Published online Nov 15, 2022. doi: 10.4251/wjgo.v14.i11.2195
Peer-review started: July 26, 2022
First decision: August 19, 2022
Revised: September 6, 2022
Accepted: October 2, 2022
Article in press: October 2, 2022
Published online: November 15, 2022
Processing time: 111 Days and 19.3 Hours
For optimizing fecal immunochemical test (FIT)-based screening programs, reducing the rate of missed colorectal cancers (CRCs) by FIT (FIT-interval CRCs) is an important aspect. Knowledge of the molecular make-up of these missed lesions could facilitate more accurate detection of all (precursor) lesions.
To compare the molecular make-up of FIT-interval CRCs to lesions that are detected by FIT [screen-detected CRCs (SD-CRCs)].
FIT-interval CRCs observed in a Dutch pilot-program of FIT-based screening were compared to a control group of SD-CRCs in a 1:2 ratio, resulting in 27 FIT-interval CRC and 54 SD-CRCs. Molecular analyses included microsatellite instability (MSI), CpG island methylator phenotype (CIMP), DNA sequence mutations and copy number alterations (CNAs).
Although no significant differences were reached, FIT-interval CRCs were more often CIMP positive and MSI positive (33% CIMP in FIT-interval CRCs vs 21% in SD-CRCs (P = 0.274); 19% MSI in FIT-interval CRCs vs 12% in SD-CRCs (P = 0.469)), and showed more often serrated pathway associated features such as BRAF (30% vs 12%, P = 0.090) and PTEN (15% vs 2.4%, P = 0.063) mutations. APC mutations, a classic feature of the adenoma-carcinoma-sequence, were more abundant in SD-CRCs (68% vs 40% in FIT-interval CRCs P = 0.035). Regarding CNAs differences between the two groups; FIT-interval CRCs less often showed gains at the regions 8p11.22-q24.3 (P = 0.009), and more often gains at 20p13-p12.1 (P = 0.039).
Serrated pathway associated molecular features seem to be more common in FIT-interval CRCs, while classic adenoma carcinoma pathway associated molecular features seem to be more common in SD-CRCs. This indicates that proximal serrated lesions may be overrepresented among FIT-interval CRCs.
Core Tip: Fecal immunochemical test (FIT) is effective in reducing colorectal cancer (CRC) but FIT testing is not perfect. FIT interval cancers, i.e. CRCs diagnosed after a negative FIT but before the next FIT invitation, still occur. Previous studies have shown that FIT sensitivity for sessile serrated lesions (SSLs) is low, but a correlation between the occurrence of FIT interval cancers and the serrated pathway has not been established. In our study, the serrated pathway-associated molecular features were more common in FIT-interval CRCs as compared to screen detected CRCs. This indicates that proximal serrated lesions may be overrepresented among FIT interval CRCs. The findings of this study can provide guidance on strategies to further improve stool-based CRC screening with incorporating biomarkers for SSLs, thereby reducing the number of screening interval CRCs.
- Citation: van der Vlugt M, Carvalho B, Fliers J, Montazeri N, Rausch C, Grobbee EJ, Engeland MV, Spaander MCW, Meijer GA, Dekker E. Missed colorectal cancers in a fecal immunochemical test-based screening program: Molecular profiling of interval carcinomas. World J Gastrointest Oncol 2022; 14(11): 2195-2207
- URL: https://www.wjgnet.com/1948-5204/full/v14/i11/2195.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i11.2195
Fecal immunochemical test (FIT)-based screening worldwide has had a major impact on reducing colorectal cancer (CRC)-related mortality. Despite this success, the issue of false negative tests giving rise to FIT interval CRCs (e.g., CRCs diagnosed after a negative FIT but before the next FIT invitation) leaves room for improvement[1]. Monitoring the incidence of FIT-interval CRCs is a key quality indicator of a FIT-based screening program, and any CRC that occurs in spite of recent screening can be regarded as an unwanted program outcome. The sensitivity of FIT for CRC is approximately 75%-85%, which indicates that still 1 in 4-5 CRCs will be missed in any single screening round[2-4]. Possible reasons for FIT-interval CRCs are the limited sensitivity of FIT for specific molecular types of CRCs that were already present at the time of FIT-screening, or rapid progression of premalignant lesions during the interval between two screening rounds. Both of these causes may be the consequence of differences in tumor biology between FIT-interval CRCs and screen-detected CRCs (SD-CRCs). Such biological differences may translate e.g., into a lower bleeding tendency of colorectal lesions or an increased progression rate.
CRC has several precursor lesions reflecting different tumor-biology. Adenomas are well-known precursors of CRC. Adenomas may follow the canonical adenoma-carcinoma sequence with APC and KRAS mutations and subsequent typical patterns of chromosomal copy number alterations (CNAs) as classic features to develop into CRC[5-7]. More recently, also sessile serrated lesions (SSLs) have been identified as precursors of CRC. SSLs may follow the serrated neoplasia pathway resulting in CRCs that are more often microsatellite instable (MSI), CpG island methylator phenotype (CIMP) high and harbor BRAF mutations[8,9]. As a consequence of a different tumor biology, colorectal lesions can have different morphology. Most SSLs are non-polypoid (flat or slightly elevated) lesions located in the proximal colon, whereas adenomas can be either non-polypoid or polypoid (e.g., stalked or sessile). Previous studies have shown that FIT sensitivity for SSLs is low[10]. Also, non-polypoid colorectal neoplasms (NP-CRNs) have a flat morphology and are associated with more aggressive biologic behavior when compared to polypoid precursor lesions[11]. These NP-CRNs are usually located in the proximal colon. As they are more challenging to detect during colonoscopy, these NP-CRNs are thought to be a major contributor to post-colonoscopy CRCs (PC-CRCs)[12]. In a study on molecular characterization, NP-CRNs were more often found to harbor 5q-loss and BRAF mutations and have less APC and KRAS mutations[11,12]. It was hypothesized that NP-CRNs bleed less intensely and/or not continuously, which could lead to a falsely negative FIT and FIT-interval CRCs.
Generating more insight in the molecular features of FIT-interval CRCs may help to optimize screening strategies, aiming to reduce their incidence. The aim of the present study therefore was to compare the molecular composition of FIT-interval CRCs to that of SD-CRCs.
From 2006 onwards, two cohort studies of biennial FIT-based CRC screening have been conducted in the southwest and northwest regions of the Netherlands. After three screening rounds, these two cohorts were combined in 2014 to conduct a fourth round of FIT screening. A threshold of 10 μg hemoglobin (Hb) per gram feces was used. Screenees with a fecal Hb concentration above this threshold were referred for colonoscopy. Colonoscopies were performed according to international quality guidelines[13]. Details about the design of this study have been reported previously[2,14,15]. After finishing the fourth screening round in 2015, the total cohort was linked to the Netherlands Cancer Registry, managed by the Netherlands Comprehensive Cancer Organization (Utrecht, The Netherlands), in order to identify CRC missed by FIT testing during the three completed screening rounds including 2 years of follow up.
All CRCs detected during colonoscopy after a positive FIT (threshold ≥ 10 μg Hb/g feces) were recorded and labeled as SD-CRC.
FIT-interval CRCs were defined as a CRC diagnosed after a negative FIT (threshold < 10 μg Hb/g feces) and before the date of the next invitation for FIT-screening[16]. If a participant had a negative FIT and was not invited for a consecutive round (for having passed the upper age limit or after moving out of the target area) but developed CRC within the 2.37 years interval (median time between invitations), this CRC was also defined as a FIT-interval CRC.
Persons who had been inconsistent in participating in FIT screening and developed CRC outside the screening interval (median 2.37 years) were not defined as an interval CRC and not included in this study. Data on tumor stage and location (at time of resection) were collected for both FIT-interval CRC and SD-CRCs. With regard to tumor location, the colon was divided into proximal (cecum, ascending, hepatic flexure, and transverse colon) and distal colon (splenic flexure, descending colon, sigmoid colon, and rectum). All cancers were staged according to the 7th edition of the American Joint Committee on Cancer[17]. In the three intervals between four screening rounds, including the 2.37 years follow up for individuals that had reached the upper age limit after any of the first three rounds, in total 27 FIT-interval CRCs were identified. Besides, a total of 116 SD-CRCs was detected in the four screening rounds. All 27 FIT-interval CRCs were included in the study, and a random sample of SD-CRCs in a 1:2 ratio to SD-CRCs, yielding a control group of 54 SD-CRCs.
All tissue samples of FIT-interval CRCs and SD-CRCs were collected from 11 departments of pathology through the Dutch National Pathology Registry[18]. DNA from formalin-fixed, paraffin-embedded material was isolated as previously described[19]. Good quality DNA could be obtained from 25 of 27 FIT-interval CRCs and 46 of 54 SD-CRCs (see Supplementary Figure 1).
CIMP status was analyzed in the Pathology Department at the University of Maastricht. The CIMP panel (CACNA1G, IGF2, NEUROG1, RUNX3 and SOCS1)[20] was determined by nested methylation-specific polymerase chain reaction (PCR) using sodium bisulfite modified genomic DNA (EZ DNA methylation kit (ZYMO research Co., Orange, CA, United States) as described before[21,22], and CIMP positive was defined when ≥ 3 of the 5 CIMP markers were methylated. In some samples DNA was no longer available, in other samples the analysis was performed but failed, leaving CIMP-analysis results available for 21 of 27 FIT-interval CRCs and 39 of 54 SD-CRCs (see Supplementary Figure 1).
MSI status analysis was performed using the multiplex marker PCR panel from Promega (MSI Multiplex System Version 1.2, Promega, Madison, WI, United States). When two or more markers were unstable, the sample was interpreted as MSI. All other samples were classified as microsatellite stable. In some samples insufficient DNA was left, while in others the results obtained did not meet the quality criteria, leaving results for MSI analysis available for 21 of 27 FIT-interval CRCs and 41 of 54 SD-CRCs (see Supplementary Figure 1).
For mutation analysis, targeted sequencing was performed. DNA libraries were prepared using the KAPA HyperPrep Kit (KAPA Biosystems, Wilmington, MA, United States) as described in the KAPA HyperPrep Kit protocol (KR0961-v5.16). Target enrichment was performed using a custom 48 gene xGen® Predesigned Gene Capture Pools (Integrated DNA Technologies, San Diego, CA, United States), as previously described[23]. In some samples, DNA was no longer available, and one sample was analyzed but sequencing was not sufficiently deep to draw conclusions for almost all genes, leaving mutation analysis results available for 20 of 27 FIT-interval CRCs and 42 of 54 SD-CRCs (see Supp
DNA CNAs were analyzed with low-coverage whole genome sequencing as described previously[23]. Briefly, DNA was fragmented by sonication (Covaris S2, Woburn, MA, United States) and run on the HiSeq 2500 (Illumina, San Diego, CA, United States) on a 65 basepairs single-read modus using the KAPA HyperPrepKit (KAPA Biosystems, KK8504, Wilmington, MA, United States). This yielded a coverage of 0.13x (IQR 0.12-0.14) genome coverage. To compare the frequencies of alterations in the two groups, the R-package CGHtest was used[25]. Good quality DNA copy number profiles were obtained for 19 of 27 FIT-interval CRCs and for 44 of 54 SD-CRCs (see Supplementary Figure 1). Raw DNA copy number data has been deposited in the EGA[24], with the study ID: EGAS00001004683.
Differences between groups were evaluated for statistical significance using the Chi square-test statistic, Fisher’s exact test statistic, linear-by-linear test or Mann-Whitney-U test where appropriate. Two-sided P values < 0.05 were considered to indicate statistically significant differences. Differences between proportions were reported as mean with 95%CI.
Ethics approval for performing FIT-based screening including linkage to the Netherlands Cancer Registry was provided by the Dutch National Health Council (WBO 2642467, 2832758, 3049078 and 161536-112008, The Hague, The Netherlands). No separate ethics approval was necessary for the additional molecular analysis, as judged by the scientific ethics board of the AMC University Hospital. Collection and use of tissue and patient data were performed in compliance with the ‘Code for Proper Secondary Use of Human Tissue in the Netherlands’ (www.federa.org). All authors had access to the study data and reviewed and approved the final manuscript.
It was not appropriate or possible to involve patients or the public in the design, or conduct, or reporting, or dissemination plans of our research.
Demographic and clinicopathological characteristics of the patients with FIT-interval CRCs and SD-CRCs are shown in Table 1. No significant differences were found between both groups. Almost 60% of all patients were men. Among the FIT-interval CRCs, 21 patients (77.8%) were estimated to have a “normal/average” socioeconomic status, compared to 33 patients (61.1%) in the SD-CRC group. In the group of FIT-interval CRCs, fecal Hb concentrations were undetectable (n = 12, 44%) or below the threshold (n = 12, 44%). For three screenees (11%), the precise level of Hb/g feces was not available. Supplementary Table 1 shows per patient and per CRC type (SD-CRC or FIT-interval CRC) how many rounds of FIT participation had been completed prior to the diagnosis of CRC.
| Characteristics | FIT-interval CRC | SD-CRC | P value |
| Patients, n | 27 | 54 | |
| Age at diagnosis, mean (min-max) | 65.9 yr (53-76) | 65.2 yr (50-75) | NS |
| Male sex, n (%) | 16 (59.3) | 33 (61.1) | NS |
| Socio-economic status, n (%) | NS | ||
| Low | 2 (7.4) | 10 (18.5) | |
| Normal | 21 (77.8) | 33 (61.1) | |
| High | 4 (14.8) | 11 (20.4) | |
| Tumor location, n (%) | |||
| Proximal | 10 (37) | 17 (31.5) | NS |
| Tumor stage, n (%) | |||
| Stage I | 8 (29.6) | 25 (46.3) | NS |
| Stage II | 6 (22.2) | 8 (14.8) | |
| Stage III | 9 (33.3) | 20 (37.0) | |
| Stage IV | 4 (14.8) | 1 (1.9) | |
| Time interval between FIT and CRC | |||
| Mean | 1.4 yr | ||
| Min-max | 0.4-2.3 yr | ||
| IQR | 0.9-2.0 yr | ||
| Mean Hb concentration last FIT in μg Hb/g feces, mean ± SD | 2.7 ± 3.4a | 167.8 ± 168.7 | < 0.001 |
MSI was more common among patients with FIT-interval CRCs compared to patients with SD-CRCs: 19% (4/21) vs 12% (5/41) [6.85% (95%CI: -12.7, 26.4); P = 0.469], respectively. Furthermore, CIMP was more prevalent in FIT-interval CRCs than in SD-CRCs, 33% (7/21) and 21% (8/39) [12.82% (95%CI: -11.36, 36.6); P = 0.274], respectively. However, differences for both variables did not reach statistical significance (Table 2).
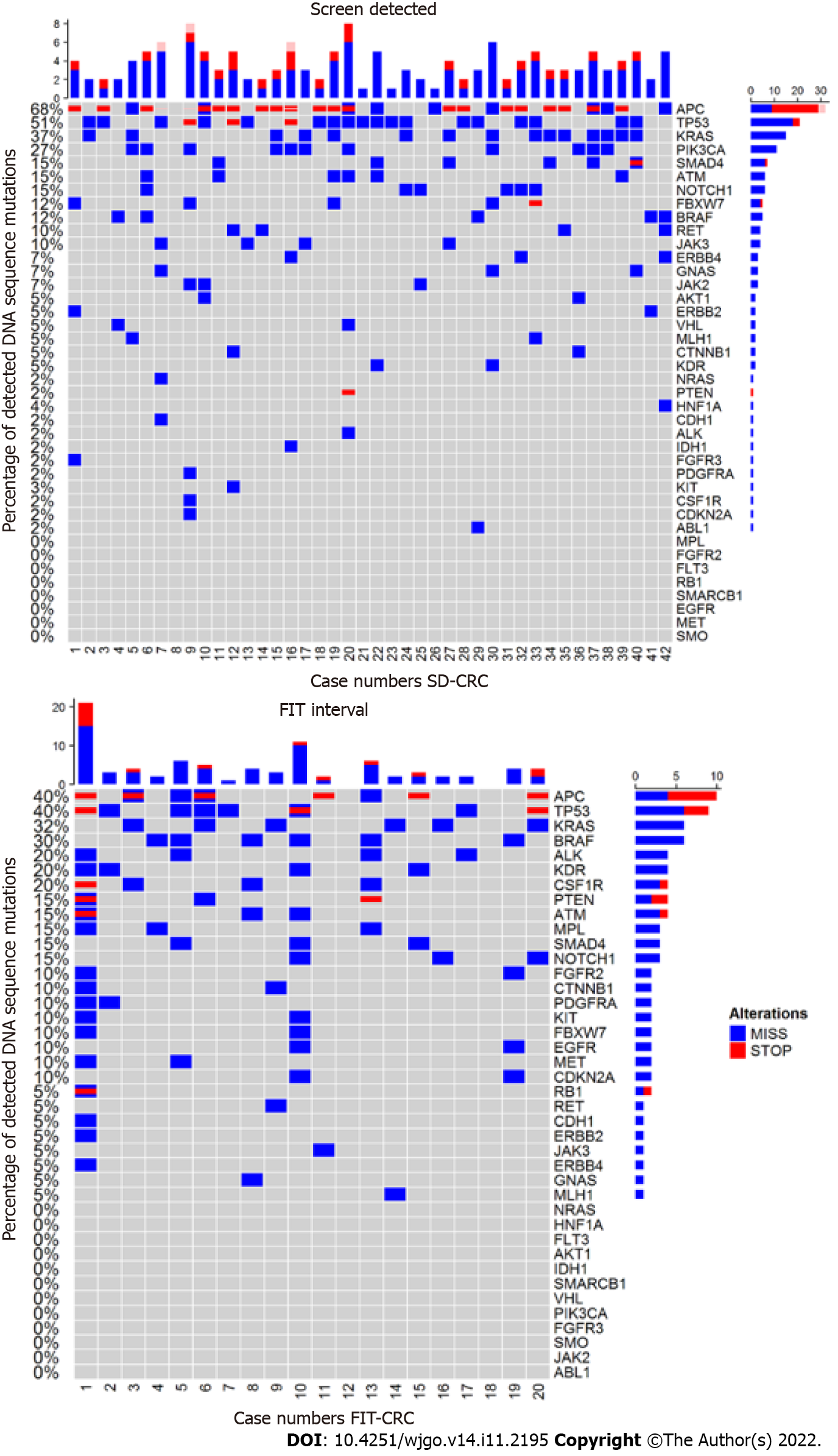
Results of the mutation analysis in FIT-interval CRCs and SD-CRCs are shown in Figure 1. Of the 48 genes tested, 37 genes were mutated in at least one sample (see Figure 1). No mutations were detected in FGFR1, FLT3, GNA11, GNAQ, HRAS, NPM1, PTPN11, SMARCB1, SMO, SRC and STK11.
APC and PIK3CA were more often mutated in SD-CRC compared to FIT-interval CRC [APC: 68% vs 40%, 28.29% (95%CI: 2.53, 54.1); P = 0.035; PIK3CA: 27% vs 0%, 26.83% (95%CI: 13.27, 40.39); P = 0.011]. KRAS was mutated in 37% of SD-CRC compared to 30% in FIT-interval CRC [5% (95%CI: -20.57, 30.58); P =0.705]. TP53 was mutated in 51% of SD-CRC compared to 40% in FIT-interval CRC [11.22% (95%CI: -15.14, 37.58); P = 0.410).
The following genes were significantly more often mutated in FIT-interval CRCs; ALK [20% vs 2.4%; 17.56% (95%CI: -0.59, 35.72); P = 0.019], CSF-1R [20% vs 2.4%; 17.56% (95%CI: -0.59, 35.72); P = 0.019], EGFR [10% vs 0%; 10% (95%CI: -3.15, 23.15); P = 0.037], FGFR2 [10% vs 0%; 10% (95%CI: -3.15, 23.15); P = 0.040], MET [10% vs 0%; 10% (95%CI: -3.15, 23.15); P = 0.040], MPL [15% vs 0%; 15% (95%CI: -0.65, 30.65); P = 0.010].
BRAF [30% vs 12%; 17.8% (95%CI: -4.64, 40.25); P = 0.090], KDR [20% vs 5%; 15.12% (95%CI: -3.61, 33.85); P = 0.063] and PTEN [15% vs 2.4%; 12.56% (95%CI: -3.78, 28.9); P = 0.063] mutations were more abundant in FIT-interval CRCs compared to SD-CRC but this difference did not reach statistical significance (see Figure 1).
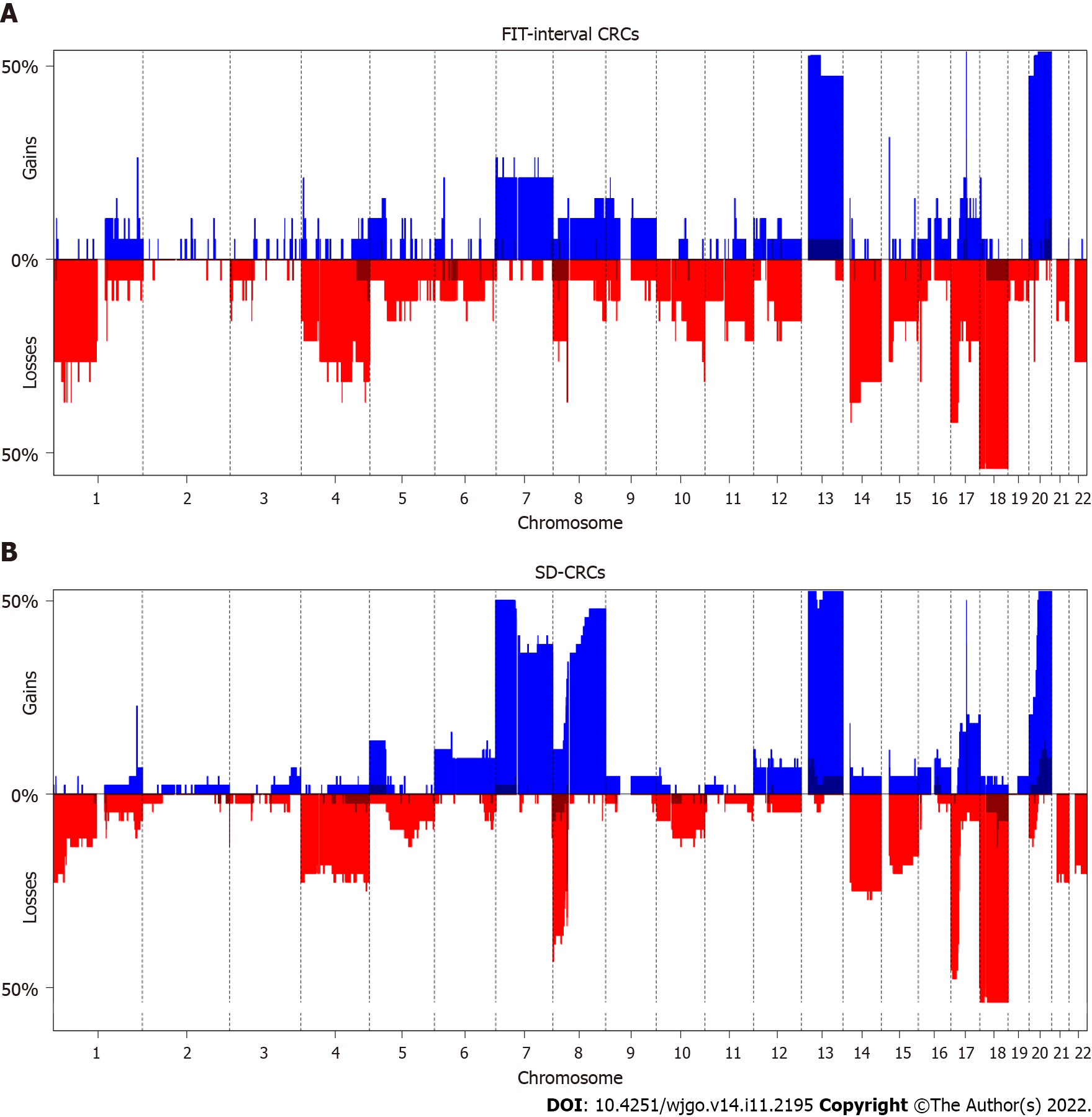
Figure 2 and Table 3 show the results of the DNA copy number analysis for FIT-interval CRC and SD-CRC. When comparing both groups, the only significantly different alterations were less frequent gains in FIT-interval CRCs at chromosome 8 region p11.22-q24.3 (P = 0.009) and more frequent gains in FIT-interval CRCs at chromosome 20 region p13-p12.1 (P = 0.039).
Of all interventions currently available, screening is one of the most powerful approaches for reducing CRC-related mortality[1]. Nevertheless, like all screening programs, CRC screening is facing the challenges of overdiagnosis and underdiagnosis. The latter mainly manifests as interval cancers. In the Dutch CRC screening program with a target population of 2.2M screenees, per screening round 544 interval cancers are observed, consistent with a FIT sensitivity of approximately 85%[26]. Theoretically, these FIT interval cancers consist of cancers that were present at the time the FIT was performed, but were missed, as well as cancer precursors missed at the time FIT was performed and that showed a rapid progression to a symptomatic cancer. On one hand sensitivity and specificity are simply determined by the cut off chosen, given the characteristics of the test. On the other hand, specific tumor characteristics driven by the underlying biology may differ between screened detected and interval cancers, a subject that so far has received little attention.
To address that question, the aim of the present study was to investigate the molecular characteristics between both categories. While we had access to a large well documented cohort of individuals followed over multiple screening rounds, the absolute number of interval cancers still was limited, which we consider to be the main reason why for most variables, differences were not statistically significant. Moreover, due to inherent formalin-fixed, paraffin-embedded associated artifacts, like DNA cross-links, the quality reads of some of the downstream analyses was poor and therefore some of the selected cases were further excluded from the final analysis (Supplementary Figure 1). Inherently these findings are exploratory in nature, yet they provide important indications on a major healthcare issue.
Indeed, the results of this exploratory study indicate that FIT-interval CRCs seem to more frequently carry the molecular features of the serrated neoplasia pathway and NP-CRNs than SD-CRCs do. FIT interval CRCs present more often CIMP high, MSI high and carry mutations like ALK, BRAF, CSF1R, EGFR, FGR2, PTEN, KDR, MET and MPL compared to SD-CRC. PTEN and KDR were previously described mutations detected in SSLs with high-grade dysplasia[27]. Furthermore, APC and KRAS mutations, which classically are part of the canonical adenoma-carcinoma sequence[5-7,27], were less frequently found in FIT-interval CRCs. Regarding DNA CNAs, FIT-interval CRCs showed very similar profiles to SD-CRCs, with only differences observed in the frequency of two genomic regions, namely, less often gains at 8p11.22-q24.3, and more often gains in FIT-interval CRCs at 20p13-p12.1. So, these results suggest that FIT-interval cancers are a mixed group of classical pathway and serrated pathway cancers, although with more commonly serrated pathway features and less commonly classical pathway features (both mutations and CNAs) in comparison with SD-CRCs. The overall pattern is striking, even if the individual variables do not reach statistical significance. Levin et al[28] also evaluated whether FIT-interval CRCs differed from FIT-positive patients with CRC by analyzing 7 KRAS mutations and 10 aberrantly methylated DNA biomarkers. They did not find any differences in their DNA profiles. In our study, however we investigated other genomic features and additional genes and did find differences as described above[29].
FIT is a good test to detect cancers. However, it does not perform so well in the detection of precursor lesions, in particular sessile serrated polyps. In a study that compared sensitivities of several FITs in a screening population, FIT sensitivity for SSLs of > 1 cm was only 5.1%[10]. One possible explanation might be the result of less bleeding tendency due to the low vessel density in serrated lesions in combination with their flat morphology and proximal location. Another explanation could be that serrated pathway lesions may show faster progression to cancer than classic adenomas. This indicates that in FIT-screening, a substantial number of serrated polyps will be missed, and therefore cancers derived from these precursor lesions might be overrepresented in FIT-interval CRCs.
NP-CRNs also show distinct molecular features compared to classical polypoid adenomas, are frequently located in the right colon and might bleed less[11,30,31]. In previous studies, NP-CRNs have been described as being less often APC and KRAS-mutated[11]. In a study comparing PC-CRCs and prevalent CRCs, KRAS mutation was inversely associated with PC-CRCs[32]. Comparably, FIT-interval CRCs were less often APC mutated in the present study (40% in FIT-interval CRCs vs 68% in SD-CRCs, P = 0.035). However, although KRAS was less often mutated in FIT-interval CRCs, the difference was not significant (30% in FIT-interval CRCs vs 37% in SD-CRCs, P = 0.705). In the previously mentioned study on PC-CRCs, BRAF-mutation was present in 28% of PC-CRCs vs 19% of prevalent CRCs (not significant)[32]. These percentages are comparable to our findings on FIT-interval CRCs (BRAF mutated in 30% of FIT-interval CRCs vs 12% of SD-CRCs, not significant). Previously, it has been shown that PC-CRCs were more likely CIMP positive and MSI than sporadic CRCs[33,34]. A separate study in a large PC-CRC cohort in the Netherlands also shows PC-CRCs to be more likely CIMP positive, MSI and BRAF mutated than prevalent CRCs[35]. This suggests that the molecular patterns observed in the interval cancers suggest that these cancers arose via non-polypoid precursors and/or serrated precursors. This may reflect that interval cancers, both FIT interval CRCs and PC-CRCs, indeed have a different biology compared to prevalent CRCs but at the same time pose technical challenges because of their morphology (difficult to be detected during colonoscopy) as well their lack of bleeding (difficult to be detected by FIT).
Some CNAs are associated to progression of adenoma to carcinoma chromosomal instability canonical pathway, and these are labeled as cancer associated events[19]. As of yet, no CNAs have been well characterized in the CRCs originating from SSLs. A study of serrated polyps, with data on a set of 38 serrated polyps (12 traditional serrated adenomas and 26 SSLs) found gains at chromosome 7, 13 and 15q[36]. The present study did not show any differences at these specific regions. However, FIT-interval CRCs had less frequent gains at chromosome 8q, and more frequent gains at chromosome 20p then SD-CRCs. As 8q is one of the genomic regions associated with the canonical adenoma-to-carcinoma progression, this finding could mean that to a certain extent, FIT-interval CRCs follow a different progression pathway.
As stated above, FIT-interval CRCs identified in multiple screening rounds represent a case-mix of cancers originated from, not only the difficult to detect NP-CRNs and sessile serrated polyps, but also classic adenomas. Precursor lesions could simply be missed by FIT just because of the low sensitivity of this test to detect advanced adenomas and not representing a different biology as reason for missing the lesion. Yet, while molecular differences were observed between FIT-interval CRCs and SD-CRCs, not all of these were statistically significant. Still these findings provide an indication that FIT-interval CRCs are a heterogeneous mixture of phenotypes and underlying molecular biology, including CRCs from flat serrated lesions, from flat adenomas, and others, that for whatever reason shed blood in a way that levels are, at least intermittently, below the limit of detection of the FIT test. In view of this, there is a clinical need to improve in screening tests for CRC early detection.
Multi-target molecular stool DNA (mt-sDNA) testing has recently been recognized as a valid CRC screening option by the American Cancer Society[37], and its test characteristics seem especially favorable for the detection of serrated lesions. In a large trial, mt-sDNA testing showed a higher detection rate of larger serrated sessile polyps than FIT (sensitivity of 42.4% for mt-sDNA-test and 5.1% for FIT, for serrated polyps > 1 cm)[10]. The combined sensitivity for advanced precancerous lesions was also higher for mt-sDNA testing than for FIT (42.4% vs 23.8%). However, although the sensitivities are better compared to FIT, the mt-sDNA test shows lower specificity, compared to FIT, which is very important in programmatic screening. Recently, a panel of protein markers detected in stool showed also a higher sensitivity for advanced adenomas without losing in specificity, in comparison to FIT. This protein-based approach would have the potential to improve effectivity of FIT screening without major impact for program logistics or cost effectivity[38]. Implementation of a more accurate test could have the potential to detect a substantial number of CRCs or precursor lesions that would otherwise result in FIT-interval CRCs.
Strengths of our study include that FIT-interval CRCs were identified over multiple rounds in a biennial FIT-based screening cohort, and tissue specimens of each tumor could be obtained. We were able to compare FIT-interval CRCs to a control group of SD-CRCs within the same screening population, region and time-span. However, an important limitation is the sample size. A total of 27 FIT-interval CRCs is still a limited number, reflecting the rarity of this entity, with inherent consequences for the statistical power of the study. To address this issue, a larger control group was composed through random selection in a 1:2 ratio. Due to budgetary and logistical constraints, we were not able to enlarge the control group. Also, we were not informed about the family history for cancer among the persons included in the study. Although rare, some of the FIT-interval CRCs might be related to familial CRC. So, although the findings of our study are suggestive for a difference in molecular make-up, further studies are needed to support our findings. We did not report tumor size and morphology of all CRCs, as this is not easy to determine in cancers (as compared to adenomas or SSLs). We were, therefore, not able to correlate size or morphology to the molecular analysis.
In conclusion, the present study provides evidence that SD-CRCs and FIT interval CRCs differ in the distribution of molecular tumor profiles. These findings can provide guidance on strategies for further improving stool-based CRC screening strategies. Future research should focus on the role of incorporating biomarkers in screening for identifying more CRCs during screening, thereby reducing the number of screening interval CRCs.
Fecal immunochemical test (FIT) is effective in reducing colorectal cancer (CRC) but FIT testing is not perfect. FIT interval cancers, i.e. CRCs diagnosed after a negative FIT but before the next FIT invitation, still occur. FIT sensitivity for CRC is approximately 75%-85% and sensitivity drops to low detection rates for adenomas and even more so for sessile serrated lesions (SSLs).
In order to lower the number of missed lesions, we need to understand which lesions are more often missed by FIT in order to improve the screening strategy. Previous studies have shown that FIT sensitivity for SSLs is low, but a correlation between the occurrence of FIT interval cancers and the serrated pathway has not been established.
Our aim was to generate more insight in the molecular features of FIT-interval CRCs, as this could help in developing a more optimal screening strategy with the aim to reduce the incidence of FIT interval cancers.
We compared the molecular make up of screen-detected CRCs (SD-CRCs) and FIT-interval CRCs, detected in a Dutch pilot-program of FIT-based screening. Molecular analyses included microsatellite instability (MSI), CpG island methylator phenotype (CIMP), DNA sequence mutations and copy number alterations.
FIT-interval CRCs were more often CIMP- and MSI-positive as compared to SD-CRCs. They also harbored more often BRAF and PTEN mutations as compared to SD-CRCs.
The serrated pathway-associated molecular features seem to be more common in FIT-interval CRCs as compared to screen detected CRCs. This might indicate that proximal serrated lesions are overrepresented among FIT interval CRCs. Further research needs to be performed. Adding molecular markers of the serrated-pathway to the FIT needs to be further explored.
These findings can provide guidance on strategies to further improve stool-based CRC screening with incorporating biomarkers for SSLs, thereby reducing the number of screening interval CRCs.
This work was performed within the framework of the COST Action (CA17118), supported by COST (European Cooperation in Science and Technology).
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Netherlands
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bustamante-Balen M, Spain; Gu GL, China S-Editor: Zhang H L-Editor: Filipodia P-Editor: Zhang H
| 1. | Malila N, Senore C, Armaroli P. International Agency for Research on C. European guidelines for quality assurance in colorectal cancer screening and diagnosis. First Edition--Organisation. Endoscopy. 2012;44:SE31-48. |
| 2. | van der Vlugt M, Grobbee EJ, Bossuyt PMM, Bos A, Bongers E, Spijker W, Kuipers EJ, Lansdorp-Vogelaar I, Spaander MCW, Dekker E. Interval Colorectal Cancer Incidence Among Subjects Undergoing Multiple Rounds of Fecal Immunochemical Testing. Gastroenterology. 2017;153:439-447.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 3. | de Wijkerslooth TR, Stoop EM, Bossuyt PM, Meijer GA, van Ballegooijen M, van Roon AH, Stegeman I, Kraaijenhagen RA, Fockens P, van Leerdam ME, Dekker E, Kuipers EJ. Immunochemical fecal occult blood testing is equally sensitive for proximal and distal advanced neoplasia. Am J Gastroenterol. 2012;107:1570-1578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 165] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 4. | van Winden O. Landelijke monitoring en evaluatie van het bevolkingsonderzoek naar darmkanker in Nederland 2014-2017. Accessed 3 March 2020. Available from: https://www.rivm.nl/sites/default/files/2019-03/monitor-evaluatie-darm-2014-2017.pdf. |
| 5. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4616] [Cited by in RCA: 4464] [Article Influence: 120.6] [Reference Citation Analysis (0)] |
| 6. | Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1202] [Cited by in RCA: 1191] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 7. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 8001] [Article Influence: 228.6] [Reference Citation Analysis (1)] |
| 8. | Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, Tanaka N, Higuchi T, Young J, Jass JR, Leggett BA. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 536] [Cited by in RCA: 566] [Article Influence: 27.0] [Reference Citation Analysis (1)] |
| 9. | O'Brien MJ, Yang S, Mack C, Xu H, Huang CS, Mulcahy E, Amorosino M, Farraye FA. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol. 2006;30:1491-1501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 381] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 10. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1233] [Article Influence: 112.1] [Reference Citation Analysis (1)] |
| 11. | Voorham QJ, Carvalho B, Spiertz AJ, van Grieken NC, Mongera S, Rondagh EJ, van de Wiel MA, Jordanova ES, Ylstra B, Kliment M, Grabsch H, Rembacken BJ, Arai T, de Bruïne AP, Sanduleanu S, Quirke P, Mulder CJ, van Engeland M, Meijer GA. Chromosome 5q loss in colorectal flat adenomas. Clin Cancer Res. 2012;18:4560-4569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 12. | le Clercq CM, Bouwens MW, Rondagh EJ, Bakker CM, Keulen ET, de Ridder RJ, Winkens B, Masclee AA, Sanduleanu S. Postcolonoscopy colorectal cancers are preventable: a population-based study. Gut. 2014;63:957-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 263] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 13. | Rembacken B, Hassan C, Riemann JF, Chilton A, Rutter M, Dumonceau JM, Omar M, Ponchon T. Quality in screening colonoscopy: position statement of the European Society of Gastrointestinal Endoscopy (ESGE). Endoscopy. 2012;44:957-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 232] [Article Influence: 17.8] [Reference Citation Analysis (1)] |
| 14. | Kapidzic A, Grobbee EJ, Hol L, van Roon AH, van Vuuren AJ, Spijker W, Izelaar K, van Ballegooijen M, Kuipers EJ, van Leerdam ME. Attendance and yield over three rounds of population-based fecal immunochemical test screening. Am J Gastroenterol. 2014;109:1257-1264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 15. | Stegeman I, van Doorn SC, Mundt MW, Mallant-Hent RC, Bongers E, Elferink MA, Fockens P, Stroobants AK, Bossuyt PM, Dekker E. Participation, yield, and interval carcinomas in three rounds of biennial FIT-based colorectal cancer screening. Cancer Epidemiol. 2015;39:388-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 16. | Sanduleanu S, le Clercq CM, Dekker E, Meijer GA, Rabeneck L, Rutter MD, Valori R, Young GP, Schoen RE; Expert Working Group on ‘Right-sided lesions and interval cancers’, Colorectal Cancer Screening Committee, World Endoscopy Organization. Definition and taxonomy of interval colorectal cancers: a proposal for standardising nomenclature. Gut. 2015;64:1257-1267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 147] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 17. | Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5537] [Cited by in RCA: 6455] [Article Influence: 430.3] [Reference Citation Analysis (0)] |
| 18. | Casparie M, Tiebosch AT, Burger G, Blauwgeers H, van de Pol A, van Krieken JH, Meijer GA. Pathology databanking and biobanking in The Netherlands, a central role for PALGA, the nationwide histopathology and cytopathology data network and archive. Cell Oncol. 2007;29:19-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 324] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 19. | Hermsen M, Postma C, Baak J, Weiss M, Rapallo A, Sciutto A, Roemen G, Arends JW, Williams R, Giaretti W, De Goeij A, Meijer G. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002;123:1109-1119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 233] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 20. | Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1423] [Cited by in RCA: 1494] [Article Influence: 78.6] [Reference Citation Analysis (0)] |
| 21. | Derks S, Postma C, Moerkerk PT, van den Bosch SM, Carvalho B, Hermsen MA, Giaretti W, Herman JG, Weijenberg MP, de Bruïne AP, Meijer GA, van Engeland M. Promoter methylation precedes chromosomal alterations in colorectal cancer development. Cell Oncol. 2006;28:247-257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 29] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 22. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821-9826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4183] [Cited by in RCA: 4247] [Article Influence: 146.4] [Reference Citation Analysis (0)] |
| 23. | van Lanschot MCJ, Carvalho B, Rausch C, Snaebjornsson P, van Engeland M, Kuipers EJ, Stoker J, Tutein Nolthenius CJ, Dekker E, Meijer GA. Molecular profiling of longitudinally observed small colorectal polyps: A cohort study. EBioMedicine. 2019;39:292-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Lappalainen I, Almeida-King J, Kumanduri V, Senf A, Spalding JD, Ur-Rehman S, Saunders G, Kandasamy J, Caccamo M, Leinonen R, Vaughan B, Laurent T, Rowland F, Marin-Garcia P, Barker J, Jokinen P, Torres AC, de Argila JR, Llobet OM, Medina I, Puy MS, Alberich M, de la Torre S, Navarro A, Paschall J, Flicek P. The European Genome-phenome Archive of human data consented for biomedical research. Nat Genet. 2015;47:692-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 259] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 25. | van de Wiel MA, Smeets SJ, Brakenhoff RH, Ylstra B. CGHMultiArray: exact P-values for multi-array comparative genomic hybridization data. Bioinformatics. 2005;21:3193-3194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Toes-Zoutendijk E, Kooyker AI, Dekker E, Spaander MCW, Opstal-van Winden AWJ, Ramakers C, Buskermolen M, van Vuuren AJ, Kuipers EJ, van Kemenade FJ, Velthuysen MF, Thomeer MGJ, van Veldhuizen H, van Ballegooijen M, Nagtegaal ID, de Koning HJ, van Leerdam ME, Lansdorp-Vogelaar I; Dutch National Colorectal Cancer Screening Working Group. Incidence of Interval Colorectal Cancer After Negative Results From First-Round Fecal Immunochemical Screening Tests, by Cutoff Value and Participant Sex and Age. Clin Gastroenterol Hepatol. 2020;18:1493-1500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 27. | Murakami T, Akazawa Y, Yatagai N, et al Molecular characterization of sessile serrated adenoma/polyps with dysplasia/carcinoma based on immunohistochemistry, next-generation sequencing, and microsatellite instability testing: a case series study. Diagn Pathol. 2018 Nov 20;13(1):88.. |
| 28. | Levin TR, Corley DA, Jensen CD, Marks AR, Zhao WK, Zebrowski AM, Quinn VP, Browne LW, Taylor WR, Ahlquist DA, Lidgard GP, Berger BM. Genetic Biomarker Prevalence Is Similar in Fecal Immunochemical Test Positive and Negative Colorectal Cancer Tissue. Dig Dis Sci. 2017;62:678-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 29. | Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6773] [Cited by in RCA: 6648] [Article Influence: 511.4] [Reference Citation Analysis (0)] |
| 30. | Nakama H, Zhang B, Kamijo N. Sensitivity of immunochemical fecal occult blood test for colorectal flat adenomas. Hepatogastroenterology. 2004;51:1333-1336. [PubMed] |
| 31. | van Doorn SC, Stegeman I, Stroobants AK, Mundt MW, de Wijkerslooth TR, Fockens P, Kuipers EJ, Bossuyt PM, Dekker E. Fecal immunochemical testing results and characteristics of colonic lesions. Endoscopy. 2015;47:1011-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | Shaukat A, Arain M, Anway R, Manaktala S, Pohlman L, Thyagarajan B. Is KRAS mutation associated with interval colorectal cancers? Dig Dis Sci. 2012;57:913-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 33. | Sawhney MS, Farrar WD, Gudiseva S, Nelson DB, Lederle FA, Rector TS, Bond JH. Microsatellite instability in interval colon cancers. Gastroenterology. 2006;131:1700-1705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 237] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 34. | Arain MA, Sawhney M, Sheikh S, Anway R, Thyagarajan B, Bond JH, Shaukat A. CIMP status of interval colon cancers: another piece to the puzzle. Am J Gastroenterol. 2010;105:1189-1195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 293] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 35. | Bogie RMM, le Clercq CMC, Voorham QJM, Cordes M, Sie D, Rausch C, van den Broek E, de Vries SDJ, van Grieken NCT, Riedl RG, Sastrowijoto P, Speel EJ, Vos R, Winkens B, van Engeland M, Ylstra B, Meijer GA, Masclee AAM, Carvalho B. Molecular pathways in post-colonoscopy versus detected colorectal cancers: results from a nested case-control study. Br J Cancer. 2022;126:865-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 36. | Gaiser T, Meinhardt S, Hirsch D, Killian JK, Gaedcke J, Jo P, Ponsa I, Miró R, Rüschoff J, Seitz G, Hu Y, Camps J, Ried T. Molecular patterns in the evolution of serrated lesion of the colorectum. Int J Cancer. 2013;132:1800-1810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 37. | Wolf AMD, Fontham ETH, Church TR, Flowers CR, Guerra CE, LaMonte SJ, Etzioni R, McKenna MT, Oeffinger KC, Shih YT, Walter LC, Andrews KS, Brawley OW, Brooks D, Fedewa SA, Manassaram-Baptiste D, Siegel RL, Wender RC, Smith RA. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin. 2018;68:250-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 945] [Cited by in RCA: 1309] [Article Influence: 187.0] [Reference Citation Analysis (0)] |
| 38. | Bosch LJW, de Wit M, Pham TV, Coupé VMH, Hiemstra AC, Piersma SR, Oudgenoeg G, Scheffer GL, Mongera S, Sive Droste JT, Oort FA, van Turenhout ST, Larbi IB, Louwagie J, van Criekinge W, van der Hulst RWM, Mulder CJJ, Carvalho B, Fijneman RJA, Jimenez CR, Meijer GA. Novel Stool-Based Protein Biomarkers for Improved Colorectal Cancer Screening: A Case-Control Study. Ann Intern Med. 2017;167:855-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (1)] |