Published online Aug 15, 2022. doi: 10.4251/wjgo.v14.i8.1562
Peer-review started: March 8, 2022
First decision: April 7, 2022
Revised: April 13, 2022
Accepted: July 18, 2022
Article in press: July 18, 2022
Published online: August 15, 2022
Processing time: 155 Days and 4 Hours
Colorectal cancer (CRC) is a highly malignant cancer with a high incidence and mortality in China. It is urgent to find a diagnostic marker with higher sensitivity and specificity than the traditional approaches for CRC diagnosis.
To provide new ideas for the diagnosis of CRC based on serum proteomics.
Specimens from 83 healthy people, 62 colon polyp (CRP) patients, and 101 CRC patients were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. The diagnostic value of the profiles of differentially expressed proteins was then analyzed.
Compared with the healthy control group, CRC patients had elevated expression of 5 proteins and reduced expression of 14 proteins. The area under the curve (AUC) for a differentially expressed protein with a mass-to-charge ratio of 2022.34 was the largest; the AUC was 0.843, which was higher than the AUC of 0.717 observed with carcinoembryonic antigen (CEA), and the sensitivity and specificity of this identified marker were 75.3% and 79.5%, respectively. After cross-validation, the accuracy of diagnosis using levels of this differentially expressed protein was 82.37%. Compared with the CRP group, the expression of 3 proteins in the serum of CRC patients was elevated and 11 proteins were expressed at reduced levels. Proteins possessing mass-to-charge ratio values of 2899.38 and 877.3 were selected to establish a classification tree model. The results showed that the accuracy of CRC diagnosis was 89.5%, the accuracy of CRP diagnosis was 81.6%, and the overall accuracy of this approach was 86.3%. The overall sensitivity and specificity of diagnosis using the proteomics approach were 81.8% and 66.75%, respectively. The sensitivities and specificities of diagnoses based on CEA and carbohydrate antigen 19-9 expression were 55.6% and 91.3% and 65.4% and 65.2%, respectively.
We demonstrated that serum proteomics may be helpful for the detection of CRC, and it may assist clinical practice for CRC diagnosis.
Core Tip: At present, the main techniques for proteomic research are mass spectrometry and two-dimensional gel electrophoresis. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry can analyze not only cells and tissues but also powders, solutions, and membranes. This technology is an ideal platform for the identification of tumor markers to be used in clinical practice. In this study, by using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, we analyzed the serum protein expression profiles of healthy controls, colorectal polyp patients, and colorectal cancer patients to find differentially expressed protein peaks, and aimed to evaluate the diagnostic value of serum proteomics for colorectal cancer.
- Citation: Wang HJ, Xie YB, Zhang PJ, Jiang T. Evaluation of the diagnostic value of serum-based proteomics for colorectal cancer. World J Gastrointest Oncol 2022; 14(8): 1562-1573
- URL: https://www.wjgnet.com/1948-5204/full/v14/i8/1562.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i8.1562
Colorectal cancer (CRC) is a highly malignant cancer with a high incidence and mortality in China[1]. Although many scholars in China and abroad have performed many studies on the pathogenesis and clinical manifestations of CRC, the CRC etiology is still not fully understood, and its pathogenesis has not been substantially elucidated. The commonly used serum tumor markers for CRC are carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA-199)[2]. Due to the low sensitivity and specificity of these tumor markers, it is urgent to find a diagnostic marker with higher sensitivity and specificity[3].
Due to the advent of the post-gene era, by analyzing the proteins and peptides of normal and cancerous cells, we can search for disease-specific markers and provide a new technical platform for theoretical and clinical research on CRC[4]. The study of altered protein and peptide expression with CRC could provide a reliable molecular theoretical basis for its early diagnosis, postoperative detection, postoperative recurrence prediction and prognosis[5]. At present, the main techniques for proteomic research are mass spectrometry and two-dimensional gel electrophoresis (2D-PAGE). Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) is a technology that was developed in the 1980s. At present, MALDI-TOF mass spectrometry technology has been widely used in the detection of sugars, nucleic acids, proteins, etc. The structural analysis and molecular weight determination of biological macromolecules and synthetic polymers have become some of the core objectives of current proteomics research[6]. This technique has many advantages, such as accurate mass-to-charge ratio calculation, low cost, large affinity surface, and good reproducibility, and MALDI-TOF-MS can analyze not only cells and tissues but also powders, solutions, and membranes. This technology was demonstrated to be a potential screening method for various diseases[7-9]. It is also a label-free detection technology, which reduces the cost of detection, and has high sensitivity and high-throughput detection capabilities[10,11]. Therefore, this technology is an ideal platform for the identification of tumor markers to be used in clinical practice. This approach to tumor biomarker discovery will play an important role in tumor screening, early diagnosis, individualized treatment and other aspects of cancer management[12].
In this study, by using MALDI-TOF mass spectrometry, we analyzed the serum protein expression profiles of healthy controls, colorectal polyp (CRP) patients, and CRC patients to find differentially expressed protein peaks. After specific marker proteins were identified, a diagnostic model based on their profiles was built to verify the clinical value of this proteomic approach, and the model’s performance was compared with that of the conventional tumor markers CEA and CA-199. Thus, we aimed to utilize a new strategy for the diagnosis of CRC and to provide evidence that this approach yields high-quality and efficient diagnostic models for clinical use.
Signed informed consent forms were obtained, and this study was approved by the Ethics Committee of the First Center of Chinese PLA General Hospital. Serum specimens in our study were all taken from the First Center of Chinese PLA General Hospital. This study included patients with precancerous lesions and colorectal tumors. The total number of specimens was 246, including samples from 83 healthy people, 62 CRP patients, and 101 CRC cancer patients. The inclusion criteria of the control group were no obvious organic lesions after physical examination, no diseases involved in this study and no other major clinically diagnosed diseases. CRC and CRP patients were diagnosed by pathologists after surgical resection of tumor tissue and endoscopic biopsy. The exclusion criteria were patients lacking confirmation by medical examination, patients undergoing chemotherapy or current acute infection[13], and sample coagulation for more than 12 h. Table 1 shows the clinical characteristics of the participants. After the whole blood samples were collected, they were centrifuged at 3500 r/min for 7 min at room temperature and immediately stored at -80 °C.
| Group | n | Ratio of gender (Male/female) | mean age |
| CRP | 62 | 1.3:1 | 58.3 |
| CRC | 101 | 1.1:1 | 59.7 |
| HC | 83 | 1:1 | 51.1 |
Blood was collected from the study subjects in the morning after fasting, and the blood was anticoagulated with EDTA and centrifuged with a clinical centrifuge at 3500 r/min for 7 min within 30 min. The serum was aliquoted at 50 μL per tube and stored in a -80 °C freezer according to the universal method. Sample freezing and thawing for more than 2 times was avoided. For the analysis of the serum samples, a 1:50 dilution of the sample in assay buffer was performed.
A maximum of 25 μL of serum or plasma was required per well. The samples used were stored in polypropylene tubes, and sample storage in glass tubes was avoided. The processing of samples with obvious hemolysis or lipemia was also avoided. It should be noted that heparin concentrations > 10 IU/mL in blood were not used as an anticoagulant because excessive heparin leads to an abnormal increase in measured values. Repeated freezing and thawing of serum samples can easily cause peptide precipitation, which will result in the loss of some peptides in the peptide MALDI-TOF MS spectrum. For this reason, repeated freezing and thawing was avoided.
The magnetic bead kit was removed from the 4 °C refrigerator, a tube of weak cation magnetic bead suspension was removed, and the tube was shaken up and down manually for 1 min to suspend the magnetic beads completely and uniformly in the liquid phase. Then, 5 μL of SPE-CM magnetic bead suspension was pipetted into a 200-μL sample tube, and 10 μL of magnetic beads was added to the sample tube and mixed by pipetting up and down to avoid foaming. Next, 5 μL of serum was added to the sample tube and mixed by pipetting up and down at least 5 times with the pipette to avoid foaming. The mixture was incubated at room temperature for 5 min, and then the sample tube was placed into the magnetic bead separator. After the magnetic beads adhered to the wall of the separator for 1 min, the magnetic beads were separated from the suspended liquid, and the color of the separated liquid was confirmed to be clear. The suspended liquid was absorbed with the sample addition gun. Care was taken to avoid the pipette tip touching the magnetic beads to prevent the magnetic beads from being aspirated. Subsequently, 100 μL of Magnetic Bead Wash Buffer was added to the sample tube. The sample tube was moved 10 times between two adjacent openings before and after being placed into the magnetic bead separator. The sample tube was then placed on the magnetic bead separator so that the magnetic beads adhered to the wall, and then the suspended liquid was absorbed with the sample addition gun. During this step, touching the magnetic beads with the pipette tip was avoided to prevent the magnetic beads from being aspirated. The suspended liquid was then completely aspirated during a final pipetting step. The sample tube was removed from the magnetic bead separator, 5 μL of magnetic bead elution buffer was added to the sample tube, and the process was repeated 10 times while avoiding foaming. The sample tube was placed into the magnetic bead separator, and the magnetic beads adhered to the wall for 2 min. After the magnetic beads were completely separated from the suspended liquid, the supernatant was transferred to a clean 0.5-mL sample tube. Five microliters of magnetic bead stabilization buffer were added to a 0.5-mL sample tube, carefully pipetted, and mixed with the sample pipette. Then, the sample was collected in a 0.5-mL tube, and the eluate with magnetic bead stabilization buffer was used immediately for mass spectrometry analysis or frozen at -20 °C for analysis within 24 h.
α-Cyano-4-hydroxycinnamic acid (1 g/L) was prepared and dissolved in acetone. The final matrix solution of 0.3 g/L, with ethanol/acetone = 2:1, was prepared. Fresh matrix solution was prepared on the same day of analysis. At room temperature, Peptide Calibration Standard (#206195) was dissolved in 125 μL of 0.1% TFA water, mixed for 1 min, and allowed to stand for 5 min. Protein Calibration Standard I (#206355) was dissolved in 125 μL of 0.1% TFA water, mixed for 1 min, and allowed to stand for 5 min. A total of 77 mg of ammonium acetate was dissolved in 100 mL of Milli-Q water to prepare 10 mmol/L ammonium acetate. A solution of 5 μL (25 μL) of Peptide Calibration Standard, 25 μL (125 μL) of Protein Calibration Standard I, and 20 μl (100 μL) of 10 mmol/L ammonium acetate were mixed well for 1 min. Solutions were aliquoted at 5 μL and stored at -20 °C for several weeks. For the list of standard products, see Table 2.
| Substance | Average mass (M + H)+ | Resolution | StDeV |
| Angiotensin II | 1047.18 | 360 | 10 |
| Angiotensin I | 1297.48 | 365 | 10 |
| Substance P | 1348.64 | 380 | 17 |
| Bombesin | 1620.86 | 420 | 23 |
| ACTH clip 1-17 | 2094.42 | 475 | 22 |
| ACTH clip 18-39 | 2466.68 | 540 | 17 |
| Somatostatin 28 | 3149.57 | 580 | 25 |
| Ubiquitin | 4283.45 | 800 | 40 |
| Insulin | 5734.56 | 580 | 25 |
| Cytochrome c | 6181.05 | 560 | 85 |
| Ubiquitin | 8565.89 | 345 | 25 |
The target surface was first rinsed with hot water. The target surface was then cleaned with dust-free paper and acetone, followed by Milli-Q water, and then with methanol and dried at room temperature. A tube of aliquoted standard and several aliquoted samples were thawed at room temperature. One microliter of standard was then mixed with 10 μL of matrix. One microliter of this solution was placed on a region of the 600-μm AnchorChip standard and dried at room temperature for several min. One microliter of the magnetic bead-treated specimen was mixed with 10 μL of matrix. The magnetic bead eluent was placed onto the AnchorChip sample position. The collection range and adjustment of laser energy were essentially identical across the collection of standard products. The same crystallization point of each sample was collected at 8 points and accumulated to 500 shots. Then, the accumulated maps were saved to a specified folder and labelled.
The acquisition range was 1-13 kDa. The laser energy was determined according to the laboratory mass spectrometer. High laser energy can be used to bombard the sample to the crystallization point, and then the energy spectrum that is 10%-20% lower than the high laser energy can be used to collect the spectrum. Data taken at the crystallization target point and different points were used for multipoint analysis, which resulted in a total of 8 crystallization point data values used to obtain the accumulated spectrum, and the average molecular weight deviation of the standard product was less than 100 ppm. The data were statistically processed by ClinProTools software[14,15]. When comparing the protein peak intensities between the two groups, P < 0.05 was considered statistically significant.
Spectra of CRC patient and healthy control (HC) samples were imported into ClinProTools software for processing and generation of serum differential protein profiles. Processes such as normalization, baseline extraction, peak definition, recalibration, and comparison of multiplex spectra were automated. The ClinProTools software in MALDI-TOF-MS was used to analyze the serum protein profiles of the CRC group and the HC group. According to the changes in the peak intensities in the two groups of data, a T test was used to calculate the P value. Nineteen of these differentially expressed protein peaks formed the basis of the auxiliary diagnostic protein profiles (Table 3). Compared with the HC group, CRC patients had elevated expression of 5 proteins with mass-to-charge ratios of 2022.34, 1866.09, 2899.72, 1778.9, and 1897.01 and reduced expression of 14 proteins with mass-to-charge ratios of 4210.57, 2932.56, 3192.08, 3883.65, 877.4, 7772.42, 3158.36, 2272.09, 4645.79, 4092.12, 4268.05, 2952.92, 3262.98, and 2660.37.
| Mass (m/z) | DAve | PTTA | PWKW | PAD | Ave1 | Ave2 |
| 4210.57 | 536.3 | 0.00000308 | 0.00000149 | 0.066 | 700.27 | 1236.57 |
| 2932.56 | 22.98 | 0.00000635 | < 0.000001 | 0.00721 | 28.61 | 51.6 |
| 2022.34 | 282.69 | 0.0000161 | < 0.000001 | < 0.000001 | 309.73 | 27.04 |
| 3192.08 | 26.53 | 0.0000333 | 0.0000105 | 0.000326 | 30.96 | 57.49 |
| 3883.65 | 31.52 | 0.000476 | 0.000129 | 0.00122 | 46.6 | 78.12 |
| 877.4 | 3.23 | 0.000645 | 0.000129 | 0.000683 | 5.93 | 9.16 |
| 1866.09 | 62.56 | 0.000826 | 0.000407 | < 0.000001 | 83.77 | 21.21 |
| 2899.72 | 13.03 | 0.000997 | 0.000189 | < 0.000001 | 38.31 | 25.28 |
| 7772.42 | 338.93 | 0.00141 | 0.00147 | 0.0503 | 594.88 | 933.81 |
| 3158.36 | 53.74 | 0.00284 | 0.000264 | < 0.000001 | 62.18 | 115.92 |
| 2272.09 | 20.34 | 0.00284 | 0.0000451 | < 0.000001 | 22.18 | 42.51 |
| 4645.79 | 22.93 | 0.00432 | 0.00483 | 0.231 | 57.18 | 80.11 |
| 4092.12 | 22.14 | 0.00577 | 0.0321 | 0.324 | 113.17 | 135.31 |
| 1778.9 | 7.03 | 0.00739 | 0.00672 | < 0.000001 | 18.66 | 11.64 |
| 4268.05 | 27.62 | 0.00795 | 0.000333 | < 0.000001 | 69.05 | 96.67 |
| 2952.92 | 27.19 | 0.0104 | 0.0265 | 0.232 | 70.78 | 97.97 |
| 3262.98 | 50 | 0.0116 | 0.00564 | < 0.000001 | 69.12 | 119.11 |
| 1897.01 | 14.98 | 0.0183 | 0.0122 | < 0.000001 | 41.59 | 26.61 |
| 2660.37 | 35.86 | 0.0197 | 0.0367 | 0.286 | 115.58 | 151.44 |
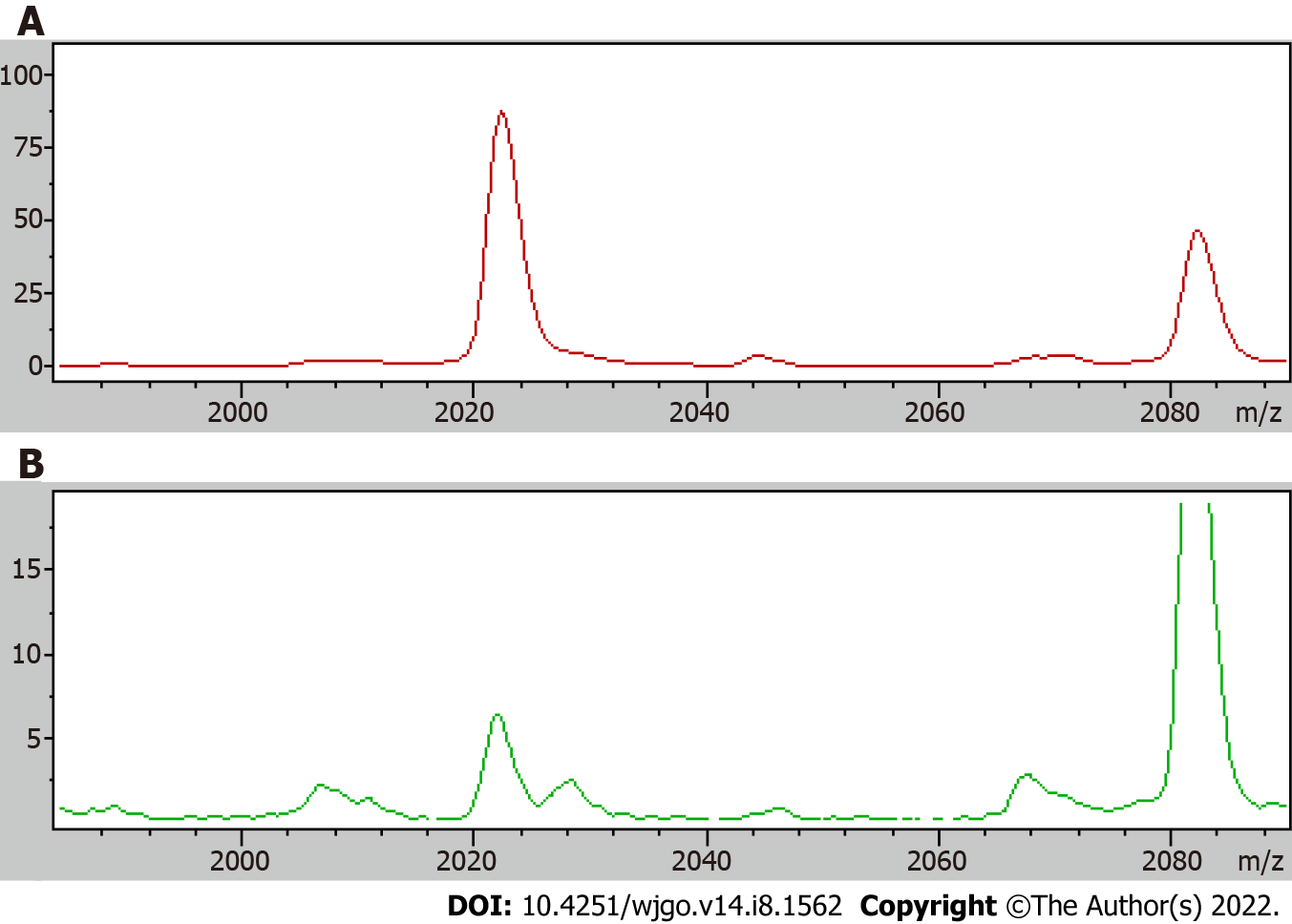
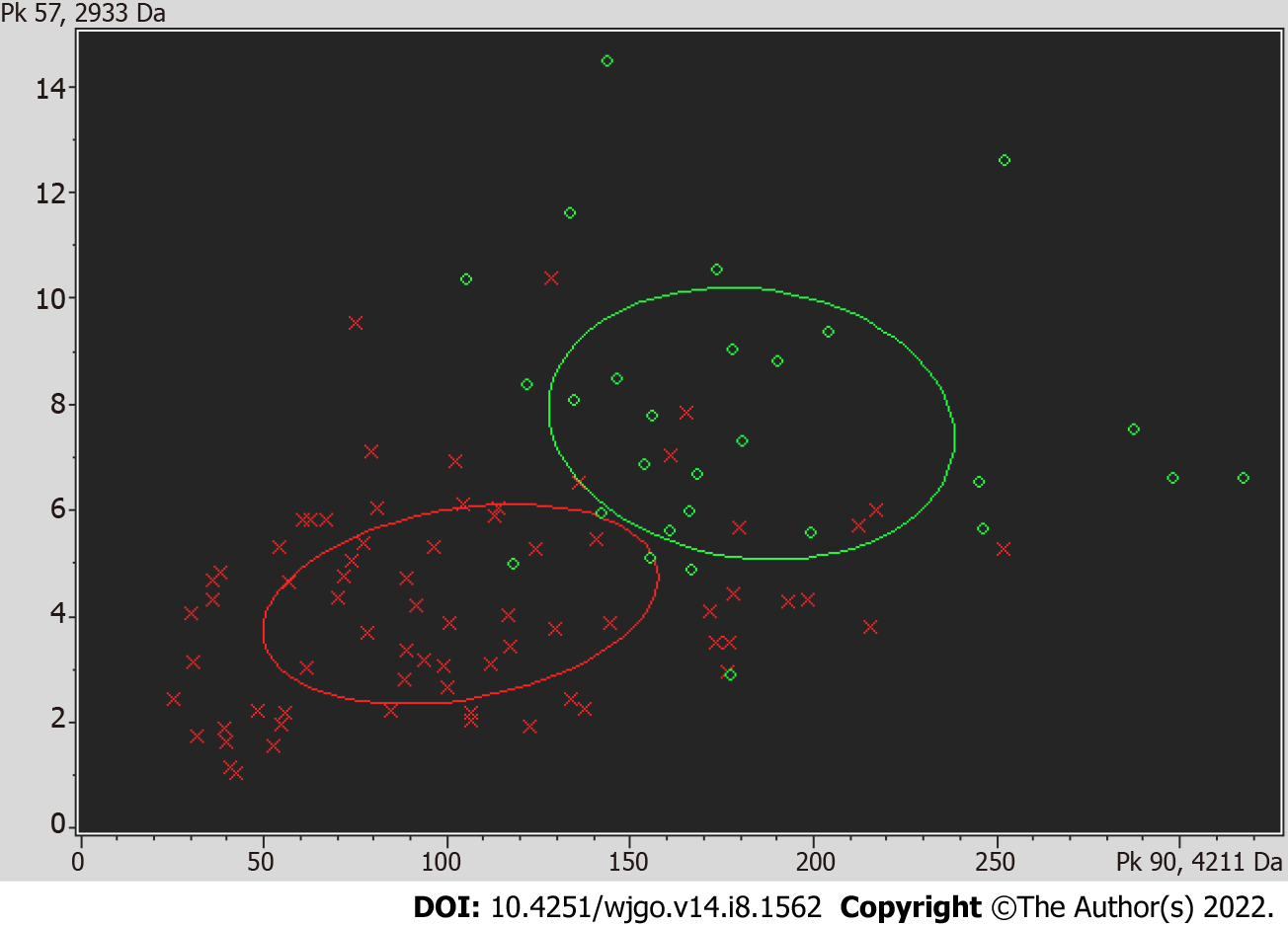
A receiver operating curve was drawn for the 19 differentially expressed proteins, and their respective area under the curves (AUCs) are shown in Table 4. The AUC of the differentially expressed protein P5 (mass-to-charge ratio of 2022.34) was the largest; the AUC was 0.843, the sensitivity was 75.3%, and the specificity was 79.5%, and thus, all of these metrics indicated high performance of protein P5-based diagnosis. The AUC obtained using the profile of the marker possessing a mass-to-charge ratio of 2022.34 was higher than the AUC of CEA-based diagnosis, which was 0.717. The comparison of the mean expression levels of the differentially expressed protein with a mass-to-charge ratio of 2022.34 in the serum of colon cancer patients and the serum of healthy groups is shown in Figure 1. Using the difference peaks obtained by the T test in ClinProTools software, the built-in Genetic Algorithm was used to calculate the cross-validation rate and identification ability. The diagnostic value of the two differentially expressed proteins possessing molecular weights of 4210.57 Da and 2932.56 Da was analyzed, as shown in Figure 2, and the cross-validation accuracy was 82.37%.
| DPP | m/z | AUC |
| P1 | 877.4 | 0.714 |
| P2 | 1778.9 | 0.595 |
| P3 | 1866.09 | 0.687 |
| P4 | 1897.01 | 0.640 |
| P5 | 2022.34 | 0.843 |
| P6 | 2272.09 | 0.705 |
| P7 | 2899.72 | 0.746 |
| P8 | 2932.56 | 0.783 |
| P9 | 2952.92 | 0.589 |
| P10 | 3158.36 | 0.759 |
| P11 | 3192.08 | 0.811 |
| P12 | 3262.98 | 0.751 |
| P13 | 3883.65 | 0.642 |
| P14 | 4092.12 | 0.655 |
| P15 | 4210.57 | 0.805 |
| P16 | 4268.05 | 0.744 |
| P17 | 4645.79 | 0.617 |
| P18 | 7772.42 | 0.714 |
| P19 | 2660.37 | 0.666 |
According to the change in the peak intensity across the two groups of data, the P value was calculated by the T test, and 14 differentially expressed protein profiles were obtained (Table 5). Compared with the CRP group, in the serum of CRC patients, 3 proteins were highly expressed, with mass-to-charge ratios of 2863.23, 2022.52, and 2899.88 m/z. Eleven proteins were expressed at reduced levels, with mass-to-charge ratios of 861.19, 4475.16, 845.16, 4210.83, 866.41, 1072.04, 2106.07, 4645.62, 2953.55, 2661.25, and 877.3.
| Mass | DAve | PTTA | PWKW | PAD | Ave1 | Ave2 |
| 861.19 | 33.51 | 0.000077 | 0.00000468 | < 0.000001 | 36.09 | 69.6 |
| 4475.16 | 87.57 | 0.000186 | 0.00000468 | 0.00000185 | 96.92 | 184.49 |
| 2863.23 | 37.14 | 0.000332 | 0.00073 | < 0.000001 | 88.71 | 51.57 |
| 845.16 | 4.89 | 0.000376 | 0.0000351 | 0.00000115 | 7.69 | 12.59 |
| 4210.83 | 278.83 | 0.000672 | 0.00073 | 0.135 | 702.87 | 981.7 |
| 2022.52 | 251.03 | 0.000672 | 0.0167 | < 0.000001 | 353.51 | 102.48 |
| 866.41 | 9.71 | 0.00209 | 0.000277 | < 0.000001 | 15.5 | 25.21 |
| 2899.88 | 12.32 | 0.00232 | 0.0000748 | < 0.000001 | 38.41 | 26.09 |
| 1072.04 | 5.17 | 0.0032 | 0.000277 | < 0.000001 | 8.66 | 13.83 |
| 2106.07 | 12.84 | 0.00374 | 0.0000351 | < 0.000001 | 35.74 | 48.59 |
| 4645.62 | 24.65 | 0.00634 | 0.00872 | 0.0751 | 65.03 | 89.68 |
| 2953.55 | 28.69 | 0.0112 | 0.0167 | 0.114 | 77.95 | 106.64 |
| 2661.25 | 41.77 | 0.0134 | 0.0137 | 0.0322 | 121.96 | 163.73 |
| 877.3 | 6.11 | 0.0278 | 0.0000078 | < 0.000001 | 5.81 | 11.92 |
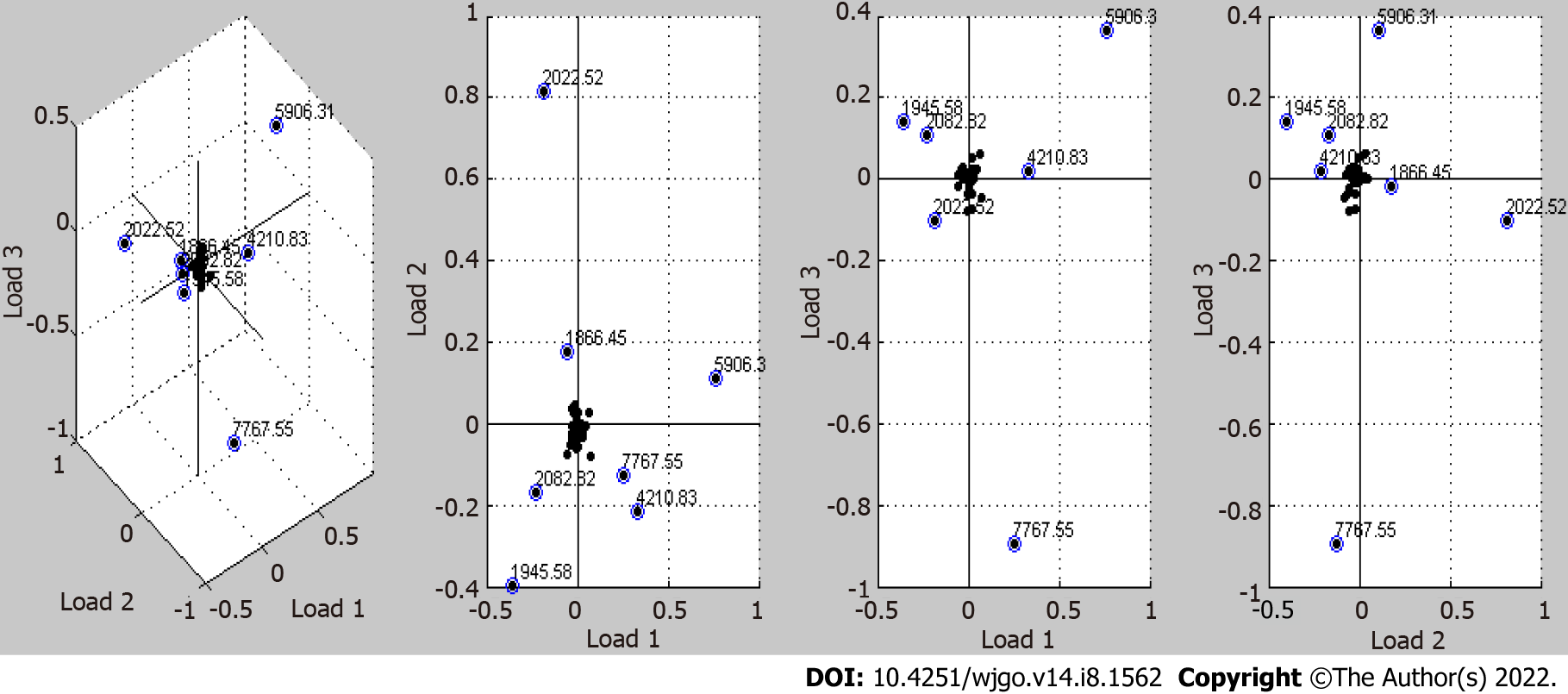
The peaks of differentially expressed proteins obtained by the T test in ClinProTools software were analyzed with the principal component analysis algorithm built in the software. As shown in Figure 3, 7 protein profile peaks (1866.45, 1945.58, 2022.52, 2082.82, 4210.83, 5906.31, and 7767.55 m/z) with relatively large relative dispersion are shown in the first three loading (Loading 1, Loading 2, and Loading 3) models. The total contribution of each peak is the cumulative sum of the three loading values of the peak multiplied by the contribution rate of the main factor. Other peaks that are located near the main axis with loading values close to 0 were ignored. The coordinate values and total contribution values of the seven peaks are shown in Table 6.
| m/z | 1866.45 | 1945.58 | 2022.52 | 2082.82 | 4210.83 | 5906.31 | 7767.55 |
| Loading 1 | -0.05 | -0.35 | -0.2 | -0.225 | 0.32 | 0.76 | 0.25 |
| Loading 2 | 0.17 | -0.4 | 0.81 | -0.17 | -0.22 | 0.11 | -0.12 |
| Loading 3 | -0.015 | 0.14 | -0.1 | 0.11 | 0.02 | 0.36 | -0.9 |
| TC | 5.45 | 25.93 | 24.36 | 15.52 | 20.08 | 44.84 | 23.07 |
The spectra of 14 differentially expressed proteins were analyzed by CLINPROT software in MALDI-TOF-MS, and a classification tree model was established. Complete random classification of the 163 collected specimens (62 colon polyp specimens, 101 colon cancer specimens) into the training group or the test group was performed. The numbers of CRC patients and CRP patients in the test and validation groups were 57 and 38 and 44 and 24, respectively. A classification tree diagnostic model was established using the 95 samples in the training group. Through this approach, it was revealed that the differentially expressed proteins with mass-to-charge ratios of 2899.38 and 877.3 were automatically selected to establish a classification tree model. The classification tree model established by these two differentially expressed proteins was then used to classify patient diagnosis. The results showed that 6 of 57 patients with CRC were missed, resulting in an accuracy of 89.5%, and 7 of 38 patients with CRP were misdiagnosed with CRC, resulting in an accuracy of 81.6% and an overall accuracy of 86.3%.
The established model was validated with the remaining samples (44 CRC and 24 CRP specimens). The levels of the differentially expressed proteins with mass-to-charge ratios of 2899.38 and 877.3 were used to establish a classification tree model, and the levels of CEA and CA-199 in the same samples (44 CRC specimens and 44 CRP specimens) were also measured. The accuracy of the proteomics-based diagnostic model was evaluated according to the sensitivity and specificity of the validation results, and these values were compared with the sensitivities and specificities of diagnosis with CEA and CA-199 Levels. The sensitivity and specificity of diagnosis using the classification tree were 81.8% and 66.75, respectively. The sensitivity and specificity values for diagnosis using CEA and CA-199 Levels were 55.6% and 91.3% and 65.4% and 65.2%, respectively, as shown in Table 7.
| Index | Sensitivity (%) | Specificity (%) |
| Classification tree | 81.8 | 66.7 |
| CEA | 55.6 | 91.3 |
| CA-199 | 65.4 | 65.2 |
MALDI-TOF mass spectrometry has been widely used in the structural analysis and molecular weight determination of sugars, nucleic acids, proteins and other biological macromolecules and synthetic polymers and has become one of the core technologies in current proteomics research[16]. This technique has many advantages, such as a precise mass-to-charge ratio, low cost, large affinity surface, and good repeatability[9,17].
MALDI-TOF mass spectrometry was used to analyze the serum protein expression profile of patients with CRC to identify differentially expressed protein peaks. At the same time, the serum proteins of patients with CRC and CRP were analyzed. Using these biological data, specific protein markers were selected, and then a CRC classification tree diagnostic model was established. The model was used to predict diagnosis with the analyzed serum protein markers of the patients to verify its clinical value and to compare the result of this approach with that of diagnoses achieved with the conventional tumor markers CEA and CA-199. This approach may thus improve diagnostic evaluation of CRC. Using MALDI-TOF mass spectrometry data from colon cancer patient serum and healthy human serum, 19 protein profiles with significant differences across the two groups were obtained. Compared with the healthy control group, there were 5 protein peaks with increased expression and 14 protein peaks with reduced expression in the serum of CRC patients. The receiver operating curves of the 19 differentially expressed proteins were drawn, and it was found that the AUC of diagnosis obtained using data from a protein with a mass-to-charge ratio of 2022.34 was the largest at 0.843, and the sensitivity and specificity were 75.3% and 79.5%, respectively. Therefore, this differentially expressed protein exhibited high diagnostic value for CRC patients. Based on these metrics, the diagnostic value of this approach was higher than that of either CEA or CA-199 alone, and this approach could be further optimized.
To this end, we used MALDI-TOF mass spectrometry technology to analyze the peripheral blood protein auxiliary diagnostic spectrum of patients with colorectal disease to find the differentially expressed proteins with high diagnostic sensitivity and specificity to verify whether it can be used as a new diagnostic biomarker of CRC. Using MALDI-TOF mass spectrometry technology to analyze the differences in protein expression in the serum of patients with CRC and patients with CRP, 14 protein peaks with significant differences were obtained. Compared with the serum of patients with CRP, the serum of patients with CRC had elevated expression of 3 proteins and reduced expression of 11 proteins. Fourteen differential peak proteins were calculated using CLINPROT software in MALDI-TOF-MS, and a classification tree model was established. Differential proteins with mass-to-charge ratios of 2899.38 and 877.3 were automatically selected to build a classification tree model. Using this classification tree model to classify patients, the results showed that 6 of 57 colon cancer patients were missed with an accuracy of 89.5%, and 7 of 38 colon polyp patients were misdiagnosed with colon cancer for an accuracy of 81.6%. The overall accuracy rate was 86.3%. Compared with diagnoses made using CEA and CA-199 Levels, this proteomics model has high sensitivity and specificity and can more effectively distinguish CRC from CRP. The high sensitivity and specificity of MALDI-TOF-based diagnostics indicated that the combination of MALDI-TOF and bioinformatics could help to improve the early diagnosis of CRC and could be used as an auxiliary diagnostic method for the treatment and prognosis of patients.
MALDI-TOF mass spectrometry technology has been widely used in clinical tumor diagnosis, treatment, and prognosis monitoring and can be used as a powerful tumor marker discovery research tool[18]. In the next few years, with the development of genomics and proteomics, an increasing number of new markers with high diagnostic sensitivity and specificity will be discovered, identified, and applied in the clinic, which will significantly improve the clinical detection rate of cancer, allow for its early diagnosis and treatment[19], and provide new methods and ideas for the study of tumorigenesis mechanisms[20].
However, there are still some limitations to this study. First, since MALDI-TOF-MS is a relatively novel protein detection method for serum samples, a standard operation protocol was not determined. This limits the utility of this approach to clinical practice. Second, the diagnostic performance of specific protein profiles in our study was assessed, but we have not confirmed these specific protein profiles in our study. Third, the relatively small sample size may somewhat bias the results.
In conclusion, we demonstrated that serum proteomics may be helpful for the detection of CRC, and it may provide a potential tool for CRC clinical management. For discriminating HC and CRC subjects using this approach, the sensitivity was 75.3%, and the specificity was 79.5%. After cross-validation, the diagnostic accuracy was 82.37%. For discriminating CRP and CRC patients, the overall accuracy of the classification tree model based on the 2899.38 m/z and 877.3 m/z protein profiles was 86.3%. The overall sensitivity and specificity of this approach were 81.8% and 66.75%, respectively.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) plays an important role in tumor research and in clinical applications and provides new ideas and approaches for tumor prevention, early diagnosis, and individualized treatment.
The clinical use of MALDI-TOF-MS serum-based biomarkers for colorectal cancer detection should be investigated.
We aimed to evaluate the diagnostic value of serum proteomics for colorectal cancer detection.
Eighty-three healthy controls, 62 colon polyp patients and 101 colorectal cancer patients were enrolled, and their serum samples were analyzed by serum proteomics. The diagnostic value of differential protein profiles was evaluated and compared with that of conventional biomarkers.
The area under the curve resulting from a diagnostic based on the levels of a differentially expressed protein with a mass-to-charge ratio of 2022.34 for discriminating healthy controls and colorectal cancer patients was 0.843, while the sensitivity was 75.3% and the specificity was 79.5%. After cross-validation, the diagnostic accuracy of this approach was 82.37%. For classification of the colorectal polyp group, proteins with mass-to-charge ratios of 2899.38 and 877.3 were automatically selected to establish a classification tree model. The sensitivity and specificity were 81.8% and 66.75%, respectively. The sensitivities and specificities of diagnostics based on the levels of carcinoembryonic antigen and carbohydrate antigen 19-9 were 55.6% and 91.3% and 65.4% and 65.2%, respectively.
We have built an assistant diagnostic method for the detection of colorectal cancer based on serum proteomics. It may be helpful for colorectal cancer clinical management.
Studies with standard detection protocols and larger sample sizes should be performed in the future, and the protein profiles should also be confirmed.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Albillos A, Spain; Kalofonos HP, Greece; Scurtu RR, Romania S-Editor: Wang JL L-Editor: A P-Editor: Wang JL
| 1. | Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet. 2005;365:153-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 862] [Cited by in RCA: 946] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 2. | Ladabaum U, Dominitz JA, Kahi C, Schoen RE. Strategies for Colorectal Cancer Screening. Gastroenterology. 2020;158:418-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 408] [Article Influence: 81.6] [Reference Citation Analysis (0)] |
| 3. | Burnett-Hartman AN, Lee JK, Demb J, Gupta S. An Update on the Epidemiology, Molecular Characterization, Diagnosis, and Screening Strategies for Early-Onset Colorectal Cancer. Gastroenterology. 2021;160:1041-1049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 172] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 4. | Yiu AJ, Yiu CY. Biomarkers in Colorectal Cancer. Anticancer Res. 2016;36:1093-1102. [PubMed] |
| 5. | Ding D, Han S, Zhang H, He Y, Li Y. Predictive biomarkers of colorectal cancer. Comput Biol Chem. 2019;83:107106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 6. | Gan X, Wang T, Chen ZY, Zhang KH. Blood-derived molecular signatures as biomarker panels for the early detection of colorectal cancer. Mol Biol Rep. 2020;47:8159-8168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 7. | Li K, Pei Y, Wu Y, Guo Y, Cui W. Performance of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) in diagnosis of ovarian cancer: a systematic review and meta-analysis. J Ovarian Res. 2020;13:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Park HG, Jang KS, Park HM, Song WS, Jeong YY, Ahn DH, Kim SM, Yang YH, Kim YG. MALDI-TOF MS-based total serum protein fingerprinting for liver cancer diagnosis. Analyst. 2019;144:2231-2238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Sun J, Yu G, Yang Y, Qiao L, Xu B, Ding C, Liu Y, Yu S. Evaluation of prostate cancer based on MALDI-TOF MS fingerprinting of nanoparticle-treated serum proteins/peptides. Talanta. 2020;220:121331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Snyder CM, Alley WR Jr, Campos MI, Svoboda M, Goetz JA, Vasseur JA, Jacobson SC, Novotny MV. Complementary Glycomic Analyses of Sera Derived from Colorectal Cancer Patients by MALDI-TOF-MS and Microchip Electrophoresis. Anal Chem. 2016;88:9597-9605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Kirana C, Peng L, Miller R, Keating JP, Glenn C, Shi H, Jordan TW, Maddern GJ, Stubbs RS. Combination of laser microdissection, 2D-DIGE and MALDI-TOF MS to identify protein biomarkers to predict colorectal cancer spread. Clin Proteomics. 2019;16:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Del Prete E, Facchiano A, Profumo A, Angelini C, Romano P. GeenaR: A Web Tool for Reproducible MALDI-TOF Analysis. Front Genet. 2021;12:635814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 13. | Ni Y, Xie G, Jia W. Metabonomics of human colorectal cancer: new approaches for early diagnosis and biomarker discovery. J Proteome Res. 2014;13:3857-3870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 137] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 14. | Chen JH, She KK, Wong OY, Teng JL, Yam WC, Lau SK, Woo PC, Cheng VC, Yuen KY. Use of MALDI Biotyper plus ClinProTools mass spectra analysis for correct identification of Streptococcus pneumoniae and Streptococcus mitis/oralis. J Clin Pathol. 2015;68:652-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 15. | Kubo Y, Ueda O, Nagamitsu S, Yamanishi H, Nakamura A, Komatsu M. Novel strategy of rapid typing of Shiga toxin-producing Escherichia coli using MALDI Biotyper and ClinProTools analysis. J Infect Chemother. 2021;27:1137-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Rodrigo MA, Zitka O, Krizkova S, Moulick A, Adam V, Kizek R. MALDI-TOF MS as evolving cancer diagnostic tool: a review. J Pharm Biomed Anal. 2014;95:245-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 17. | Karpova MA, Moshkovskii SA, Toropygin IY, Archakov AI. Cancer-specific MALDI-TOF profiles of blood serum and plasma: biological meaning and perspectives. J Proteomics. 2010;73:537-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Cho YT, Su H, Wu WJ, Wu DC, Hou MF, Kuo CH, Shiea J. Biomarker Characterization by MALDI-TOF/MS. Adv Clin Chem. 2015;69:209-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 19. | Froehlich BC, Popp R, Sobsey CA, Ibrahim S, LeBlanc A, Mohammed Y, Buchanan M, Aguilar-Mahecha A, Pötz O, Chen MX, Spatz A, Basik M, Batist G, Zahedi RP, Borchers CH. A multiplexed, automated immuno-matrix assisted laser desorption/ionization mass spectrometry assay for simultaneous and precise quantitation of PTEN and p110α in cell lines and tumor tissues. Analyst. 2021;146:6566-6575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Petre G, Durand H, Pelletier L, Poulenard M, Nugue G, Ray PF, Rendu J, Coutton C, Berger F, Bidart M. Rapid Proteomic Profiling by MALDI-TOF Mass Spectrometry for Better Brain Tumor Classification. Proteomics Clin Appl. 2020;14:e1900116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |