Published online Sep 15, 2022. doi: 10.4251/wjgo.v14.i9.1665
Peer-review started: March 20, 2022
First decision: April 17, 2022
Revised: May 9, 2022
Accepted: July 31, 2022
Article in press: July 31, 2022
Published online: September 15, 2022
Processing time: 173 Days and 4.6 Hours
Colorectal cancer (CRC) constitutes the third most frequently reported malig
Core Tip: Colorectal cancer (CRC) constitutes the third most frequent malignancy. CRC is a complex, multistep process. The impact of environmental factors as well as the disturbance of the gut microbial ecosystem is associated with CRC development. There is a strong relationship between the gut micro
- Citation: Koustas E, Trifylli EM, Sarantis P, Papadopoulos N, Aloizos G, Tsagarakis A, Damaskos C, Garmpis N, Garmpi A, Papavassiliou AG, Karamouzis MV. Implication of gut microbiome in immunotherapy for colorectal cancer. World J Gastrointest Oncol 2022; 14(9): 1665-1674
- URL: https://www.wjgnet.com/1948-5204/full/v14/i9/1665.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i9.1665
Colorectal cancer (CRC) constitutes the third most frequently reported malignancy in the male po
The relationship between the intestinal microbiome and disease development, including carcinogenesis, was relatively undervalued in the last decade. However, the interrelation of gut microbiota with the main functions of the host has recently been in the spotlight[4]. The digestive tract contains the largest amount of microbiota colonization among other anatomical regions, accounting for approximately 70% of the human microbiota make-up[5], including viral and bacterial microorganisms, archaea and fungi[6,7]. The proximal parts of the GI tract, including the stomach and small intestine, present few microbiota species whereas the distal part, the colon, presents the largest number of species (microorganisms) in the colonic substance[7]. The six main phyla of the gut microbiome (90% of the population) include[8]: Bacteroidetes, Actinobacteria, Firmicutes, Proteobacteria, Verrucomicrobia, and Euryachaeota[9]. Of all the genera found in the human gut, Bacteroides makes up the majority of the population (30%)[10], implying its significant effecton the human functional system. Additionally, many genera from the Firmicutes phylum compose a high amount of the intestinal substance, such as lactobacillus, Clostridium, Faecalibacterium, Eubacterium and Ruminococcus[11]. The application of metagenomics on fecal specimens has given the opportunity for microbiome quantification and analysis, and potentially its use as a potent diagnostic tool[12].
PubMed was searched to identify studies on gut microbiome, immunotherapy and CRC. PubMed and Reference Citation Analysis (https://www.referencecitationanalysis.com/) were searched to identify studies on gut microbiome, immunotherapy and CRC. The literature review was completed on February 28, 2022. The following search terms were applied: “Colorectal cancer”, “Immunotherapy”, “Checkpoint inhibitors,” “Tumor microenvironment,” and “Gut microbiome”. The reference lists of all related articles were screened for other potentially relevant studies. The search citation analysis is presented in the reference list. Finally, the authors similarly reviewed the reference lists of eligible articles to identify further eligible articles, books and other forms of publication. Publications that are written in any other language other than English were excluded. Publications of abstracts were also excluded.
Gut microbiota exhibits diverse functions in the human organism and are responsible for many metabolic processes and biosynthesis. Vitamin synthesis constitutes one of the key roles of gut microbiota, such as riboflavin, vitamin B1, biotin, vitamin K and cobalamin[13]. They also have a crucial role in non-digestible carbohydrate metabolism; to transform them into short-chain fatty acids (SCFAs), such as butyric acid, acetic acid and propionic[14], which are produced by the main phyla of bacteriome, this includes Bacteroidetes and Firmicutes[15]. Alteration of the above metabolic process leads to modification of the fatty acid production and overall metabolic imbalance[16]. Along with their involvement in vitamin and short fatty acids synthesis, they take part in bile acid production[17]. Neuromodulators are also produced by gut microbiota, with a significant implication for the gut-brain axis, which includes the peripheral and central nervous systems as well as the enteric nervous system[18]. Many neurological and psychiatric disorders are closely connected with the gut microbiome. This can occur because they are responsible for synthesizing many pro-inflammatory cytokines, amyloids and liposaccharides[18]. Based on metagenomics, genome disturbance and dysbiotic flora can cause a predisposition to develop a number of malignancies[19], including non-neoplastic disorders, such as atopy, functional intestinal disturbances, like irritable bowel syndrome (IBS), inflammatory bowel disease (IBD) and metabolic syndrome[20,21].
There is a strong relationship between gut microbiome dysbiosis and bowel pathogenicity. In the case of the bowel, functional disorders such as IBS have many studies illustrating an altered consistency of the bacteriome, with both an increase or decrease in the quantity of many bacteria. It is specifically observed as an aberrant increase of Ruminococcus, Firmicutes, and Clostridium spp. with an abnormal decrease of Ruminococcusalbus and callidus, Bacteroidesfragilis and bulgatus[18]. Additionally, the overproduction of SCFAs that deregulate the secretion of serotonin from the enteroendocrine cells leads to increased bowel movements and fermentation. This causes the symptomatology associated with meteorism[22]. Patients who suffer from organic bowel diseases, such as IBD, Ulcerative colitis and Crohn’s disease (CD) have been observed to have an altered microbiome. The modification of the gut microbiome is closely associated to dietary habits[23]. Patients with CD specifically demonstrate increased amounts of Neisseria caeacorrodens, E. coli and proteobacteria[24], while enhanced amounts of fungal species such as Candida albicans, Cyberlindnera jadinii and Saccharomyces cerevisiaecan also be observed[25]. In addition, a decreased number of some bacterial taxa, such as Firmicutes, Faecalibacterium prausnitzii, Bacteroidetes and Roseburia, is observed[26]. Dietary habits that include a high amount of fruit and vegetable consumption can lower the risk for developing CD[27].
Intestinal epithelial cell sare closely interrelated with the immune system via the existence of goblet and Paneth cells and their products. Goblet cells are located in intestinal mucosa and have a crucial role in producing mucus. Paneth cells are located in the crypts of Lieberkühn, secreting various immuno
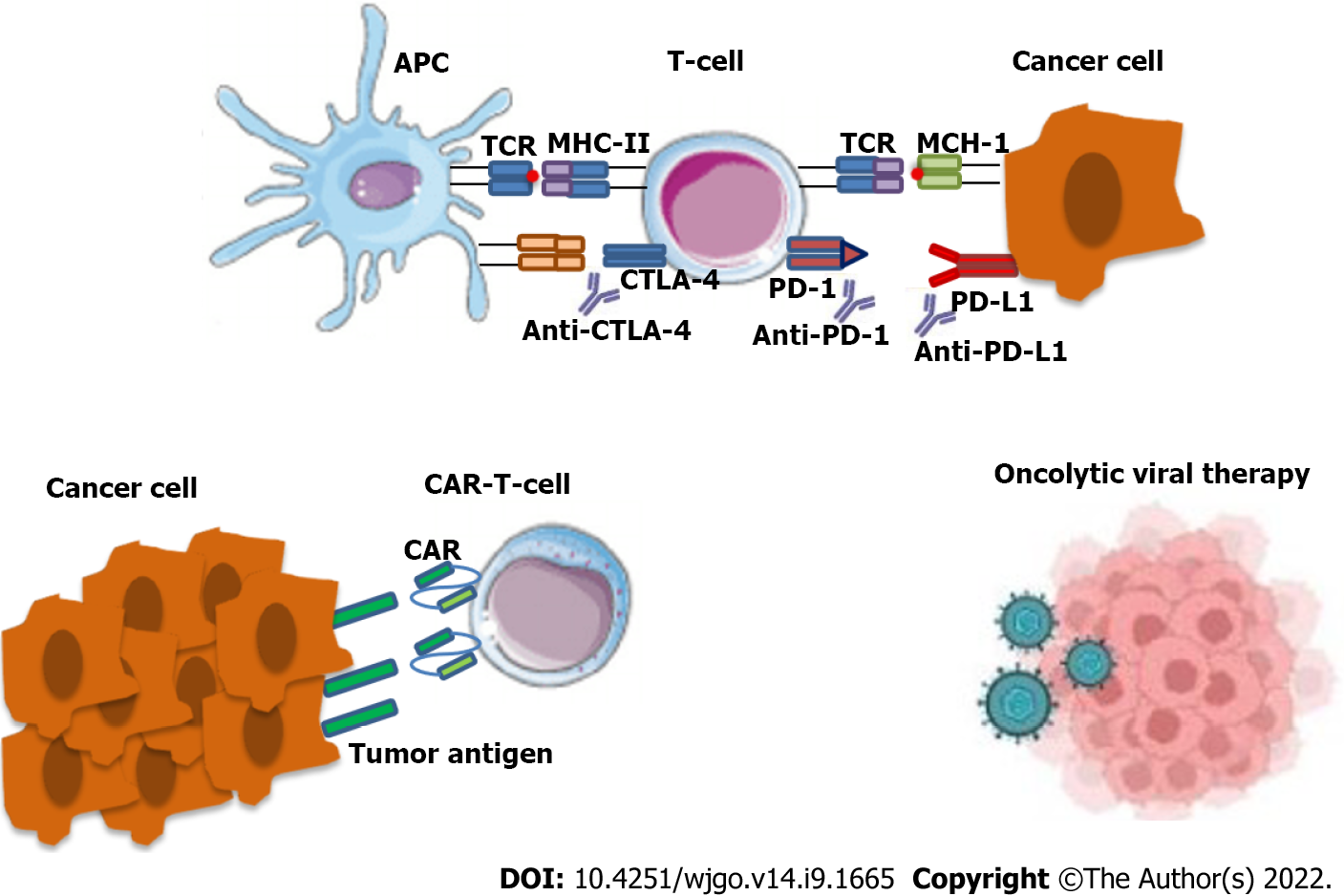
Immunotherapy constitutes a significant therapeutic option, including immune checkpoint inhibitors, cancer vaccines and chimeric antigen receptor-T cells[30]. This treatment modality makes use of the immune responses to create an anti-neoplastic effect. The main therapeutic agents include the following monoclonal antibodies: (1) Anti-cytotoxic TT-lymphocyte antigen-4 (anti-CTLA-4); (2) Anti-program
This therapeutic modality is currently selected as an anti-cancer treatment specifically in cases of tumors that are characterized by high microsatellite instability (MSI-H)[32]. Tumors that present MSI-H arise from a defective DNA mismatch repair (MMR) mechanism that leads to the accumulation of genetic mutations. This can be seen in the case of mutant MSH2, PMS2, MSH6 and MLH1. Or by epigenetic aberration, such as genome hyper-methylation[33]. There are many reports that gut mi
There are many studies about the implication of gut microbiota in immunotherapeutic agents including immune checkpoint inhibitors for melanoma. Fewer studies exist about its role in CRC treatment management.
Modifications in the gut microbiome and microbial metabolites have been involved in many pathological processes and diseases, including colon carcinogenesis[36]. Many intestinal bacterial products have been implicated in malignant states in the intestinal tract[37]. Several studies demonstrate the presence of an altered microbiome either in CRC patients’ fecal specimens or in malignant tissues compared to healthy patients[38]. These alterations in the microbiome which take place in the initial steps of CRC development can be utilized as predictive biomarkers as well as microbial diagnostic gene markers. This can be utilized in patients with an increased risk of developing colon adenomas that can potentially lead to CRC[39].
Environmental factors have a high influence on the gut microbiome along with idiosyncratic factors[40] which subsequently induce carcinogenesis and CRC development via the overgrowth of particular microbial species in the flora[41]. The formulation of colonic microbial substances is closely related to modifiable factors such as eating behavior and style of living[42]. While there is a key role in the metabolism of nutrients[43], there is also a diversity of environmental risk factors that are associated with colorectal carcinogenesis such as obesity, tobacco use, alcohol consumption and prepared meat products[44].
Many studies demonstrate the implication of specific bacterial taxa in carcinogenesis, such as Enterococcus faecalis, Helicobacter hepaticus, Bacteroides fragilis and Fusobacterium nucleatum. The products of the previously mentioned microbes lead to genomic alterations[45]. While in the case of the Fusobacterium nucleatum, the carcinogenesis indirectly occurs via the perpetual secretion of pro-inflammatory cytokines[46]. This phenomenon implies the close interrelation of the microbiome with immune response and metabolic processes[47].
There is a notable reduction of genera from the Firmicutes phylum, which produce a significant metabolite, the alleged butyrate. An enhanced reproduction of specific phyla, such as Bacteroides fragilis, Peptostreptococcus stomatis as well as Tarvi monas micra, Fusobacterium nucleatum[48] and Solobacterium moorei[49]. Additionally, there are reports that show an increased amount of Enterococcus, Escherichia coli, Klebsiella and Streptococcus, as well as a decrease in Rothia[2].
There is considerable evidence that CRC development is closely associated with the presence of Fusobacteriaceae family members, such as Fusobacterium nucleatum, necrophorum and mortiferum[37] via a mechanism that was reportedly observed in mice[50].
Generally, dysbiosis which includes the modification of microbial taxa in the gut ecosystem leads either to a limited variety of microbiota or the overgrowth of microbes. This can further lead to the development of opportunistic infections[51], destruction of the intestinal epithelial barrier, bacterial translocation to the mesenteric lymph nodes or the circulatory system, ultimately leading to a local and systemic inflammatory response[52].
Recruitment of T lymphocytes is observed in CRC malignant tissues[53] via the secretion of chemotactic cytokines. This is further related to an abundance in proteobacteria Ruminococcaceae, B. fragilis and E.coli. Alternatively, a high number of Fusobacteriais is associated with a dismal prognosis. In in vitro it has been observed to express an increased number of recruited T cells and inflammatory modulators [interleukin (IL)-6, IL-8, IL-1][54], an inhibitory effect on Natural killer cells, as well as tumor-infiltrating lymphocytes[55]. Although Fusobacterium nucleatum is normally associated with a worse prognosis, it constitutes a promoter for differentiation in regulatory T cells leading to a decrease in expression of scurfin or forkheadbox P3 which is correlated to prolonged survival[56].
The therapeutic management of CRC is considered quite challenging due to the complex molecular basis including genetic and epigenetic alterations[57]. In recent years, immunotherapeutic agents are utilized for tumors that present high MSI-H which results from a defective DNA MMR or epigenetic modification[33]. An epigenetic aberration is genome hyper-methylation in addition tomutant genes such as PMS2, MLH1 as well as MSH2 and MSH6[58]. In the case of MSI-H colorectal tumors, there is evident methylation of CpG islands in the promoter of the BRAF proto-oncogene[59]. It is observed that patients with BRAF and RAS genetic mutations present resistance to immunotherapeutic treatments with a limited enhancement of survival[60]. It can occur in cases of epidermal growth factor receptor inhibitors, like cetuximab, as well as Panitumumab[61]. In comparison with MSI tumors, the mi
Tumor microenvironment (TME) includes multiple types of cells, such as fibroblasts, immune cells, endothelial and stromal cells[66]. TME demonstrates a significant role in immune responses, particularly in CRC, and constitutes as a therapeutic target for many anti-cancer agents[67]. The stroma around the tumor has a key role in resistance to chemotherapy due to the fact that it includes a heterogeneous population of cells with various levels of differentiation. This contributes to invasive tumor behavior and dissemination. This is shown in the case of tumor-associated macrophages and cancer-associated fibroblasts. Both of these are related to a dismal prognosis and neoangiogenesis[68,69], as well as Myeloid-derived suppressor cells which are also implicated in tumor progression and invasion. Their effect is under the regulation of tumoral products like chemokine (C-C motif) ligand 2 and 5 (CCL2 and CCL5)[70].
It was previously stated that the gut microbiota exhibited various effects on the differentiation mechanism and tumor development. While they influence the tumor response to immunotherapeutics[71], the existence of intra-tumoral bacteria is reported in many solid tumors, especially in breast cancer. It was demonstrated that the microbiome is particular for each kind of malignant tumor presenting distinct metabolic functions[72]. Based on data that was collected by whole-transcriptome analysis, there is a distinct microbiome correlated with different malignant tumors, implying a specific microbial profile for each type of cancer[73]. Additionally, TME has a crucial role in the existence and multi
Resistance to immunotherapy is difficult to overcome in clinical practice[31]. Manipulation of gut microbiota constitutes a promising method for reducing the resistance to therapeutic agents. This is implied by the notable effect of intestinal microbial products on the malignant tumor where they could also be considered cancer-driving molecules[75].
Experimental studies on mice have shown that bacteria have a crucial role in the anti-cancer immune response. While the response was limited in the case of germ-free mice[28], it was primarily reported that intestinal microbiota have a significant role in the response especially to immune checkpoint inhibitors. However, the previous observation was also demonstrated in humans when an immune checkpoint blockade was applied[28]. In mouse-model studies, fecal microbial transplantation (FMT) from mice that presented immune-responsive microbiota, to germ-free mice, provided a better anti-neoplastic response and tumor growth management. This result is associated with an increased amount of cytotoxic T lymphocytes (CD8+) in TME[76]. Whereas the transfer of fecal samples, including microbiota prone for carcinogenesis, provides the opposite results to physiological mice[77]. However, the correlation of the anti-tumor response with external factors must be taken into consideration.
Alterations in the consistency of bacteriome were reported in cases of patients with an active response to PD-1 inhibitors. More specifically, these patients presented a higher amount of Enterococcus faecium, Bifidobacterium longum and Collinsella aerofaciens. Fecal specimens that presented the above microbial taxa were characterized as “responder” stool samples and were transferred via FMT to germ-free mice. Subsequently, the germ-free mice started to express the stool phenotype of the responders[28].
Based on various human and animal-model cohort studies, intestinal microbiota could not only have been beneficial but also toxic effects on immune checkpoint inhibition[78]. Reduced toxicity was observed in specimens where Bacteroidetes genera were in abundance. Although they relate to unresponsiveness to immune checkpoint inhibitors (ICIs), in contrast to Firmicutes, and especially in the case of Ruminococaceae, they were not only responsive to ICIs but also presented toxic effects. In cases of overgrown Faecalibacterium prausnitzii, patients had an increased risk of presenting colitis related with CTLA-4 inhibitors[79,80].
Based on all the characteristics of the intestinal microbiota, they can either promote the anti-neoplastic response or induce inflammation and carcinogenesis[81]. A reduced anti-cancer response in the host was observed in germ-free mice or with antibiotic administration (broad-spectrum)[28,35]. In cases with urinary tract malignancies and lung cancer, antibiotics had a harmful effect on anti-PD1/PD-L1 treatment[35] in comparison to cyclophosphamide which presented a promoting effect on the overgrowth of Barnesiella intestine hominis in the intestinal tract and a stimulatory effect on anti-cancer immune response[82].
However, the manipulation of microbiota and utilization of antibiotics for the killing of bacteria is detrimental to the response to immunotherapeutic agents. This method includes the risk of killing favorable bacterial species. To avoid the non-elective effect of antibiotics, bacteriophage therapy is administered which permits a selective elimination of unfavorable bacteria[83].
Lastly, environmental and lifestyle habits could potentially alter the gut microbiome. These include physical exercise, proper dietary habits, sleep patterns, as well as via the utilization of FMT[84]. Bacteriotherapy or FMT includes the transferring of beneficial bacterial species such as Bacteroides, Bifidobacteria, E. hirae and Akkermansia mucini philia[85].
The relationship between the intestinal microbiome and disease development, such as carcinogenesis, was underestimated in the last decades. Nevertheless, the crucial role of intestinal microbiota has been in the spotlight as of recent years. Not only for their significant influence on the main metabolic functions of the host but also on the immune and anti-tumor responses. Immunotherapeutic agents are commonly used specifically for cases with chemo-resistant advanced colorectal malignant tumors. The implication of gut microbiota in the anti-cancer immune response is still under research. However, there are many reports supporting that the lack of specific bacterial taxa in CRC patients leads to a limited response to immunotherapy or complete unresponsiveness with the presence of specific phyla that could promote the anti-cancer response. Based on various human and animal-model cohort studies, intestinal microbiota could not only have beneficial effects on immune checkpoint inhibition but also have detrimental effects. The aforementioned phenomenon illustrates the necessity for the manipulation of intestinal microbiota. Specifically for the highest anti-neoplastic immune response, either via bacteriophage therapy or lifestyle habits modifications as well as FMT. Further research regarding the implication of gut microbiome on immunotherapy responses is needed for the identification of additional druggable targets, along with the manipulation of intestinal microbiota to achieve an optimal therapeutic response personalized for each patient.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Greece
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Manojlovic N, Serbia; Popovic LS, Serbia S-Editor: Fan JR L-Editor: Filipodia P-Editor: Fan JR
| 1. | Song M, Chan AT, Sun J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology. 2020;158:322-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 464] [Cited by in RCA: 513] [Article Influence: 102.6] [Reference Citation Analysis (2)] |
| 2. | Quigley EM. Gut bacteria in health and disease. Gastroenterol Hepatol (N Y). 2013;9:560-569. [PubMed] |
| 3. | Gagnière J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, Bringer MA, Pezet D, Bonnet M. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 2016;22:501-518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 613] [Cited by in RCA: 558] [Article Influence: 62.0] [Reference Citation Analysis (5)] |
| 4. | Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell. 2018;33:954-964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 548] [Article Influence: 78.3] [Reference Citation Analysis (0)] |
| 5. | Saus E, Iraola-Guzmán S, Willis JR, Brunet-Vega A, Gabaldón T. Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Mol Aspects Med. 2019;69:93-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 233] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 6. | Passos MDCF, Moraes-Filho JP. Intestinal microbiota in digestive diseases. Arq Gastroenterol. 2017;54:255-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 7. | Shapira M. Gut Microbiotas and Host Evolution: Scaling Up Symbiosis. Trends Ecol Evol. 2016;31:539-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 267] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 8. | Feng Q, Chen WD, Wang YD. Gut Microbiota: An Integral Moderator in Health and Disease. Front Microbiol. 2018;9:151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 273] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 9. | Gao R, Gao Z, Huang L, Qin H. Gut microbiota and colorectal cancer. Eur J Clin Microbiol Infect Dis. 2017;36:757-769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 144] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 10. | Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787-8803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1421] [Cited by in RCA: 1839] [Article Influence: 183.9] [Reference Citation Analysis (58)] |
| 11. | Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, Mele MC. What is the Healthy Gut Microbiota Composition? Microorganisms. 2019;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 969] [Cited by in RCA: 2077] [Article Influence: 346.2] [Reference Citation Analysis (0)] |
| 12. | Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Böhm J, Brunetti F, Habermann N, Hercog R, Koch M, Luciani A, Mende DR, Schneider MA, Schrotz-King P, Tournigand C, Tran Van Nhieu J, Yamada T, Zimmermann J, Benes V, Kloor M, Ulrich CM, von Knebel Doeberitz M, Sobhani I, Bork P. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 651] [Cited by in RCA: 859] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 13. | LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 790] [Cited by in RCA: 960] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 14. | Makki K, Deehan EC, Walter J, Bäckhed F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe. 2018;23:705-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 898] [Cited by in RCA: 1558] [Article Influence: 222.6] [Reference Citation Analysis (0)] |
| 15. | Pushpanathan P, Mathew GS, Selvarajan S, Seshadri KG, Srikanth P. Gut microbiota and its mysteries. Indian J Med Microbiol. 2019;37:268-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 16. | Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature. 2016;534:213-217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 724] [Cited by in RCA: 951] [Article Influence: 105.7] [Reference Citation Analysis (1)] |
| 17. | Fiorucci S, Carino A, Baldoni M, Santucci L, Costanzi E, Graziosi L, Distrutti E, Biagioli M. Bile Acid Signaling in Inflammatory Bowel Diseases. Dig Dis Sci. 2021;66:674-693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 142] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 18. | Gomaa EZ. Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek. 2020;113:2019-2040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 701] [Article Influence: 140.2] [Reference Citation Analysis (4)] |
| 19. | Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, Andoh A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018;11:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 905] [Article Influence: 113.1] [Reference Citation Analysis (3)] |
| 20. | Temraz S, Nassar F, Nasr R, Charafeddine M, Mukherji D, Shamseddine A. Gut Microbiome: A Promising Biomarker for Immunotherapy in Colorectal Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 21. | Dabke K, Hendrick G, Devkota S. The gut microbiome and metabolic syndrome. J Clin Invest. 2019;129:4050-4057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 450] [Article Influence: 90.0] [Reference Citation Analysis (0)] |
| 22. | Kadooka Y, Sato M, Imaizumi K, Ogawa A, Ikuyama K, Akai Y, Okano M, Kagoshima M, Tsuchida T. Regulation of abdominal adiposity by probiotics (Lactobacillus gasseri SBT2055) in adults with obese tendencies in a randomized controlled trial. Eur J Clin Nutr. 2010;64:636-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 469] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 23. | Younis N, Zarif R, Mahfouz R. Inflammatory bowel disease: between genetics and microbiota. Mol Biol Rep. 2020;47:3053-3063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 24. | Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Büttner L, de Lima Romão E, Behrendt CL, Lopez CA, Sifuentes-Dominguez L, Huff-Hardy K, Wilson RP, Gillis CC, Tükel Ç, Koh AY, Burstein E, Hooper LV, Bäumler AJ, Winter SE. Precision editing of the gut microbiota ameliorates colitis. Nature. 2018;553:208-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 405] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 25. | Lane ER, Zisman TL, Suskind DL. The microbiota in inflammatory bowel disease: current and therapeutic insights. J Inflamm Res. 2017;10:63-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 168] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 26. | Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Doré J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 857] [Cited by in RCA: 912] [Article Influence: 57.0] [Reference Citation Analysis (0)] |
| 27. | Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn's disease. Lancet. 2017;389:1741-1755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1121] [Cited by in RCA: 1806] [Article Influence: 225.8] [Reference Citation Analysis (111)] |
| 28. | Li W, Deng Y, Chu Q, Zhang P. Gut microbiome and cancer immunotherapy. Cancer Lett. 2019;447:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 178] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 29. | Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012;12:821-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 484] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 30. | Dahiya DS, Kichloo A, Singh J, Albosta M, Lekkala M. Current immunotherapy in gastrointestinal malignancies A Review. J Investig Med. 2021;69:689-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 31. | Jacob JB, Jacob MK, Parajuli P. Review of immune checkpoint inhibitors in immuno-oncology. Adv Pharmacol. 2021;91:111-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 32. | Lichtenstern CR, Ngu RK, Shalapour S, Karin M. Immunotherapy, Inflammation and Colorectal Cancer. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 202] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 33. | Gologan A, Sepulveda AR. Microsatellite instability and DNA mismatch repair deficiency testing in hereditary and sporadic gastrointestinal cancers. Clin Lab Med. 2005;25:179-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 42] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 34. | Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017;17:271-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 653] [Article Influence: 81.6] [Reference Citation Analysis (0)] |
| 35. | Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragón L, Jacquelot N, Qu B, Ferrere G, Clémenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2493] [Cited by in RCA: 3804] [Article Influence: 475.5] [Reference Citation Analysis (0)] |
| 36. | Ashktorab H, Kupfer SS, Brim H, Carethers JM. Racial Disparity in Gastrointestinal Cancer Risk. Gastroenterology. 2017;153:910-923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 195] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 37. | Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25:968-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 880] [Article Influence: 146.7] [Reference Citation Analysis (0)] |
| 38. | Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, Shi Y, Song M, da Silva A, Gu M, Li W, Hamada T, Kosumi K, Hanyuda A, Liu L, Kostic AD, Giannakis M, Bullman S, Brennan CA, Milner DA, Baba H, Garraway LA, Meyerhardt JA, Garrett WS, Huttenhower C, Meyerson M, Giovannucci EL, Fuchs CS, Nishihara R, Ogino S. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol. 2016;7:e200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 226] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 39. | Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, Wang X, Xu X, Chen N, Wu WK, Al-Aama J, Nielsen HJ, Kiilerich P, Jensen BA, Yau TO, Lan Z, Jia H, Li J, Xiao L, Lam TY, Ng SC, Cheng AS, Wong VW, Chan FK, Yang H, Madsen L, Datz C, Tilg H, Wang J, Brünner N, Kristiansen K, Arumugam M, Sung JJ. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 917] [Cited by in RCA: 785] [Article Influence: 98.1] [Reference Citation Analysis (0)] |
| 40. | Song M, Chan AT. Environmental Factors, Gut Microbiota, and Colorectal Cancer Prevention. Clin Gastroenterol Hepatol. 2019;17:275-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 229] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 41. | Clay SL, Fonseca-Pereira D, Garrett WS. Colorectal cancer: the facts in the case of the microbiota. J Clin Invest. 2022;132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 89] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 42. | Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu J, Wijmenga C, Zhernakova A, Elinav E, Segal E. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1342] [Cited by in RCA: 1842] [Article Influence: 263.1] [Reference Citation Analysis (0)] |
| 43. | Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1039] [Cited by in RCA: 1355] [Article Influence: 150.6] [Reference Citation Analysis (0)] |
| 44. | Hills RD Jr, Pontefract BA, Mishcon HR, Black CA, Sutton SC, Theberge CR. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 323] [Cited by in RCA: 671] [Article Influence: 111.8] [Reference Citation Analysis (0)] |
| 45. | Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19:55-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 2676] [Article Influence: 535.2] [Reference Citation Analysis (1)] |
| 46. | Chattopadhyay I, Dhar R, Pethusamy K, Seethy A, Srivastava T, Sah R, Sharma J, Karmakar S. Exploring the Role of Gut Microbiome in Colon Cancer. Appl Biochem Biotechnol. 2021;193:1780-1799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 79] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 47. | Cheng Y, Ling Z, Li L. The Intestinal Microbiota and Colorectal Cancer. Front Immunol. 2020;11:615056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 329] [Article Influence: 65.8] [Reference Citation Analysis (0)] |
| 48. | Loftus M, Hassouneh SA, Yooseph S. Bacterial community structure alterations within the colorectal cancer gut microbiome. BMC Microbiol. 2021;21:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 49. | Yang Y, Cai Q, Shu XO, Steinwandel MD, Blot WJ, Zheng W, Long J. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int J Cancer. 2019;144:2381-2389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 50. | Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, Li H, Guo B, Zhu Q, Wei Q, Moyer MP, Wang P, Cai S, Goel A, Qin H, Ma Y. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152:851-866.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 455] [Cited by in RCA: 716] [Article Influence: 89.5] [Reference Citation Analysis (1)] |
| 51. | Frosali S, Pagliari D, Gambassi G, Landolfi R, Pandolfi F, Cianci R. How the Intricate Interaction among Toll-Like Receptors, Microbiota, and Intestinal Immunity Can Influence Gastrointestinal Pathology. J Immunol Res. 2015;2015:489821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 174] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 52. | Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 724] [Cited by in RCA: 1084] [Article Influence: 135.5] [Reference Citation Analysis (0)] |
| 53. | Ruan H, Leibowitz BJ, Zhang L, Yu J. Immunogenic cell death in colon cancer prevention and therapy. Mol Carcinog. 2020;59:783-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 54. | Proença MA, Biselli JM, Succi M, Severino FE, Berardinelli GN, Caetano A, Reis RM, Hughes DJ, Silva AE. Relationship between Fusobacterium nucleatum, inflammatory mediators and microRNAs in colorectal carcinogenesis. World J Gastroenterol. 2018;24:5351-5365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 62] [Cited by in RCA: 86] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 55. | Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 978] [Article Influence: 97.8] [Reference Citation Analysis (0)] |
| 56. | Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, Nagase H, Nishimura J, Yamamoto H, Takiguchi S, Tanoue T, Suda W, Morita H, Hattori M, Honda K, Mori M, Doki Y, Sakaguchi S. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 660] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 57. | Sideris M, Papagrigoriadis S. Molecular biomarkers and classification models in the evaluation of the prognosis of colorectal cancer. Anticancer Res. 2014;34:2061-2068. [PubMed] |
| 58. | Fishel R. Mismatch repair. J Biol Chem. 2015;290:26395-26403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 174] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 59. | Li WQ, Kawakami K, Ruszkiewicz A, Bennett G, Moore J, Iacopetta B. BRAF mutations are associated with distinctive clinical, pathological and molecular features of colorectal cancer independently of microsatellite instability status. Mol Cancer. 2006;5:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 137] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 60. | Thiel A, Ristimäki A. Toward a Molecular Classification of Colorectal Cancer: The Role of BRAF. Front Oncol. 2013;3:281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 61. | Koustas E, Karamouzis MV, Mihailidou C, Schizas D, Papavassiliou AG. Co-targeting of EGFR and autophagy signaling is an emerging treatment strategy in metastatic colorectal cancer. Cancer Lett. 2017;396:94-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 62. | Koustas E, Papavassiliou AG, Karamouzis MV. The role of autophagy in the treatment of BRAF mutant colorectal carcinomas differs based on microsatellite instability status. PLoS One. 2018;13:e0207227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 63. | André T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P, de la Fouchardiere C, Rivera F, Elez E, Bendell J, Le DT, Yoshino T, Van Cutsem E, Yang P, Farooqui MZH, Marinello P, Diaz LA Jr; KEYNOTE-177 Investigators. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N Engl J Med. 2020;383:2207-2218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 962] [Cited by in RCA: 1813] [Article Influence: 362.6] [Reference Citation Analysis (0)] |
| 64. | Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LA Jr. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16:361-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1114] [Cited by in RCA: 1229] [Article Influence: 204.8] [Reference Citation Analysis (0)] |
| 65. | Raman SS, Hecht JR, Chan E. Talimogene laherparepvec: review of its mechanism of action and clinical efficacy and safety. Immunotherapy. 2019;11:705-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 66. | Wang DK, Zuo Q, He QY, Li B. Targeted Immunotherapies in Gastrointestinal Cancer: From Molecular Mechanisms to Implications. Front Immunol. 2021;12:705999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 67. | Grizzi F, Basso G, Borroni EM, Cavalleri T, Bianchi P, Stifter S, Chiriva-Internati M, Malesci A, Laghi L. Evolving notions on immune response in colorectal cancer and their implications for biomarker development. Inflamm Res. 2018;67:375-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 68. | Koustas E, Sarantis P, Kyriakopoulou G, Papavassiliou AG, Karamouzis MV. The Interplay of Autophagy and Tumor Microenvironment in Colorectal Cancer-Ways of Enhancing Immunotherapy Action. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 69. | Yang X, Li Y, Zou L, Zhu Z. Role of Exosomes in Crosstalk Between Cancer-Associated Fibroblasts and Cancer Cells. Front Oncol. 2019;9:356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 144] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 70. | Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, Kaiser EA, Snyder LA, Pollard JW. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222-225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2327] [Cited by in RCA: 2228] [Article Influence: 159.1] [Reference Citation Analysis (0)] |
| 71. | Ge Y, Wang X, Guo Y, Yan J, Abuduwaili A, Aximujiang K, Wu M. Correction to: Gut microbiota influence tumor development and Alter interactions with the human immune system. J Exp Clin Cancer Res. 2021;40:334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 72. | Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, Meltser A, Douglas GM, Kamer I, Gopalakrishnan V, Dadosh T, Levin-Zaidman S, Avnet S, Atlan T, Cooper ZA, Arora R, Cogdill AP, Khan MAW, Ologun G, Bussi Y, Weinberger A, Lotan-Pompan M, Golani O, Perry G, Rokah M, Bahar-Shany K, Rozeman EA, Blank CU, Ronai A, Shaoul R, Amit A, Dorfman T, Kremer R, Cohen ZR, Harnof S, Siegal T, Yehuda-Shnaidman E, Gal-Yam EN, Shapira H, Baldini N, Langille MGI, Ben-Nun A, Kaufman B, Nissan A, Golan T, Dadiani M, Levanon K, Bar J, Yust-Katz S, Barshack I, Peeper DS, Raz DJ, Segal E, Wargo JA, Sandbank J, Shental N, Straussman R. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368:973-980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 490] [Cited by in RCA: 1464] [Article Influence: 292.8] [Reference Citation Analysis (0)] |
| 73. | Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, Kosciolek T, Janssen S, Metcalf J, Song SJ, Kanbar J, Miller-Montgomery S, Heaton R, Mckay R, Patel SP, Swafford AD, Knight R. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579:567-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 741] [Cited by in RCA: 718] [Article Influence: 143.6] [Reference Citation Analysis (0)] |
| 74. | Qiu Q, Lin Y, Ma Y, Li X, Liang J, Chen Z, Liu K, Huang Y, Luo H, Huang R, Luo L. Exploring the Emerging Role of the Gut Microbiota and Tumor Microenvironment in Cancer Immunotherapy. Front Immunol. 2020;11:612202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 85] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 75. | Pitt JM, Vétizou M, Daillère R, Roberti MP, Yamazaki T, Routy B, Lepage P, Boneca IG, Chamaillard M, Kroemer G, Zitvogel L. Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity. 2016;44:1255-1269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 584] [Cited by in RCA: 800] [Article Influence: 100.0] [Reference Citation Analysis (0)] |
| 76. | Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1390] [Cited by in RCA: 2134] [Article Influence: 304.9] [Reference Citation Analysis (1)] |
| 77. | Dzutsev A, Badger JH, Perez-Chanona E, Roy S, Salcedo R, Smith CK, Trinchieri G. Microbes and Cancer. Annu Rev Immunol. 2017;35:199-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 204] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 78. | Zhou CB, Zhou YL, Fang JY. Gut Microbiota in Cancer Immune Response and Immunotherapy. Trends Cancer. 2021;7:647-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 200] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 79. | Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, Boselli L, Routier E, Cassard L, Collins M, Vaysse T, Marthey L, Eggermont A, Asvatourian V, Lanoy E, Mateus C, Robert C, Carbonnel F. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2019;30:2012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 80. | Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, Koh AY. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia. 2017;19:848-855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 307] [Cited by in RCA: 498] [Article Influence: 62.3] [Reference Citation Analysis (0)] |
| 81. | Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14:356-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 451] [Cited by in RCA: 664] [Article Influence: 83.0] [Reference Citation Analysis (0)] |
| 82. | Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene. 2020;39:4925-4943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 382] [Article Influence: 76.4] [Reference Citation Analysis (0)] |
| 83. | Zhang J, Hong Y, Harman NJ, Das A, Ebner PD. Genome sequence of a salmonella phage used to control salmonella transmission in Swine. Genome Announc. 2014;2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 84. | Schwartz DJ, Rebeck ON, Dantas G. Complex interactions between the microbiome and cancer immune therapy. Crit Rev Clin Lab Sci. 2019;56:567-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 85. | Wang JW, Kuo CH, Kuo FC, Wang YK, Hsu WH, Yu FJ, Hu HM, Hsu PI, Wang JY, Wu DC. Fecal microbiota transplantation: Review and update. J Formos Med Assoc. 2019;118 Suppl 1:S23-S31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 333] [Article Influence: 55.5] [Reference Citation Analysis (0)] |