Published online Sep 15, 2022. doi: 10.4251/wjgo.v14.i9.1856
Peer-review started: January 20, 2022
First decision: April 17, 2022
Revised: April 26, 2022
Accepted: August 7, 2022
Article in press: August 7, 2022
Published online: September 15, 2022
Processing time: 232 Days and 10.9 Hours
Hepatitis B virus (HBV) is a cause of hepatocellular carcinoma (HCC). Interestingly, this process is not necessarily mediated through cirrhosis and may in fact involve oncogenic processes. Prior studies have suggested specific oncogenic gene expression pathways were affected by viral regulatory proteins. Thus, identifying these genes and associated pathways could highlight predictive factors for HCC transformation and has implications in early diagnosis and treatment.
To elucidate HBV oncogenesis in HCC and identify potential therapeutic targets.
We employed our Search, Tag, Analyze, Resource platform to conduct a meta-analysis of public data from National Center for Biotechnology Information’s Gene Expression Omnibus. We performed meta-analysis consisting of 155 tumor samples compared against 185 adjacent non-tumor samples and analyzed results with ingenuity pathway analysis.
Our analysis revealed liver X receptors/retinoid X receptor (RXR) activation and farnesoid X receptor/RXR activation as top canonical pathways amongst others. Top upstream regulators identified included the Ras family gene rab-like protein 6 (RABL6). The role of RABL6 in oncogenesis is beginning to unfold but its specific role in HBV-related HCC remains undefined. Our causal analysis suggests RABL6 mediates pathogenesis of HBV-related HCC through promotion of genes related to cell division, epigenetic regulation, and Akt signaling. We conducted survival analysis that demonstrated increased mortality with higher RABL6 expression. Additionally, homeobox A10 (HOXA10) was a top upstream regulator and was strongly upregu
This meta-analysis describes possible roles of RABL6 and HOXA10 in the pathogenesis of HBV-related HCC. RABL6 and HOXA10 represent potential therapeutic targets and warrant further investigation.
Core Tip: Hepatitis B virus (HBV) is a cause of hepatocellular carcinoma (HCC). Interestingly, this process is not necessarily mediated through cirrhosis and may in fact involve oncogenic processes. Prior studies have suggested specific oncogenic gene expression pathways were affected by viral regulatory proteins. Thus, identifying these genes and associated pathways could highlight predictive factors for HCC transformation, and has implications in early diagnosis and treatment. Our manuscript leverages big data to offer key insights to oncogenesis of HBV infection in HCC. We were able to dissect key genetic drivers to disease and namely demonstrate a newfound role for rab-like protein 6 and homeobox A10.
- Citation: Aljabban J, Rohr M, Syed S, Cohen E, Hashi N, Syed S, Khorfan K, Aljabban H, Borkowski V, Segal M, Mukhtar M, Mohammed M, Boateng E, Nemer M, Panahiazar M, Hadley D, Jalil S, Mumtaz K. Dissecting novel mechanisms of hepatitis B virus related hepatocellular carcinoma using meta-analysis of public data. World J Gastrointest Oncol 2022; 14(9): 1856-1873
- URL: https://www.wjgnet.com/1948-5204/full/v14/i9/1856.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i9.1856
Hepatitis B virus (HBV) is a major cause of liver disease, significantly contributing to global morbidity and mortality. Recent estimates show there are approximately 240-300 million people chronically infected with HBV worldwide[1,2]. Chronic HBV infection leads to cirrhosis in 30% of patients, of which 53% later develop hepatocellular carcinoma (HCC)[3]. The Global Burden of Disease Study estimated that HBV-related cirrhosis and liver cancer annually causes 786000 deaths worldwide[4]. HBV utilizes both direct and indirect means to promote HCC. For example, HBV-induced HCC, without cirrhosis suggests involvement of oncogenic pathways independent of chronic liver inflammation. Some studies have implicated viral regulatory proteins such as HBV X protein affecting gene expression pathways[5]. In addition, mutations resulting in growth advantages may be conferred to infected cells by virtue of host chromosomal HBV DNA integration - a phenomenon known as insertional mutagenesis[5]. In the decades since Garcia et al[6] and Wang et al[7] identified cyclin (CCN) A and retinoic acid receptor-beta genes, respectively, as targets of HBV integration, other points of vulnerability have been identified. Li et al[8] found HBV integration into telomerase reverse transcriptase-promoter genes results in sex hormone-dependent responsiveness, providing a possible explanation for the threefold male-to-female preponderance of HBV-related HCC[6-8].
Despite advancements in nucleoside nucleotide analog (NA)-based treatment, and adequate viral suppression resulting in undetectable HBV DNA, patients are still at risk for developing HCC. This may be due to NAs ability to suppress viremia but not eliminate infection and leading to oncogenesis. Importantly, chronic HBV infection, irrespective of cirrhosis represents a lifetime risk for HCC deve
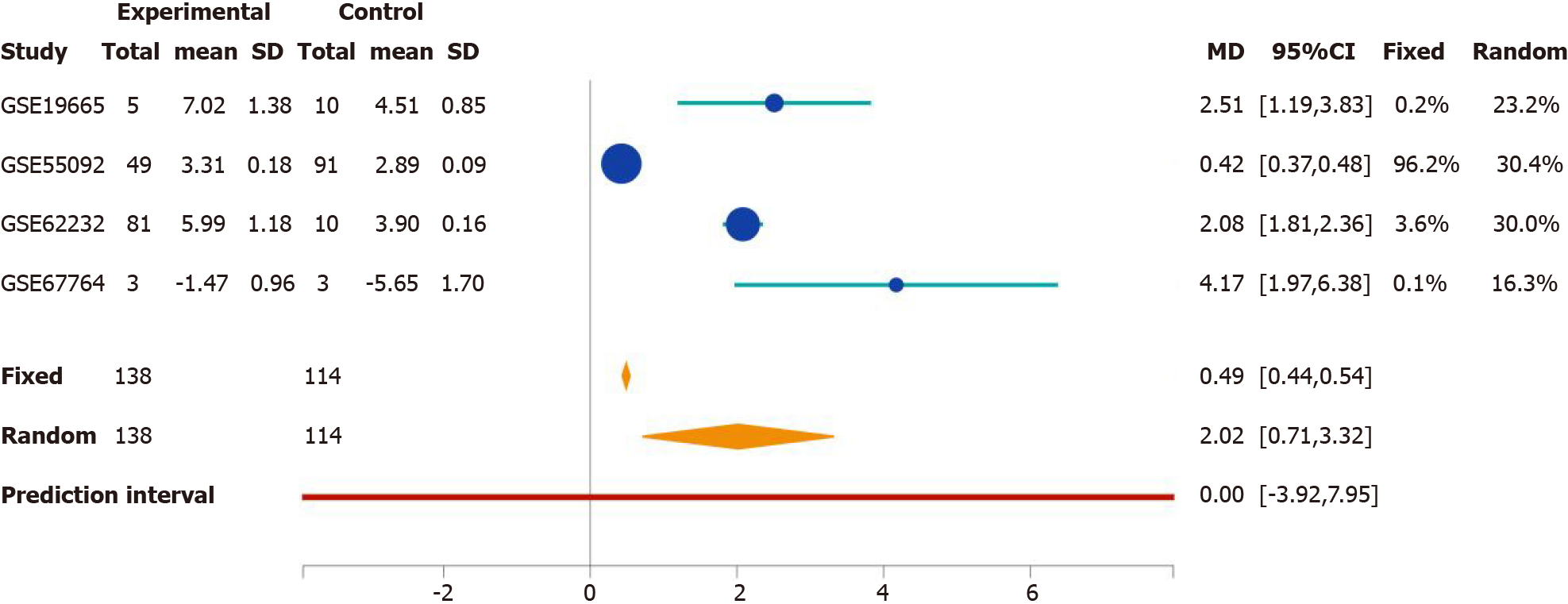
We developed the Search Tag Analyze Resource (STARGEO) platform to utilize the wealth of genomics data featured in the National Center for Biotechnology Information’s Gene Expression Omnibus. STARGEO allows for meta-analysis of transcriptomic signatures between sample sets, such as between disease and normal tissue, through tagging of biological samples from public data. Briefly, through stargeo.org, we searched for studies that studied HBV-related HCC. We then manually curated samples through the “Tagging” interface built into STARGEO based on interactive regular expressions. We gathered liver tumor samples under the “HBV_HCC” tag and control samples from adjacent tumor samples under the “HCC_Control” tag. More information on STARGEO and can be found in our previous paper[10]. To investigate HBV-specific HCC, we tagged 155 tumor and 185 adjacent non-tumor samples as a control. Samples were paired 1:1 within each study. Data was sourced from the GSE19665, GSE44074, GSE55092, GSE62232, and GSE67764 series[10-15]. HCC patients in these studies had confirmed chronic HBV infection with no other co-morbid hepatic infections and had biopsied taken at time of diagnosis. Genetic analysis focused on hepatocytes. Stargeo.org mappings are based on mygene.info gene annotation service to map all probe identifiers to Entrez gene identifiers[16]. The mean difference of contrasts for expressions and the standard deviation of that mean difference were calculate for each gene in every study. Standard meta-analysis with fixed and random effects model were used to combine these estimates across studies to generate both meta P values and effects size across studies. Study weight percentages were calculated using the inverse variance method via the DerSimonian-Laird estimate[17]. We use Python to achieve the analyses explained above in stargeo.org. More information on this particular analysis of HBV-related HCC can be found on http://stargeo.org/analysis/669/. Individual genes can be searched and the number of patient samples in which we observed change in genetic expression is available, along with other information (see Figure 1). Lastly, to best contextualize our results we cited high quality articles in Reference Citation Analysis (https://www.referencecitationanalysis.com).
In order to dissect potential mechanism of disease, potential biomarkers, and therapeutic targets, we extracted more than 21000 genes for our meta-analysis and analyzed the output using the ingenuity pathway analysis (IPA) tool[18]. Analysis was restricted to genes that showed statistical significance (P < 0.05) in both fixed and random effects models with an absolute experimental log ratio greater than 0.7 between experimental (HCC) and control samples. A total of 1035 genes were included in the IPA analysis. Top up- and downregulated genes determined by STARGEO are featured in Table 1. Genes analyzed by IPA are summarized in Supplementary Table 1 with P values and experimental log ratios.
| Top upregulated | Top downregulated | ||
| GPC3 | 2.426 | CLEC4G | -4.212 |
| ANLN | 2.398 | CLEC1B | -4.122 |
| CCNB1 | 2.239 | LINC01092 | -3.834 |
| ASPM | 2.220 | SLCO1B3 | -3.788 |
| CDK1 | 2.215 | CLEC4M | -3.689 |
| NEK2 | 2.121 | HAMP | -3.657 |
| EPS8L3 | 2.116 | STAB2 | -3.604 |
| PBK | 2.017 | OIT3 | -3.575 |
| DTL | 2.010 | MT1M | -3.508 |
| PRC1 | 1.993 | HHIP | -3.417 |
HCC is a complex disease that is a result of several pathological drivers. We used IPA Upstream Regulator analysis to elucidate upstream transcription regulators that best reflect our observed genetic expression dataset[18]. The P values are based on the degree of overlap of known effector targets and our gene list submission. The activation z-score illustrates the upstream regulator activation state, the magnitude of which represents likely activation states of upstream regulators.
The association between rab-like protein 6 (RABL6) expression in HCC patients and survival was found using GPEIA2[19]. GPEIA2 uses samples from The Cancer Genome Atlas (TCGA) HCC cohort and analyzes survival based on the Kaplan-Meier survival method. Expression was split into the top 75% and bottom 25% of RABL6 expression. There were no interactions with human subjects or interventions involved in this study. Additionally, as all content presented is sourced from publicly available data and no patient-protected information was used. Therefore, no institutional review board approval was deemed necessary.
IPA analysis demonstrated liver X receptor (LXR)/retinoid x receptor (RXR) activation, lipopolysaccharide-interleukin-1 (LPS-IL-1) mediated RXR inhibition, acetone degradation, melatonin degradation, and farnesoid X receptor (FXR)/RXR activation as top canonical pathways in HBV-associated HCC (see Supplementary Table 2 for more information on top canonical pathways). FXR (NR1H4) was downregulated in our analysis suggesting its inhibition (Supplementary Table 2).
Top upregulated genes in our analysis are implicated in several signaling processes (Table 1). For example, glypican-3 (GPC3), a cell surface glycoprotein that interacts with Wnt/β-catenin, Yes associated protein, and Hedgehog pathways[20]. Additionally, Akt signaling was activated through the epidermal growth factor receptor kinase substrate 8-like protein 3, a substrate for the epidermal growth factor receptor[21]. Furthermore, we found upregulation of the serine/threonine kinase PDZ-binding kinase (PBK), a mitotic kinase related to mitogen-activated protein kinase kinase (MAPKK)[22]. Other top regulated genes are involved in cell cycle division and control including anilin, CCNB1, assembly factor for microtubules, cyclin dependent kinase 1, and protein regulator of cytokinesis 1. Moreover, we found upregulation of the serine/threonine kinase NIMA related kinase a mediator of centrosome separation in mitosis and meiosis, and deniticleless, an adaptor of the E3-ubiquitin ligase that targets p21 to drive cell division[23].
Top downregulated genes are involved in liver metabolism and other processes. Several members of the C-type lectin family CLEC4G, CLEC1B, and CLEC4M were found among the most repressed genes. C-type lectins participate in adhesion and can act as signaling receptors for inflammation and immune-related processes[24]. Other downregulated genes are indicative of impaired liver function including the liver specific anion transporter solute carrier organic anion transporter family member 1B3, iron regulator hepcidin (HAMP), and the metallothionen (MT1M)[25]. Lastly, we found downregulation of Hh-interacting protein (HHIP), a negative regulator of Hedgehog signaling[26].
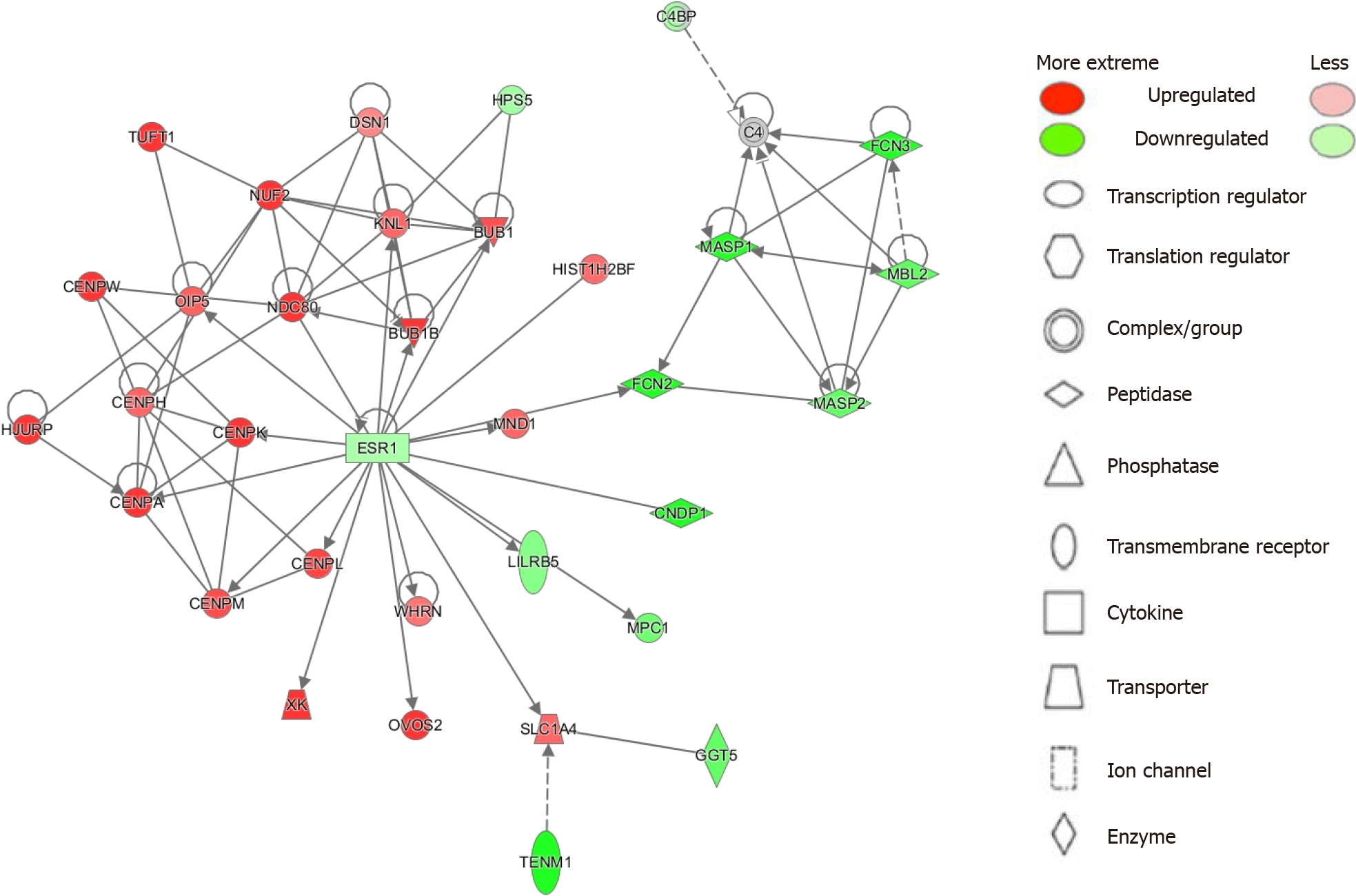
To elucidate top disease functions from our results we employed the IPA Network analysis function[18]. IPA ranks networks from the Global Molecular Network based on the number of focus genes from given networks that match with our analysis. Significance is given by the p-score [p-score = -log10(P value)]. We identified 25 networks with most being involved in cancer, cell cycle, gastrointestinal disease, and other disease functions. The top 6 networks are summarized in Table 2 and the top network is illustrated in Figure 2 (see Supplementary Table 3 for all networks).
| Top disease and function | P-score | Focus genes | Genes in network |
| Cell cycle; cellular assembly and organization; DNA replication, recombination and repair | 46 | 33 | BUB1, BUB1B, C4, C4BP, CENPA, CENPH, CENPK, CENPL, CENPM, CENPW, CNDP1, DSN1, ESR1, FCN2, FCN3, GGT5, HIST1H2BF, HJURP, HPS5, KNL1, LILRB5, MASP1, MASP2, MBL2, MND1, MPC1, NDC80, NUF2, OIP5, OVOS2, SLC1A4, TENM1, TUFT1, WHRN, XK |
| Cancer; cell death and survival; organismal injury and abnormalities | 42 | 31 | ANKS6, Ap1, AURKA, B9D1, BCKDHB, BOLA2/BOLA2B, CA5A, CBX5, CCT3, CDK1, CDKN2A, CEMIP, EPB41L5, estrogenreceptor, ETFRF1, EZH2, H2AFX, HIST1H2AM, HMGA1, KIF11, MCM2, MFAP4, MKI67, NAT2, NGFR, NT5DC2, PRKDC, Rnr, SETDB1, Smad2/3, TCF19, TK1, TMEM131L, TUBE1, ZSWIM5 |
| Cancer; cell cycle; cellular movement | 42 | 31 | ATAD2, ATPase, BMP, BMP5, CCBE1, CEP55, CTH, DTL, ECT2, GORASP2, IGF2BP1, IL12 (family), IL18R1, IL1RAP, IPO9, KIF14, KIF23, LUM, MAP1LC3, MSH2, NAAA, NUP62, OLA1, PBLD, PLSCR4, RAD54B, SIGLEC1, STAU2, TEX37, TRAF5, TRIP13, VSIG2, VWA8, WDYHV1, XPO5 |
| Cell morphology, cellular assembly and organization, DNA replication, recombination, and repair | 39 | 30 | ACAA2, ARMC6, BCHE, C4BPA, CDCA3, CDCA8, CENPE, CENPF, Ciap, CRNDE, ENO3, Enolase, FOXM1, KALRN, KIF20A, KIF2C, KIF4A, LRAT, MAGEA3/MAGEA6, MZT1, NAV1, NEB, PRC1, RAS, RASGRP2, RASSF4, SESTD1, SGO2, SRD5A1, SRD5A2, Steroid 5 alpha-Reductase, TARBP1, TGM3, transglutaminase, TRIO |
| Cancer; organismal injury and abnormalities; reproductive system disease | 39 | 30 | ANGPTL6, BMPER, Cysteine Protease, DPF3, EGLN, FNIP2, GCDH, GDF2, Granzyme B-Perforin-SRGN, GREM2, HMGCL, HOXA13, KIF15, LYVE1, MS4A7, MT1G, NOSTRIN, PCDH9, PDE7B, PLVAP, RNF125, RNF165, RRAGD, SERPINB9, SESN3, SLC7A2, Smad1/5/8, SPARCL1, SRGN, STC1, TPX2, TRIM16, Vegf, VSIG4, ZFP |
| Cancer, gastrointestinal disease, hepatic system disease | 37 | 29 | AKR1D1, ALDH, ALDH1A3, ALDH6A1, ALDH8A1, ANK3, CA2, COBLL1, CYP39A1, ENAH, ESM1, FOS, GBA, GLS2, GPM6A, GPSM2, GRHPR, GUCY1A1, HIST1H3H, histone-lysine N-methyltransferase, HOOK1, MECOM, NCKAP1L, NSD2, PALM2, PXMP2, Rab11, RAB11FIP4, sGC, SLC1A1, SMYD3, Sos, TSKU, UXS1 |
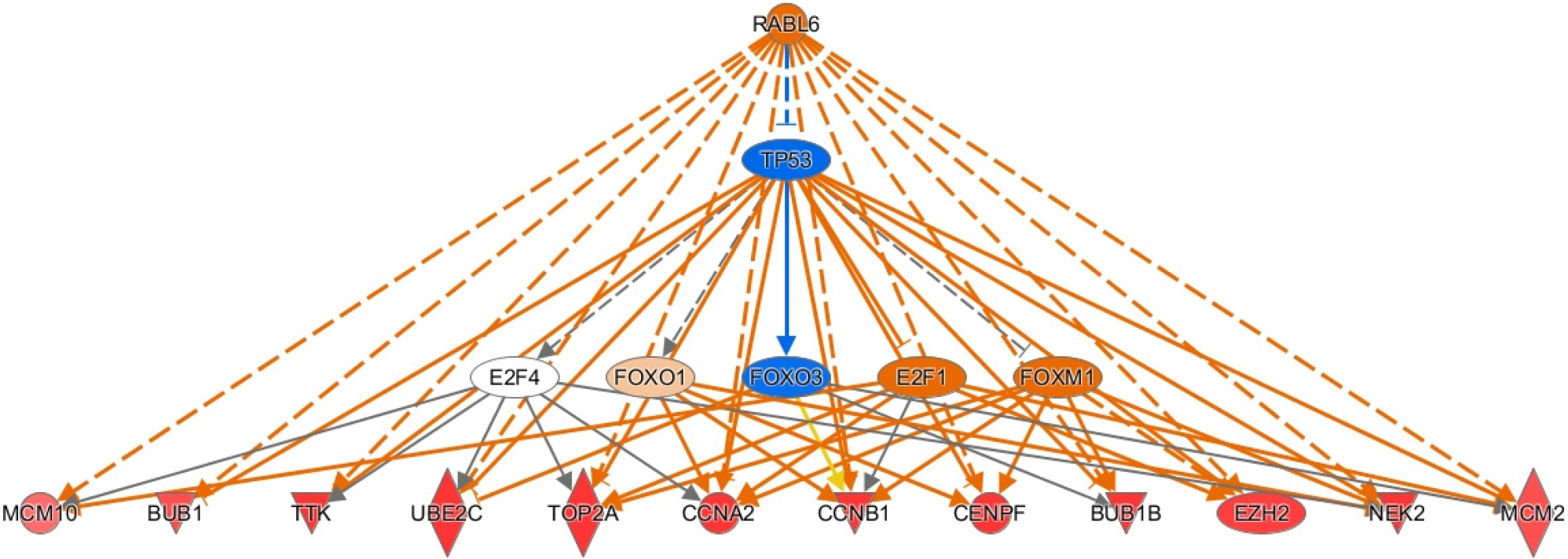
To investigate genes that most greatly influenced our gene dataset and oncogenesis we used IPA to identify top upstream regulators (see methods). We searched for genes that were the identified as the most activated up-regulators in IPA and were upregulated in our dataset (see Supplementary Table 4). The most activated upstream regulator was RABL6, a member of the Ras family of GTPases[27]. We also found activation of transcription factors T-box transcription factor 2 (TBX2) involved in hepatocyte proliferation, migration, and invasion[28]. We also noted activation of E2F transcription factor 1 (E2F1), which has a mixed but predominantly proliferative role in HCC[29]. Additionally, we found activation of the transcription factor forkhead box M1 (FOXM1), which promotes cell turnover through CCNB1 and CCND1 upregulation and through other mechanisms[30,31]. Other activated transcription factors implicated in tumor activity included RUNX transcription factor 1, melanocyte inducing transcription factor, and homeobox A10 (HOXA10)[32-35]. Moreover, other top upstream regulators demonstrated varied biologic activity. For example, E1A binding protein P400 (EP400), a component of the NuA4 histone acetyltransferase complex, is associated with epigenetic activity[36]. Other upstream regulators are proteins involved in signaling pathways including actin like 6a, an actin binding protein involved in Notch1/SOX2 signaling, LIN9-MYBL2 [interacts with the tumor suppressors such as retinoblastoma (Rb) protein], SHC adaptor protein 1 (SHC1, a signaling adaptor for growth factor receptors), protein inhibitor of activated STAT 4 (PIAS4, a protein inhibitor of STAT), and Tumor necrosis factor (TNF) receptor associated factor 2 [TRAF2, role in TNF and nuclear factor-kappaB (NF-κB)][37-42]. We also found activation of RNA binding proteins including ELAVL1, a regulator of ferroptosis in hepatic stellate cells, as well as oncogenic members of the negative elongation factor family NELFE, NELFA, and NELFCD[43]. Lastly, we found activation of oncogenesis-promoting kinases such as mitogen activated kinase 4 and neurotrophic receptor kinase 2 (NTRK2), which aid in cell adhesion[44,45].
We next focused on inhibited upstream regulators (Supplementary Table 5), several of which are implicated in the innate immune response including the innate receptor DExD/H-Box helicase 58 (DDX58), or RIG-I, and the pattern recognition and toll-like receptors (TLR)2, TLR3, TLR4, and TLR9[46,47]. There was predicted inhibition and downregulation of inflammatory mediators NF-κB and interleukins interleukin (IL)-5, IL-12, IL-18, and IL-33. We also found downregulation and predicted inhibition of the immune co-stimulatory molecule CD86, as well as forkhead head transcription factor FOXO3, a mediator of the antioxidant response and autophagy[48]. Several other inhibited upstream regulators are well-described tumor suppressors such as TP53, CDKN1A, Rb gene, and Rb transcriptional suppressor type 2[49]. Additionally, we found predicted inhibition of hepatocyte nuclear factors HNF4 and HNF41A, and the LXR NR1H3, which have been shown to exhibit anti-tumor activity[50-53]. We also found downregulation and predicted inhibition of the transcription factor CCAAT enhancer binding protein delta (CEBPD), a regulator of apoptosis and potential tumor suppressor[54]. Lastly, we observed downregulation and predicted inhibition of the SAM domain, SH3 domain, and nuclear localization signals 1 (SAMSN1), a lung cancer tumor suppressor that is hypermethylated in HCC[55,56].
To elucidate the pathologic potential of the upstream regulators described above, we assessed downstream effector genes through IPA. We focused on upstream regulators with high activation z-scores. We first investigated RABL6 as it is the most activated upstream regulator in our analysis and its target genes play important roles in HBV-HCC (see Supplementary Table 4). Most of the activated genes are involved in promoting cell division. For example, the mitotic spindle checkpoint genes BUB1 and BUB1B, several cyclins including CCNA2, CCNB2, and CCNE2, and the M-phase inducer CDC25C were all upregulated[57]. Likewise, we observed upregulation of centromere protein F, helicase RAD54B, and topoisomerase TOP2A. We also found upregulation of the mitogen PBK, NEK2 (a regulator of mitotic progression), and the kinase TTK, all of which promote HCC cell proliferation and migration via Akt signaling[21,22,58]. Similarly, we found upregulation of minichromosome maintenance family (MCMs) members MCM2 and MCM10, which also promote cell division[59-61]. There are also downstream genes of RABL6 with recently described roles in cancer. For example, we found upregulation of the ubiquitin-conjugating enzyme E2C. Knockdown of this gene has been shown to suppress cellular proliferation, migration, and invasion in HCC[62]. Lastly, RABL6 may mediate HCC progression through downregulation of enhancer of zeste homolog 2, which regulates histone and DNA methylation and silences tumor suppressors[63,64]. Thus, our analysis suggests RABL6 promotes HCC through several pro-oncogenic mechanisms. A summary of the downstream effects of RABL6 are presented in Figure 3.
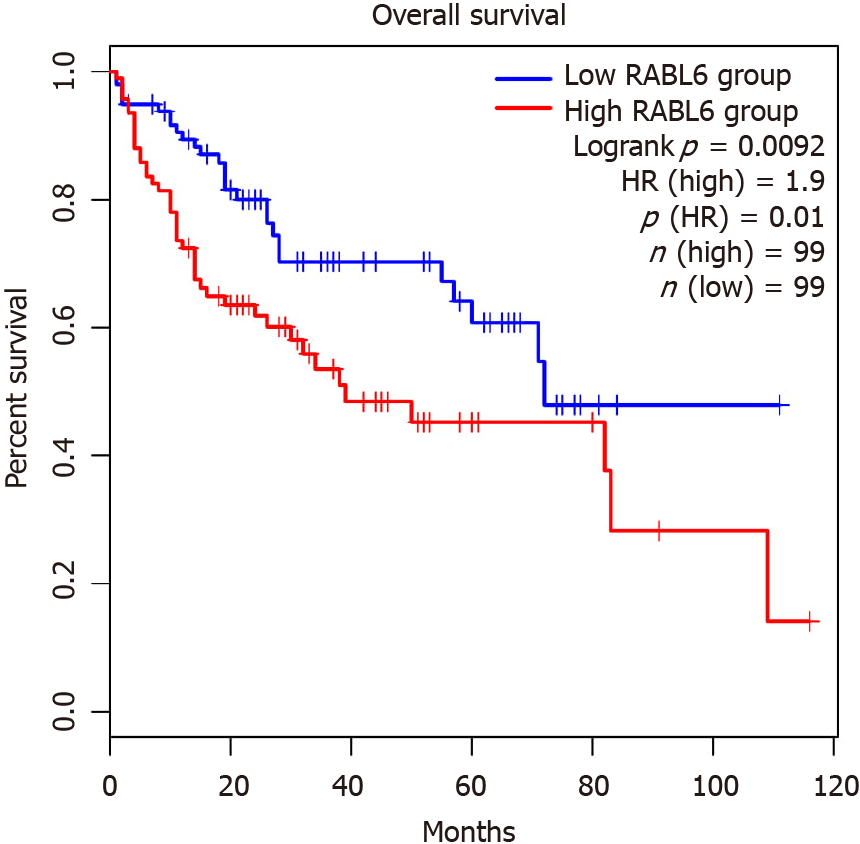
Since RABL6 has not been described in HCC, we conducted survival analysis using the GEP1A2 platform. GEP1A2 is a website that uses patient samples from TCGA, which is used for bioinformatic analysis on genes of interest among differ cancer types[19]. We examined prognosis based on quartile expression of RABL6 (upper 75% vs lower 25%) in HCC patients. We found a statistically lower chance of survival with higher expression of RABL6.
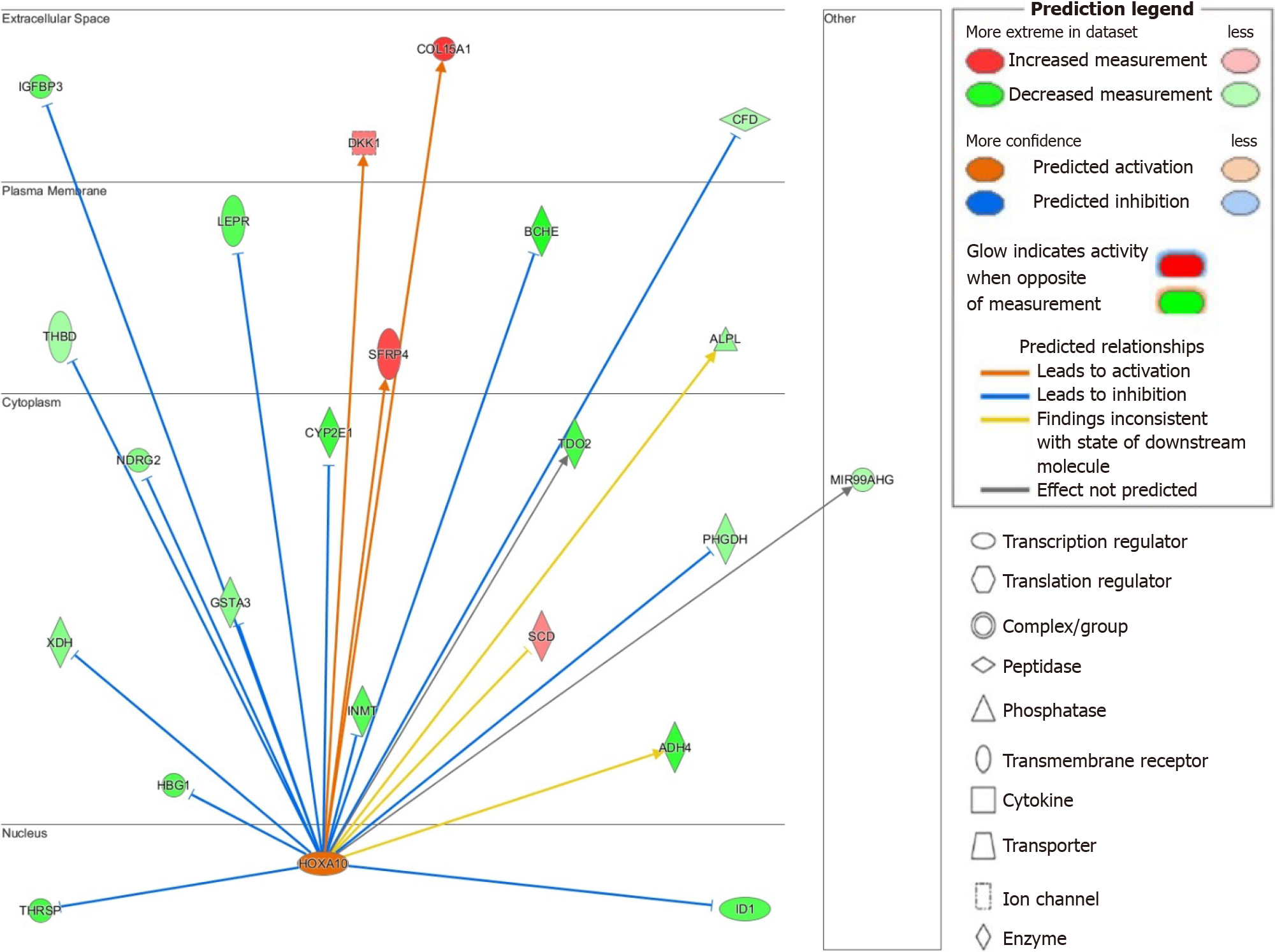
The upstream regulators TBX2, E2F1, FOXM1, and EP400 share similar activated genes to those described for RABL6 (see Supplementary material), such as HOXA10, and thus may act synergistically to promote HBV-HCC. This prompted us to next study activated genes downstream of HOXA10 due to its stark upregulation and high activation z-score (see Supplementary Table 6). While role of HOXA10 is not well-defined in HCC, knockdown model has been recently shown to inhibit HCC cell tumorigenesis[35]. Additionally, another study by Shao et al[65] demonstrated the involvement of HOXA10 in the renewal and survival of liver tumor initiation cells. IPA identified several genes downstream of HOXA10 that may explain its pathogenic activity. For example, HOXA10 may induce tumor progression through downregulation of the tumor suppressor gene NDRG2 as well as glutathione S-transferase A3 or GSTA3, whose inactivity results in hepatocyte oxidative stress and liver injury[66,67]. We also found upregulation of dickkopf-1, a negative regulator of Wnt signaling and negative prognosticator for HCC[68]. Insulin-like growth factor binding protein-3 is a potential mediator of growth suppression signals and is downregulated in our dataset[69]. The hepatic enzyme CYP2E1 was similarly downregulated and is known to be repressed in HCC and linked with a poor prognosis[70]. Lastly, xanthine dehydrogenase, a rate-limiting enzyme in purine metabolism, was downregulated and its suppression has been linked to enhanced cancer stem-cell activity in HCC[71]. A suggested model of the potential multifactorial role HOXA10 and its interplay in HCC is shown in Figure 4.
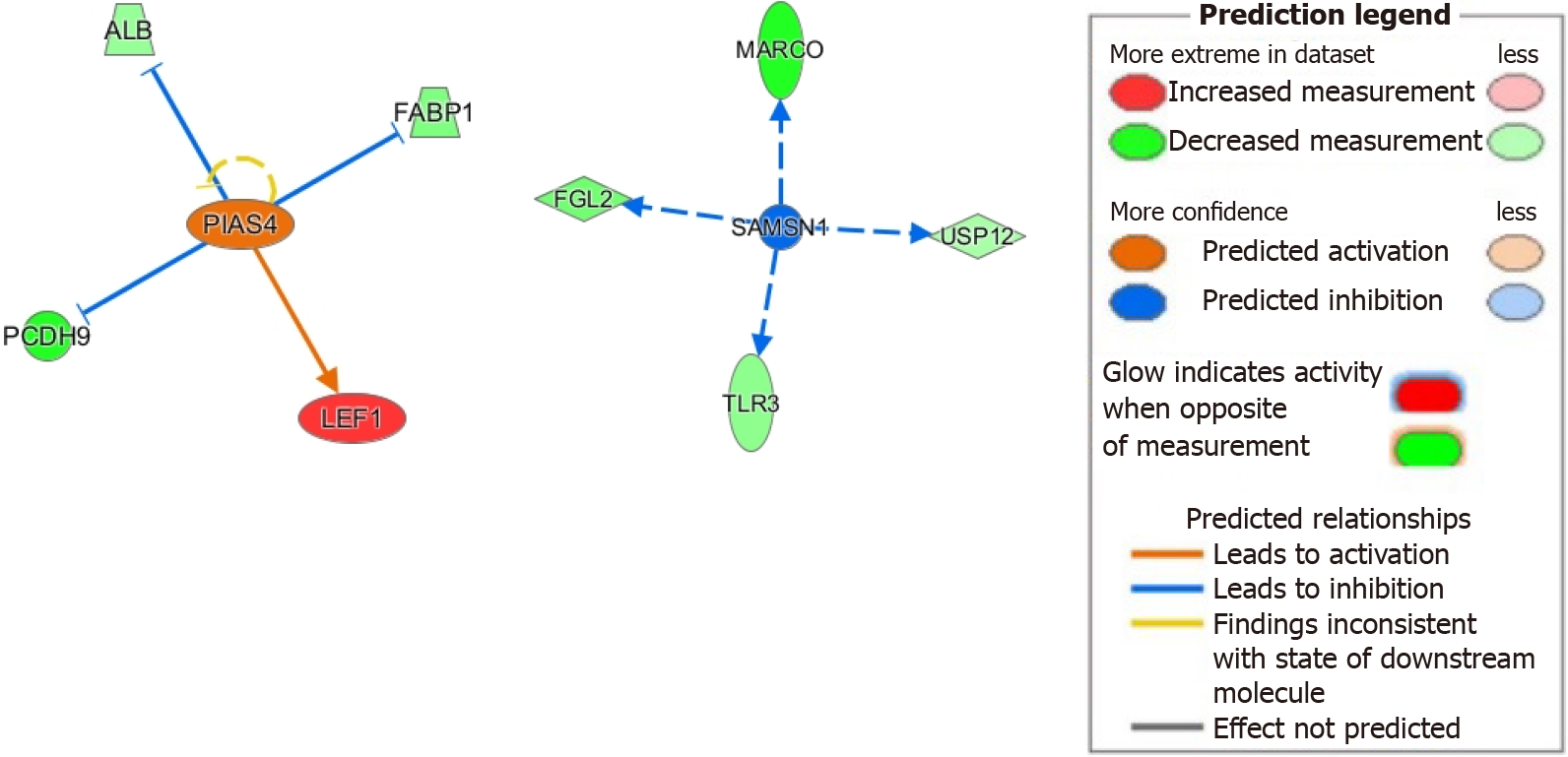
Next, we investigated the downstream signaling of PIAS4 given its significant upregulation and activation z-score (see Supplementary Table 4). PIAS4 involvement in HCC has been recently described[41]. Downstream of PIAS4, we found upregulation of lymphoid enhancer factor 1 and downregulation of protocadherin 9, both of which promote epithelial-mesenchymal transition in HCC[72-74]. We also found downregulation of fatty acid binding protein 1, which has been shown to reduce oxidative stress, a major contributor to HCC development[75]. Its loss may also lead to microsatellite instability in colorectal carcinomas and may have similar effects in HCC[76]. Furthermore, we found downregulation of albumin, with evidence showing albumin itself suppressing HCC cellular proliferation[77]. These results suggest a mechanistic role for PIAS4 in HCC progression (Figure 5).
Lastly, we investigated how inhibition of SAMSN1 may contribute to HCC. As mentioned previously, SAMSN1 inhibition is linked to malignant HCC tumorigenesis[55]. From our IPA analysis, inhibition of SAMSN1 has immunologic implications including downregulation of the pattern recognition receptor TLR3 and macrophage receptor with collagenous structure. These changes are known to negatively impact HCC prognosis[46,78]. Furthermore, we found downregulation of the de-ubiquitinase USP12, which complexes with WD repeat protein WDR48 to suppress Akt signaling and tumor cell survival[79]. Our suggested model of molecular mechanisms linking SAMSN1 inhibition with HCC is showed in Figure 5.
Despite robust vaccination strategies in some countries, hepatitis B infection remains a leading global cause of liver cancer[1,3]. Therapeutic options for HBV-related HCC remain poor owing to an overall lack of understanding of pathways involved in HBV oncogenesis. Current literature suggests HCC development is a result of aberrant activation of cellular signaling processes such as Wnt/FZD/β-catenin, PI3K/Akt/mTOR, IRS1/IGF, and Ras/Raf/MAPK[80]. Even with such knowledge, directed therapy for HBV-related HCC cases requires a more detailed understanding of the interactome. In this meta-analysis, we found potential underlying cellular pathways that define HBV-related HCC and its disease mechanisms. Our results build upon known contributors to HBV-related HCC including LXR/FXR/RXR signaling, Akt signaling, and immunological changes within the tumor microenvironment that are favorable for both HBV infection and tumor progression. We also illustrate the possible activities of upstream regulators, whose role in HBV-related HCC are not well described, such as RABL6, HOXA10, PIAS4, and SAMSN1.
We began our analysis by studying the top canonical pathways identified by IPA, which included LXR/RXR activation, LPS-IL-1 mediated RXR inhibition, acetone degradation, melatonin degradation, and FXR/RXR activation. LXR heterodimerizes with RXR and bind to LXR response elements, directly regulating gene expression[81]. LXR primarily regulates expression of genes essential for lipid metabolism and is a known HCC tumor-suppressor[82]. Similarly, the bile acid regulator FXR (NR1H4) suppresses hepatocarcinogenesis and was starkly downregulated in our analysis[83]. In addition, inhibition of the acetone degradation pathway in HCC suggests alteration hepatic ketone metabolism, suggesting an increase in acetone levels may serve as a disease biomarker[84]. Lastly, we saw predicted inhibition of the melatonin degradation pathway, which would presumably lead to a rise in melatonin. Melatonin has been demonstrated to inhibit HCC progression through let7i-3p-mediated RAF1 suppression[85]. Overall, the top canonical pathways above reinforce the roles of LXR/RXR/FXR signaling in HCC pathogenesis and suggests a role for melatonin degradation in HBV-related HCC.
Top up- and downregulated genes identified in our analysis are implicated in oncogenic cellular signaling and other pathologic processes. For example, GPC3, the most upregulated gene in our analysis, functions as a co-receptor for Wnt proteins[86]. Wnt signaling is vital for hepatobiliary function and cell differentiation. Therefore it’s no surprise that aberrations in activity are major contributors to HCC tumorigenesis and other liver disorders[87]. GPC3 is also implicated in hedgehog signaling[20], another important regulator of cell growth and differentiation; overactivation of which is associated with multiple cancer types including HCC[88]. In vitro studies suggest GPC3 mediates cell proliferation through hedgehog signaling[89]. Additionally, the negative hedgehog regulator HHIP was one of the most downregulated genes in our dataset[26]. This change may act synergistically with GPC3 to further promote hedgehog signaling and cellular proliferation. Lastly, we found the kinase PBK as one of the most upregulated genes. This kinase is related to the MAPKK family and has been recently been shown to promote HCC metastasis through ETV4a-uPar signaling[22,90]. ETV4 is part of the ETS family of transcription factors and directly regulates cell division to promote pancreatic cancer and other cancer types[91,92].
In addition to activation of cellular signaling pathways described above, our results also highlight changes in the tumor microenvironment and the immune response that may confer advantages to HBV infection and tumor progression. Our most downregulated genes included several members of the C-type lectin family including CLEC4G, CLEC1B, and CLEC4M. C-type lectins function in both the innate and adaptive immune response and downregulation of has been demonstrated in HCC[24,93]. Lower expression of CLEC1B is associated with poorer outcomes in HCC[94]. In addition, we found downregulation of pattern recognition receptors, like TLR3 and TLR2, implying impairment of the innate immune response. For example, TLR3, a receptor that recognizes viral components and double-stranded RNA[95], has been associated with control of HBV infection and apoptosis of HCC cells[46]. Likewise, TLR2, whose activity limits HCC cellular proliferation, was downregulated[47]. We also noted down
Our analysis revealed FOXM1, E2F1, and EP400 as activated top upstream regulators in HBV-related HCC, each of which play a prominent role in facilitating cancer proliferation. FOXM1 has previously been shown to promote HCC progression via expression of genes KIF4A and CCNB1[22]. Additionally, FOXM1 promotes tumor cell proliferation via increasing expression of CCNB1 and CCND1, and decreasing expression of cell cycle checkpoint molecules p27 and p21[100]. E2F1 is a transcription factor that has been shown to have both proliferative and apoptotic effects in HCC although the proliferative effects seem to be more prominent[29]. E2F1 activates MYBL2 (another upregulated transcription factor in our analysis) and is involved in cell cycle progression[101]. EP400 is a component of NuA4 histone acetyltransferase complex and is associated with activation of various genes. Recent studies have revealed it to be a critical transcription factor associated with greater HCC relapse and lower overall survival[36].
In addition, our results showed RABL6, ESR1, NR0B2, and CEBPD as top upstream regulators that negatively regulate HCC tumor suppression. RABL6 is a member of Ras GTPase family that is overexpressed in HCC[102]. Survival analysis suggests overexpression leads to a poorer prognosis (Figure 6). ESR1 was downregulated in our analysis, concordant with prior findings implicating ESR1 as a potential HCC tumor suppressor gene[103]. Moreover, our results showed downregulation of NR0B2 and CEBPD. NR0B2 is a nuclear receptor and tumor suppressor; downregulation of which is associated with HCC and renal cell carcinoma[104,105]. Similarly, CEBPD has also been posited as a candidate HCC tumor suppressor gene primarily through modulating IL-1 signaling[106].
Interestingly, our results also revealed that HBV-related HCC progression may be intrinsically linked with repression of inflammatory and innate immune responses. Our analysis showed stark inhibition of the NF-κB pathway, a Myc-dependent driver of HCC tumorigenesis[107]. Indeed, other studies have proposed the NF-κB pathway is an important mediator of hepatic fibrosis and disease progression, especially when inhibition is pronounced[108]. While our meta-analysis allows for robust results of larger datasets, it has certain limitations. Annotations of public samples are limited and can introduce confounding variables to our analysis. For one, samples were taken from patients at different stages of HBV-related HCC. There may be significant differences in genetic aberrations based on tumor stage and grade. While samples were taken at the time of diagnosis, patient characteristics, such as ethnicity, comorbidities, and medications, were not clarified and may affect the results. Additionally, there may be differences in how samples were processed and how omics were performed between the studies included in our analysis. Of note, the studies in our meta-analysis did not clarify if changes in gene expression were attributable to HBV-DNA integration to the hepatocyte genome. Analysis focused on hepatocytes and the genes identified have links to HBV infection epiphonema as detailed above so a portion of the changes we have seen are associated to HBV. As explained, analysis focused on hepatocytes so gene changes would not be related to infiltrates. We did identify several genes involved in cell cycle regulation, but cellularity is important to function of these genes and could not be described in this approach. For future directions, we aim to validate our top genetic candidates using patient samples and control for such factors as stage of disease. We also hope to compare results between different etiologies of HCC such as HCV and alcohol. Doing so will elucidate the similarities and differences between these etiologies, allowing for a greater understanding of the oncogenic process while aiding the development of directed therapeutics for patient-specific treatment.
HBV is a leading cause of HCC and treatment options are still limited. In this meta-analysis based on public data, we studied the pathogenesis of HBV and pave the way for novel therapeutic avenues. We illustrated genetic changes that contribute to pro-oncogenic signaling through such pathways as the Akt, hedgehog, ETV4, and Wnt pathways. We also illustrated changes in the tumor microenvironment and immune response that are contributory to HBV infection and tumor progression. Additionally, we clarify the role of key upstream regulators such as RABL6, HOXA10, PIAS4, and SAMNS1 and describe how their downstream effects contribute to disease. These observations need to be further confirmed in prospective studies on oncogenesis. There is also need for investigating HBV-related cirrhosis and progressive changes of HBV-related HCC to assess the stepwise activity that define HBV oncogenesis.
Hepatitis B virus (HBV) is a major cause of hepatocellular carcinoma (HCC) through several mecha
Studying HBV related HCC will offer novel insights to viral hepatic oncogenesis. It will also potentially lead to more directed therapy in HBV-related HCC.
Identify genetic changes and pathways that define HBV-related HCC. Identify novel therapeutic targets.
Used our novel Search Tag Analyze Resource platform to mine liver biopsies from HBV-related HCC patients and used ingenuity pathway analysis to study the results of our meta-analysis.
Our meta-analysis highlighted several genes and pathways with oncogenic potential. Of note, we describe two potential novel mediators of oncogenesis in rab-like protein 6 (RABL6) and homeobox A10 (HOXA10).
This meta-analysis describes possible roles of RABL6 and HOXA10 in the pathogenesis of HBV-related HCC. RABL6 and HOXA10 represent potential therapeutic targets and warrant further investigation.
The next steps to our research is to validate RABL6 and HOXA10 relevance in HBV-related HCC using clinical samples and establish its mechanistic underpinnings in an animal model.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: American Association for the Study of Liver Diseases; American Gastroenterological Association; American College of Gastroenterology.
Specialty type: Oncology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bredt LC, Brazil; Kao JT, Taiwan S-Editor: Wang JJ L-Editor: A P-Editor: Yuan YY
| 1. | Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11:97-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1734] [Cited by in RCA: 1750] [Article Influence: 83.3] [Reference Citation Analysis (0)] |
| 2. | Alberts CJ, Clifford GM, Georges D, Negro F, Lesi OA, Hutin YJ, de Martel C. Worldwide prevalence of hepatitis B virus and hepatitis C virus among patients with cirrhosis at country, region, and global levels: a systematic review. Lancet Gastroenterol Hepatol. 2022;7:724-735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 115] [Article Influence: 38.3] [Reference Citation Analysis (1)] |
| 3. | Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1764] [Cited by in RCA: 1843] [Article Influence: 97.0] [Reference Citation Analysis (0)] |
| 4. | Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo JP, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KM, Nasseri K, Norman P, O'Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA 3rd, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De León FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh PH, Yip P, Zabetian A, Zheng ZJ, Lopez AD, Murray CJ, AlMazroa MA, Memish ZA. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095-2128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9500] [Cited by in RCA: 9579] [Article Influence: 736.8] [Reference Citation Analysis (0)] |
| 5. | Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64:S84-S101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 710] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 6. | Garcia M, de Thé H, Tiollais P, Samarut J, Dejean A. A hepatitis B virus pre-S-retinoic acid receptor beta chimera transforms erythrocytic progenitor cells in vitro. Proc Natl Acad Sci U S A. 1993;90:89-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 25] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 7. | Wang J, Chenivesse X, Henglein B, Bréchot C. Hepatitis B virus integration in a cyclin A gene in a hepatocellular carcinoma. Nature. 1990;343:555-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 489] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 8. | Li CL, Li CY, Lin YY, Ho MC, Chen DS, Chen PJ, Yeh SH. Androgen Receptor Enhances Hepatic Telomerase Reverse Transcriptase Gene Transcription After Hepatitis B Virus Integration or Point Mutation in Promoter Region. Hepatology. 2019;69:498-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 9. | Yoo J, Hann HW, Coben R, Conn M, DiMarino AJ. Update Treatment for HBV Infection and Persistent Risk for Hepatocellular Carcinoma: Prospect for an HBV Cure. Diseases. 2018;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Hadley D, Pan J, El-Sayed O, Aljabban J, Aljabban I, Azad TD, Hadied MO, Raza S, Rayikanti BA, Chen B, Paik H, Aran D, Spatz J, Himmelstein D, Panahiazar M, Bhattacharya S, Sirota M, Musen MA, Butte AJ. Precision annotation of digital samples in NCBI's gene expression omnibus. Sci Data. 2017;4:170125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 11. | Deng YB, Nagae G, Midorikawa Y, Yagi K, Tsutsumi S, Yamamoto S, Hasegawa K, Kokudo N, Aburatani H, Kaneda A. Identification of genes preferentially methylated in hepatitis C virus-related hepatocellular carcinoma. Cancer Sci. 2010;101:1501-1510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 82] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 12. | Ueda T, Honda M, Horimoto K, Aburatani S, Saito S, Yamashita T, Sakai Y, Nakamura M, Takatori H, Sunagozaka H, Kaneko S. Gene expression profiling of hepatitis B- and hepatitis C-related hepatocellular carcinoma using graphical Gaussian modeling. Genomics. 2013;101:238-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 13. | Melis M, Diaz G, Kleiner DE, Zamboni F, Kabat J, Lai J, Mogavero G, Tice A, Engle RE, Becker S, Brown CR, Hanson JC, Rodriguez-Canales J, Emmert-Buck M, Govindarajan S, Kew M, Farci P. Viral expression and molecular profiling in liver tissue versus microdissected hepatocytes in hepatitis B virus-associated hepatocellular carcinoma. J Transl Med. 2014;12:230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 14. | Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud AL, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc JF, Mazzaferro V, Calvo F, Villanueva A, Nault JC, Bioulac-Sage P, Stratton MR, Llovet JM, Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505-511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1006] [Cited by in RCA: 1341] [Article Influence: 134.1] [Reference Citation Analysis (0)] |
| 15. | Lin X, Yang Y, Guo Y, Liu H, Jiang J, Zheng F, Wu B. PTTG1 is involved in TNF-α-related hepatocellular carcinoma via the induction of c-myc. Cancer Med. 2019;8:5702-5715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 16. | Wu C, Macleod I, Su AI. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41:D561-D565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 237] [Cited by in RCA: 265] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 17. | DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26739] [Cited by in RCA: 30408] [Article Influence: 779.7] [Reference Citation Analysis (0)] |
| 18. | Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523-530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2902] [Cited by in RCA: 4208] [Article Influence: 350.7] [Reference Citation Analysis (0)] |
| 19. | Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1991] [Cited by in RCA: 3071] [Article Influence: 511.8] [Reference Citation Analysis (0)] |
| 20. | Kolluri A, Ho M. The Role of Glypican-3 in Regulating Wnt, YAP, and Hedgehog in Liver Cancer. Front Oncol. 2019;9:708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 21. | Li P, Hu T, Wang H, Tang Y, Ma Y, Wang X, Xu Y, Chen G. Upregulation of EPS8L3 is associated with tumorigenesis and poor prognosis in patients with liver cancer. Mol Med Rep. 2019;20:2493-2499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 22. | Yang QX, Zhong S, He L, Jia XJ, Tang H, Cheng ST, Ren JH, Yu HB, Zhou L, Zhou HZ, Ren F, Hu ZW, Gong R, Huang AL, Chen J. PBK overexpression promotes metastasis of hepatocellular carcinoma via activating ETV4-uPAR signaling pathway. Cancer Lett. 2019;452:90-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 23. | Zhang Y, Wang W, Wang Y, Huang X, Zhang Z, Chen B, Xie W, Li S, Shen S, Peng B. NEK2 promotes hepatocellular carcinoma migration and invasion through modulation of the epithelial-mesenchymal transition. Oncol Rep. 2018;39:1023-1033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 24. | Brown GD, Willment JA, Whitehead L. C-type lectins in immunity and homeostasis. Nat Rev Immunol. 2018;18:374-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 473] [Article Influence: 78.8] [Reference Citation Analysis (0)] |
| 25. | Kanda M, Nomoto S, Okamura Y, Nishikawa Y, Sugimoto H, Kanazumi N, Takeda S, Nakao A. Detection of metallothionein 1G as a methylated tumor suppressor gene in human hepatocellular carcinoma using a novel method of double combination array analysis. Int J Oncol. 2009;35:477-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Tada M, Kanai F, Tanaka Y, Tateishi K, Ohta M, Asaoka Y, Seto M, Muroyama R, Fukai K, Imazeki F, Kawabe T, Yokosuka O, Omata M. Down-regulation of hedgehog-interacting protein through genetic and epigenetic alterations in human hepatocellular carcinoma. Clin Cancer Res. 2008;14:3768-3776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 77] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 27. | Montalbano J, Lui K, Sheikh MS, Huang Y. Identification and characterization of RBEL1 subfamily of GTPases in the Ras superfamily involved in cell growth regulation. J Biol Chem. 2009;284:18129-18142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 28. | Liu X, Miao Z, Wang Z, Zhao T, Xu Y, Song Y, Huang J, Zhang J, Xu H, Wu J. TBX2 overexpression promotes proliferation and invasion through epithelial-mesenchymal transition and ERK signaling pathway. Exp Ther Med. 2019;17:723-729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | Farra R, Grassi G, Tonon F, Abrami M, Grassi M, Pozzato G, Fiotti N, Forte G, Dapas B. The Role of the Transcription Factor E2F1 in Hepatocellular Carcinoma. Curr Drug Deliv. 2017;14:272-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 30. | Chen X, Müller GA, Quaas M, Fischer M, Han N, Stutchbury B, Sharrocks AD, Engeland K. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol Cell Biol. 2013;33:227-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 189] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 31. | Hu G, Yan Z, Zhang C, Cheng M, Yan Y, Wang Y, Deng L, Lu Q, Luo S. FOXM1 promotes hepatocellular carcinoma progression by regulating KIF4A expression. J Exp Clin Cancer Res. 2019;38:188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 122] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 32. | Li Q, Lai Q, He C, Fang Y, Yan Q, Zhang Y, Wang X, Gu C, Wang Y, Ye L, Han L, Lin X, Chen J, Cai J, Li A, Liu S. RUNX1 promotes tumour metastasis by activating the Wnt/β-catenin signalling pathway and EMT in colorectal cancer. J Exp Clin Cancer Res. 2019;38:334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 141] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 33. | Nooron N, Ohba K, Takeda K, Shibahara S, Chiabchalard A. Dysregulated Expression of MITF in Subsets of Hepatocellular Carcinoma and Cholangiocarcinoma. Tohoku J Exp Med. 2017;242:291-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 34. | Li J, Ye M, Zhou C. Expression Profile and Prognostic Values of HOXA Family Members in Laryngeal Squamous Cell Cancer. Front Oncol. 2020;10:368. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 35. | Zhang Y, Chen J, Wu SS, Lv MJ, Yu YS, Tang ZH, Chen XH, Zang GQ. HOXA10 knockdown inhibits proliferation, induces cell cycle arrest and apoptosis in hepatocellular carcinoma cells through HDAC1. Cancer Manag Res. 2019;11:7065-7076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 36. | Hong W, Hu Y, Fan Z, Gao R, Yang R, Bi J, Hou J. In silico identification of EP400 and TIA1 as critical transcription factors involved in human hepatocellular carcinoma relapse. Oncol Lett. 2020;19:952-964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Xiao S, Chang RM, Yang MY, Lei X, Liu X, Gao WB, Xiao JL, Yang LY. Actin-like 6A predicts poor prognosis of hepatocellular carcinoma and promotes metastasis and epithelial-mesenchymal transition. Hepatology. 2016;63:1256-1271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 112] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 38. | Zeng Z, Yang H, Xiao S. ACTL6A expression promotes invasion, metastasis and epithelial mesenchymal transition of colon cancer. BMC Cancer. 2018;18:1020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 39. | Calvisi DF, Simile MM, Ladu S, Frau M, Evert M, Tomasi ML, Demartis MI, Daino L, Seddaiu MA, Brozzetti S, Feo F, Pascale RM. Activation of v-Myb avian myeloblastosis viral oncogene homolog-like2 (MYBL2)-LIN9 complex contributes to human hepatocarcinogenesis and identifies a subset of hepatocellular carcinoma with mutant p53. Hepatology. 2011;53:1226-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 40. | Huang P, Feng X, Zhao Z, Yang B, Fang T, Guo M, Xia J. p66Shc promotes HCC progression in the tumor microenvironment via STAT3 signaling. Exp Cell Res. 2019;383:111550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 41. | Liu Q, Zhou B, Liao R, Zhou X, Yan X. PIAS4, upregulated in hepatocellular carcinoma, promotes tumorigenicity and metastasis. J Cell Biochem. 2020;121:3372-3381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 42. | Schneider AT, Gautheron J, Feoktistova M, Roderburg C, Loosen SH, Roy S, Benz F, Schemmer P, Büchler MW, Nachbur U, Neumann UP, Tolba R, Luedde M, Zucman-Rossi J, Panayotova-Dimitrova D, Leverkus M, Preisinger C, Tacke F, Trautwein C, Longerich T, Vucur M, Luedde T. RIPK1 Suppresses a TRAF2-Dependent Pathway to Liver Cancer. Cancer Cell. 2017;31:94-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 126] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 43. | Dang H, Takai A, Forgues M, Pomyen Y, Mou H, Xue W, Ray D, Ha KCH, Morris QD, Hughes TR, Wang XW. Oncogenic Activation of the RNA Binding Protein NELFE and MYC Signaling in Hepatocellular Carcinoma. Cancer Cell. 2017;32:101-114.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 118] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 44. | Feng XJ, Pan Q, Wang SM, Pan YC, Wang Q, Zhang HH, Zhu MH, Zhang SH. MAP4K4 promotes epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma. Tumour Biol. 2016;37:11457-11467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 45. | Lee SJ, Kim NKD, Lee SH, Kim ST, Park SH, Park JO, Park YS, Lim HY, Kang WK, Park WY, Bang HJ, Kim KM, Park K, Lee J. NTRK gene amplification in patients with metastatic cancer. Precis Futur Med. 2017;1. [RCA] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 46. | Chen XL, Xu YY, Chen L, Wang GL, Shen Y. TLR3 Plays Significant Roles against HBV-Associated HCC. Gastroenterol Res Pract. 2015;2015:572171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 47. | Chen Y, Huang Z, Chen X, Ye H. Activation of the Toll-like receptor 2 signaling pathway inhibits the proliferation of HCC cells in vitro. Oncol Rep. 2019;42:2267-2278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 48. | Tikhanovich I, Cox J, Weinman SA. Forkhead box class O transcription factors in liver function and disease. J Gastroenterol Hepatol. 2013;28 Suppl 1:125-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 141] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 49. | Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 720] [Cited by in RCA: 786] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 50. | Ning BF, Ding J, Yin C, Zhong W, Wu K, Zeng X, Yang W, Chen YX, Zhang JP, Zhang X, Wang HY, Xie WF. Hepatocyte nuclear factor 4 alpha suppresses the development of hepatocellular carcinoma. Cancer Res. 2010;70:7640-7651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 197] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 51. | Takashima Y, Horisawa K, Udono M, Ohkawa Y, Suzuki A. Prolonged inhibition of hepatocellular carcinoma cell proliferation by combinatorial expression of defined transcription factors. Cancer Sci. 2018;109:3543-3553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 52. | Long H, Guo X, Qiao S, Huang Q. Tumor LXR Expression is a Prognostic Marker for Patients with Hepatocellular Carcinoma. Pathol Oncol Res. 2018;24:339-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 53. | Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1110] [Cited by in RCA: 1138] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 54. | Pan YC, Li CF, Ko CY, Pan MH, Chen PJ, Tseng JT, Wu WC, Chang WC, Huang AM, Sterneck E, Wang JM. CEBPD reverses RB/E2F1-mediated gene repression and participates in HMDB-induced apoptosis of cancer cells. Clin Cancer Res. 2010;16:5770-5780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 55. | Sueoka S, Kanda M, Sugimoto H, Shimizu D, Nomoto S, Oya H, Takami H, Ezaka K, Hashimoto R, Tanaka Y, Okamura Y, Yamada S, Fujii T, Nakayama G, Koike M, Fujiwara M, Kodera Y. Suppression of SAMSN1 Expression is Associated with the Malignant Phenotype of Hepatocellular Carcinoma. Ann Surg Oncol. 2015;22 Suppl 3:S1453-S1460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 56. | Yamada H, Yanagisawa K, Tokumaru S, Taguchi A, Nimura Y, Osada H, Nagino M, Takahashi T. Detailed characterization of a homozygously deleted region corresponding to a candidate tumor suppressor locus at 21q11-21 in human lung cancer. Genes Chromosomes Cancer. 2008;47:810-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 73] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 57. | Yang WX, Pan YY, You CG. CDK1, CCNB1, CDC20, BUB1, MAD2L1, MCM3, BUB1B, MCM2, and RFC4 May Be Potential Therapeutic Targets for Hepatocellular Carcinoma Using Integrated Bioinformatic Analysis. Biomed Res Int. 2019;2019:1245072. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 58. | Liu X, Liao W, Yuan Q, Ou Y, Huang J. TTK activates Akt and promotes proliferation and migration of hepatocellular carcinoma cells. Oncotarget. 2015;6:34309-34320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 76] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 59. | Liao X, Liu X, Yang C, Wang X, Yu T, Han C, Huang K, Zhu G, Su H, Qin W, Huang R, Yu L, Deng J, Zeng X, Ye X, Peng T. Distinct Diagnostic and Prognostic Values of Minichromosome Maintenance Gene Expression in Patients with Hepatocellular Carcinoma. J Cancer. 2018;9:2357-2373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 60. | Liu Z, Li J, Chen J, Shan Q, Dai H, Xie H, Zhou L, Xu X, Zheng S. MCM family in HCC: MCM6 indicates adverse tumor features and poor outcomes and promotes S/G2 cell cycle progression. BMC Cancer. 2018;18:200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 61. | Karavias D, Maroulis I, Papadaki H, Gogos C, Kakkos S, Karavias D, Bravou V. Overexpression of CDT1 Is a Predictor of Poor Survival in Patients with Hepatocellular Carcinoma. J Gastrointest Surg. 2016;20:568-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 62. | Xiong Y, Lu J, Fang Q, Lu Y, Xie C, Wu H, Yin Z. UBE2C functions as a potential oncogene by enhancing cell proliferation, migration, invasion, and drug resistance in hepatocellular carcinoma cells. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 63. | Au SL, Wong CC, Lee JM, Fan DN, Tsang FH, Ng IO, Wong CM. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. 2012;56:622-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 240] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 64. | Chen S, Pu J, Bai J, Yin Y, Wu K, Wang J, Shuai X, Gao J, Tao K, Wang G, Li H. EZH2 promotes hepatocellular carcinoma progression through modulating miR-22/galectin-9 axis. J Exp Clin Cancer Res. 2018;37:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 65. | Shao M, Yang Q, Zhu W, Jin H, Wang J, Song J, Kong Y, Lv X. LncHOXA10 drives liver TICs self-renewal and tumorigenesis via HOXA10 transcription activation. Mol Cancer. 2018;17:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 66. | Zheng J, Li Y, Yang J, Liu Q, Shi M, Zhang R, Shi H, Ren Q, Ma J, Guo H, Tao Y, Xue Y, Jiang N, Yao L, Liu W. NDRG2 inhibits hepatocellular carcinoma adhesion, migration and invasion by regulating CD24 expression. BMC Cancer. 2011;11 251:1-251:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 67. | Crawford DR, Ilic Z, Guest I, Milne GL, Hayes JD, Sell S. Characterization of liver injury, oval cell proliferation and cholangiocarcinogenesis in glutathione S-transferase A3 knockout mice. Carcinogenesis. 2017;38:717-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 68. | Yu B, Yang X, Xu Y, Yao G, Shu H, Lin B, Hood L, Wang H, Yang S, Gu J, Fan J, Qin W. Elevated expression of DKK1 is associated with cytoplasmic/nuclear beta-catenin accumulation and poor prognosis in hepatocellular carcinomas. J Hepatol. 2009;50:948-957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 119] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 69. | Hanafusa T, Yumoto Y, Nouso K, Nakatsukasa H, Onishi T, Fujikawa T, Taniyama M, Nakamura S, Uemura M, Takuma Y, Yumoto E, Higashi T, Tsuji T. Reduced expression of insulin-like growth factor binding protein-3 and its promoter hypermethylation in human hepatocellular carcinoma. Cancer Lett. 2002;176:149-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 92] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 70. | Ho JC, Cheung ST, Leung KL, Ng IO, Fan ST. Decreased expression of cytochrome P450 2E1 is associated with poor prognosis of hepatocellular carcinoma. Int J Cancer. 2004;111:494-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 71. | Chen GL, Ye T, Chen HL, Zhao ZY, Tang WQ, Wang LS, Xia JL. Xanthine dehydrogenase downregulation promotes TGFβ signaling and cancer stem cell-related gene expression in hepatocellular carcinoma. Oncogenesis. 2017;6:e382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 72. | Zhu P, Lv J, Yang Z, Guo L, Zhang L, Li M, Han W, Chen X, Zhuang H, Lu F. Protocadherin 9 inhibits epithelial-mesenchymal transition and cell migration through activating GSK-3β in hepatocellular carcinoma. Biochem Biophys Res Commun. 2014;452:567-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 73. | Lv J, Zhu P, Zhang X, Zhang L, Chen X, Lu F, Yu Z, Liu S. PCDH9 acts as a tumor suppressor inducing tumor cell arrest at G0/G1 phase and is frequently methylated in hepatocellular carcinoma. Mol Med Rep. 2017;16:4475-4482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 74. | Chen CL, Tsai YS, Huang YH, Liang YJ, Sun YY, Su CW, Chau GY, Yeh YC, Chang YS, Hu JT, Wu JC. Lymphoid Enhancer Factor 1 Contributes to Hepatocellular Carcinoma Progression Through Transcriptional Regulation of Epithelial-Mesenchymal Transition Regulators and Stemness Genes. Hepatol Commun. 2018;2:1392-1407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 75. | Wang G, Bonkovsky HL, de Lemos A, Burczynski FJ. Recent insights into the biological functions of liver fatty acid binding protein 1. J Lipid Res. 2015;56:2238-2247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 171] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 76. | Wood SM, Gill AJ, Brodsky AS, Lu S, Friedman K, Karashchuk G, Lombardo K, Yang D, Resnick MB. Fatty acid-binding protein 1 is preferentially lost in microsatellite instable colorectal carcinomas and is immune modulated via the interferon γ pathway. Mod Pathol. 2017;30:123-133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 77. | Nojiri S, Joh T. Albumin suppresses human hepatocellular carcinoma proliferation and the cell cycle. Int J Mol Sci. 2014;15:5163-5174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 82] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 78. | Sun H, Song J, Weng C, Xu J, Huang M, Huang Q, Sun R, Xiao W, Sun C. Association of decreased expression of the macrophage scavenger receptor MARCO with tumor progression and poor prognosis in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2017;32:1107-1114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 79. | Gangula NR, Maddika S. WD repeat protein WDR48 in complex with deubiquitinase USP12 suppresses Akt-dependent cell survival signaling by stabilizing PH domain leucine-rich repeat protein phosphatase 1 (PHLPP1). J Biol Chem. 2013;288:34545-34554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 80. | Torresi J, Tran BM, Christiansen D, Earnest-Silveira L, Schwab RHM, Vincan E. HBV-related hepatocarcinogenesis: the role of signalling pathways and innovative ex vivo research models. BMC Cancer. 2019;19:707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 81. | Repa JJ, Mangelsdorf DJ. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu Rev Cell Dev Biol. 2000;16:459-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 549] [Cited by in RCA: 578] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 82. | Ducheix S, Montagner A, Theodorou V, Ferrier L, Guillou H. The liver X receptor: a master regulator of the gut-liver axis and a target for non alcoholic fatty liver disease. Biochem Pharmacol. 2013;86:96-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 83. | Yang F, Huang X, Yi T, Yen Y, Moore DD, Huang W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67:863-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 391] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 84. | Liu Y, Hong Z, Tan G, Dong X, Yang G, Zhao L, Chen X, Zhu Z, Lou Z, Qian B, Zhang G, Chai Y. NMR and LC/MS-based global metabolomics to identify serum biomarkers differentiating hepatocellular carcinoma from liver cirrhosis. Int J Cancer. 2014;135:658-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 105] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 85. | Wang TH, Hsueh C, Chen CC, Li WS, Yeh CT, Lian JH, Chang JL, Chen CY. Melatonin Inhibits the Progression of Hepatocellular Carcinoma through MicroRNA Let7i-3p Mediated RAF1 Reduction. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 86. | Capurro MI, Xiang YY, Lobe C, Filmus J. Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 2005;65:6245-6254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 386] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 87. | Bengochea A, de Souza MM, Lefrançois L, Le Roux E, Galy O, Chemin I, Kim M, Wands JR, Trepo C, Hainaut P, Scoazec JY, Vitvitski L, Merle P. Common dysregulation of Wnt/Frizzled receptor elements in human hepatocellular carcinoma. Br J Cancer. 2008;99:143-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 175] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 88. | Matsui WH. Cancer stem cell signaling pathways. Medicine (Baltimore). 2016;95:S8-S19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 172] [Cited by in RCA: 217] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 89. | Wang S, Chen N, Chen Y, Sun L, Li L, Liu H. Elevated GPC3 level promotes cell proliferation in liver cancer. Oncol Lett. 2018;16:970-976. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 90. | Gaudet S, Branton D, Lue RA. Characterization of PDZ-binding kinase, a mitotic kinase. Proc Natl Acad Sci U S A. 2000;97:5167-5172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 181] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 91. | Tyagi N, Deshmukh SK, Srivastava SK, Azim S, Ahmad A, Al-Ghadhban A, Singh AP, Carter JE, Wang B, Singh S. ETV4 Facilitates Cell-Cycle Progression in Pancreatic Cells through Transcriptional Regulation of Cyclin D1. Mol Cancer Res. 2018;16:187-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 92. | Qi M, Liu Z, Shen C, Wang L, Zeng J, Wang C, Li C, Fu W, Sun Y, Han B. Overexpression of ETV4 is associated with poor prognosis in prostate cancer: involvement of uPA/uPAR and MMPs. Tumour Biol. 2015;36:3565-3572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 93. | Chen J, Qian Z, Li F, Li J, Lu Y. Integrative Analysis of Microarray Data to Reveal Regulation Patterns in the Pathogenesis of Hepatocellular Carcinoma. Gut Liver. 2017;11:112-120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 94. | Hu K, Wang ZM, Li JN, Zhang S, Xiao ZF, Tao YM. CLEC1B Expression and PD-L1 Expression Predict Clinical Outcome in Hepatocellular Carcinoma with Tumor Hemorrhage. Transl Oncol. 2018;11:552-558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 95. | Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, Davies DR. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379-381. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 585] [Cited by in RCA: 558] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 96. | Liu Z, Dou C, Jia Y, Li Q, Zheng X, Yao Y, Liu Q, Song T. RIG-I suppresses the migration and invasion of hepatocellular carcinoma cells by regulating MMP9. Int J Oncol. 2015;46:1710-1720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 97. | Hou Z, Zhang J, Han Q, Su C, Qu J, Xu D, Zhang C, Tian Z. Hepatitis B virus inhibits intrinsic RIG-I and RIG-G immune signaling via inducing miR146a. Sci Rep. 2016;6:26150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 98. | Chen L, Ashe S, Brady WA, Hellström I, Hellström KE, Ledbetter JA, McGowan P, Linsley PS. Costimulation of antitumor immunity by the B7 counterreceptor for the T lymphocyte molecules CD28 and CTLA-4. Cell. 1992;71:1093-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 751] [Cited by in RCA: 772] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 99. | Markowitz GJ, Yang P, Fu J, Michelotti GA, Chen R, Sui J, Yang B, Qin WH, Zhang Z, Wang FS, Diehl AM, Li QJ, Wang H, Wang XF. Inflammation-Dependent IL18 Signaling Restricts Hepatocellular Carcinoma Growth by Enhancing the Accumulation and Activity of Tumor-Infiltrating Lymphocytes. Cancer Res. 2016;76:2394-2405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 100. | Yu M, Tang Z, Meng F, Tai M, Zhang J, Wang R, Liu C, Wu Q. Elevated expression of FoxM1 promotes the tumor cell proliferation in hepatocellular carcinoma. Tumour Biol. 2016;37:1289-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 101. | Nakajima T, Yasui K, Zen K, Inagaki Y, Fujii H, Minami M, Tanaka S, Taniwaki M, Itoh Y, Arii S, Inazawa J, Okanoue T. Activation of B-Myb by E2F1 in hepatocellular carcinoma. Hepatol Res. 2008;38:886-895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 102. | Petrizzo A, Caruso FP, Tagliamonte M, Tornesello ML, Ceccarelli M, Costa V, Aprile M, Esposito R, Ciliberto G, Buonaguro FM, Buonaguro L. Identification and Validation of HCC-specific Gene Transcriptional Signature for Tumor Antigen Discovery. Sci Rep. 2016;6:29258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 103. | Hishida M, Nomoto S, Inokawa Y, Hayashi M, Kanda M, Okamura Y, Nishikawa Y, Tanaka C, Kobayashi D, Yamada S, Nakayama G, Fujii T, Sugimoto H, Koike M, Fujiwara M, Takeda S, Kodera Y. Estrogen receptor 1 gene as a tumor suppressor gene in hepatocellular carcinoma detected by triple-combination array analysis. Int J Oncol. 2013;43:88-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 104. | Park YY, Choi HS, Lee JS. Systems-level analysis of gene expression data revealed NR0B2/SHP as potential tumor suppressor in human liver cancer. Mol Cells. 2010;30:485-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 105. | Prestin K, Olbert M, Hussner J, Isenegger TL, Gliesche DG, Böttcher K, Zimmermann U, Meyer Zu Schwabedissen HE. Modulation of expression of the nuclear receptor NR0B2 (small heterodimer partner 1) and its impact on proliferation of renal carcinoma cells. Onco Targets Ther. 2016;9:4867-4878. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 106. | Liu P, Cao W, Ma B, Li M, Chen K, Sideras K, Duitman JW, Sprengers D, Khe Tran TC, Ijzermans JNM, Biermann K, Verheij J, Spek CA, Kwekkeboom J, Pan Q, Peppelenbosch MP. Action and clinical significance of CCAAT/enhancer-binding protein delta in hepatocellular carcinoma. Carcinogenesis. 2019;40:155-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 107. | He J, Gerstenlauer M, Chan LK, Leithäuser F, Yeh MM, Wirth T, Maier HJ. Block of NF-kB signaling accelerates MYC-driven hepatocellular carcinogenesis and modifies the tumor phenotype towards combined hepatocellular cholangiocarcinoma. Cancer Lett. 2019;458:113-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 108. | Luedde T, Schwabe RF. NF-κB in the liver--linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:108-118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1094] [Cited by in RCA: 1087] [Article Influence: 77.6] [Reference Citation Analysis (0)] |