Published online Sep 15, 2021. doi: 10.4251/wjgo.v13.i9.995
Peer-review started: February 21, 2021
First decision: May 8, 2021
Revised: May 30, 2021
Accepted: July 27, 2021
Article in press: July 27, 2021
Published online: September 15, 2021
Processing time: 200 Days and 18.3 Hours
MicroRNAs (miRNAs) are non-coding RNA molecules composed of 19–25 nucleotides that regulate gene expression and play a central role in the regulation of several immune-mediated disorders, including inflammatory bowel diseases (IBD). IBD, represented by ulcerative colitis and Crohn’s disease, is characterized by chronic intestinal inflammation associated with an increased risk of colorectal cancer (CRC). CRC is one of the most prevalent tumors in the world, and its main risk factors are obesity, physical inactivity, smoking, alcoholism, advanced age, and some eating habits, in addition to chronic intestinal inflammatory processes and the use of immunosuppressants administered to IBD patients. Recent studies have identified miRNAs associated with an increased risk of developing CRC in this population. The identification of miRNAs involved in this tumorigenic process could be useful to stratify cancer risk development for patients with IBD and to monitor and assess prognosis. Thus, the present review aimed to summarize the role of miRNAs as biomarkers for the diagnosis and prognosis of IBD-associated CRC. In the future, therapies based on miRNA modulation could be used both in clinical practice to achieve remission of the disease and restore the quality of life for patients with IBD, and to identify the patients with IBD at high risk for tumor development.
Core Tip: Inflammatory bowel diseases (IBD), represented by ulcerative colitis and Crohn's disease, are characterized by recurrent chronic intestinal inflammation associated with an increased risk of colorectal cancer. MicroRNAs (miRNAs) are small non-coding RNA molecules, composed of 19 to 25 nucleotides, which can regulate gene expression and play an important role in regulating cellular processes. Altered expression of these molecules is related to the progression of inflammation and an increased risk of colorectal cancer (CRC). Thus, the aim of the present review was to evaluate the role of miRNAs as biomarkers for the diagnosis and prognosis of IBD-associated CRC.
- Citation: Grillo TG, Quaglio AEV, Beraldo RF, Lima TB, Baima JP, Di Stasi LC, Sassaki LY. MicroRNA expression in inflammatory bowel disease-associated colorectal cancer. World J Gastrointest Oncol 2021; 13(9): 995-1016
- URL: https://www.wjgnet.com/1948-5204/full/v13/i9/995.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i9.995
MicroRNAs (miRNAs) are non-coding RNA molecules composed of approximately 19–25 nucleotides that are capable of regulating gene expression[1]. MiRNAs are involved in biological functions such as embryonic development, proliferation, cell differentiation, metabolism, apoptosis, and stress response[2,3]. The isolation of miRNAs is possible using various biological materials such as tissues, cells, and body fluids (tears, urine, serum, and plasma)[4]. Polymerase chain reaction, in situ hybridization, microarrays, and RNA sequencing are the main methods used to detect miRNA expression[1].
MiRNAs can downregulate mRNA[5] by binding to the 3′ untranslated region of the target mRNAs[6]. As only one miRNA can regulate many mRNA targets, any minimal structural change can lead to major changes in cell homeostasis[7], disease evolution[8], and predisposition to neoplastic and inflammatory conditions[9-11]. Alterations in miRNA expression have been described in several tumors, including colorectal cancer (CRC) acting as oncogenes or tumor suppressors[12]. This expression is specific according to the tumor type and the surrounding tissue; thus, the study of tumor miRNAs helps to differentiate normal and tumor tissues and, in addition, reflects the degree of tumor differentiation[13].
Inflammatory bowel diseases (IBD), such as ulcerative colitis (UC) and Crohn’s disease (CD), are characterized by chronic intestinal inflammation associated with an increased risk of CRC. Tumor development is one of the most feared long-term disease complications, accounting for 15% of deaths in patients with IBD[14]. UC patients are approximately 30 times more likely to develop CRC than the general population[15], the main risk factors being the extension and duration of the disease[16], family history of CRC[17], and the presence of primary sclerosing cholangitis (PSC)[18]. Likewise, patients with CD are at a higher risk of small bowel cancer[19].
In IBD studies, miRNAs have been found to be involved in pathogenesis and can serve as diagnostic biomarkers and therapeutic targets. Recent studies have identified miRNAs associated with an increased risk of developing CRC in this population. The identification of miRNAs involved in this tumorigenic process could be useful to stratify cancer risk development for patients with IBD and to monitor and assess prognosis. Thus, this review article aimed to characterize the miRNAs expressed in IBD, CRC, and IBD-related CRC to better understand their role in the diagnosis and prognosis of these diseases, in addition to analyzing their potential as therapeutic targets.
IBD is a chronic disease involving the gastrointestinal tract, the pathophysiology of which is complex, encompassing environmental, genetic factors and the immune response[20], and comprises two specific diseases, UC and CD[20-22]. There is no specific examination to establish the diagnosis of CD and UC, which requires the association of clinical, biochemical, endoscopic, radiological, and histologic data[23]. Thus, a distinction between these two diseases can be challenging when inflammation is limited to the colon, and 10%–15% of IBD patients are classified as having indeterminate colitis[24].
In recent years, new biomarkers associated with easy application have been identified, such as miRNAs, to facilitate disease diagnosis and prognosis[25]. However, the real role of miRNAs in IBD is not fully understood, and it is believed that some miRNAs might be common to both diseases, whereas others are unique to each, depending on the severity of disease activity and the tissue analyzed[26]. Changes in miRNA expression in patients with IBD were first described by Wu et al[27] in 2008. The authors showed an increase in the expression of eight miRNAs (Let-7f, miR-16, miR-21, miR-23a, miR-24, miR-29a, miR-126, and miR-195) and a decrease in the expression of three miRNAs (miR-192, miR-375, and miR-422b) in colonic tissue samples from patients with active UC. In 2010, the same group[28] observed increased expression of miR-23b, miR106a, and miR-191 and decreased expression of miR-19b and miR-629 in colonic samples of patients with colonic CD and increased expression of miR-16, miR-21, miR-223, and miR-594 in terminal ileum samples of CD patients with active disease. In UC patients, miR-19a, miR-21, miR-31, miR-146a and miR-375 levels were found increased when compared with CD patients indicating these miRNAs as potential differential biomarkers for CD and UC[29,30]. In addition to these studies, others studies have demonstrated changes in miRNA expression in the colonic tissue of patients with IBD[31,32].
Guo et al[31] evaluated the differential expression of miRNA in inflamed or non-inflamed ileum mucosa of patients with CD and found decreased expression of miR-192-5p in those with inflammation. Among the alterations in the expression of miRNAs already observed, miR-10a, miR-192, and miR-320 seem to negatively regulate the inflammatory response by inhibiting the expression of NOD2 mRNA (domain 2 of nucleotide-binding oligomerization)[33-35]. In contrast, miR-155, by activating the nuclear factor kappa B (NF-κB) signaling pathway, plays an important role in the progression of intestinal inflammation[36]. Besides that, Lu et al[37] showed that miR-155 was capable to decrease SH-2 containing inositol 5’ polyphosphatase 1 (SHIP-1), an important phosphatase correlated with membrane trafficking, contributing to inflammation pathogenesis.
Shi et al[38] demonstrated that the miR-31/interleukin (IL)-25 signaling axis can regulate the Th1/Th17 IL-12/23-mediated inflammatory response in experimental colitis, indicating that a decrease in miR-31 expression with a consequent increase in IL-25 levels could be an alternative treatment for IBD. This pathway is also intricately linked to miR-155 and miR-223[39,40].
Pre-clinical studies have also been performed to evaluate the role of miRNAs in intestinal inflammation. Nata et al[41] administered miR-146b intraperitoneally to mice with dextran sodium sulfate (DSS)-induced intestinal inflammation and observed an improvement in the inflammatory process and intestinal barrier, demonstrating a potential use of miRNAs for IBD treatment[41]. Additionally, Huang et al[42] observed the regulation of leukocyte infiltration and consequently a reduction in the inflammatory process with a intracolonic injection of miR-141 in mice[42]. On the other hand, the use of an antagomir for miR-155, a small synthetic RNA complementary to miR-155 used to silence this miRNA, in the DSS-induced intestinal inflammation model improved intestinal inflammation indicating miR-155 as a possible target for IBD treatment[37]. Jin et al[43] using miRNA mimics, namely miR-133a to target UCP2 (mitochondrial uncoupling protein 2) observed a reduction in the severity of DSS-induced intestinal inflammation, suggesting that miRNA mimics are another therapeutic option for IBD patients[43]. In another study with the DSS-model of intestinal inflammation, Tian et al[44] found a super expression of miR-31, results similar to what was found in inflamed mucosa from patients with CD or UC.
Besides that, levels of miR-301a were also increased in intestinal epithelial cells from patients with active IBD reducing the expression of BTG anti-proliferation factor 1 (BTG1) and promoting Th17 cell differentiation through downregulation of Smad Nuclear Interacting Protein 1 (SNIP1). BTG1 reduces epithelial integrity and promote inflammation in mouse colon and leading to tumorigenesis. This way, blockade of miR-301a in vivo may serve as a novel therapeutic approach in the treatment of IBD and colitis associated-CRC[45,46].
Some miRNAs act on the same inflammatory pathways as drugs currently used to treat IBD. MiR-29 has been described to comprise a family of miRNAs with the ability to decrease IL-23 levels[47,48], effects similar to ustekinumab and others anti-interleukin 12/23 used for the treatment of moderate to severe IBD. The blockade of integrin α4β7 in T helper lymphocytes has anti-inflammatory activity through the inhibition of leukocyte adhesion to endothelial cells. Harris et al[49] observed that endogenous miR-126 could inhibit this adhesion through the regulation of VCAM-1 adherence[49,50], effects similar to those found with the use of vedolizumab, a mono
In addition to their possible application in clinical practice, some miRNAs can be used as predictors of the response to clinical treatment. Morilla et al[52] evaluated patients with severe UC and their response to corticosteroids, or infliximab and cyclosporine in those corticosteroid-refractory UC patients. The authors identified 15 miRNAs associated with the response to corticosteroids, six with the response to infliximab, and four with the response to cyclosporine, indicating that miRNAs can be used to screen patients according to the probability of responding to a specific medication. Cordes et al[53] evaluated the potential of miR-320a to monitor disease activity and predict the course of disease in patients with IBD, and found that blood levels of miR-320a were significantly increased in patients with active CD and UC when compared to healthy controls assessing the role of miRNA in monitoring inflammatory activity. Moreover, miR-320a levels were strongly correlated with endoscopic disease activity in both CD and UC patients, highlighting the use of miRNA as a non-invasive tool useful in monitoring disease activity in these patients.
New studies have evaluated the role miRNAs in fecal samples of IBD patients. Verdier et al[54] analyzed more than 800 miRNAs in fecal samples from control individuals and patients with IBD with levels of miR-223 and miR-1246 significantly increased in fecal samples from patients with active IBD. Furthermore, the miRNAs were correlated with clinical disease activity scores, such as the Mayo score, and fecal calprotectin levels in these patients, indicating that miRNAs from fecal samples might be a new noninvasive and easy fecal biomarker for monitoring IBD activity. MiR-223 was also identified as a fecal marker in a study by Schönauen et al[25], in addition to miR-155 and miR-16. A summary of miRNAs found altered in each disease is shown in Table 1[55-61] and Table 2[62-69].
| MiRNA | Expression | Ref. |
| let-7i | Increased | [55] |
| miR-7 | Increased | [32] |
| miR-9 | Increased | [32] |
| miR-16 | Increased | [27,28] |
| miR-19a | Increased | [29] |
| miR-21 | Increased | [9,28,32] |
| miR-22 | Increased | [32] |
| miR-23b | Increased | [28] |
| miR-29a | Increased | [27] |
| miR-29b | Increased | [32] |
| miR-29c | Increased | [32] |
| miR-30a | Increased | [32] |
| miR-30b | Increased | [32] |
| miR-30c | Increased | [32] |
| miR-31 | Increased | [32,56] |
| miR-34c-5p | Increased | [32] |
| miR-101 | Increased | [29] |
| miR-106a | Increased | [28,32] |
| miR-126 | Increased | [32] |
| miR-127-3p | Increased | [32] |
| miR-130a | Increased | [32] |
| miR-133b | Increased | [32] |
| miR-141 | Increased | [42] |
| miR-146a | Increased | [29,32] |
| miR-146b-5p | Increased | [32] |
| miR-150 | Increased | [32] |
| miR-155 | Increased | [32,56] |
| miR-181c | Increased | [32] |
| miR-191 | Increased | [28] |
| miR-196 | Increased | [57] |
| miR-196a | Increased | [32] |
| miR-206 | Increased | [58] |
| miR-223 | Increased | [28,32] |
| miR-301a | Increased | [45] |
| miR-324-3p | Increased | [32] |
| miR-328 | Increased | [59] |
| miR-422a | Increased | [59] |
| miR-449b | Increased | [55] |
| miR-594 | Increased | [28] |
| miR 663 | Increased | [58] |
| miR 885-5p | Increased | [59] |
| let-7b | Decreased | [59] |
| miR-10a | Decreased | [33] |
| miR-18a | Decreased | [59] |
| miR-19b | Decreased | [28] |
| miR-26a | Decreased | [32] |
| miR-140-3p | Decreased | [59] |
| miR-143 | Decreased | [60] |
| miR-192-5p | Decreased | [31] |
| miR-194b | Decreased | [58] |
| miR-203 | Decreased | [61] |
| miR-216b | Decreased | [58] |
| miR-320a | Decreased | [34] |
| miR-320b | Decreased | [34] |
| miR-320c | Decreased | [34] |
| miR-375 | Decreased | [27] |
| miR-548e | Decreased | [58] |
| miR-559 | Decreased | [58] |
| miR-629 | Decreased | [28] |
| MiRNA | Expression | Ref. |
| let-7e | Increased | [62] |
| let-7f | Increased | [27] |
| miR-7 | Increased | [32] |
| miR-16 | Increased | [27,28] |
| miR-19a | Increased | [29] |
| miR-20b | Increased | [62] |
| miR-21 | Increased | [9,32] |
| miR-23a | Increased | [27] |
| miR-24 | Increased | [27] |
| miR-29a | Increased | [27] |
| miR-29b | Increased | [32] |
| miR-31 | Increased | [32,56] |
| miR-98 | Increased | [62] |
| miR-101 | Increased | [29] |
| miR-125b-1 | Increased | [62] |
| miR-126 | Increased | [27,63] |
| miR-127-3p | Increased | [32] |
| miR-135b | Increased | [32] |
| miR-146a | Increased | [29,32] |
| miR-150 | Increased | [64] |
| miR-155 | Increased | [32,56] |
| miR-195 | Increased | [27] |
| miR-196a | Increased | [32] |
| miR-206 | Increased | [58] |
| miR-214 | Increased | [65] |
| miR-223 | Increased | [28,32] |
| miR-301a | Increased | [45] |
| miR-324-3p | Increased | [32] |
| miR-548a-3p | Increased | [59] |
| miR-650 | Increased | [59] |
| miR-663 | Increased | [58] |
| miR-10a | Decreased | [33] |
| miR-26a | Decreased | [32] |
| miR-124 | Decreased | [66] |
| miR-143 | Decreased | [60] |
| miR-145 | Decreased | [60] |
| miR-188-5p | Decreased | [32] |
| miR-192 | Decreased | [27] |
| miR-194 | Decreased | [67] |
| miR-194b | Decreased | [58] |
| miR-196b | Decreased | [59] |
| miR-200b | Decreased | [68] |
| miR-215 | Decreased | [32] |
| miR-216b | Decreased | [58] |
| miR-320a | Decreased | [34] |
| miR-320b | Decreased | [34] |
| miR-320c | Decreased | [34] |
| miR-346 | Decreased | [32] |
| miR-375 | Decreased | [27] |
| miR-422b | Decreased | [27] |
| miR-489 | Decreased | [59] |
| miR-548e | Decreased | [58] |
| miR-559 | Decreased | [58] |
| miR-630 | Decreased | [59] |
| miR-4284 | Decreased | [69] |
The future treatment of IBD involves the application of pharmacological strategies to control or even stop the progression of inflammation and to improve sensitivity to the therapy. This could occur, for example, through anti-miRNA oligonucleotides to inactivate miRNAs association with increased expression in the inflammatory process or increase the expression of suppressor miRNAs[70]. The alteration of immune system cells by miRNAs is also a factor for inflammation[71]. Thus, tracking the immune status in IBD based on miRNA alterations may be powerful for designing individualized therapies[72].
Taken together, these differences in the expression of miRNAs in UC and CD patients are relevant from the moment they lead to the emergence of biomarkers for diagnosis and therapeutic targets, aiming to improve the management of IBD; however, larger and more consistent studies are necessary for their implementation in clinical practice[60,72].
CRC is the second most common cancer in women and the third in men, with higher rates in developed countries, and is responsible for approximately 900000 deaths each year[73]. An increase in the global incidence to 2.5 million new cases is expected in 2035[74,75], mainly due to an increase in exposure to risk factors. Obesity, physical inactivity, smoking, alcoholism, aging, and eating habits are some of the main risk factors for the appearance of tumors[73]. Genetic factors are also involved, such as the presence of a positive family history in 10%–20% of the cases[76] and hereditary syndromes in 5%–7% of the cases[77]. Patients with long-standing IBD constitute a risk group due to the presence of the inflammatory processes and the use of immunosuppressive drugs, with CRC being responsible for 15% of deaths in this population[14].
The development of CRC results from an evolutionary period of approximately 10–15 years and originates, in most cases, from alterations to the crypt pattern that evolves to a pre-neoplastic lesion (polyp) and later to a tumor[78,79]. Precursor lesions appear in two ways. The first is via adenoma-carcinoma, responsible for 70%–90% of tumors and related to an adenomatous polyposis coli (APC) gene mutation with the subsequent activation of RAS and loss of tumor suppressor p53 (TP53) function; the second is through a serrated neoplasia, which is responsible for 10%–20% of cases[73,80] and is mainly related to RAS and RAF mutations[73].
It has also been noted that changes in cell homeostasis due to genetic changes lead to the activation of oncogenes and inactivation of tumor suppressor genes[78,79]. WNT signaling pathways, epidermal growth factor (EGFR), the TP53 complex, and transforming growth factor beta (TGF-β) are implicated in the carcinogenesis of CRC[81]. The WNT pathway is related to the regulation of stem cell activity in intestinal crypts, and inadvertent signaling by this pathway leads to the inhibition of cell differentiation and death, leading to the development of polyps and consequently carcinoma[82,83].
It was noted that marked expression of miR-135b, which activates the WNT pathway is involved in sporadic and inflammatory CRC and is related to the tumor stage and a worse prognosis[84]. The EGFR signaling pathway is also responsible for cellular activities and is related to certain oncogenes, predominantly Kirsten rat sarcoma viral oncogene homolog (KRAS), for which mutations are present in approximately 30%–40% of CRC cases, resulting in worse prognosis[85]. It is also important to note that KRAS seems to be regulated by isoforms of the let-7 family, such as let-7a, which when deregulated, contributes to colorectal carcinogenesis[86].
Senescence, cell cycle arrest, apoptosis, invasion, and metastasis are related to TP53 when cells are subjected to stress[87]. Some of the miRNAs that participate in the TP53 pathway include let-7i, miR-20a, miR-21, miR-25, miR-34a/b/c, miR-145, miR-181b, miR183, miR-195, miR-215, and miR-451 with a special attention with miR-34 family. Activation of TP53 by miR-34a has already been observed in several types of cancer, especially CRC, with a overexpression of this miRNA in those patients[88]. Finally, the TGF-β pathway regulates activities such as proliferation, differentiation, and apoptosis, and miRNAs that regulate the TGF- β receptor 2 (TGFBR2) have been identified, including miR-17-5p, miR-20a, miR-21, miR-23b, miR-106a, and miR-301a[89].
There are specific molecular expression profiles in CRC cells compared to those in non-tumor cells. Among the overexpressed miRNAs, miR-106, miR-31, miR-21, miR-25, miR-20a, miR-93, miR-183, and miR-203 have been identified, whereas those with reduced expression include miR-1, miR-126, miR-30a, miR-143, miR-145, miR-191, and miR-192[81]. A reduced expression of miR-192 seems to be related to an increase in tumor size[90], and a reduced expression of miR-145 was determined to be related to invasion, metastasis, degree of differentiation, and tumor size[91], demonstrating the relationship between specific miRNAs and tumor behavior.
It is believed that there is a difference in miRNA expression based on the stage of the tumor. For example, overexpression of miR-92a can be a biomarker for the early diagnosis of CRC[92], whereas the overexpression of miR-21 and miR-31 is associated with advanced CRC[93,94]. According to that, Tsukamoto et al[95], demonstrated that the overexpression of exosomal miR-21 showed a significant association with liver metastasis and TNM stage in CRC patients being associated with a decrease in the overall survival and disease-free survival rates. Besides that, as a proangiogenic miRNA, miR-21 targets the programmed cell death protein 4 (PDCD4) gene enhancing invasion, intravasation and metastasis[96].
Giráldez et al[97], in turn, showed a positive correlation of appearance of distant metastasis in advanced CRC patients and miR-103 overexpression. Besides that, miR-29a also presented overexpressed in metastatic CRC patients when compared to non-metastatic ones[98], and plasmatic expression of miR-203 and miR-141 could help in the differentiation of early and advanced CRC as demonstrated by Sun et al[99] In addition to these alterations correlated with stage of the tumor, there are also changes related to the response to treatment. MiR-1914-3p and miR-1915-3p were found downregulated in plasma samples from patients with chemo resistant CRC. This way, up-regulation of miR-1914-3p and -1915-3p reduces the chemoresistance abilities of chemo resistant CRC cells and may represent a possible therapy and diagnosis tool in CRC[100].
Alteration in angiogenesis is a contributing factor to tumor development, supporting proliferation, growth, dissemination, and metastasis[101,102]. MiRNAs are thought to participate as regulators of angiogenesis, acting both as antiangiogenic and proangiogenic[103,104], directly influencing endothelial cells or indirectly modulating protein expression[104], which makes them an interesting pathway in antiangiogenic therapies[105]. Non-responding bevacizumab (antibody anti-vascular endothelial growth factor A (VEGF-A) patients had increased levels of miR-126 correlated with an increase in tumor size[106]. On the other hand, miR-140-5p showed a tumor suppression role in CRC, targeting VEGF-A/MMP-2 pathway, and leading to inhibition of tumor progression and angiogenesis[107]. Additionally, miR-497 also blocks VEGF-A/ERK/MMP-9 pathway with reduction on angiogenesis, invasion, and metastasis in CRC[108].
Tumor growth depends on angiogenesis and the formation of new blood vessels, to ensure a continuous supply of oxygen and nutrients. This way, antiangiogenic agents are used to treat cancers, either alone or in combination[109]. However, the mechanistic details of how these combination therapies work are far from clear, and the lack of validated prognostic and predictive biomarkers represents one of the greatest obstacles in determining treatment outcomes and optimal response[109,110]. Based on that, miRNAs could serve as new predictive biomarker, therapeutic targets as an antiangiogenic therapy, or screening tools for immune-based therapies[111].
MiRNAs stability and predictive property make them ideal serum and plasma biomarkers in cancer patients, and they may be useful in predicting patterns of sensitivity and resistance to anti-cancer drugs[112]. This has brought attention to future personalized treatment strategies targeting miRNA expression in these patients. Thus, the identification of miRNAs as a tool for the early detection, prognostic evaluation, and treatment of CRC has gained clinical importance in recent years[81].
A summary of miRNAs found altered in CRC is shown in Table 3[113-141].
| MiRNA | Ref. |
| Let-7f | [27,113] |
| miR-1 | [114,115] |
| miR-101 | [29,116] |
| miR-103 | [117] |
| miR-106a | [28,32,117] |
| miR-106b | [118,119] |
| miR-107 | [117,120] |
| miR-10a | [33,117] |
| miR-122 | [115,121] |
| miR-126 | [27,32,122] |
| miR-1288 | [114,121] |
| miR-1305 | [114,123] |
| miR-133b | [32,124] |
| miR-135b | [32,121] |
| miR-138 | [116,123,125] |
| miR-139 | [117] |
| miR-139-5p | [113,121] |
| miR-141 | [42,121] |
| miR-143 | [60,126,127] |
| miR-145 | [60,128] |
| miR-147 | [117] |
| miR-148a | [113,117] |
| miR-150 | [32,114] |
| miR-155 | [32,56,120] |
| miR-16 | [27,28,116,124] |
| miR-17 | [124] |
| miR-181a | [117] |
| miR-181c | [32,117] |
| miR-183 | [117,121,124] |
| miR-18a | [59,129] |
| miR-191 | [28,117] |
| miR-192 | [27,90] |
| miR-194 | [67,117,121] |
| miR-195 | [27,117,130] |
| miR-196a | [32,124] |
| miR-19a | [29,117,118] |
| miR-19b | [28,113] |
| miR-200a | [117,121] |
| miR-200b | [68,121] |
| miR-200c | [117,121] |
| miR-203 | [61,124] |
| miR-20a | [113,119,131] |
| miR-21 | [9,32,132] |
| miR-210 | [133] |
| miR-212 | [133] |
| miR-214 | [65,123] |
| miR-215 | [32,90,121] |
| miR-218 | [115] |
| miR-222 | [117] |
| miR-223 | [28,32,120,121] |
| miR-224 | [117,121] |
| miR-23b | [28,114] |
| miR-25 | [117,124] |
| miR-27a | [117] |
| miR-27b | [115] |
| miR-29a | [27,114,117] |
| miR-29b | [29,114] |
| miR-30a | [32,115] |
| miR-30c | [32,115,120] |
| miR-31 | [32,56,128] |
| miR-32 | [120,134] |
| miR-320a | [34,135] |
| miR-328 | [59,134] |
| miR-34a | [136] |
| miR-375 | [27,137] |
| miR-422a | [59,113,116] |
| miR-422b | [27,138] |
| miR-424 | [118,123] |
| miR-451 | [114,116] |
| miR-490-3p | [115,121,125] |
| miR-497 | [115] |
| miR-506 | [139] |
| miR-552 | [121,134,140] |
| miR-650 | [59] |
| miR-7 | [32,123,140] |
| miR-885-5p | [59] |
| miR-892b | [115,121] |
| miR-92a | [119,141] |
| miR-93 | [113,124] |
| miR-95 | [117,123] |
| miR-96 | [121] |
| miR-98 | [62,113,117] |
Chronic inflammation is a contributing factor to carcinogenesis; therefore, patients with IBD, and especially those with colonic involvement, are at an increased risk for CRC, which is responsible for approximately 15% of deaths in this population[14]. However, the exact mechanisms underlying this relationship have not yet been fully elucidated[20,142]. Among the risk factors involved, we can emphasize a positive family history of CRC, long disease duration, colonic involvement, the presence of PSC, and the presence of disease activity[142]. For this reason, screening colonoscopy should be performed 8 years after the onset of symptoms in all patients with IBD to detect dysplasia in the early stages, with subsequent surveillance ranging from 1–5 years according to individual risk stratification[16]. Patients with IBD-associated PSC must undergo annual endoscopic surveillance after the diagnosis of PSC because of the increased risk of developing CRC[16].
The accumulation of reactive oxidative species from continuous cycles of inflammation and tissue repair results in damage to DNA, proteins, and lipids, leading to tumor development in patients with UC[143]. The progression of colitis-associated CRC occurs through processes that are perpetuated by the absence of dysplasia, undefined dysplasia, and low-and high-grade dysplasia until it progresses definitively to cancer[144]. Therefore, despite adequate surveillance, detecting dysplasia in these patients is difficult owing to difficulties in distinguishing lesions from the adjacent inflamed mucosa and foci of constant tissue regeneration and repair caused by the inflammatory process[145]. In 2010, O’ Connor et al[146] observed that chronic inflammation in these patients exposed them to tumorigenic traits based on the NF-κB pathways and inflammatory mediators such IL-6 and tumor necrosis factor-alpha (TNF-α). Oxidative stress, the activation of survival pathways, apoptosis, and the formation of a tumorigenic environment have been found to be involved in carcinogenesis. Changes in P53 gene occur early and can be detected before the onset of dysplasia, being identified in 47–85% of patients with colitis-associated CRC[146,147].
The release of cytokines resulting from chronic inflammation is related to the development of all stages of cancer, such as initiation, promotion, angiogenesis, and metastasis, and the transcription factor NF-κB is a primary factor in the inflammation/cancer cascade[146]. In addition to the involvement of molecular mediators in the inflammation/cancer relationship, such as cytokines, growth factors, Toll-like receptors (TLRs), Pl3K/MAPK signaling, and transcription factors (NF-κB/STAT3, P53, c-Myc, and Wnt/β-catenin), among others[148], it is worth noting that cellular changes caused by a chronic inflammatory status in patients with IBD also contribute to the development of the tumor[149].
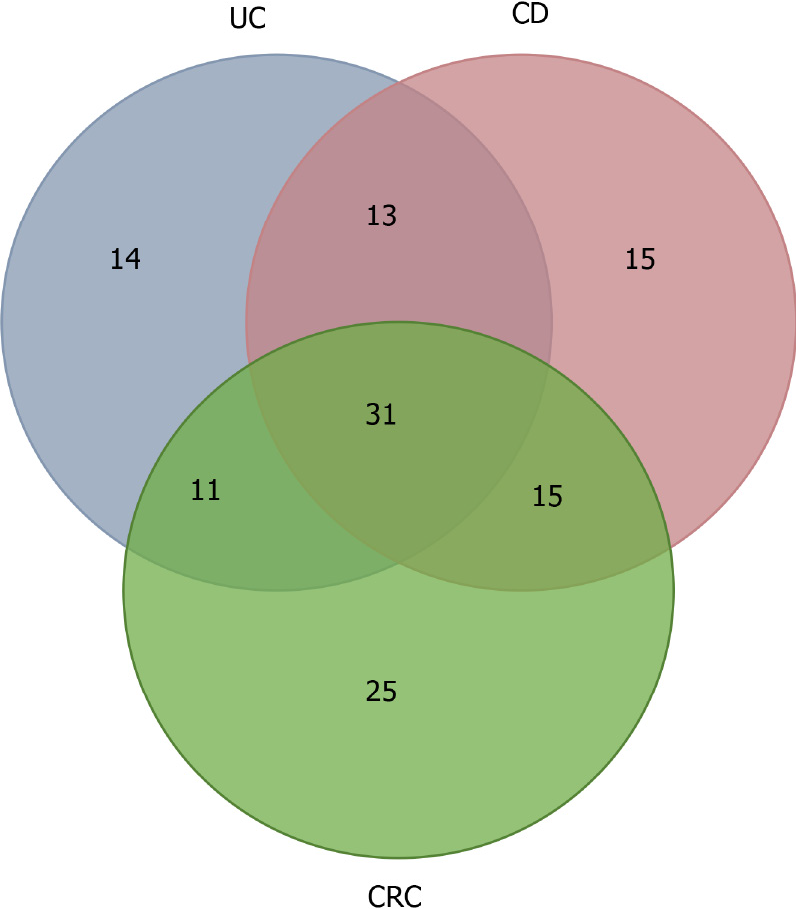
MiRNAs are believed to participate in the intestinal inflammation of IBD patients and contribute to the inflammation/tumor process. The identification of miRNAs involved in this tumorigenic process could be useful to stratify the risk of tumor development in patients with IBD and monitor and assess the prognosis of cases. Figure 1 and Table 4 shows the miRNAs found altered in UC, CD, CRC, and IBD-associated CRC summarizing the alterations described for each condition and the relationship between them.
| Disease | Altered miRNAs |
| UC | miR-489 miR-630 miR-125b-1 let-7f miR-188-5p miR-196b miR-20b miR-346 miR-24 miR-548a-3p let-7e miR-23a miR-124 miR-4284 |
| CD | let-7i miR-34c-5p miR-22 miR-9 miR-140-3p miR-196 miR-130a miR-30b miR-594 let-7b miR-29c miR-629 miR-449b miR-192-5p miR-146b-5p |
| CRC | miR-497 miR-27b miR-92a miR-451 miR-20a miR-1 miR-506 Let-7f miR-181a miR-218 miR-32 miR-34a miR-106b miR-210 miR-93 miR-148a miR-17 miR-25 miR-222 miR-103 miR-147 miR-212 miR-27a miR-107 miR-139 |
| UC – CD | miR-320c miR-559 miR-216b miR-301a miR-320b miR-548e miR-146a miR-127-3p miR-324-3p miR-663 miR-194b miR-206 miR-26a |
| UC – CRC | miR-200b miR-145 miR-98 miR-214 miR-194 miR-195 miR-135b miR-215 miR-422b miR-650 miR-192 |
| CD – CRC | miR-18a miR-19b miR-122 miR-328 miR-30a miR-106a miR-30c miR-23b miR-181c miR-422a miR-203 miR-141 miR-191 miR-133b miR-885-5p |
| IBD – CRC | miR-21 miR-155 miR-200c miR-139-5p miR-223 miR-892b miR-7 miR-143 miR-150 miR-320a miR-10a miR-29b miR-196a miR-95 miR-490-3p miR-16 miR-19a miR-126 miR-101 miR-1305 miR-424 miR-1288 miR-183 miR-96 miR-138 miR-375 miR-200a miR-29a miR-31 miR-224 miR-552 |
The first study on the differential expression of miRNAs in IBD and CRC-progression from non-neoplastic mucosa to dysplasia and invasive cancer was published in 2012[150]. Using naïve immunotherapy patients from CD or UC and tree types of tissue (non-neoplastic, dysplastic and neoplastic) from each patient, the authors observed that five miRNAs (miR-193b, miR-373, let-7e, miR-15b, and miR-372) were significantly downregulated in both diseases, correlated with the progression from non-neoplastic tissue to dysplasia and from dysplasia to cancer[150]. In CD patients, during non-neoplastic to dysplasia progression, miR-181a, miR-146b-5p, let-7e, and miR-17 were found to be upregulated, on the other hand, during the progression from dysplasia to cancer, let-7e, miR-17, and miR-143 were downregulated. From the deregulated miRNAs, let-7e, miR-15b, miR-17, miR-122, miR-124, and miR-372 had a tumorigenic effect on the TP53 pathway[150].
Bai et al[151], integrated genome-wide gene expression profiles and biological pathway information to explore the associations among UC, CD and CRC at function and gene level, and found 34, 20 and 47 risk pathways for UC, CD and CRC, respectively. Furthermore, the authors found that UC and CD share 16 pathways, indicating that the two inflammatory diseases are strikingly linked with each other at the biological pathway level. On the other hand, more pathways were shared between CRC and UC compared to CRC and CD, which might suggest that UC has a potential functional link with CRC. Pathways for UC and CRC were mainly related to the immune system and metabolism like the Intestinal immune network for IgA production[151].
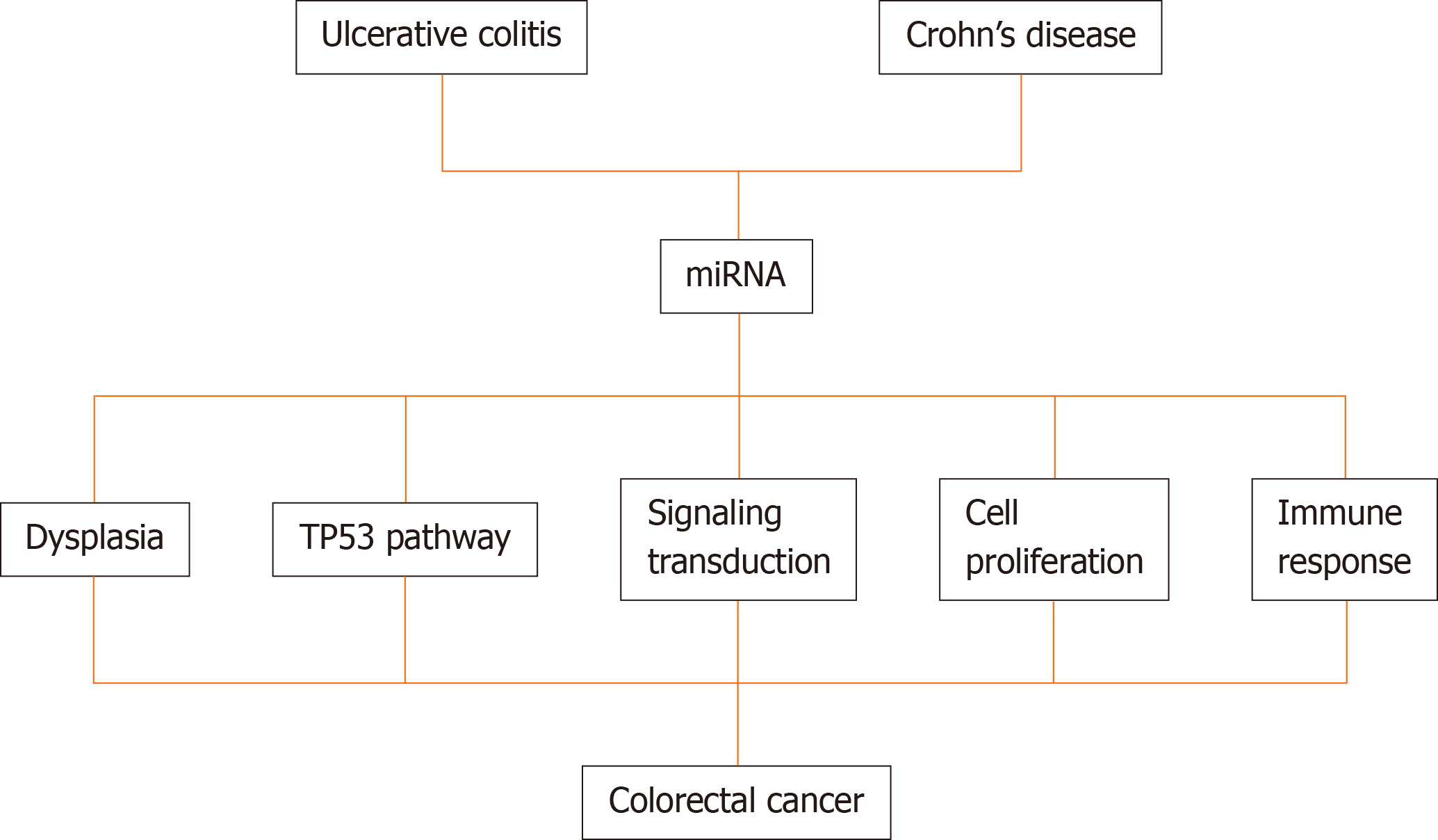
When analyzing the correlation between miRNA and the risk pathways, four miRNAs participate in all three diseases (miR-146a, miR-335, miR-26b and miR-124). Targets of these four miRNAs were mainly associated with “signaling transduction”, “cell proliferation” and “immune responses”. The authors concluded that miRNAs, genes and pathways are connected and there is a crosstalk between different pathways, and the miRNAs might mediate pathway crosstalk via regulating the corresponding gene[151].
A study conducted by Olaru et al[123] evaluated fragments of colon tissue from healthy people and compared with fragments of non-inflamed mucosa, inflamed mucosa of patients with IBD, IBD-dysplasia tissue and IBD-associated cancer tissue. The authors identified five upregulated miRNAs in IBD-related cancer, namely, miR-31, miR-135b, miR-200a, miR-224, and miR-552. MiR-224 was expressed at the highest-level during differentiation between IBD patients with tumors and patients with IBD without tumors.
Several miRNAs are being correlated with the transition of normal tissue to dysplasia or neoplasia. In CD, miR-196 is a marker of dysplasia[57]. Further, miR-124a, a tumor-suppressive miRNA, undergoes methylation during exposure to chronic inflammation leading to the emergence of dysplasia and then neoplastic tissues in UC patients[152]. Wan et al[153] revealed that miR-155 is related to the involvement of cancer cells and worse prognosis. Additionally, Fang et al[154] analyzed patients with colorectal disease and healthy controls and determined that miR-24, miR-320a, and miR-423-5p, which were aberrantly expressed, were associated with high sensitivity for the detection of early CRC and could be promising biomarkers for IBD. In a recent study, Al-Mustanjid et al[155] used a system biology approach to identify common molecular signatures and pathways that interact between IBD and CRC and found that mir-335-5p, mir-26b-5p, mir-124-3p, mir-16-5p, mir-192-5p, mir-548c-3p, mir-29b-3p, mir-155-5p, mir-21-5p, mir-15a-5p are related with the 177 common differentially expressed genes between IBD and CRC. A schematic diagram showing the pathways modulated by miRNAs in IBD-related CRC progression is shown in Figure 2.
Due to the role of inflammation in CRC carcinogenesis, prevention methods targeting pro-inflammatory pathways have been studied. Among these pathways are the NF-κB pathway, which regulates innate and adaptive immune functions[156], and the phosphatidylinositol 3 kinase pathways (Pl3K), TLR, Janus kinase, and the activating factor transcript 3 (JAK/STAT3), which are also involved in the inflammation/cancer cascade[157-159]. Thus, the development of drugs that regulate miRNAs and these pro-inflammatory pathways has become an important field to prevent the development of CRC in these patients[20].
After predicting the target genes for the IBD-associated CRC miRNAs[160] were found 8939 possible targets for the selected miRNAs (Supporting material). The enrichment analysis performed showed large modulation of signaling pathways that participate in the pathophysiology of both IBD and CRC, pathways such as the VEGFA, TGF- β and EGFR pathways that have already been discussed in this review[161-163] (Table 5). The participation of these pathways in both diseases could help to explain the correlation between these two conditions and why IBD patients are more likely to develop CRC.
| Pathway | P value | Adjusted P value |
| VEGFA-VEGFR2 signaling pathway | 2.577e-31 | 1.203e-28 |
| TGF-beta signaling pathway | 1.823e-23 | 4.257e-21 |
| EGF/EGFR signaling pathway | 6.399e-20 | 9.961e-18 |
| DNA damage response (only ATM dependent) | 8.953e-20 | 1.045e-17 |
| Oncostatin M signaling pathway | 1.426e-18 | 1.322e-16 |
| Integrated breast cancer pathway | 1.699e-18 | 1.322e-16 |
| Insulin signaling | 4.429e-18 | 2.955e-16 |
| Focal adhesion | 2.120e-17 | 1.238e-15 |
| Signaling pathways in glioblastoma | 4.047e-17 | 2.100e-15 |
| ErbB signaling pathway | 4.559e-16 | 2.129e-14 |
Inflammatory processes and immunosuppressive drugs are the main risk factors for the development of tumors in patients with IBD. The identification of miRNAs as diagnostic biomarkers can revolutionize the screening of high-risk patients, allowing for personalized surveillance according to individual risk and the early diagnosis of lesions, directly affecting the treatment and prognosis of the patient. Further studies are needed to clarify the role of miRNAs in disease pathogenesis and evolution in patients with IBD, in addition to the identification of miRNAs related to the therapeutic response and disease prognosis. In the future, therapies based on miRNA modulation could be used in clinical practice to achieve remission of the disease and restore the quality of life for patients with IBD.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Raoul W S-Editor: Ma YJ L-Editor: A P-Editor: Li JH
| 1. | Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 853] [Cited by in RCA: 1821] [Article Influence: 227.6] [Reference Citation Analysis (0)] |
| 2. | Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848-5856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 727] [Cited by in RCA: 789] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 3. | Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67:129-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 505] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 4. | Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733-1741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 2086] [Article Influence: 139.1] [Reference Citation Analysis (0)] |
| 5. | Saito Y, Suzuki H, Hibi T. The role of microRNAs in gastrointestinal cancers. J Gastroenterol. 2009;44 Suppl 19:18-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16085] [Article Influence: 1005.3] [Reference Citation Analysis (2)] |
| 7. | Baz M, Ibrahim T. Role of microRNAs in the predisposition to gastrointestinal malignancies. World J Clin Cases. 2020;8:1580-1585. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 8. | Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, Goel A. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766-1774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 251] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 9. | Shi C, Liang Y, Yang J, Xia Y, Chen H, Han H, Yang Y, Wu W, Gao R, Qin H. MicroRNA-21 knockout improve the survival rate in DSS induced fatal colitis through protecting against inflammation and tissue injury. PLoS One. 2013;8:e66814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 123] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 10. | Gracias DT, Stelekati E, Hope JL, Boesteanu AC, Doering TA, Norton J, Mueller YM, Fraietta JA, Wherry EJ, Turner M, Katsikis PD. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol. 2013;14:593-602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 223] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 11. | Singh UP, Murphy AE, Enos RT, Shamran HA, Singh NP, Guan H, Hegde VL, Fan D, Price RL, Taub DD, Mishra MK, Nagarkatti M, Nagarkatti PS. miR-155 deficiency protects mice from experimental colitis by reducing T helper type 1/type 17 responses. Immunology. 2014;143:478-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 102] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 12. | Yang L, Belaguli N, Berger DH. MicroRNA and colorectal cancer. World J Surg. 2009;33:638-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 100] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 13. | Cowland JB, Hother C, Grønbaek K. MicroRNAs and cancer. APMIS. 2007;115:1090-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 135] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 14. | Munkholm P. Review article: the incidence and prevalence of colorectal cancer in inflammatory bowel disease. Aliment Pharmacol Ther. 2003;18 Suppl 2:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 395] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 15. | Viscido A, Bagnardi V, Sturniolo GC, Annese V, Frieri G, D'Arienzo A, Papi C, Riegler G, Corrao G, Caprilli R; GISC: Italian Group for the Study of the Colon and Rectum. Survival and causes of death in Italian patients with ulcerative colitis. A GISC nationwide study. Dig Liver Dis. 2001;33:686-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | Magro F, Gionchetti P, Eliakim R, Ardizzone S, Armuzzi A, Barreiro-de Acosta M, Burisch J, Gecse KB, Hart AL, Hindryckx P, Langner C, Limdi JK, Pellino G, Zagórowicz E, Raine T, Harbord M, Rieder F; European Crohn’s and Colitis Organisation [ECCO]. Third European Evidence-based Consensus on Diagnosis and Management of Ulcerative Colitis. Part 1: Definitions, Diagnosis, Extra-intestinal Manifestations, Pregnancy, Cancer Surveillance, Surgery, and Ileo-anal Pouch Disorders. J Crohns Colitis. 2017;11:649-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1446] [Cited by in RCA: 1298] [Article Influence: 162.3] [Reference Citation Analysis (0)] |
| 17. | Askling J, Dickman PW, Karlén P, Broström O, Lapidus A, Löfberg R, Ekbom A. Family history as a risk factor for colorectal cancer in inflammatory bowel disease. Gastroenterology. 2001;120:1356-1362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 283] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 18. | Zheng HH, Jiang XL. Increased risk of colorectal neoplasia in patients with primary sclerosing cholangitis and inflammatory bowel disease: a meta-analysis of 16 observational studies. Eur J Gastroenterol Hepatol. 2016;28:383-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 106] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 19. | Burisch J, Munkholm P. The epidemiology of inflammatory bowel disease. Scand J Gastroenterol. 2015;50:942-951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 247] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 20. | Feng Y, Zhang Y, Zhou D, Chen G, Li N. MicroRNAs, intestinal inflammatory and tumor. Bioorg Med Chem Lett. 2019;29:2051-2058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | Magro F, Langner C, Driessen A, Ensari A, Geboes K, Mantzaris GJ, Villanacci V, Becheanu G, Borralho Nunes P, Cathomas G, Fries W, Jouret-Mourin A, Mescoli C, de Petris G, Rubio CA, Shepherd NA, Vieth M, Eliakim R; European Society of Pathology (ESP); European Crohn's and Colitis Organisation (ECCO). European consensus on the histopathology of inflammatory bowel disease. J Crohns Colitis. 2013;7:827-851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 540] [Cited by in RCA: 467] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 22. | de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13:13-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1185] [Cited by in RCA: 1120] [Article Influence: 124.4] [Reference Citation Analysis (1)] |
| 23. | Maaser C, Sturm A, Vavricka SR, Kucharzik T, Fiorino G, Annese V, Calabrese E, Baumgart DC, Bettenworth D, Borralho Nunes P, Burisch J, Castiglione F, Eliakim R, Ellul P, González-Lama Y, Gordon H, Halligan S, Katsanos K, Kopylov U, Kotze PG, Krustinš E, Laghi A, Limdi JK, Rieder F, Rimola J, Taylor SA, Tolan D, van Rheenen P, Verstockt B, Stoker J; European Crohn’s and Colitis Organisation [ECCO] and the European Society of Gastrointestinal and Abdominal Radiology [ESGAR]. ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 1: Initial diagnosis, monitoring of known IBD, detection of complications. J Crohns Colitis. 2019;13:144-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1242] [Cited by in RCA: 1164] [Article Influence: 194.0] [Reference Citation Analysis (0)] |
| 24. | Guindi M, Riddell RH. Indeterminate colitis. J Clin Pathol. 2004;57:1233-1244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 115] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 25. | Schönauen K, Le N, von Arnim U, Schulz C, Malfertheiner P, Link A. Circulating and Fecal microRNAs as Biomarkers for Inflammatory Bowel Diseases. Inflamm Bowel Dis. 2018;24:1547-1557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 26. | Whiteoak SR, Felwick R, Sanchez-Elsner T, Fraser Cummings JR. MicroRNAs in inflammatory bowel diseases: paradoxes and possibilities. Inflamm Bowel Dis. 2015;21:1160-1165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 27. | Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624-1635.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 404] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 28. | Wu F, Zhang S, Dassopoulos T, Harris ML, Bayless TM, Meltzer SJ, Brant SR, Kwon JH. Identification of microRNAs associated with ileal and colonic Crohn's disease. Inflamm Bowel Dis. 2010;16:1729-1738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 236] [Cited by in RCA: 239] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 29. | Schaefer JS, Attumi T, Opekun AR, Abraham B, Hou J, Shelby H, Graham DY, Streckfus C, Klein JR. MicroRNA signatures differentiate Crohn's disease from ulcerative colitis. BMC Immunol. 2015;16:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 30. | Thorlacius-Ussing G, Schnack Nielsen B, Andersen V, Holmstrøm K, Pedersen AE. Expression and Localization of miR-21 and miR-126 in Mucosal Tissue from Patients with Inflammatory Bowel Disease. Inflamm Bowel Dis. 2017;23:739-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 31. | Guo Z, Wu R, Gong J, Zhu W, Li Y, Wang Z, Li N, Li J. Altered microRNA expression in inflamed and non-inflamed terminal ileal mucosa of adult patients with active Crohn's disease. J Gastroenterol Hepatol. 2015;30:109-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 32. | Fasseu M, Tréton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, Aparicio T, Daniel F, Soulé JC, Moreau R, Bouhnik Y, Laburthe M, Groyer A, Ogier-Denis E. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 2010;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 223] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 33. | Wu W, He C, Liu C, Cao AT, Xue X, Evans-Marin HL, Sun M, Fang L, Yao S, Pinchuk IV, Powell DW, Liu Z, Cong Y. miR-10a inhibits dendritic cell activation and Th1/Th17 cell immune responses in IBD. Gut. 2015;64:1755-1764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 121] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 34. | Pierdomenico M, Cesi V, Cucchiara S, Vitali R, Prete E, Costanzo M, Aloi M, Oliva S, Stronati L. NOD2 Is Regulated By Mir-320 in Physiological Conditions but this Control Is Altered in Inflamed Tissues of Patients with Inflammatory Bowel Disease. Inflamm Bowel Dis. 2016;22:315-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 35. | Chuang AY, Chuang JC, Zhai Z, Wu F, Kwon JH. NOD2 expression is regulated by microRNAs in colonic epithelial HCT116 cells. Inflamm Bowel Dis. 2014;20:126-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 77] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 36. | Min M, Peng L, Yang Y, Guo M, Wang W, Sun G. MicroRNA-155 is involved in the pathogenesis of ulcerative colitis by targeting FOXO3a. Inflamm Bowel Dis. 2014;20:652-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 37. | Lu ZJ, Wu JJ, Jiang WL, Xiao JH, Tao KZ, Ma L, Zheng P, Wan R, Wang XP. MicroRNA-155 promotes the pathogenesis of experimental colitis by repressing SHIP-1 expression. World J Gastroenterol. 2017;23:976-985. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 42] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 38. | Shi T, Xie Y, Fu Y, Zhou Q, Ma Z, Ma J, Huang Z, Zhang J, Chen J. The signaling axis of microRNA-31/interleukin-25 regulates Th1/Th17-mediated inflammation response in colitis. Mucosal Immunol. 2017;10:983-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 39. | Wang H, Chao K, Ng SC, Bai AH, Yu Q, Yu J, Li M, Cui Y, Chen M, Hu JF, Zhang S. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease. Genome Biol. 2016;17:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 140] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 40. | Hou J, Hu X, Chen B, Chen X, Zhao L, Chen Z, Liu F, Liu Z. miR-155 targets Est-1 and induces ulcerative colitis via the IL-23/17/6-mediated Th17 pathway. Pathol Res Pract. 2017;213:1289-1295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 41. | Nata T, Fujiya M, Ueno N, Moriichi K, Konishi H, Tanabe H, Ohtake T, Ikuta K, Kohgo Y. MicroRNA-146b improves intestinal injury in mouse colitis by activating nuclear factor-κB and improving epithelial barrier function. J Gene Med. 2013;15:249-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 42. | Huang Z, Shi T, Zhou Q, Shi S, Zhao R, Shi H, Dong L, Zhang C, Zeng K, Chen J, Zhang J. miR-141 Regulates colonic leukocytic trafficking by targeting CXCL12β during murine colitis and human Crohn's disease. Gut. 2014;63:1247-1257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 119] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 43. | Jin X, Chen D, Zheng RH, Zhang H, Chen YP, Xiang Z. miRNA-133a-UCP2 pathway regulates inflammatory bowel disease progress by influencing inflammation, oxidative stress and energy metabolism. World J Gastroenterol. 2017;23:76-86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 46] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 44. | Tian Y, Xu J, Li Y, Zhao R, Du S, Lv C, Wu W, Liu R, Sheng X, Song Y, Bi X, Li G, Li M, Wu X, Lou P, You H, Cui W, Sun J, Shuai J, Ren F, Zhang B, Guo M, Hou X, Wu K, Xue L, Zhang H, Plikus MV, Cong Y, Lengner CJ, Liu Z, Yu Z. MicroRNA-31 Reduces Inflammatory Signaling and Promotes Regeneration in Colon Epithelium, and Delivery of Mimics in Microspheres Reduces Colitis in Mice. Gastroenterology. 2019;156:2281-2296.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 158] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 45. | He C, Shi Y, Wu R, Sun M, Fang L, Wu W, Liu C, Tang M, Li Z, Wang P, Cong Y, Liu Z. miR-301a promotes intestinal mucosal inflammation through induction of IL-17A and TNF-α in IBD. Gut. 2016;65:1938-1950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 133] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 46. | He C, Yu T, Shi Y, Ma C, Yang W, Fang L, Sun M, Wu W, Xiao F, Guo F, Chen M, Yang H, Qian J, Cong Y, Liu Z. MicroRNA 301A Promotes Intestinal Inflammation and Colitis-Associated Cancer Development by Inhibiting BTG1. Gastroenterology. 2017;152:1434-1448.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 118] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 47. | Bae HJ, Noh JH, Kim JK, Eun JW, Jung KH, Kim MG, Chang YG, Shen Q, Kim SJ, Park WS, Lee JY, Nam SW. MicroRNA-29c functions as a tumor suppressor by direct targeting oncogenic SIRT1 in hepatocellular carcinoma. Oncogene. 2014;33:2557-2567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 116] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 48. | Chapman CG, Pekow J. The emerging role of miRNAs in inflammatory bowel disease: a review. Therap Adv Gastroenterol. 2015;8:4-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 126] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 49. | Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105:1516-1521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 737] [Cited by in RCA: 797] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 50. | Fedyk ER, Wyant T, Yang LL, Csizmadia V, Burke K, Yang H, Kadambi VJ. Exclusive antagonism of the α4 β7 integrin by vedolizumab confirms the gut-selectivity of this pathway in primates. Inflamm Bowel Dis. 2012;18:2107-2119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 51. | Pathak S, Grillo AR, Scarpa M, Brun P, D'Incà R, Nai L, Banerjee A, Cavallo D, Barzon L, Palù G, Sturniolo GC, Buda A, Castagliuolo I. MiR-155 modulates the inflammatory phenotype of intestinal myofibroblasts by targeting SOCS1 in ulcerative colitis. Exp Mol Med. 2015;47:e164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 92] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 52. | Morilla I, Uzzan M, Laharie D, Cazals-Hatem D, Denost Q, Daniel F, Belleannee G, Bouhnik Y, Wainrib G, Panis Y, Ogier-Denis E, Treton X. Colonic MicroRNA Profiles, Identified by a Deep Learning Algorithm, That Predict Responses to Therapy of Patients With Acute Severe Ulcerative Colitis. Clin Gastroenterol Hepatol. 2019;17:905-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (1)] |
| 53. | Cordes F, Demmig C, Bokemeyer A, Brückner M, Lenze F, Lenz P, Nowacki T, Tepasse P, Schmidt HH, Schmidt MA, Cichon C, Bettenworth D. MicroRNA-320a Monitors Intestinal Disease Activity in Patients With Inflammatory Bowel Disease. Clin Transl Gastroenterol. 2020;11:e00134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 54. | Verdier J, Breunig IR, Ohse MC, Roubrocks S, Kleinfeld S, Roy S, Streetz K, Trautwein C, Roderburg C, Sellge G. Faecal Micro-RNAs in Inflammatory Bowel Diseases. J Crohns Colitis. 2020;14:110-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 55. | Ben-Shachar S, Yanai H, Sherman Horev H, Elad H, Baram L, Isakov O, Tulchinsky H, Pasmanik-Chor M, Shomron N, Dotan I. Correction: MicroRNAs Expression in the Ileal Pouch of Patients with Ulcerative Colitis Is Robustly Up-Regulated and Correlates with Disease Phenotypes. PLoS One. 2016;11:e0165220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 56. | Gwiggner M, Martinez-Nunez RT, Whiteoak SR, Bondanese VP, Claridge A, Collins JE, Cummings JRF, Sanchez-Elsner T. MicroRNA-31 and MicroRNA-155 Are Overexpressed in Ulcerative Colitis and Regulate IL-13 Signaling by Targeting Interleukin 13 Receptor α-1. Genes (Basel). 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 57. | Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hébuterne X, Harel-Bellan A, Mograbi B, Darfeuille-Michaud A, Hofman P. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet. 2011;43:242-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 428] [Cited by in RCA: 458] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 58. | Lin J, Welker NC, Zhao Z, Li Y, Zhang J, Reuss SA, Zhang X, Lee H, Liu Y, Bronner MP. Novel specific microRNA biomarkers in idiopathic inflammatory bowel disease unrelated to disease activity. Mod Pathol. 2014;27:602-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 59. | Iborra M, Bernuzzi F, Correale C, Vetrano S, Fiorino G, Beltrán B, Marabita F, Locati M, Spinelli A, Nos P, Invernizzi P, Danese S. Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin Exp Immunol. 2013;173:250-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 60. | Pekow JR, Dougherty U, Mustafi R, Zhu H, Kocherginsky M, Rubin DT, Hanauer SB, Hart J, Chang EB, Fichera A, Joseph LJ, Bissonnette M. miR-143 and miR-145 are downregulated in ulcerative colitis: putative regulators of inflammation and protooncogenes. Inflamm Bowel Dis. 2012;18:94-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 61. | Peck BC, Weiser M, Lee SE, Gipson GR, Iyer VB, Sartor RB, Herfarth HH, Long MD, Hansen JJ, Isaacs KL, Trembath DG, Rahbar R, Sadiq TS, Furey TS, Sethupathy P, Sheikh SZ. MicroRNAs Classify Different Disease Behavior Phenotypes of Crohn's Disease and May Have Prognostic Utility. Inflamm Bowel Dis. 2015;21:2178-2187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 62. | Coskun M, Bjerrum JT, Seidelin JB, Troelsen JT, Olsen J, Nielsen OH. miR-20b, miR-98, miR-125b-1*, and let-7e* as new potential diagnostic biomarkers in ulcerative colitis. World J Gastroenterol. 2013;19:4289-4299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 67] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 63. | Feng X, Wang H, Ye S, Guan J, Tan W, Cheng S, Wei G, Wu W, Wu F, Zhou Y. Up-regulation of microRNA-126 may contribute to pathogenesis of ulcerative colitis via regulating NF-kappaB inhibitor IκBα. PLoS One. 2012;7:e52782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 64. | Bian Z, Li L, Cui J, Zhang H, Liu Y, Zhang CY, Zen K. Role of miR-150-targeting c-Myb in colonic epithelial disruption during dextran sulphate sodium-induced murine experimental colitis and human ulcerative colitis. J Pathol. 2011;225:544-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 65. | Polytarchou C, Hommes DW, Palumbo T, Hatziapostolou M, Koutsioumpa M, Koukos G, van der Meulen-de Jong AE, Oikonomopoulos A, van Deen WK, Vorvis C, Serebrennikova OB, Birli E, Choi J, Chang L, Anton PA, Tsichlis PN, Pothoulakis C, Verspaget HW, Iliopoulos D. MicroRNA214 Is Associated With Progression of Ulcerative Colitis, and Inhibition Reduces Development of Colitis and Colitis-Associated Cancer in Mice. Gastroenterology. 2015;149:981-92.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 111] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 66. | Koukos G, Polytarchou C, Kaplan JL, Morley-Fletcher A, Gras-Miralles B, Kokkotou E, Baril-Dore M, Pothoulakis C, Winter HS, Iliopoulos D. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. 2013;145:842-52.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 191] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 67. | Zahm AM, Hand NJ, Tsoucas DM, Le Guen CL, Baldassano RN, Friedman JR. Rectal microRNAs are perturbed in pediatric inflammatory bowel disease of the colon. J Crohns Colitis. 2014;8:1108-1117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 68. | Shen Y, Zhou M, Yan J, Gong Z, Xiao Y, Zhang C, Du P, Chen Y. miR-200b inhibits TNF-α-induced IL-8 secretion and tight junction disruption of intestinal epithelial cells in vitro. Am J Physiol Gastrointest Liver Physiol. 2017;312:G123-G132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 69. | Koukos G, Polytarchou C, Kaplan JL, Oikonomopoulos A, Ziring D, Hommes DW, Wahed R, Kokkotou E, Pothoulakis C, Winter HS, Iliopoulos D. A microRNA signature in pediatric ulcerative colitis: deregulation of the miR-4284/CXCL5 pathway in the intestinal epithelium. Inflamm Bowel Dis. 2015;21:996-1005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 70. | Fisher K, Lin J. MicroRNA in inflammatory bowel disease: Translational research and clinical implication. World J Gastroenterol. 2015;21:12274-12282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 46] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 71. | Xu XM, Zhang HJ. miRNAs as new molecular insights into inflammatory bowel disease: Crucial regulators in autoimmunity and inflammation. World J Gastroenterol. 2016;22:2206-2218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 58] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (1)] |
| 72. | James JP, Riis LB, Malham M, Høgdall E, Langholz E, Nielsen BS. MicroRNA Biomarkers in IBD-Differential Diagnosis and Prediction of Colitis-Associated Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 73. | Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1570] [Cited by in RCA: 3024] [Article Influence: 504.0] [Reference Citation Analysis (3)] |
| 74. | Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3058] [Cited by in RCA: 3306] [Article Influence: 413.3] [Reference Citation Analysis (3)] |
| 75. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55828] [Article Influence: 7975.4] [Reference Citation Analysis (132)] |
| 76. | Henrikson NB, Webber EM, Goddard KA, Scrol A, Piper M, Williams MS, Zallen DT, Calonge N, Ganiats TG, Janssens AC, Zauber A, Lansdorp-Vogelaar I, van Ballegooijen M, Whitlock EP. Family history and the natural history of colorectal cancer: systematic review. Genet Med. 2015;17:702-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 102] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 77. | Syngal S, Brand RE, Church JM, Giardiello FM, Hampel HL, Burt RW; American College of Gastroenterology. ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol. 2015;110:223-262; quiz 263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 957] [Cited by in RCA: 1090] [Article Influence: 109.0] [Reference Citation Analysis (0)] |
| 78. | Medema JP. Cancer stem cells: the challenges ahead. Nat Cell Biol. 2013;15:338-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 516] [Cited by in RCA: 559] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 79. | Nassar D, Blanpain C. Cancer Stem Cells: Basic Concepts and Therapeutic Implications. Annu Rev Pathol. 2016;11:47-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 405] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 80. | Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6773] [Cited by in RCA: 6673] [Article Influence: 513.3] [Reference Citation Analysis (0)] |
| 81. | Mohammadi A, Mansoori B, Baradaran B. The role of microRNAs in colorectal cancer. Biomed Pharmacother. 2016;84:705-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 129] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 82. | Hofacker IL. How microRNAs choose their targets. Nat Genet. 2007;39:1191-1192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 83. | Chivukula RR, Shi G, Acharya A, Mills EW, Zeitels LR, Anandam JL, Abdelnaby AA, Balch GC, Mansour JC, Yopp AC, Maitra A, Mendell JT. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell. 2014;157:1104-1116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 84. | Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795-5802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 376] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 85. | Dinu D, Dobre M, Panaitescu E, Bîrlă R, Iosif C, Hoara P, Caragui A, Boeriu M, Constantinoiu S, Ardeleanu C. Prognostic significance of KRAS gene mutations in colorectal cancer--preliminary study. J Med Life. 2014;7:581-587. [PubMed] |
| 86. | Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 471] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 87. | Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 713] [Cited by in RCA: 714] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 88. | Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 924] [Cited by in RCA: 1054] [Article Influence: 95.8] [Reference Citation Analysis (0)] |
| 89. | Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136:3699-3714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 626] [Cited by in RCA: 654] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 90. | Chiang Y, Song Y, Wang Z, Liu Z, Gao P, Liang J, Zhu J, Xing C, Xu H. microRNA-192, -194 and -215 are frequently downregulated in colorectal cancer. Exp Ther Med. 2012;3:560-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 89] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 91. | Liu Q, Yang W, Luo Y, Hu S, Zhu L. Correlation between miR-21 and miR-145 and the incidence and prognosis of colorectal cancer. J BUON. 2018;23:29-35. [PubMed] |
| 92. | Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 750] [Cited by in RCA: 750] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 93. | Wu Y, Song Y, Xiong Y, Wang X, Xu K, Han B, Bai Y, Li L, Zhang Y, Zhou L. MicroRNA-21 (Mir-21) Promotes Cell Growth and Invasion by Repressing Tumor Suppressor PTEN in Colorectal Cancer. Cell Physiol Biochem. 2017;43:945-958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 146] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 94. | Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, Chen HY, Sun XF. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers. 2009;26:27-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 114] [Reference Citation Analysis (0)] |
| 95. | Tsukamoto M, Iinuma H, Yagi T, Matsuda K, Hashiguchi Y. Circulating Exosomal MicroRNA-21 as a Biomarker in Each Tumor Stage of Colorectal Cancer. Oncology. 2017;92:360-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 160] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 96. | Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128-2136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1350] [Cited by in RCA: 1458] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 97. | Giráldez MD, Lozano JJ, Ramírez G, Hijona E, Bujanda L, Castells A, Gironella M. Circulating microRNAs as biomarkers of colorectal cancer: results from a genome-wide profiling and validation study. Clin Gastroenterol Hepatol. 2013;11:681-8.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 139] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 98. | Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012;36:e61-e67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 166] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 99. | Sun Y, Liu Y, Cogdell D, Calin GA, Sun B, Kopetz S, Hamilton SR, Zhang W. Examining plasma microRNA markers for colorectal cancer at different stages. Oncotarget. 2016;7:11434-11449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 100. | Hu J, Cai G, Xu Y, Cai S. The Plasma microRNA miR-1914* and -1915 Suppresses Chemoresistant in Colorectal Cancer Patients by Down-regulating NFIX. Curr Mol Med. 2016;16:70-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 101. | Lee SL, Rouhi P, Dahl Jensen L, Zhang D, Ji H, Hauptmann G, Ingham P, Cao Y. Hypoxia-induced pathological angiogenesis mediates tumor cell dissemination, invasion, and metastasis in a zebrafish tumor model. Proc Natl Acad Sci USA. 2009;106:19485-19490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 189] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 102. | Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20:409-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 1016] [Article Influence: 127.0] [Reference Citation Analysis (0)] |
| 103. | Goradel NH, Mohammadi N, Haghi-Aminjan H, Farhood B, Negahdari B, Sahebkar A. Regulation of tumor angiogenesis by microRNAs: State of the art. J Cell Physiol. 2019;234:1099-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 104. | Wang Y, Wang L, Chen C, Chu X. New insights into the regulatory role of microRNA in tumor angiogenesis and clinical implications. Mol Cancer. 2018;17:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 136] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 105. | Leone P, Buonavoglia A, Fasano R, Solimando AG, De Re V, Cicco S, Vacca A, Racanelli V. Insights into the Regulation of Tumor Angiogenesis by Micro-RNAs. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 106. | Hansen TF, Carlsen AL, Heegaard NH, Sørensen FB, Jakobsen A. Changes in circulating microRNA-126 during treatment with chemotherapy and bevacizumab predicts treatment response in patients with metastatic colorectal cancer. Br J Cancer. 2015;112:624-629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 99] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 107. | Zhang W, Zou C, Pan L, Xu Y, Qi W, Ma G, Hou Y, Jiang P. MicroRNA-140-5p inhibits the progression of colorectal cancer by targeting VEGFA. Cell Physiol Biochem. 2015;37:1123-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 108. | Qiu Y, Yu H, Shi X, Xu K, Tang Q, Liang B, Hu S, Bao Y, Xu J, Cai J, Peng W, Cao Q, Yin P. microRNA-497 inhibits invasion and metastasis of colorectal cancer cells by targeting vascular endothelial growth factor-A. Cell Prolif. 2016;49:69-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 109. | Casanovas O. Cancer: Limitations of therapies exposed. Nature. 2012;484:44-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 110. | Jászai J, Schmidt MHH. Trends and Challenges in Tumor Anti-Angiogenic Therapies. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 111. | Romano G, Kwong LN. Diagnostic and therapeutic applications of miRNA-based strategies to cancer immunotherapy. Cancer Metastasis Rev. 2018;37:45-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 112. | Ma R, Jiang T, Kang X. Circulating microRNAs in cancer: origin, function and application. J Exp Clin Cancer Res. 2012;31:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 143] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 113. | Chang KH, Miller N, Kheirelseid EA, Lemetre C, Ball GR, Smith MJ, Regan M, McAnena OJ, Kerin MJ. MicroRNA signature analysis in colorectal cancer: identification of expression profiles in stage II tumors associated with aggressive disease. Int J Colorectal Dis. 2011;26:1415-1422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 86] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 114. | Slattery ML, Wolff E, Hoffman MD, Pellatt DF, Milash B, Wolff RK. MicroRNAs and colon and rectal cancer: differential expression by tumor location and subtype. Genes Chromosomes Cancer. 2011;50:196-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 117] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 115. | Mosakhani N, Sarhadi VK, Borze I, Karjalainen-Lindsberg ML, Sundström J, Ristamäki R, Osterlund P, Knuutila S. MicroRNA profiling differentiates colorectal cancer according to KRAS status. Genes Chromosomes Cancer. 2012;51:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 116. | Faltejskova P, Svoboda M, Srutova K, Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K, Kiss I, Vyzula R, Slaby O. Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med. 2012;16:2655-2666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 117] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 117. | Monzo M, Navarro A, Bandres E, Artells R, Moreno I, Gel B, Ibeas R, Moreno J, Martinez F, Diaz T, Martinez A, Balagué O, Garcia-Foncillas J. Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell Res. 2008;18:823-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 148] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 118. | Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis. 2010;11:50-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 180] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 119. | Nishida N, Nagahara M, Sato T, Mimori K, Sudo T, Tanaka F, Shibata K, Ishii H, Sugihara K, Doki Y, Mori M. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res. 2012;18:3054-3070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 140] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 120. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4471] [Cited by in RCA: 4523] [Article Influence: 238.1] [Reference Citation Analysis (0)] |
| 121. | Olaru AV, Selaru FM, Mori Y, Vazquez C, David S, Paun B, Cheng Y, Jin Z, Yang J, Agarwal R, Abraham JM, Dassopoulos T, Harris M, Bayless TM, Kwon J, Harpaz N, Livak F, Meltzer SJ. Dynamic changes in the expression of MicroRNA-31 during inflammatory bowel disease-associated neoplastic transformation. Inflamm Bowel Dis. 2011;17:221-231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 122. | Li XM, Wang AM, Zhang J, Yi H. Down-regulation of miR-126 expression in colorectal cancer and its clinical significance. Med Oncol. 2011;28:1054-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 123. | Olaru AV, Yamanaka S, Vazquez C, Mori Y, Cheng Y, Abraham JM, Bayless TM, Harpaz N, Selaru FM, Meltzer SJ. MicroRNA-224 negatively regulates p21 expression during late neoplastic progression in inflammatory bowel disease. Inflamm Bowel Dis. 2013;19:471-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 124. | Earle JS, Luthra R, Romans A, Abraham R, Ensor J, Yao H, Hamilton SR. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn. 2010;12:433-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 135] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 125. | Balaguer F, Moreira L, Lozano JJ, Link A, Ramirez G, Shen Y, Cuatrecasas M, Arnold M, Meltzer SJ, Syngal S, Stoffel E, Jover R, Llor X, Castells A, Boland CR, Gironella M, Goel A. Colorectal cancers with microsatellite instability display unique miRNA profiles. Clin Cancer Res. 2011;17:6239-6249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 93] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 126. | Kulda V, Pesta M, Topolcan O, Liska V, Treska V, Sutnar A, Rupert K, Ludvikova M, Babuska V, Holubec L Jr, Cerny R. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet. 2010;200:154-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 119] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 127. | Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845-850. [PubMed] |
| 128. | Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 543] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 129. | Diosdado B, van de Wiel MA, Terhaar Sive Droste JS, Mongera S, Postma C, Meijerink WJ, Carvalho B, Meijer GA. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101:707-714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 209] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 130. | Wang X, Wang J, Ma H, Zhang J, Zhou X. Downregulation of miR-195 correlates with lymph node metastasis and poor prognosis in colorectal cancer. Med Oncol. 2012;29:919-927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 100] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 131. | Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, Sugihara K, Mori M. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 166] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 132. | Xu XM, Qian JC, Deng ZL, Cai Z, Tang T, Wang P, Zhang KH, Cai JP. Expression of miR-21, miR-31, miR-96 and miR-135b is correlated with the clinical parameters of colorectal cancer. Oncol Lett. 2012;4:339-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 133. | Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y, Zhu L, Wang J, Yang R, Zhang Y, Ren Z, Zen K, Zhang J, Zhang CY. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385-1392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 450] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 134. | Sarver AL, French AJ, Borralho PM, Thayanithy V, Oberg AL, Silverstein KA, Morlan BW, Riska SM, Boardman LA, Cunningham JM, Subramanian S, Wang L, Smyrk TC, Rodrigues CM, Thibodeau SN, Steer CJ. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2009;9:401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 252] [Cited by in RCA: 261] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 135. | Zhang Y, He X, Liu Y, Ye Y, Zhang H, He P, Zhang Q, Dong L, Dong J. microRNA-320a inhibits tumor invasion by targeting neuropilin 1 and is associated with liver metastasis in colorectal cancer. Oncol Rep. 2012;27:685-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 136. | Roy S, Levi E, Majumdar AP, Sarkar FH. Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. J Hematol Oncol. 2012;5:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 133] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 137. | Dai X, Chiang Y, Wang Z, Song Y, Lu C, Gao P, Xu H. Expression levels of microRNA-375 in colorectal carcinoma. Mol Med Rep. 2012;5:1299-1304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 138. | Arndt GM, Dossey L, Cullen LM, Lai A, Druker R, Eisbacher M, Zhang C, Tran N, Fan H, Retzlaff K, Bittner A, Raponi M. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer. 2009;9:374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 207] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 139. | Zhang Y, Lin C, Liao G, Liu S, Ding J, Tang F, Wang Z, Liang X, Li B, Wei Y, Huang Q, Li X, Tang B. MicroRNA-506 suppresses tumor proliferation and metastasis in colon cancer by directly targeting the oncogene EZH2. Oncotarget. 2015;6:32586-32601. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 140. | Hamfjord J, Stangeland AM, Hughes T, Skrede ML, Tveit KM, Ikdahl T, Kure EH. Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS One. 2012;7:e34150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 131] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 141. | Wu CW, Ng SS, Dong YJ, Ng SC, Leung WW, Lee CW, Wong YN, Chan FK, Yu J, Sung JJ. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut. 2012;61:739-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 216] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 142. | Lichtenstein GR, Loftus EV, Isaacs KL, Regueiro MD, Gerson LB, Sands BE. ACG Clinical Guideline: Management of Crohn's Disease in Adults. Am J Gastroenterol. 2018;113:481-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 612] [Cited by in RCA: 932] [Article Influence: 133.1] [Reference Citation Analysis (0)] |
| 143. | Roessner A, Kuester D, Malfertheiner P, Schneider-Stock R. Oxidative stress in ulcerative colitis-associated carcinogenesis. Pathol Res Pract. 2008;204:511-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 202] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 144. | Bressenot A, Cahn V, Danese S, Peyrin-Biroulet L. Microscopic features of colorectal neoplasia in inflammatory bowel diseases. World J Gastroenterol. 2014;20:3164-3172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (1)] |
| 145. | Collins PD. Strategies for detecting colon cancer and dysplasia in patients with inflammatory bowel disease. Inflamm Bowel Dis. 2013;19:860-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 146. | O'Connor PM, Lapointe TK, Beck PL, Buret AG. Mechanisms by which inflammation may increase intestinal cancer risk in inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:1411-1420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 147. | Itzkowitz SH. Molecular biology of dysplasia and cancer in inflammatory bowel disease. Gastroenterol Clin North Am. 2006;35:553-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 125] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 148. | Josse C, Bours V. MicroRNAs and Inflammation in Colorectal Cancer. Adv Exp Med Biol. 2016;937:53-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 149. | Choi CR, Bakir IA, Hart AL, Graham TA. Clonal evolution of colorectal cancer in IBD. Nat Rev Gastroenterol Hepatol. 2017;14:218-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 124] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 150. | Kanaan Z, Rai SN, Eichenberger MR, Barnes C, Dworkin AM, Weller C, Cohen E, Roberts H, Keskey B, Petras RE, Crawford NP, Galandiuk S. Differential microRNA expression tracks neoplastic progression in inflammatory bowel disease-associated colorectal cancer. Hum Mutat. 2012;33:551-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 100] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 151. | Bai J, Li Y, Shao T, Zhao Z, Wang Y, Wu A, Chen H, Li S, Jiang C, Xu J, Li X. Integrating analysis reveals microRNA-mediated pathway crosstalk among Crohn's disease, ulcerative colitis and colorectal cancer. Mol Biosyst. 2014;10:2317-2328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (1)] |
| 152. | Ueda Y, Ando T, Nanjo S, Ushijima T, Sugiyama T. DNA methylation of microRNA-124a is a potential risk marker of colitis-associated cancer in patients with ulcerative colitis. Dig Dis Sci. 2014;59:2444-2451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 153. | Wan J, Xia L, Xu W, Lu N. Expression and Function of miR-155 in Diseases of the Gastrointestinal Tract. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 154. | Fang Z, Tang J, Bai Y, Lin H, You H, Jin H, Lin L, You P, Li J, Dai Z, Liang X, Su Y, Hu Q, Wang F, Zhang ZY. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res. 2015;34:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 121] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 155. | Al-Mustanjid M, Mahmud SMH, Royel MRI, Rahman MH, Islam T, Rahman MR, Moni MA. Detection of molecular signatures and pathways shared in inflammatory bowel disease and colorectal cancer: A bioinformatics and systems biology approach. Genomics. 2020;112:3416-3426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 156. | Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3245] [Cited by in RCA: 3804] [Article Influence: 223.8] [Reference Citation Analysis (0)] |
| 157. | Wang SW, Sun YM. The IL-6/JAK/STAT3 pathway: potential therapeutic strategies in treating colorectal cancer (Review). Int J Oncol. 2014;44:1032-1040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 256] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 158. | Shi M, Chen X, Ye K, Yao Y, Li Y. Application potential of toll-like receptors in cancer immunotherapy: Systematic review. Medicine (Baltimore). 2016;95:e3951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 159. | Liu X, Xu Y, Zhou Q, Chen M, Zhang Y, Liang H, Zhao J, Zhong W, Wang M. PI3K in cancer: its structure, activation modes and role in shaping tumor microenvironment. Future Oncol. 2018;14:665-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 160. | Huang HY, Lin YC, Li J, Huang KY, Shrestha S, Hong HC, Tang Y, Chen YG, Jin CN, Yu Y, Xu JT, Li YM, Cai XX, Zhou ZY, Chen XH, Pei YY, Hu L, Su JJ, Cui SD, Wang F, Xie YY, Ding SY, Luo MF, Chou CH, Chang NW, Chen KW, Cheng YH, Wan XH, Hsu WL, Lee TY, Wei FX, Huang HD. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;48:D148-D154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 293] [Cited by in RCA: 664] [Article Influence: 132.8] [Reference Citation Analysis (0)] |
| 161. | Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3294] [Cited by in RCA: 5052] [Article Influence: 421.0] [Reference Citation Analysis (0)] |
| 162. | Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90-W97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4582] [Cited by in RCA: 6916] [Article Influence: 768.4] [Reference Citation Analysis (0)] |
| 163. | Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Jeon M, Ma'ayan A. Gene Set Knowledge Discovery with Enrichr. Curr Protoc. 2021;1:e90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 1951] [Article Influence: 487.8] [Reference Citation Analysis (0)] |