Published online Nov 15, 2022. doi: 10.4251/wjgo.v14.i11.2183
Peer-review started: August 21, 2022
First decision: September 13, 2022
Revised: September 28, 2022
Accepted: October 27, 2022
Article in press: October 27, 2022
Published online: November 15, 2022
Processing time: 85 Days and 19.2 Hours
Gastric cancer (GC) is considered a major global health problem. The role of TRIM55, a member of the three-domain protein family, in GC is unknown.
To determine the expression of TRIM55 in GC tissues and its relationship with clinicopathological characteristics, and to investigate the effects of TRIM55 on the malignant biological behavior of GC cells.
Differential expression of TRIM55 in GC and para-cancer tissues was detected by immunohistochemistry, and the relationship between TRIM55 level and clini
TRIM55 expression was significantly increased in GC tissues compared with adjacent normal tissues. High expression of TRIM55 was associated with advanced pathological stage and poor prognosis. Overexpression of TRIM55 promoted invasion and metastasis of GC cells in vitro by regulating epithelial-mesenchymal transition (EMT), whereas knockdown of TRIM55 had the opposite effect. Our data showed that TRIM55 is highly expressed in GC tissues, and is associated with poor prognosis. TRIM55 plays the role of an oncogene in GC, and it promotes metastasis of GC through the regulation of EMT.
TRIM55 may be a possible target for the diagnosis and prognosis of GC patients.
Core Tip: TRIM55 expression was elevated in gastric cancer (GC) cancer tissues. Depletion of TRIM55 in GC cells suppressed proliferation, migration and invasion of cells. Knockdown of TRIM55 affected the expression of cell epithelial-mesenchymal transition-related proteins. TRIM55 may serve as an oncogene in GC.
- Citation: Li WW, Yuan H, Kong S, Tian SB. E3 ubiquitin ligase TRIM55 promotes metastasis of gastric cancer cells by mediating epithelial-mesenchymal transition. World J Gastrointest Oncol 2022; 14(11): 2183-2194
- URL: https://www.wjgnet.com/1948-5204/full/v14/i11/2183.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i11.2183
Gastric cancer (GC) is one of the most common tumors and seriously affects patients’ health[1]. Comprehensive treatment based on surgery is the preferred strategy for GC[2]. Postoperative metastasis is the leading cause of death from GC; however, the mechanisms underlying the occurrence and the development of GC have not been fully elucidated, and there is a lack of effective markers for early diagnosis[3]. Therefore, identifying molecular markers of GC is important for the early diagnosis, treatment, and prognostic evaluation of GC. The three-domain protein (TRIM) family is composed of many members, including the tripartite motif, which consists of a RING domain, 1 or 2 Box motifs, and a coiled-coil region[4]. Some TRIM proteins are involved in the regulation of cellular transcription, cell proliferation, and tumor development; thus, they play a role in either promoting or inhibiting cancer[5]. The structural diversity of TRIM family proteins underpins their functional diversity. TRIM55, also known as muscle-specific RING zinc finger protein 2, maintains muscle development and cardiac function. TRIM55 plays an important role in early skeletal muscle differentiation and the generation of muscle fibers[6]. Studies have shown that mir-30-5p can inhibit muscle cell differentiation and regulate the alternative splicing of TRIM55 by targeting Muscleblind-like Protein[7]. TRIM55 can regulate the TNF-α-CCL2 pathway and promote an inflammatory response in the development of mesangial proliferative glomerulonephritis[8]; however, the role of TRIM55 in GC has not been fully elucidated.
In this study, we determined the differential expression of TRIM55 in GC patients and investigated the relationship between TRIM55 and clinicopathological characteristics. We found that TRIM55 induced proliferation, migration, and invasion of GC cells. We also investigated the mechanism underlying these effects.
Tissues were obtained from 91 GC patients admitted to the Department of Gastrointestinal Surgery of Shandong Provincial Hospital Affiliated to Shandong First Medical University between July 2014 and December 2015. The tumor tissue samples and adjacent normal gastric mucosal tissue samples were validated by pathologists. Of the 91 patients, 61 were male and 30 were female, with an average age of 63.6 years. Data such as gender and age of the patient, tumor location, tumor pathological stage, and lymph node metastasis, were also collected. In addition, five fresh GC tissue samples and matched adjacent normal gastric tissues were obtained from our hospital. The research protocols were approved by the Ethics Committee of the Provincial Hospital Affiliated to Shandong First Medical University, and all the patients signed an informed consent form before surgery.
Human GC cell lines (AGS, MKN28, MGC803, SGC7901, HGC27, and MKN45) and the immortalized gastric mucosa cell line (GES-1) were provided by the Cell Center of the Chinese Academy of Medical Sciences. All the cell lines were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS, Gibco, United States). Cells were cultured in an incubator at 37 °C and 5% CO2. According to the CDS coding sequence of TRIM55, small interfering RNAs (siRNA) to knock down TRIM55 were designed and synthesized by RiboBio (Guangzhou, China), and the scramble nonsense sequence was used as the negative control (si-control). The targeting TRIM55 siRNA sequence was as follows: TRIM55 siRNA, 5’-AUCAAACUUCUCACAAAGCUC-3’. The control siRNA was not homologous to any human genome sequence. For the overexpression assay, the coding sequence of TRIM55 was amplified and cloned into a pcDNA3.1-HA vector to construct the TRIM55 overexpression system (TRIM55 plasmid). The pcDNA3.1-HA empty vector was used as the negative control in HGC27 cells.
The GC tissue and adjacent tissue were embedded in paraffin and sliced into 4-μm-thick sections. Xylene was used for dewaxing and citrate buffer was used for antigen repair. After washing with PBS 4 times, the samples were sealed with BSA blocking solution at 37 °C. TRIM55 primary antibody (Novus, United States) was added and incubated overnight at 4 °C. According to the instructions of the im
Total RNA was extracted using TRIzol reagent (Invitrogen) and reverse transcribed to cDNA using a reverse transcription kit (Takara, Dalian, China) following the manufacturer’s instructions. The quantitative real-time polymerase chain reaction three-step method was used, and the 2-ΔΔCt method was used to calculate the relative expression levels of TRIM55. GAPDH was used as the endogenous control. The upstream primer sequence of TRIM55 was 5 '-GGTTTTGGATAGACATGGGGT-3', and the downstream primer sequence was 5 '-TTCTCCTCTTGGGTTCGGGT-3'.
Cells were seeded in 96-well plates (2000 cells/well) and placed into an incubator for further culture. Ten microliters of cell counting kit-8 (CCK-8) reagent (Dojindo, Mashikimachi, Japan) was added to each well on days 1, 2, 3, and 4. After 4 h incubation, the absorbance value at 450 nm (OD450) was measured with a microplate reader. The growth curve of cells was drawn according to the OD value. Three independent assays were performed for each time point.
Two thousand cells/well were plated in a six-well plate and allowed to grow in complete growth medium for 14 d. The colonies were then fixed with 4% paraformaldehyde for 15 min, followed by staining with 0.1% crystal violet solution for 10 min. The number of colonies was counted, and the average value was calculated from three independent experiments.
GC cells at the logarithmic growth stage were trypsinized and suspended in serum-free media at a concentration of 1 × 106 cells/mL. Two hundred microliters of the cell suspension was added to the upper transwell chamber. Culture medium containing 10% FBS was added to the lower chamber. For the invasion assay, the transwell membrane was coated with Matrigel. After 48 h incubation, the cells that did not pass through the membrane in the upper chamber were removed with cotton swabs. The migrated/invaded cells were fixed with paraformaldehyde for 30 min and stained with 0.1% crystal violet solution. The cells were observed under a microscope and five random fields were selected to count the number of migrated or invaded cells.
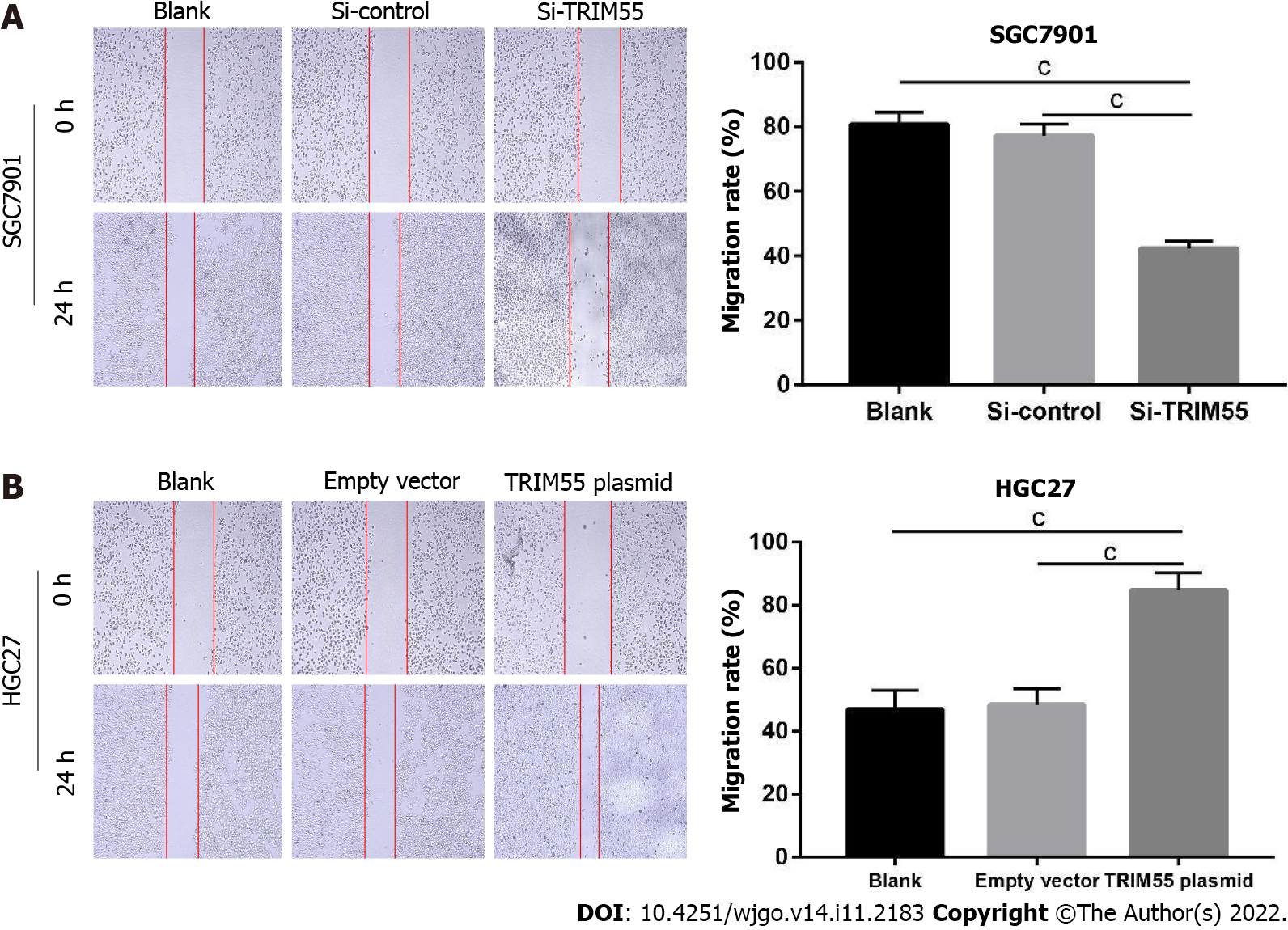
GC cells were inoculated into a six-well plate at a density of 5 × 105 cells/well until 90% confluence was reached. Then, a sterile 100-μL pipette tip was used to scrape the bottom surface of the plate to form a wound vertically. The cells were washed with 1× PBS, and complete growth medium was added. The wound margins were observed at 0 and 24 h in 5 randomly selected microscopic regions, and the mean cell spacing was calculated.
GC cells were collected and lysed in immunoprecipitation lysis buffer. After protein quantification by the BCA method, 30 μg of protein sample was used for electrophoresis. After transfer and blocking, the membrane was incubated overnight at 4 °C with the primary antibody. Horseradish peroxidase-labeled secondary antibody was added and incubated for 1 h. The bands were visualized using chemiluminescence reagent.
Statistical analyses were performed using GraphPad Prism software version 7.0 (CA, United States). All the data are represented as mean ± SD from three independent experiments. Differences among groups were compared using student’s t-test and one-way analysis of variance. The prognostic factors were analyzed by univariate and multivariate Cox regression models. The Kaplan-Meier method was used to calculate the survival rate, and the log-rank test was used to compare different survival curves. A value of P < 0.05 was considered to indicate a statistically significant difference.
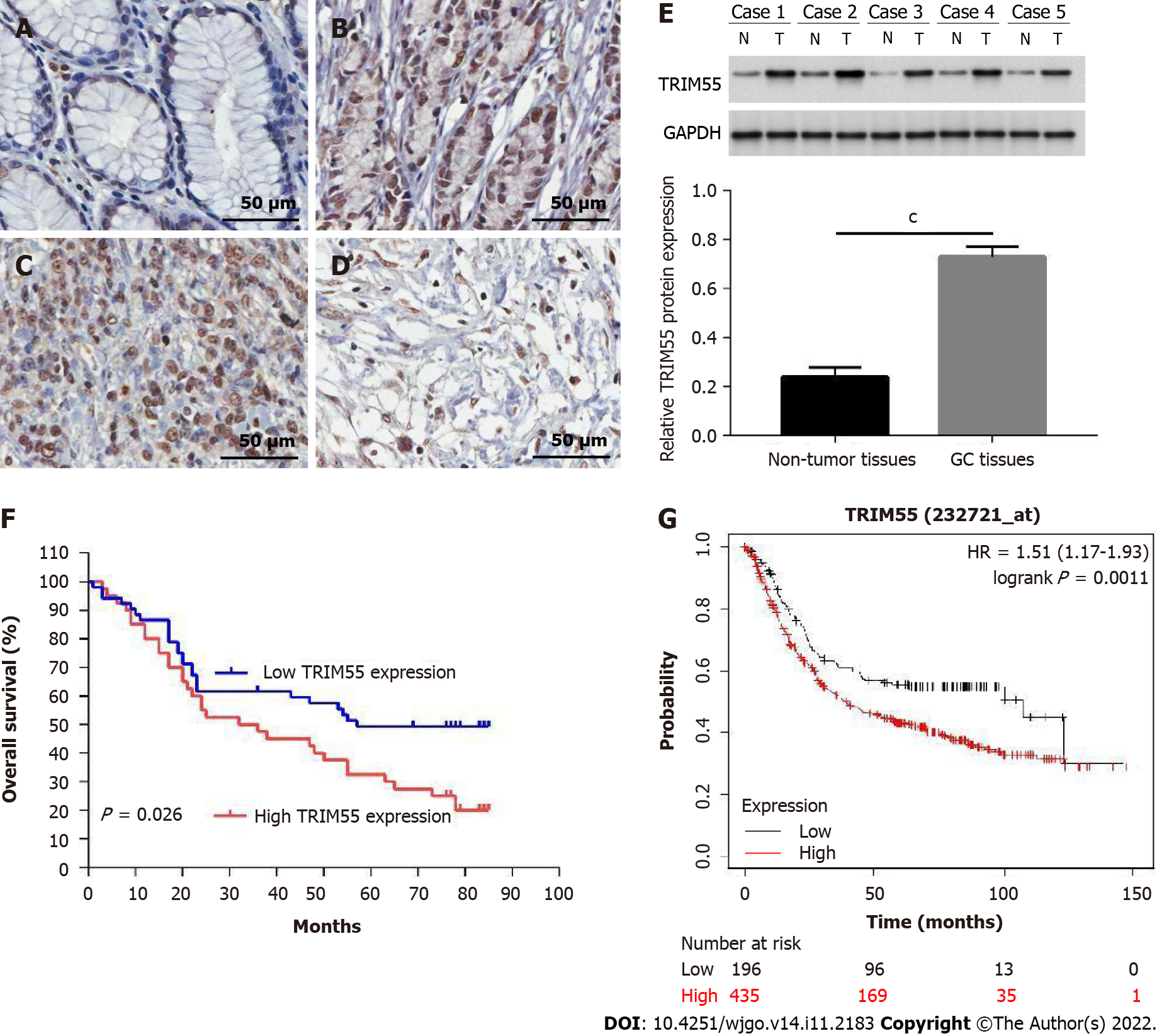
Immunohistochemical results showed that TRIM55 was mainly expressed in the nucleus. Figure 1A-D shows the representative immunohistochemical staining images of TRIM55 in normal gastric mucosa tissue and cancer tissue. The expression of TRIM55 was significantly high in 91 GC tissues and low in para-cancer tissues (Table 1, P < 0.001). There was no significant correlation between TRIM55 expression and age at diagnosis, gender, tumor location, or Lauren type (P > 0.05, Table 2). However, a significant relationship was found between TRIM55 expression and tumor stage, lymph node metastasis, and T stage (P < 0.05, Table 2).
| Tissue | N | High expression | Low expression | P value |
| Gastric cancer | 91 | 40 | 51 | < 0.001 |
| Normal | 91 | 3 | 89 |
| Variables | No. of case | TRIM55 expression | χ2 | P value | |
| High (n = 40) | Low (n = 51) | ||||
| Gender | 0.284 | 0.594 | |||
| Male | 61 | 28 | 33 | ||
| Female | 30 | 12 | 18 | ||
| Age (n) | 0.216 | 0.642 | |||
| ≤ 65 | 48 | 20 | 28 | ||
| > 65 | 43 | 20 | 23 | ||
| Tumor location | 0.008 | 0.928 | |||
| Proximal | 14 | 6 | 8 | ||
| Dismatal | 77 | 34 | 43 | ||
| pT stage | 6.232 | 0.013 | |||
| T1 + T2 | 43 | 13 | 30 | ||
| T3 + T4 | 48 | 27 | 21 | ||
| Lymph node metastasis | 5.506 | 0.019 | |||
| Negative | 49 | 16 | 33 | ||
| Positive | 42 | 24 | 18 | ||
| TNM stage | 5.146 | 0.023 | |||
| Ⅰ + Ⅱ | 53 | 18 | 35 | ||
| Ⅲ | 38 | 22 | 16 | ||
| Lauren histotype | 3.366 | 0.067 | |||
| Intestinal | 47 | 25 | 22 | ||
| Diffuse | 44 | 15 | 29 | ||
To further confirm the results, we performed western blotting using five GC specimens and matched normal tissues. The results showed that protein levels of TRIM55 in GC tissues were up-regulated compared to non-tumor tissues (Figure 1E).
The 5-year survival rate of patients with high TRIM55 expression was lower than that of those with low expression (P = 0.026, Figure 1F). The median overall survival of patients with high and low expression of TRIM55 was 34 mo [95% confidence interval (CI): 13.3–62.8] and 57 mo (95%CI: 25.5–84.5), res
| Variable | Univariate analysis | Multivariate analysis | ||||
| HR | 95%CI | P value | HR | 95%CI | P value | |
| Gender | 0.79 | 0.12–1.38 | 0.463 | |||
| Age | 0.56 | 0.38–1.50 | 0.517 | |||
| Tumor location | 1.67 | 0.53–2.27 | 0.770 | |||
| pT stage | 0.85 | 0.61–0.93 | 0.041 | |||
| Lymph node metastasis | 3.29 | 2.07–5.44 | 0.005 | 2.64 | 1.35–3.08 | 0.021 |
| TNM stage | 2.18 | 1.32–2.89 | 0.016 | 1.20 | 1.06–1.88 | 0.028 |
| Lauren histotype | 1.45 | 0.39–2.46 | 0.433 | |||
| TRIM55 | 2.37 | 1.53–3.81 | 0.028 | 1.48 | 1.17–1.92 | 0.035 |
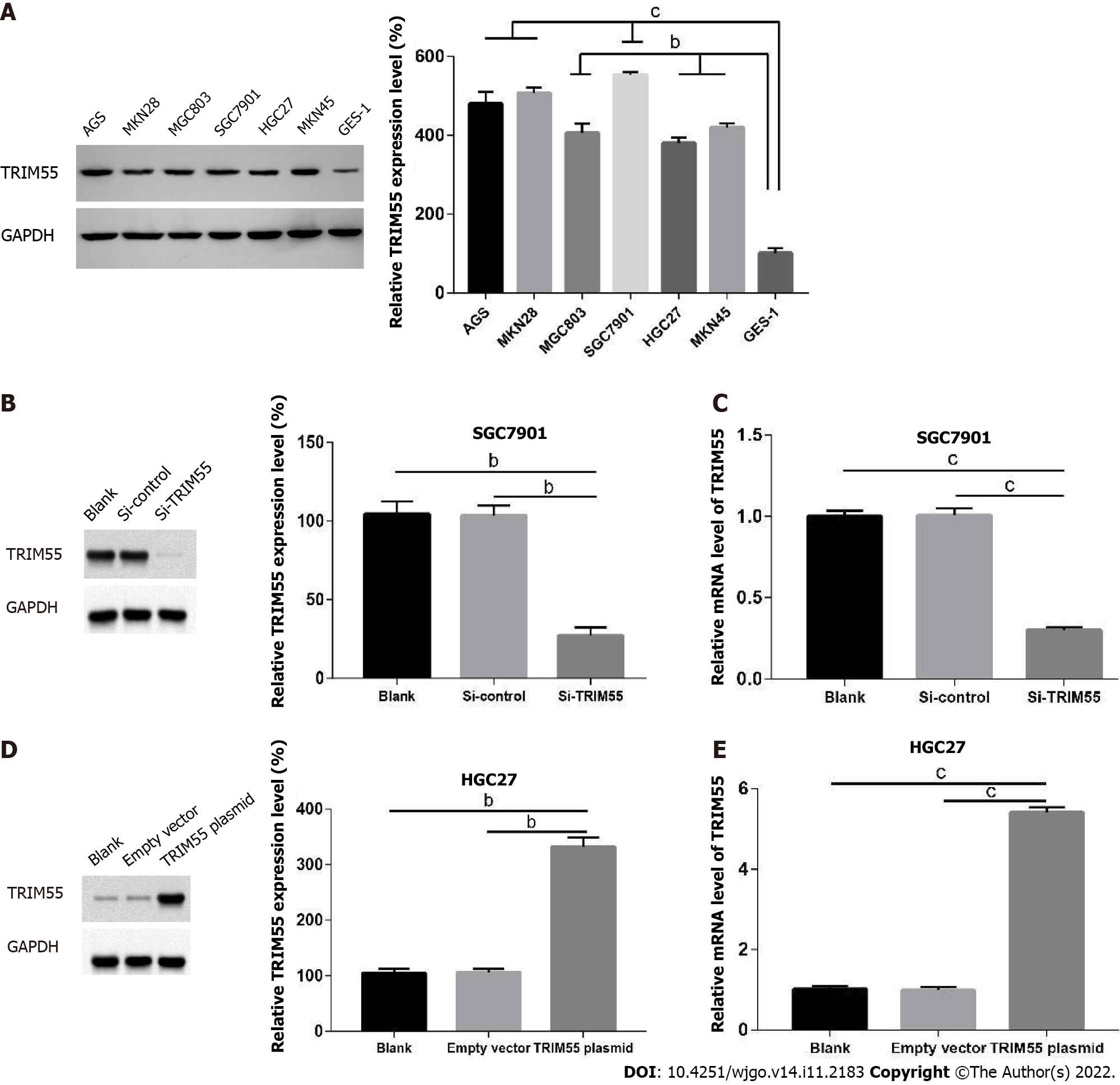
The protein level of TRIM55 was analyzed in the immortalized gastric mucosal epithelial cell line GES-1 and six GC cell lines (AGS, MKN28, MGC803, SGC7901, HGC27, and MKN45). The results showed that the expression level of TRIM55 in GC cell lines was significantly higher than that in GES-1 cells (Figure 2A). Among them, the expression level of TRIM55 was highest in SGC7901 and lowest in the HGC27 cell line.
To analyze the effects of TRIM55 knockdown, SGC7901 (cells with high TRIM55 expression) GC cells were transfected with TRIM55 siRNA. The results showed that both mRNA and protein expressions of TRIM55 were significantly lower in cells transfected with TRIM55 siRNA compared to those in the control group (Figure 2B and C). HGC27 cells with low endogenous TRIM55 expression were trans
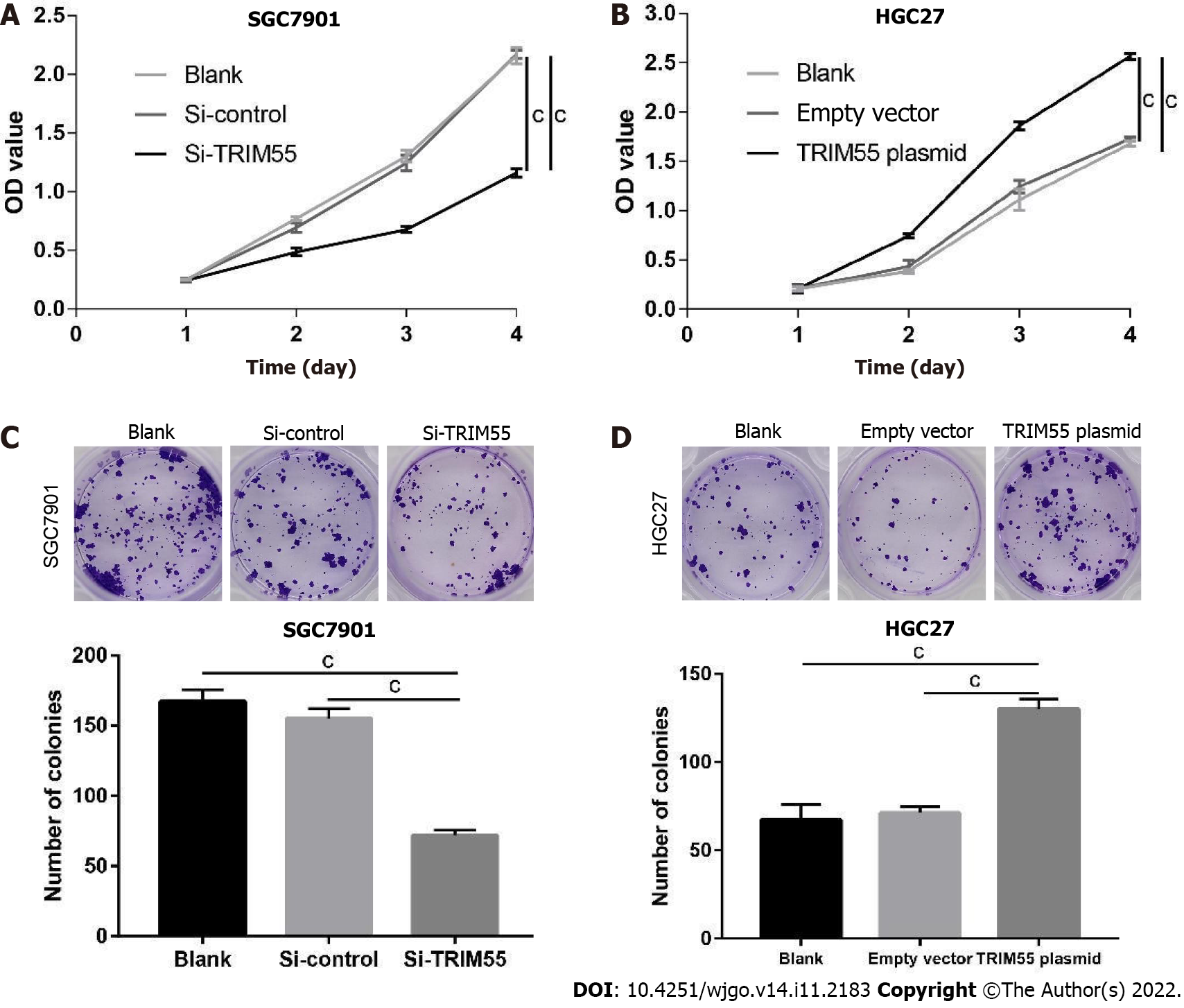
The CCK-8 assay was used to determine the proliferation of GC cells. The results showed that SGC7901 cell proliferation was significantly inhibited on days 2, 3, and 4 after TRIM55 knockdown (P < 0.001, Figure 3A). However, the proliferation of HGC27 cells was significantly increased on days 2, 3, and 4 (P < 0.001, Figure 3B) when TRIM55 was overexpressed in these cells. These results suggest that TRIM55 can promote the proliferation of GC cells.
The colony formation assay showed that the colony number was significantly reduced after siRNA transfection in SGC7901 cells (Figure 3C). Similarly, overexpression of TRIM55 in HGC27 cells enhanced the ability of cells to form colonies (Figure 3D).
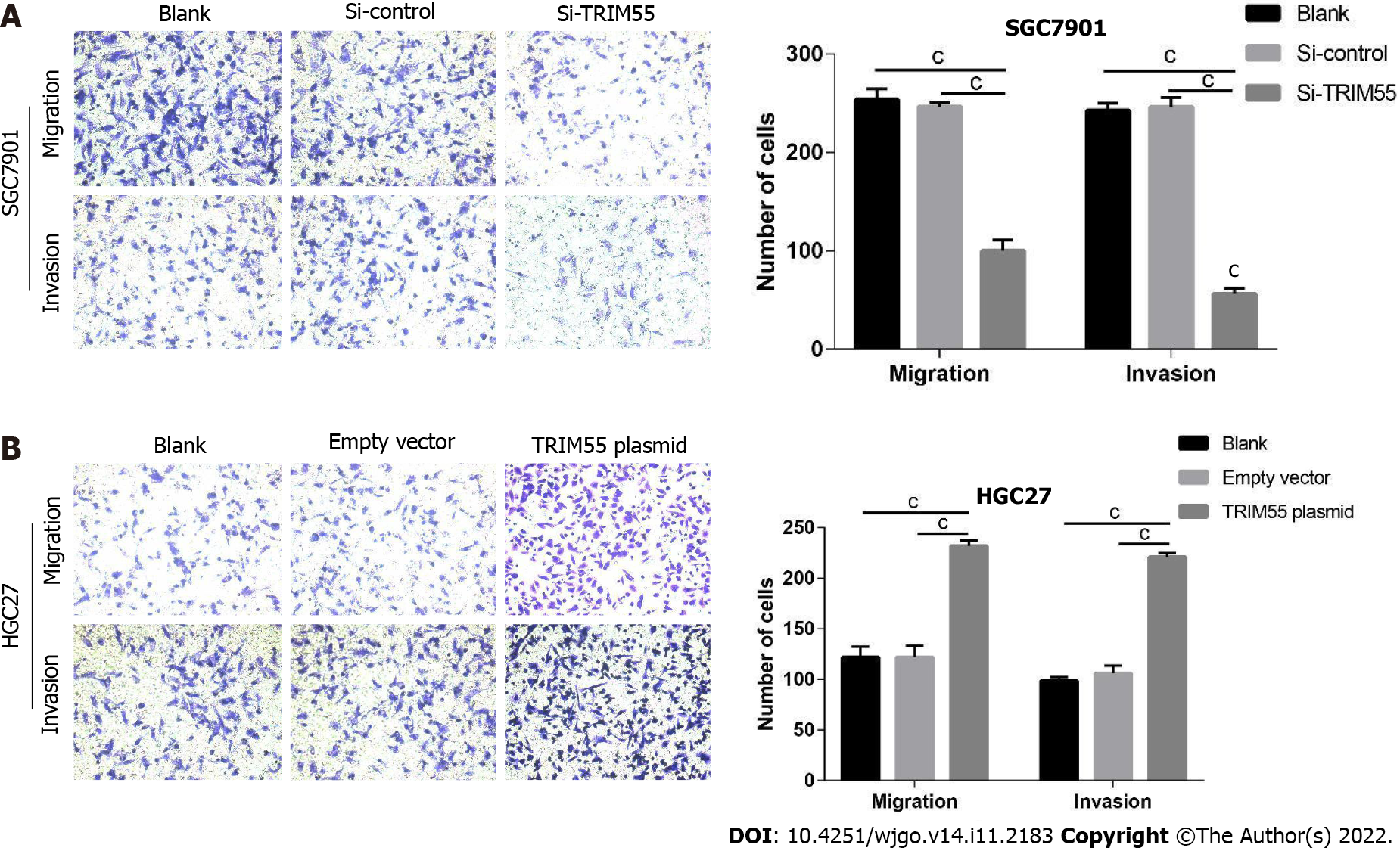
The transwell migration and invasion assay results showed that TRIM55 knockdown significantly reduced the migration and invasion of SGC7901 cells (Figure 4A). Furthermore, the number of migrated and invaded HGC27 cells was significantly increased after TRIM55 overexpression (Figure 4B). The wound-healing assay revealed that knockdown of TRIM55 significantly impaired the migration ability of SGC7901 cells (Figure 5A), while HGC27 migration ability was enhanced after overexpression of TRIM55 (Figure 5B). These results suggest that TRIM55 could promote the invasion and migration of GC cells.
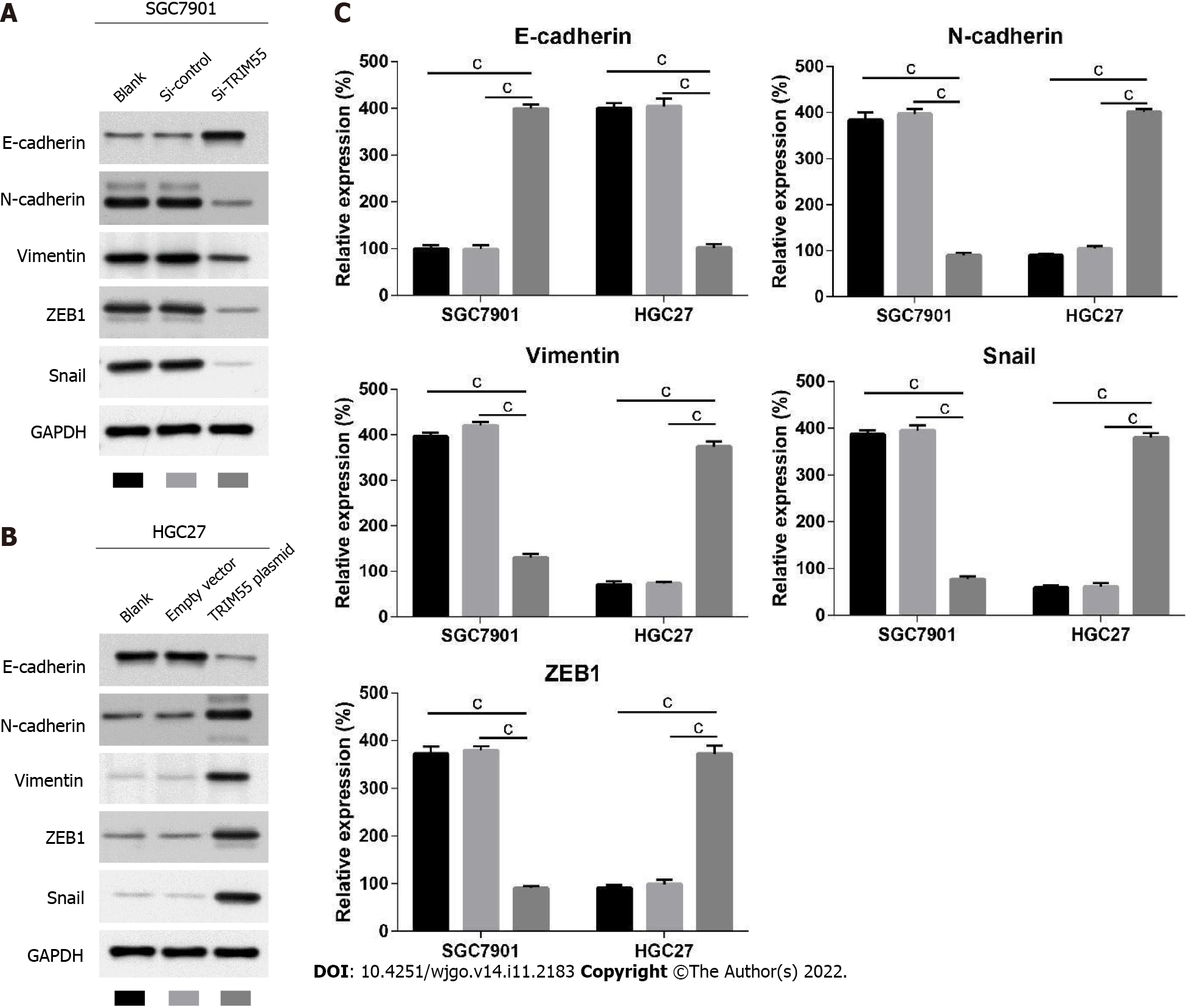
To further explore the potential mechanism of how TRIM55 promotes the progression of GC, we analyzed the expression of epithelial-mesenchymal transition (EMT)-related proteins by western blot analysis (Figure 6). The results showed that in the SGC7901 cell line, E-cadherin expression was significantly up-regulated after TRIM55 knockdown, while N-cadherin, Vimentin, ZEB1, and Snail expression were significantly down-regulated compared with the control group (Figure 6A). Fur
TRIM55 belongs to the TRIM protein family, which plays an important role in the development and progression of tumors. TRIM proteins have three characteristic domains and function as an E3 ligase; they also mediate ubiquitination and regulate intracellular pathophysiological and tumor-related processes by degrading target molecules. TRIM family members exhibit oncogenic and tumor-suppressive capacities in different human cancer types by regulating signal transduction pathways[9,10]. EMT is a transitional process in which epithelial cells lose intercellular adhesion and polarity, and they acquire mesenchymal cell characteristics, enhanced cell motility, and migration ability[11,12].
The progression of EMT is regulated by translational factors and epigenetic modification[13]. Also, microRNAs and long non-coding RNAs are also involved in EMT regulation as post-translational regulators[14]. The tumor cells or other stromal cells can secret exosomes. Exosomes are extracellular vesicles with a lipid bilayer containing proteins, lipids and functional RNAs, which can transfer information between tumor cells or between tumor cells and the tumor microenvironment, thereby regulating the EMT process[15,16]. As EMT plays essential physiological roles, EMT-targeted therapy combined with conventional chemotherapy can improve the sensitivity of tumor cells to drugs.
TRIM proteins were found to be associated with EMT in various types of cancer. TRIM11 protein was upregulated in GC tissue and cell lines, and it could promote cell proliferation, migration, invasion, and EMT of GC by activating β-catenin signaling[17]. TRIM47 mainly influenced the EMT signaling pathway, was highly expressed in GC, and was associated with poor prognosis of patients[18]. In GC, TRIM44 expression was also increased in GC tissues and cell lines, and it regulated GC cell metastasis by altering the expression of EMT-associated factors[19].
Previous studies have demonstrated that the role of TRIM55 in tumors is tissue specific. For example, a study revealed that TRIM55 expression was significantly suppressed in lung adenocarcinoma tissues and tumor cells. TRIM55 exerted its tumor-suppressive effect by increasing the degradation of Snail protein via ubiquitination[20]. TRIM55 was downregulated in hepatocellular carcinoma (HCC) tissue and associated with tumor stage and poor prognosis. Overexpression of TRIM55 can suppress the migration and invasion of HCC cells through EMT and the MMP2 pathways[21].
To the best of our knowledge, our study is the first to determine the expression levels and biological functions of TRIM55 in GC. We demonstrated that TRIM55 could be a potential new biomarker for diagnosing and evaluating GC patients. First, we performed immunohistochemical staining, and the results showed that TRIM55 was highly expressed in GC tissues and that TRIM55 expression was related to the T stage, lymph node metastasis, and TNM stage of GC. Survival analysis showed that the 5-year survival rate of GC patients with high expression of TRIM55 was significantly reduced. Cox regression analysis also confirmed that TRIM55 expression was an independent prognostic factor in GC. These results suggest that TRIM55 may be involved in the occurrence and development of GC. Furthermore, to confirm the biological functions of TRIM55 in GC, we performed a gain and loss of function experiment in vitro. Results from the CCK-8 assay, colony formation assay, transwell, and wound healing assays indicated that inhibition of TRIM55 decreases the proliferation and invasion of GC cells, whereas the overexpression of TRIM55 promotes these processes in GC cells. Finally, we used western blot analysis to confirm that knockdown or overexpression of TRIM55 could alter the EMT-related protein, suggesting that TRIM55 could regulate the EMT process. TRIM proteins could serve the ubiquitination function to stabilize or dislocate target proteins in various cellular compartments[4]. Ubiquitination is a post-transcriptional modification that labels the target proteins to be degraded at the proteasome level. Thus, TRIM family members determine both tumor suppressor and oncogenic roles by affecting the signal pathways in cancer development and progression. For example, TRIM29 and TRIM8 exhibited contextual function in different cancers[22-24]. They negatively or positively regulate tumorigenesis and tumor progression by affecting pathways. In our study, we showed that TRIM55 is highly expressed in gastric tumors and cultured tumor cells. TRIM55 has E3 ubiquitin ligase activity and whether it can regulate the EMT-related proteins through ubiquitination requires further investigation.
However, some limitations exist in our study. First, the detection of TRIM55 was based on a single-center clinical cohort, and the functional experiments were only performed in vitro. Future studies should enroll more patients and utilize animal models to confirm our conclusions. Second, TRIM55 has E3 ubiquitin ligase activity and whether it can regulate the EMT-related proteins through ubiquitination requires further study.
In summary, our study analyzed the expression of TRIM55 in GC and demonstrated that TRIM55 could promote GC cell proliferation, migration, and invasion via the EMT process. Overexpression of TRIM55 could be an independent factor predicting poor survival, and TRIM55 may serve as a potential therapeutic target for GC. In addition, whether TRIM55 can affect EMT through other molecular mechanisms remains to be examined in future studies.
TRIM55 plays important role in hepatocellular carcinoma and lung adenocarcinoma. However, little is known about the role of TRIM55 in gastric cancer (GC).
To discover the targets for the diagnosis, treatment and prognosis prediction of GC.
To explore the biological function of TRIM55 and its underlying molecular mechanism in GC.
The expression of TRIM55 was determined by quantitative real-time polymerase chain reaction and Western blot. Cell counting kit-8 assay, colony formation, wound healing assay and transwell assay were used to investigate the TRIM55 function.
TRIM55 expression levels were significantly increased in GC cell lines and tissues. High expression of TRIM55 was correlated with poor prognosis of GC patients. Knockdown of TRIM55 in GC cell lines inhibited proliferation, colony formation, migration and invasion in vitro. TRIM55 can regulate the expression of epithelial-mesenchymal transition-related proteins in GC cells.
TRIM55 functions as an oncogene through promoting cell proliferation, migration and invasion in GC.
TRIM55 may be a new potential target in GC treatment.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Liu YQ, United States; Qin Y, China S-Editor: Fan JR L-Editor: A P-Editor: Yuan YY
| 1. | Rahman R, Asombang AW, Ibdah JA. Characteristics of gastric cancer in Asia. World J Gastroenterol. 2014;20:4483-4490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 265] [Cited by in RCA: 315] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 2. | Tung I, Sahu A. The treatment of resectable gastric cancer: a literature review of an evolving landscape. J Gastrointest Oncol. 2022;13:871-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 3. | Xia JY, Aadam AA. Advances in screening and detection of gastric cancer. J Surg Oncol. 2022;125:1104-1109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 78] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 4. | Mohammadi A, Pour Abbasi MS, Khorrami S, Khodamoradi S, Mohammadi Goldar Z, Ebrahimzadeh F. The TRIM proteins in cancer: from expression to emerging regulatory mechanisms. Clin Transl Oncol. 2022;24:460-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 5. | Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792-804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 630] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 6. | Perera S, Holt MR, Mankoo BS, Gautel M. Developmental regulation of MURF ubiquitin ligases and autophagy proteins nbr1, p62/SQSTM1 and LC3 during cardiac myofibril assembly and turnover. Dev Biol. 2011;351:46-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 7. | Zhang BW, Cai HF, Wei XF, Sun JJ, Lan XY, Lei CZ, Lin FP, Qi XL, Plath M, Chen H. miR-30-5p Regulates Muscle Differentiation and Alternative Splicing of Muscle-Related Genes by Targeting MBNL. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 8. | Lu Y, Mei Y, Chen L, Wu L, Wang X, Zhang Y, Fu B, Chen X, Xie Y, Cai G, Bai X, Li Q. The role of transcriptional factor D-site-binding protein in circadian CCL2 gene expression in anti-Thy1 nephritis. Cell Mol Immunol. 2019;16:735-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 9. | Cambiaghi V, Giuliani V, Lombardi S, Marinelli C, Toffalorio F, Pelicci PG. TRIM proteins in cancer. Adv Exp Med Biol. 2012;770:77-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 10. | Mantovani F, Walerych D, Sal GD. Targeting mutant p53 in cancer: a long road to precision therapy. FEBS J. 2017;284:837-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 11. | Xu R, Won JY, Kim CH, Kim DE, Yim H. Roles of the Phosphorylation of Transcriptional Factors in Epithelial-Mesenchymal Transition. J Oncol. 2019;2019:5810465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 12. | Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11:28-39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 348] [Cited by in RCA: 523] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 13. | Lu W, Kang Y. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev Cell. 2019;49:361-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 706] [Article Influence: 141.2] [Reference Citation Analysis (0)] |
| 14. | Huang Y, Hong W, Wei X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J Hematol Oncol. 2022;15:129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 489] [Reference Citation Analysis (0)] |
| 15. | Jiang J, Li J, Zhou X, Zhao X, Huang B, Qin Y. Exosomes Regulate the Epithelial-Mesenchymal Transition in Cancer. Front Oncol. 2022;12:864980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 35] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 16. | Kim H, Lee S, Shin E, Seong KM, Jin YW, Youn H, Youn B. The Emerging Roles of Exosomes as EMT Regulators in Cancer. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 17. | Lan Q, Tan X, He P, Li W, Tian S, Dong W. TRIM11 Promotes Proliferation, Migration, Invasion and EMT of Gastric Cancer by Activating β-Catenin Signaling. Onco Targets Ther. 2021;14:1429-1440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 18. | Xia Y, Wei Z, Huang W, Wei X, He Y. Trim47 overexpression correlates with poor prognosis in gastric cancer. Neoplasma. 2021;68:307-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 19. | Wang F, Ruan L, Yang J, Zhao Q, Wei W. TRIM14 promotes the migration and invasion of gastric cancer by regulating epithelialtomesenchymal transition via activation of AKT signaling regulated by miR1955p. Oncol Rep. 2018;40:3273-3284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 20. | Guo T, Zhang Z, Zhu L, Chen W, Ding Y, Li W, Huang Y, Huang J, Pan X. TRIM55 suppresses malignant biological behavior of lung adenocarcinoma cells by increasing protein degradation of Snail1. Cancer Biol Ther. 2022;23:17-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 21. | Li X, Huang L, Gao W. Overexpression of Tripartite Motif Conaining 55 (TRIM55) Inhibits Migration and Invasion of Hepatocellular Carcinoma (HCC) Cells via Epithelial-Mesenchymal Transition and Matrix Metalloproteinase-2 (MMP2). Med Sci Monit. 2019;25:771-777. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 22. | Esposito JE, De Iuliis V, Avolio F, Liberatoscioli E, Pulcini R, Di Francesco S, Pennelli A, Martinotti S, Toniato E. Dissecting the Functional Role of the TRIM8 Protein on Cancer Pathogenesis. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 23. | Deng X, Fu X, Teng H, Fang L, Liang B, Zeng R, Chen L, Zou Y. E3 ubiquitin ligase TRIM29 promotes pancreatic cancer growth and progression via stabilizing Yes-associated protein 1. J Transl Med. 2021;19:332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 24. | Xu M, Hu J, Zhou B, Zhong Y, Lin N, Xu R. TRIM29 prevents hepatocellular carcinoma progression by inhibiting Wnt/β-catenin signaling pathway. Acta Biochim Biophys Sin (Shanghai). 2019;51:68-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |