostasomes) in human semen. Prostate. 2009;69:159-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 254] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 98. | Cossetti C, Iraci N, Mercer TR, Leonardi T, Alpi E, Drago D, Alfaro-Cervello C, Saini HK, Davis MP, Schaeffer J, Vega B, Stefanini M, Zhao C, Muller W, Garcia-Verdugo JM, Mathivanan S, Bachi A, Enright AJ, Mattick JS, Pluchino S. Extracellular vesicles from neural stem cells transfer IFN-γ via Ifngr1 to activate Stat1 signaling in target cells. Mol Cell. 2014;56:193-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 239] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 99. | Choi D, Montermini L, Kim DK, Meehan B, Roth FP, Rak J. The Impact of Oncogenic EGFRvIII on the Proteome of Extracellular Vesicles Released from Glioblastoma Cells. Mol Cell Proteomics. 2018;17:1948-1964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 100. | Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M, Hladik F. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290-7304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 446] [Cited by in RCA: 444] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 101. | Kanaoka R, Iinuma H, Dejima H, Sakai T, Uehara H, Matsutani N, Kawamura M. Usefulness of Plasma Exosomal MicroRNA-451a as a Noninvasive Biomarker for Early Prediction of Recurrence and Prognosis of Non-Small Cell Lung Cancer. Oncology. 2018;94:311-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 79] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 102. | Fortunato O, Gasparini P, Boeri M, Sozzi G. Exo-miRNAs as a New Tool for Liquid Biopsy in Lung Cancer. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 103. | Endzeliņš E, Berger A, Melne V, Bajo-Santos C, Soboļevska K, Ābols A, Rodriguez M, Šantare D, Rudņickiha A, Lietuvietis V, Llorente A, Linē A. Detection of circulating miRNAs: comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer. 2017;17:730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 201] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 104. | Puik JR, Meijer LL, Le Large TY, Prado MM, Frampton AE, Kazemier G, Giovannetti E. miRNA profiling for diagnosis, prognosis and stratification of cancer treatment in cholangiocarcinoma. Pharmacogenomics. 2017;18:1343-1358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 105. | Sun ZP, Li AQ, Jia WH, Ye S, Van Eps G, Yu JM, Yang WJ. MicroRNA expression profiling in exosomes derived from gastric cancer stem-like cells. Oncotarget. 2017;8:93839-93855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 106. | Tang Y, Cui Y, Li Z, Jiao Z, Zhang Y, He Y, Chen G, Zhou Q, Wang W, Zhou X, Luo J, Zhang S. Radiation-induced miR-208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J Exp Clin Cancer Res. 2016;35:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 141] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 107. | Wei F, Ma C, Zhou T, Dong X, Luo Q, Geng L, Ding L, Zhang Y, Zhang L, Li N, Li Y, Liu Y. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol Cancer. 2017;16:132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 203] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 108. | Zhao W, Shan B, He D, Cheng Y, Li B, Zhang C, Duan C. Recent Progress in Characterizing Long Noncoding RNAs in Cancer Drug Resistance. J Cancer. 2019;10:6693-6702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 109. | Wang M, Zhou L, Yu F, Zhang Y, Li P, Wang K. The functional roles of exosomal long non-coding RNAs in cancer. Cell Mol Life Sci. 2019;76:2059-2076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 107] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 110. | Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M, De Leo G, Alessandro R. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 383] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 111. | Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3273] [Cited by in RCA: 3137] [Article Influence: 522.8] [Reference Citation Analysis (0)] |
| 112. | Cheng X, Zhang L, Zhang K, Zhang G, Hu Y, Sun X, Zhao C, Li H, Li YM, Zhao J. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis. 2018;77:770-779. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 239] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 113. | Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X, Li X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 292] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 114. | Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4:594-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1514] [Cited by in RCA: 1696] [Article Influence: 62.8] [Reference Citation Analysis (0)] |
| 115. | Raimondo S, Saieva L, Corrado C, Fontana S, Flugy A, Rizzo A, De Leo G, Alessandro R. Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism. Cell Commun Signal. 2015;13:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 153] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 116. | Takahashi A, Okada R, Nagao K, Kawamata Y, Hanyu A, Yoshimoto S, Takasugi M, Watanabe S, Kanemaki MT, Obuse C, Hara E. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2017;8:15287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 395] [Cited by in RCA: 587] [Article Influence: 73.4] [Reference Citation Analysis (0)] |
| 117. | Yang Y, Li CW, Chan LC, Wei Y, Hsu JM, Xia W, Cha JH, Hou J, Hsu JL, Sun L, Hung MC. Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell Res. 2018;28:862-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 389] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 118. | Demory Beckler M, Higginbotham JN, Franklin JL, Ham AJ, Halvey PJ, Imasuen IE, Whitwell C, Li M, Liebler DC, Coffey RJ. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12:343-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 417] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 119. | Shimoda M, Principe S, Jackson HW, Luga V, Fang H, Molyneux SD, Shao YW, Aiken A, Waterhouse PD, Karamboulas C, Hess FM, Ohtsuka T, Okada Y, Ailles L, Ludwig A, Wrana JL, Kislinger T, Khokha R. Loss of the Timp gene family is sufficient for the acquisition of the CAF-like cell state. Nat Cell Biol. 2014;16:889-901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 165] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 120. | Achreja A, Zhao H, Yang L, Yun TH, Marini J, Nagrath D. Exo-MFA - A 13C metabolic flux analysis framework to dissect tumor microenvironment-secreted exosome contributions towards cancer cell metabolism. Metab Eng. 2017;43:156-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 121. | Cho JA, Park H, Lim EH, Lee KW. Exosomes from breast cancer cells can convert adipose tissue-derived mesenchymal stem cells into myofibroblast-like cells. Int J Oncol. 2012;40:130-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 183] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 122. | Baroni S, Romero-Cordoba S, Plantamura I, Dugo M, D'Ippolito E, Cataldo A, Cosentino G, Angeloni V, Rossini A, Daidone MG, Iorio MV. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. 2016;7:e2312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 245] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 123. | Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M, Lochnit G, Preissner KT, Zöller M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010;70:1668-1678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 453] [Cited by in RCA: 532] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 124. | Gesierich S, Berezovskiy I, Ryschich E, Zöller M. Systemic induction of the angiogenesis switch by the tetraspanin D6.1A/CO-029. Cancer Res. 2006;66:7083-7094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 164] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 125. | Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC, Tsai PH, Wu CY, Kuo PL. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36:4929-4942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 463] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 126. | Ridder K, Keller S, Dams M, Rupp AK, Schlaudraff J, Del Turco D, Starmann J, Macas J, Karpova D, Devraj K, Depboylu C, Landfried B, Arnold B, Plate KH, Höglinger G, Sültmann H, Altevogt P, Momma S. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 2014;12:e1001874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 249] [Cited by in RCA: 326] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 127. | Kurywchak P, Tavormina J, Kalluri R. The emerging roles of exosomes in the modulation of immune responses in cancer. Genome Med. 2018;10:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 84] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 128. | Tkach M, Kowal J, Zucchetti AE, Enserink L, Jouve M, Lankar D, Saitakis M, Martin-Jaular L, Théry C. Qualitative differences in T-cell activation by dendritic cell-derived extracellular vesicle subtypes. EMBO J. 2017;36:3012-3028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 277] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 129. | Kitai Y, Kawasaki T, Sueyoshi T, Kobiyama K, Ishii KJ, Zou J, Akira S, Matsuda T, Kawai T. DNA-Containing Exosomes Derived from Cancer Cells Treated with Topotecan Activate a STING-Dependent Pathway and Reinforce Antitumor Immunity. J Immunol. 2017;198:1649-1659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 164] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 130. | Torralba D, Baixauli F, Villarroya-Beltri C, Fernández-Delgado I, Latorre-Pellicer A, Acín-Pérez R, Martín-Cófreces NB, Jaso-Tamame ÁL, Iborra S, Jorge I, González-Aseguinolaza G, Garaude J, Vicente-Manzanares M, Enríquez JA, Mittelbrunn M, Sánchez-Madrid F. Priming of dendritic cells by DNA-containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat Commun. 2018;9:2658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 277] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 131. | Montermini L, Meehan B, Garnier D, Lee WJ, Lee TH, Guha A, Al-Nedawi K, Rak J. Inhibition of oncogenic epidermal growth factor receptor kinase triggers release of exosome-like extracellular vesicles and impacts their phosphoprotein and DNA content. J Biol Chem. 2015;290:24534-24546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 132. | Chennakrishnaiah S, Meehan B, D'Asti E, Montermini L, Lee TH, Karatzas N, Buchanan M, Tawil N, Choi D, Divangahi M, Basik M, Rak J. Leukocytes as a reservoir of circulating oncogenic DNA and regulatory targets of tumor-derived extracellular vesicles. J Thromb Haemost. 2018;16:1800-1813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 133. | Ding G, Zhou L, Qian Y, Fu M, Chen J, Xiang J, Wu Z, Jiang G, Cao L. Pancreatic cancer-derived exosomes transfer miRNAs to dendritic cells and inhibit RFXAP expression via miR-212-3p. Oncotarget. 2015;6:29877-29888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 236] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 134. | Ying X, Wu Q, Wu X, Zhu Q, Wang X, Jiang L, Chen X. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget. 2016;7:43076-43087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 291] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 135. | Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382-386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1806] [Cited by in RCA: 2049] [Article Influence: 292.7] [Reference Citation Analysis (0)] |
| 136. | Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, Montabana E, Lang UE, Fu Q, Fong L, Blelloch R. Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory. Cell. 2019;177:414-427.e13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 822] [Cited by in RCA: 992] [Article Influence: 165.3] [Reference Citation Analysis (0)] |
| 137. | Skokos D, Le Panse S, Villa I, Rousselle JC, Peronet R, David B, Namane A, Mécheri S. Mast cell-dependent B and T lymphocyte activation is mediated by the secretion of immunologically active exosomes. J Immunol. 2001;166:868-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 264] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 138. | Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N, Levy S, Zhang B, Coffey RJ, Patton JG. KRAS-dependent sorting of miRNA to exosomes. Elife. 2015;4:e07197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 263] [Cited by in RCA: 289] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 139. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8847] [Cited by in RCA: 9280] [Article Influence: 464.0] [Reference Citation Analysis (0)] |
| 140. | Michael MZ, O' Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882-891. [PubMed] |
| 141. | Bartley AN, Yao H, Barkoh BA, Ivan C, Mishra BM, Rashid A, Calin GA, Luthra R, Hamilton SR. Complex patterns of altered MicroRNA expression during the adenoma-adenocarcinoma sequence for microsatellite-stable colorectal cancer. Clin Cancer Res. 2011;17:7283-7293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 142. | Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016;30:836-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1100] [Cited by in RCA: 1486] [Article Influence: 165.1] [Reference Citation Analysis (0)] |
| 143. | Syn N, Wang L, Sethi G, Thiery JP, Goh BC. Exosome-Mediated Metastasis: From Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharmacol Sci. 2016;37:606-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 386] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 144. | Whiteside TL. The role of tumor-derived exosomes in epithelial mesenchymal transition (EMT). Transl Cancer Res. 2017;6:S90-S92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 145. | Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621-9630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 671] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 146. | Cao H, Xu E, Liu H, Wan L, Lai M. Epithelial-mesenchymal transition in colorectal cancer metastasis: A system review. Pathol Res Pract. 2015;211:557-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 289] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 147. | Siemens H, Jackstadt R, Hünten S, Kaller M, Menssen A, Götz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256-4271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 479] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 148. | Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR, Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315-1326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 451] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 149. | Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu Z, Li X, Wu M. MiR-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting Onecut2 in colorectal carcinoma. Mol Cell Biochem. 2014;390:19-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 121] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 150. | Sun Z, Zhang Z, Liu Z, Qiu B, Liu K, Dong G. MicroRNA-335 inhibits invasion and metastasis of colorectal cancer by targeting ZEB2. Med Oncol. 2014;31:982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 151. | Zheng YB, Luo HP, Shi Q, Hao ZN, Ding Y, Wang QS, Li SB, Xiao GC, Tong SL. miR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2. World J Gastroenterol. 2014;20:6515-6522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 83] [Cited by in RCA: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 152. | Geng L, Chaudhuri A, Talmon G, Wisecarver JL, Are C, Brattain M, Wang J. MicroRNA-192 suppresses liver metastasis of colon cancer. Oncogene. 2014;33:5332-5340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 153. | Solanes-Casado S, Cebrián A, Rodríguez-Remírez M, Mahíllo I, García-García L, Río-Vilariño A, Baños N, de Cárcer G, Monfort-Vengut A, Castellano V, Fernández-Aceñero MJ, García-Foncillas J, Del Puerto-Nevado L. Overcoming PLK1 inhibitor resistance by targeting mevalonate pathway to impair AXL-TWIST axis in colorectal cancer. Biomed Pharmacother. 2021;144:112347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 154. | Shen X, Jiang H, Chen Z, Lu B, Zhu Y, Mao J, Chai K, Chen W. MicroRNA-145 Inhibits Cell Migration and Invasion in Colorectal Cancer by Targeting TWIST. Onco Targets Ther. 2019;12:10799-10809. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 155. | Lu MH, Huang CC, Pan MR, Chen HH, Hung WC. Prospero homeobox 1 promotes epithelial-mesenchymal transition in colon cancer cells by inhibiting E-cadherin via miR-9. Clin Cancer Res. 2012;18:6416-6425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 156. | Vishnubalaji R, Hamam R, Yue S, Al-Obeed O, Kassem M, Liu FF, Aldahmash A, Alajez NM. MicroRNA-320 suppresses colorectal cancer by targeting SOX4, FOXM1, and FOXQ1. Oncotarget. 2016;7:35789-35802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 157. | Zhang GJ, Li Y, Zhou H, Xiao HX, Zhou T. miR20a is an independent prognostic factor in colorectal cancer and is involved in cell metastasis. Mol Med Rep. 2014;10:283-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 158. | Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L, Zhang Y, Huang X. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci Rep. 2017;7:4150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 146] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 159. | Zhao S, Sun H, Jiang W, Mi Y, Zhang D, Wen Y, Cheng D, Tang H, Wu S, Yu Y, Liu X, Cui W, Zhang M, Sun X, Zhou Z, Peng Z, Yan D. miR-4775 promotes colorectal cancer invasion and metastasis via the Smad7/TGFβ-mediated epithelial to mesenchymal transition. Mol Cancer. 2017;16:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 160. | Bu P, Wang L, Chen KY, Rakhilin N, Sun J, Closa A, Tung KL, King S, Kristine Varanko A, Xu Y, Huan Chen J, Zessin AS, Shealy J, Cummings B, Hsu D, Lipkin SM, Moreno V, Gümüş ZH, Shen X. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-β. Nat Commun. 2015;6:6879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 161. | Hahn S, Jackstadt R, Siemens H, Hünten S, Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32:3079-3095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 162. | Kim NH, Kim HS, Kim NG, Lee I, Choi HS, Li XY, Kang SE, Cha SY, Ryu JK, Na JM, Park C, Kim K, Lee S, Gumbiner BM, Yook JI, Weiss SJ. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci Signal. 2011;4:ra71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 246] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 163. | Sanchez-Mejias A, Kwon J, Chew XH, Siemens A, Sohn HS, Jing G, Zhang B, Yang H, Tay Y. A novel SOCS5/miR-18/miR-25 axis promotes tumorigenesis in liver cancer. Int J Cancer. 2019;144:311-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 164. | Subramanian M, Rao SR, Thacker P, Chatterjee S, Karunagaran D. MiR-29b downregulates canonical Wnt signaling by suppressing coactivators of β-catenin in human colorectal cancer cells. J Cell Biochem. 2014;115:1974-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 165. | Jolly MK, Tripathi SC, Somarelli JA, Hanash SM, Levine H. Epithelial/mesenchymal plasticity: how have quantitative mathematical models helped improve our understanding? Mol Oncol. 2017;11:739-754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 166. | Cui F, Wang S, Lao I, Zhou C, Kong H, Bayaxi N, Li J, Chen Q, Zhu T, Zhu H. miR-375 inhibits the invasion and metastasis of colorectal cancer via targeting SP1 and regulating EMT-associated genes. Oncol Rep. 2016;36:487-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 167. | Cai HK, Chen X, Tang YH, Deng YC. MicroRNA-194 modulates epithelial-mesenchymal transition in human colorectal cancer metastasis. Onco Targets Ther. 2017;10:1269-1278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 168. | Xu T, Jing C, Shi Y, Miao R, Peng L, Kong S, Ma Y, Li L. microRNA-20a enhances the epithelial-to-mesenchymal transition of colorectal cancer cells by modulating matrix metalloproteinases. Exp Ther Med. 2015;10:683-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 169. | Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792-3801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 719] [Cited by in RCA: 784] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 170. | Rana S, Malinowska K, Zöller M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia. 2013;15:281-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 288] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 171. | Liu Y, Cao X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell. 2016;30:668-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 826] [Article Influence: 91.8] [Reference Citation Analysis (0)] |
| 172. | Takano Y, Masuda T, Iinuma H, Yamaguchi R, Sato K, Tobo T, Hirata H, Kuroda Y, Nambara S, Hayashi N, Iguchi T, Ito S, Eguchi H, Ochiya T, Yanaga K, Miyano S, Mimori K. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget. 2017;8:78598-78613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 139] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 173. | Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, Zhang Z, Cai S, Xu Y, Li X, He X, Zhong X, Li G, Chen Z, Li D. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol. 2020;13:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 532] [Article Influence: 106.4] [Reference Citation Analysis (0)] |
| 174. | Wang D, Wang X, Si M, Yang J, Sun S, Wu H, Cui S, Qu X, Yu X. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020;474:36-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 247] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 175. | Yamada N, Kuranaga Y, Kumazaki M, Shinohara H, Taniguchi K, Akao Y. Colorectal cancer cell-derived extracellular vesicles induce phenotypic alteration of T cells into tumor-growth supporting cells with transforming growth factor-β1-mediated suppression. Oncotarget. 2016;7:27033-27043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 116] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 176. | Whiteside TL. Exosomes and tumor-mediated immune suppression. J Clin Invest. 2016;126:1216-1223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 445] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 177. | Wang J, De Veirman K, De Beule N, Maes K, De Bruyne E, Van Valckenborgh E, Vanderkerken K, Menu E. The bone marrow microenvironment enhances multiple myeloma progression by exosome-mediated activation of myeloid-derived suppressor cells. Oncotarget. 2015;6:43992-44004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 139] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 178. | Huang Y, Luo Y, Ou W, Wang Y, Dong D, Peng X. Exosomal lncRNA SNHG10 derived from colorectal cancer cells suppresses natural killer cell cytotoxicity by upregulating INHBC. Cancer Cell Int. 2021;21:528. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 179. | Liu J, Chen Z, Li Y, Zhao W, Wu J, Zhang Z. PD-1/PD-L1 Checkpoint Inhibitors in Tumor Immunotherapy. Front Pharmacol. 2021;12:731798. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 216] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 180. | Sun W, Cui J, Ge Y, Wang J, Yu Y, Han B, Liu B. Tumor stem cell-derived exosomal microRNA-17-5p inhibits anti-tumor immunity in colorectal cancer via targeting SPOP and overexpressing PD-L1. Cell Death Discov. 2022;8:223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 181. | Høye AM, Erler JT. Structural ECM components in the premetastatic and metastatic niche. Am J Physiol Cell Physiol. 2016;310:C955-C967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 82] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 182. | Najafi M, Farhood B, Mortezaee K. Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J Cell Biochem. 2019;120:2782-2790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 468] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 183. | Dai G, Yao X, Zhang Y, Gu J, Geng Y, Xue F, Zhang J. Colorectal cancer cell-derived exosomes containing miR-10b regulate fibroblast cells via the PI3K/Akt pathway. Bull Cancer. 2018;105:336-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 184. | Masamune A, Yoshida N, Hamada S, Takikawa T, Nabeshima T, Shimosegawa T. Exosomes derived from pancreatic cancer cells induce activation and profibrogenic activities in pancreatic stellate cells. Biochem Biophys Res Commun. 2018;495:71-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 185. | Sánchez CA, Andahur EI, Valenzuela R, Castellón EA, Fullá JA, Ramos CG, Triviño JC. Exosomes from bulk and stem cells from human prostate cancer have a differential microRNA content that contributes cooperatively over local and pre-metastatic niche. Oncotarget. 2016;7:3993-4008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 172] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 186. | Guo Y, Ji X, Liu J, Fan D, Zhou Q, Chen C, Wang W, Wang G, Wang H, Yuan W, Ji Z, Sun Z. Effects of exosomes on pre-metastatic niche formation in tumors. Mol Cancer. 2019;18:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 314] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 187. | Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883-899. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8437] [Cited by in RCA: 8170] [Article Influence: 544.7] [Reference Citation Analysis (0)] |
| 188. | Lust JA, Lacy MQ, Zeldenrust SR, Dispenzieri A, Gertz MA, Witzig TE, Kumar S, Hayman SR, Russell SJ, Buadi FK, Geyer SM, Campbell ME, Kyle RA, Rajkumar SV, Greipp PR, Kline MP, Xiong Y, Moon-Tasson LL, Donovan KA. Induction of a chronic disease state in patients with smoldering or indolent multiple myeloma by targeting interleukin 1{beta}-induced interleukin 6 production and the myeloma proliferative component. Mayo Clin Proc. 2009;84:114-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 204] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 189. | Hong DS, Angelo LS, Kurzrock R. Interleukin-6 and its receptor in cancer: implications for translational therapeutics. Cancer. 2007;110:1911-1928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 313] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 190. | Pucci M, Raimondo S, Urzì O, Moschetti M, Di Bella MA, Conigliaro A, Caccamo N, La Manna MP, Fontana S, Alessandro R. Tumor-Derived Small Extracellular Vesicles Induce Pro-Inflammatory Cytokine Expression and PD-L1 Regulation in M0 Macrophages via IL-6/STAT3 and TLR4 Signaling Pathways. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 191. | Guillerey C, Nakamura K, Vuckovic S, Hill GR, Smyth MJ. Immune responses in multiple myeloma: role of the natural immune surveillance and potential of immunotherapies. Cell Mol Life Sci. 2016;73:1569-1589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 95] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 192. | Shi C, Yang Y, Xia Y, Okugawa Y, Yang J, Liang Y, Chen H, Zhang P, Wang F, Han H, Wu W, Gao R, Gasche C, Qin H, Ma Y, Goel A. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut. 2016;65:1470-1481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 121] [Article Influence: 13.4] [Reference Citation Analysis (1)] |
| 193. | Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9:361-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1478] [Cited by in RCA: 1409] [Article Influence: 88.1] [Reference Citation Analysis (0)] |
| 194. | Patten DA, Hussein E, Davies SP, Humphreys PN, Collett A. Commensal-derived OMVs elicit a mild proinflammatory response in intestinal epithelial cells. Microbiology (Reading). 2017;163:702-711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 195. | Cao Y, Wang Z, Yan Y, Ji L, He J, Xuan B, Shen C, Ma Y, Jiang S, Ma D, Tong T, Zhang X, Gao Z, Zhu X, Fang JY, Chen H, Hong J. Enterotoxigenic Bacteroidesfragilis Promotes Intestinal Inflammation and Malignancy by Inhibiting Exosome-Packaged miR-149-3p. Gastroenterology. 2021;161:1552-1566.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 189] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 196. | Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110-E2116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1064] [Cited by in RCA: 1258] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 197. | Katoh M. Therapeutics targeting angiogenesis: genetics and epigenetics, extracellular miRNAs and signaling networks (Review). Int J Mol Med. 2013;32:763-767. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 119] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 198. | Momeny M, Sabourinejad Z, Zarrinrad G, Moghaddaskho F, Eyvani H, Yousefi H, Mirshahvaladi S, Poursani EM, Barghi F, Poursheikhani A, Dardaei L, Bashash D, Ghazi-Khansari M, Tavangar SM, Dehpour AR, Yaghmaie M, Alimoghaddam K, Ghavamzadeh A, Ghaffari SH. Anti-tumour activity of tivozanib, a pan-inhibitor of VEGF receptors, in therapy-resistant ovarian carcinoma cells. Sci Rep. 2017;7:45954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 199. | Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470-1476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4100] [Cited by in RCA: 3940] [Article Influence: 231.8] [Reference Citation Analysis (0)] |
| 200. | Zhang Q, Peng C. Cancer-associated fibroblasts regulate the biological behavior of cancer cells and stroma in gastric cancer. Oncol Lett. 2018;15:691-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 201. | Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36:1770-1778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 400] [Cited by in RCA: 589] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 202. | Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, Marini JC, Tudawe T, Seviour EG, San Lucas FA, Alvarez H, Gupta S, Maiti SN, Cooper L, Peehl D, Ram PT, Maitra A, Nagrath D. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 505] [Cited by in RCA: 722] [Article Influence: 80.2] [Reference Citation Analysis (0)] |
| 203. | Safaei R, Larson BJ, Cheng TC, Gibson MA, Otani S, Naerdemann W, Howell SB. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther. 2005;4:1595-1604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 415] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 204. | Shedden K, Xie XT, Chandaroy P, Chang YT, Rosania GR. Expulsion of small molecules in vesicles shed by cancer cells: association with gene expression and chemosensitivity profiles. Cancer Res. 2003;63:4331-4337. [PubMed] |
| 205. | Bebawy M, Combes V, Lee E, Jaiswal R, Gong J, Bonhoure A, Grau GE. Membrane microparticles mediate transfer of P-glycoprotein to drug sensitive cancer cells. Leukemia. 2009;23:1643-1649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 262] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 206. | Yang SJ, Wang DD, Li J, Xu HZ, Shen HY, Chen X, Zhou SY, Zhong SL, Zhao JH, Tang JH. Predictive role of GSTP1-containing exosomes in chemotherapy-resistant breast cancer. Gene. 2017;623:5-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 207. | Wilson TR, Johnston PG, Longley DB. Anti-apoptotic mechanisms of drug resistance in cancer. Curr Cancer Drug Targets. 2009;9:307-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 160] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 208. | Zhang S, Zhang Y, Qu J, Che X, Fan Y, Hou K, Guo T, Deng G, Song N, Li C, Wan X, Qu X, Liu Y. Exosomes promote cetuximab resistance via the PTEN/Akt pathway in colon cancer cells. Braz J Med Biol Res. 2017;51:e6472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 209. | McCubrey JA, Abrams SL, Fitzgerald TL, Cocco L, Martelli AM, Montalto G, Cervello M, Scalisi A, Candido S, Libra M, Steelman LS. Roles of signaling pathways in drug resistance, cancer initiating cells and cancer progression and metastasis. Adv Biol Regul. 2015;57:75-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 210. | Zheng P, Luo Q, Wang W, Li J, Wang T, Wang P, Chen L, Zhang P, Chen H, Liu Y, Dong P, Xie G, Ma Y, Jiang L, Yuan X, Shen L. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis. 2018;9:434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 282] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 211. | Hu YB, Yan C, Mu L, Mi YL, Zhao H, Hu H, Li XL, Tao DD, Wu YQ, Gong JP, Qin JC. Exosomal Wnt-induced dedifferentiation of colorectal cancer cells contributes to chemotherapy resistance. Oncogene. 2019;38:1951-1965. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 157] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 212. | Qin X, Yu S, Zhou L, Shi M, Hu Y, Xu X, Shen B, Liu S, Yan D, Feng J. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int J Nanomedicine. 2017;12:3721-3733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 212] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 213. | Wang J, Zheng Y, Zhao M. Exosome-Based Cancer Therapy: Implication for Targeting Cancer Stem Cells. Front Pharmacol. 2016;7:533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 148] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 214. | Hu Y, Yan C, Mu L, Huang K, Li X, Tao D, Wu Y, Qin J. Fibroblast-Derived Exosomes Contribute to Chemoresistance through Priming Cancer Stem Cells in Colorectal Cancer. PLoS One. 2015;10:e0125625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 280] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 215. | Seo M, Kim SM, Woo EY, Han KC, Park EJ, Ko S, Choi EW, Jang M. Stemness-Attenuating miR-503-3p as a Paracrine Factor to Regulate Growth of Cancer Stem Cells. Stem Cells Int. 2018;2018:4851949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 216. | Fatima F, Nawaz M. Stem cell-derived exosomes: roles in stromal remodeling, tumor progression, and cancer immunotherapy. Chin J Cancer. 2015;34:541-553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 217. | Paolillo M, Schinelli S. Integrins and Exosomes, a Dangerous Liaison in Cancer Progression. Cancers (Basel). 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 218. | Norcic G. Liquid Biopsy in Colorectal Cancer-Current Status and Potential Clinical Applications. Micromachines (Basel). 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 219. | Duffy MJ, van Dalen A, Haglund C, Hansson L, Holinski-Feder E, Klapdor R, Lamerz R, Peltomaki P, Sturgeon C, Topolcan O. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer. 2007;43:1348-1360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 342] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 220. | Bach DH, Hong JY, Park HJ, Lee SK. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer. 2017;141:220-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 213] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 221. | Umwali Y, Yue CB, Gabriel ANA, Zhang Y, Zhang X. Roles of exosomes in diagnosis and treatment of colorectal cancer. World J Clin Cases. 2021;9:4467-4479. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 222. | Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733-1741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 2083] [Article Influence: 138.9] [Reference Citation Analysis (0)] |
| 223. | Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int. 2015;2015:125094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 223] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 224. | Chen B, Xia Z, Deng YN, Yang Y, Zhang P, Zhu H, Xu N, Liang S. Emerging microRNA biomarkers for colorectal cancer diagnosis and prognosis. Open Biol. 2019;9:180212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 225. | Wang J, Huang SK, Zhao M, Yang M, Zhong JL, Gu YY, Peng H, Che YQ, Huang CZ. Identification of a circulating microRNA signature for colorectal cancer detection. PLoS One. 2014;9:e87451. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 136] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 226. | Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 557] [Cited by in RCA: 644] [Article Influence: 58.5] [Reference Citation Analysis (1)] |
| 227. | Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg. 2014;259:735-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 244] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 228. | Min L, Chen L, Liu S, Yu Y, Guo Q, Li P, Zhu S. Loss of Circulating Exosomal miR-92b is a Novel Biomarker of Colorectal Cancer at Early Stage. Int J Med Sci. 2019;16:1231-1237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 229. | Baassiri A, Nassar F, Mukherji D, Shamseddine A, Nasr R, Temraz S. Exosomal Non Coding RNA in LIQUID Biopsies as a Promising Biomarker for Colorectal Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 230. | Fu F, Jiang W, Zhou L, Chen Z. Circulating Exosomal miR-17-5p and miR-92a-3p Predict Pathologic Stage and Grade of Colorectal Cancer. Transl Oncol. 2018;11:221-232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 163] [Article Influence: 23.3] [Reference Citation Analysis (1)] |
| 231. | Liu C, Eng C, Shen J, Lu Y, Takata Y, Mehdizadeh A, Chang GJ, Rodriguez-Bigas MA, Li Y, Chang P, Mao Y, Hassan MM, Wang F, Li D. Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget. 2016;7:76250-76260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 96] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 232. | Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X, Deng Z, Kumar A, Zhang L, Merchant ML, Yan J, Miller DM, Zhang HG. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8:14448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 373] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 233. | Peng ZY, Gu RH, Yan B. Downregulation of exosome-encapsulated miR-548c-5p is associated with poor prognosis in colorectal cancer. J Cell Biochem. 2019;120:1457-1463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 78] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 234. | Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics. 2017;7:789-804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 870] [Cited by in RCA: 1385] [Article Influence: 173.1] [Reference Citation Analysis (1)] |
| 235. | Johnstone RM, Bianchini A, Teng K. Reticulocyte maturation and exosome release: transferrin receptor containing exosomes shows multiple plasma membrane functions. Blood. 1989;74:1844-1851. [PubMed] |
| 236. | Lathe GH, Ruthven CR. The separation of substances on the basis of their molecular weights, using columns of starch and water. Biochem J. 1955;60:xxxiv. [PubMed] |
| 237. | Lv LL, Cao Y, Liu D, Xu M, Liu H, Tang RN, Ma KL, Liu BC. Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int J Biol Sci. 2013;9:1021-1031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 154] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 238. | Thakur A, Xu C, Li WK, Qiu G, He B, Ng SP, Wu CL, Lee Y. In vivo liquid biopsy for glioblastoma malignancy by the AFM and LSPR based sensing of exosomal CD44 and CD133 in a mouse model. Biosens Bioelectron. 2021;191:113476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 239. | Tao L, Chen K, Chen Z, Cong C, Qiu C, Chen J, Wang X, Chen H, Yu T, Xie W, Deng S, Xu JB. 1T' Transition Metal Telluride Atomic Layers for Plasmon-Free SERS at Femtomolar Levels. J Am Chem Soc. 2018;140:8696-8704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 131] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 240. | Fu JH, Zhong Z, Xie D, Guo YJ, Kong DX, Zhao ZX, Li M. SERS-Active MIL-100(Fe) Sensory Array for Ultrasensitive and Multiplex Detection of VOCs. Angew Chem Int Ed Engl. 2020;59:20489-20498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 241. | Kırbaş OK, Bozkurt BT, Asutay AB, Mat B, Ozdemir B, Öztürkoğlu D, Ölmez H, İşlek Z, Şahin F, Taşlı PN. Optimized Isolation of Extracellular Vesicles From Various Organic Sources Using Aqueous Two-Phase System. Sci Rep. 2019;9:19159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 242. | Fais S, O'Driscoll L, Borras FE, Buzas E, Camussi G, Cappello F, Carvalho J, Cordeiro da Silva A, Del Portillo H, El Andaloussi S, Ficko Trček T, Furlan R, Hendrix A, Gursel I, Kralj-Iglic V, Kaeffer B, Kosanovic M, Lekka ME, Lipps G, Logozzi M, Marcilla A, Sammar M, Llorente A, Nazarenko I, Oliveira C, Pocsfalvi G, Rajendran L, Raposo G, Rohde E, Siljander P, van Niel G, Vasconcelos MH, Yáñez-Mó M, Yliperttula ML, Zarovni N, Zavec AB, Giebel B. Evidence-Based Clinical Use of Nanoscale Extracellular Vesicles in Nanomedicine. ACS Nano. 2016;10:3886-3899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 368] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 243. | Karimi N, Cvjetkovic A, Jang SC, Crescitelli R, Hosseinpour Feizi MA, Nieuwland R, Lötvall J, Lässer C. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol Life Sci. 2018;75:2873-2886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 356] [Cited by in RCA: 392] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 244. | Zaharie F, Muresan MS, Petrushev B, Berce C, Gafencu GA, Selicean S, Jurj A, Cojocneanu-Petric R, Lisencu CI, Pop LA, Pileczki V, Eniu D, Muresan MA, Zaharie R, Berindan-Neagoe I, Tomuleasa C, Irimie A. Exosome-Carried microRNA-375 Inhibits Cell Progression and Dissemination via Bcl-2 Blocking in Colon Cancer. J Gastrointestin Liver Dis. 2015;24:435-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 245. | Liu D, Chen C, Cui M, Zhang H. miR-140-3p inhibits colorectal cancer progression and its liver metastasis by targeting BCL9 and BCL2. Cancer Med. 2021;10:3358-3372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 246. | Yan S, Ren X, Yang J, Wang J, Zhang Q, Xu D. Exosomal miR-548c-5p Regulates Colorectal Cancer Cell Growth and Invasion Through HIF1A/CDC42 Axis. Onco Targets Ther. 2020;13:9875-9885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 247. | Hu JL, He GY, Lan XL, Zeng ZC, Guan J, Ding Y, Qian XL, Liao WT, Ding YQ, Liang L. Inhibition of ATG12-mediated autophagy by miR-214 enhances radiosensitivity in colorectal cancer. Oncogenesis. 2018;7:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 248. | Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M. A comprehensive overview of exosomes as drug delivery vehicles - endogenous nanocarriers for targeted cancer therapy. Biochim Biophys Acta. 2014;1846:75-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 386] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 249. | Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6:287-296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 903] [Cited by in RCA: 972] [Article Influence: 108.0] [Reference Citation Analysis (0)] |
| 250. | Gilligan KE, Dwyer RM. Engineering Exosomes for Cancer Therapy. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 211] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 251. | Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33:941-951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3958] [Cited by in RCA: 4582] [Article Influence: 458.2] [Reference Citation Analysis (1)] |
| 252. | Srivastava A, Amreddy N, Razaq M, Towner R, Zhao YD, Ahmed RA, Munshi A, Ramesh R. Exosomes as Theranostics for Lung Cancer. In: Broome AM, editor Cancer Nanotechnology-Book, 2018: 1-33. |
| 253. | Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, Wei J, Nie G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35:2383-2390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 987] [Cited by in RCA: 1346] [Article Influence: 112.2] [Reference Citation Analysis (0)] |
| 254. | Yang T, Martin P, Fogarty B, Brown A, Schurman K, Phipps R, Yin VP, Lockman P, Bai S. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm Res. 2015;32:2003-2014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 539] [Cited by in RCA: 741] [Article Influence: 74.1] [Reference Citation Analysis (0)] |
| 255. | Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498-503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1167] [Cited by in RCA: 1898] [Article Influence: 237.3] [Reference Citation Analysis (1)] |
| 256. | Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, Inskoe E, Piroyan A, Sokolsky M, Okolie O, Hingtgen SD, Kabanov AV, Batrakova EV. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2016;12:655-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 615] [Cited by in RCA: 1079] [Article Influence: 107.9] [Reference Citation Analysis (0)] |
| 257. | Batrakova EV, Kim MS. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J Control Release. 2015;219:396-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 510] [Cited by in RCA: 786] [Article Influence: 78.6] [Reference Citation Analysis (1)] |
| 258. | Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3883] [Cited by in RCA: 3576] [Article Influence: 255.4] [Reference Citation Analysis (0)] |
| 259. | Luan X, Sansanaphongpricha K, Myers I, Chen H, Yuan H, Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol Sin. 2017;38:754-763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 513] [Cited by in RCA: 852] [Article Influence: 106.5] [Reference Citation Analysis (0)] |
| 260. | Lu B, Huang X, Mo J, Zhao W. Drug Delivery Using Nanoparticles for Cancer Stem-Like Cell Targeting. Front Pharmacol. 2016;7:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 261. | Liu Y, Gu Y, Han Y, Zhang Q, Jiang Z, Zhang X, Huang B, Xu X, Zheng J, Cao X. Tumor Exosomal RNAs Promote Lung Pre-metastatic Niche Formation by Activating Alveolar Epithelial TLR3 to Recruit Neutrophils. Cancer Cell. 2016;30:243-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 395] [Cited by in RCA: 512] [Article Influence: 56.9] [Reference Citation Analysis (0)] |
| 262. | Valencia K, Luis-Ravelo D, Bovy N, Antón I, Martínez-Canarias S, Zandueta C, Ormazábal C, Struman I, Tabruyn S, Rebmann V, De Las Rivas J, Guruceaga E, Bandrés E, Lecanda F. miRNA cargo within exosome-like vesicle transfer influences metastatic bone colonization. Mol Oncol. 2014;8:689-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 143] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 263. | Bourderioux M, Nguyen-Khoa T, Chhuon C, Jeanson L, Tondelier D, Walczak M, Ollero M, Bekri S, Knebelmann B, Escudier E, Escudier B, Edelman A, Guerrera IC. A new workflow for proteomic analysis of urinary exosomes and assessment in cystinuria patients. J Proteome Res. 2015;14:567-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 264. | Morse MA, Garst J, Osada T, Khan S, Hobeika A, Clay TM, Valente N, Shreeniwas R, Sutton MA, Delcayre A, Hsu DH, Le Pecq JB, Lyerly HK. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med. 2005;3:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 678] [Cited by in RCA: 882] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 265. | Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, Li G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16:782-790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 529] [Cited by in RCA: 639] [Article Influence: 37.6] [Reference Citation Analysis (0)] |