Published online Sep 15, 2021. doi: 10.4251/wjgo.v13.i9.1164
Peer-review started: March 4, 2021
First decision: April 6, 2021
Revised: April 9, 2021
Accepted: July 5, 2021
Article in press: July 5, 2021
Published online: September 15, 2021
Processing time: 190 Days and 8.6 Hours
Pancreatic cancer is a highly malignant tumor of the gastrointestinal system whose emerging resistance to chemotherapy has necessitated the development of novel antitumor treatments. Scoparone, a traditional Chinese medicine monomer with a wide range of pharmacological properties, has attracted considerable attention for its antitumor activity.
To explore the potential antitumor effect of scoparone on pancreatic cancer and the possible molecular mechanism of action.
The target genes of scoparone were determined using both the bioinformatics and multiplatform analyses. The effect of scoparone on pancreatic cancer cell proliferation, migration, invasion, cell cycle, and apoptosis was detected in vitro. The expression of hub genes was tested using quantitative reverse transcription polymerase chain reaction (qRT-PCR), and the molecular mechanism was analyzed using Western blot. The in vivo effect of scoparone on pancreatic cancer cell proliferation was detected using a xenograft tumor model in nude mice as well as immunohistochemistry.
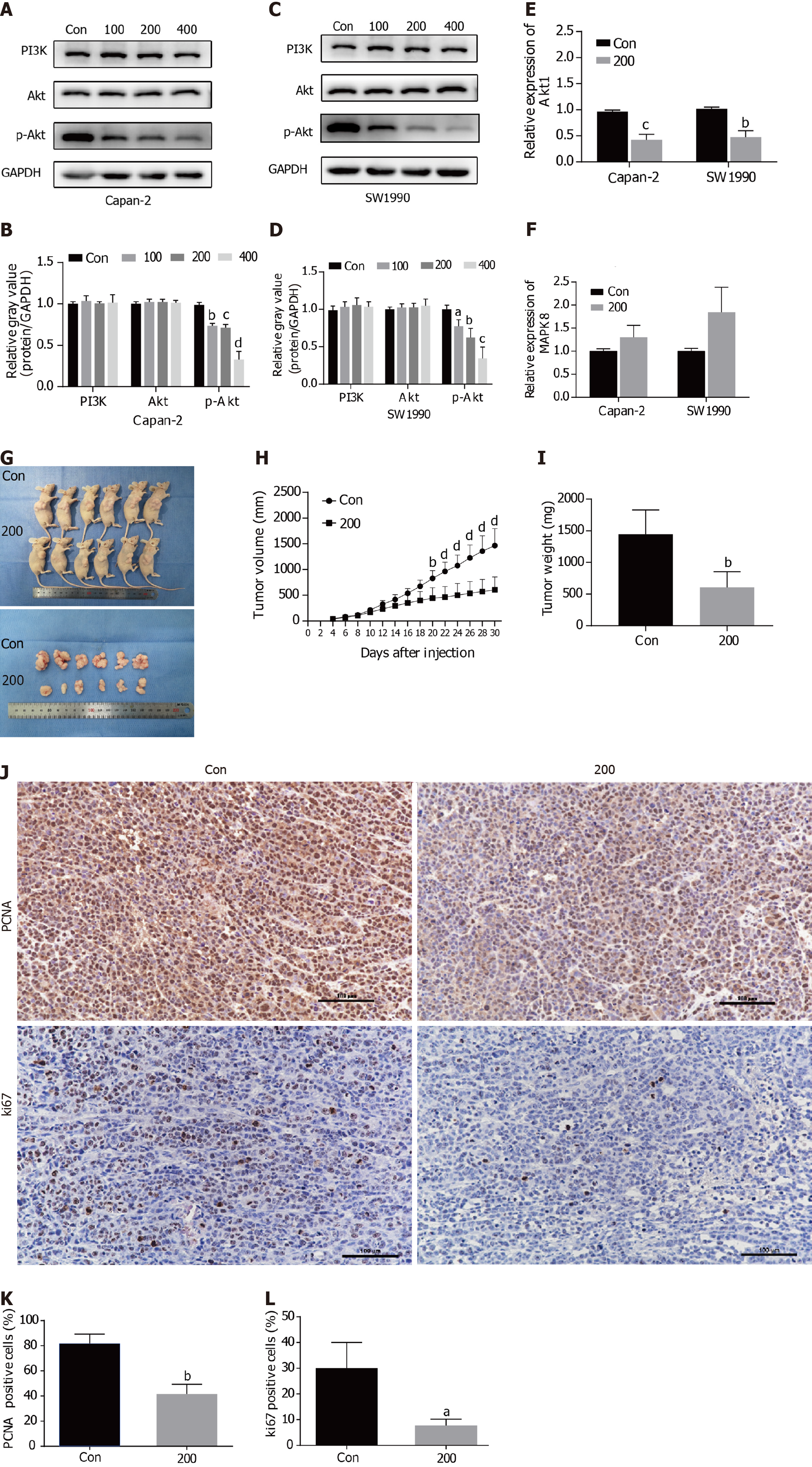
The hub genes involved in the suppression of pancreatic cancer by scoparone were obtained by network bioinformatics analyses using publicly available databases and platforms, including SwissTargetPrediction, STITCH, GeneCards, CTD, STRING, WebGestalt, Cytoscape, and Gepia; AKT1 was confirmed using qRT-PCR to be the hub gene. Cell Counting Kit-8 assay revealed that the viability of Capan-2 and SW1990 cells was significantly reduced by scoparone treatment exhibiting IC50 values of 225.2 μmol/L and 209.1 μmol/L, respectively. Wound healing and transwell assays showed that scoparone inhibited the migration and invasion of pancreatic cancer cells. Additionally, flow cytometry confirmed that scoparone caused cell cycle arrest and induced apoptosis. Scoparone also increased the expression levels of Bax and cleaved caspase-3, decreased the levels of MMP9 and Bcl-2, and suppressed the phosphorylation of Akt without affecting total PI3K and Akt. Moreover, compared with the control group, xenograft tumors, in the 200 μmol/L scoparone treatment group, were smaller in volume and lighter in weight, and the percentages of Ki65- and PCNA-positive cells were decreased.
Our findings indicate that scoparone inhibits pancreatic cancer cell proliferation in vitro and in vivo, inhibits migration and invasion, and induces cycle arrest and apoptosis in vitro through the PI3K/Akt signaling pathway.
Core Tip: To explore the potential antitumor effect of scoparone on pancreatic cancer and the possible molecular mechanism, target genes of scoparone were determined using the bioinformatics and multiplatform analyses. The effect of scoparone on pancreatic cancer cell proliferation, migration, invasion, cell cycle, and apoptosis was detected. The expression of hub genes was detected using quantitative reverse transcription polymerase chain reaction, and Western blot was used to analyze the molecular mechanism. Xenograft tumor model and immunohistochemistry were used to detect cell proliferation in vivo. Our findings indicated that scoparone inhibits pancreatic cancer cell proliferation, migration, and invasion, and induced cell cycle arrest and apoptosis through the PI3K/Akt signaling pathway.
- Citation: Li N, Yang F, Liu DY, Guo JT, Ge N, Sun SY. Scoparone inhibits pancreatic cancer through PI3K/Akt signaling pathway. World J Gastrointest Oncol 2021; 13(9): 1164-1183
- URL: https://www.wjgnet.com/1948-5204/full/v13/i9/1164.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i9.1164
Pancreatic cancer is a lethal malignancy with occult onset, difficult early diagnosis, rapid progression, poor therapeutic response, short median survival time, and poor prognosis[1,2]. According to the National Cancer Center of China, the incidence rate of pancreatic cancer is 7/100000, accounting for 2.42% of the cases of malignant tumors in China, and its 5-year survival rate is only 7.2%, which is the lowest survival rate among all malignant tumors[3,4]. Pancreatic cancer is the seventh leading cause of death worldwide[5], with an increasing trend. It is estimated that pancreatic cancer will become the second leading cause of malignant tumor-related deaths by 2030[6]. At present, surgical resection is the primary treatment method for pancreatic cancer; however, only 15%-20% of patients are eligible for surgical resection because of late diagnosis[7]. In recent years, with the development of endoscopic ultrasonography and pathology, early diagnosis of pancreatic cancer has been improved[8-10]; however, there are still only few cases that can be surgically removed. Moreover, pancreatic cancer has a strong tendency to metastasize and a high recurrence rate following surgery. Although gemcitabine, 5-fluorouracil, oxaliplatin, and other chemotherapeutic agents can effectively improve the prognosis of patients with pancreatic cancer, the emergence of drug resistance has lowered the efficacy of these treatments[11]. Meanwhile, the toxicity, negative side effects, and associated costs of chemotherapy remain a challenge. Therefore, the development of novel anti-pancreatic cancer drugs is essential.
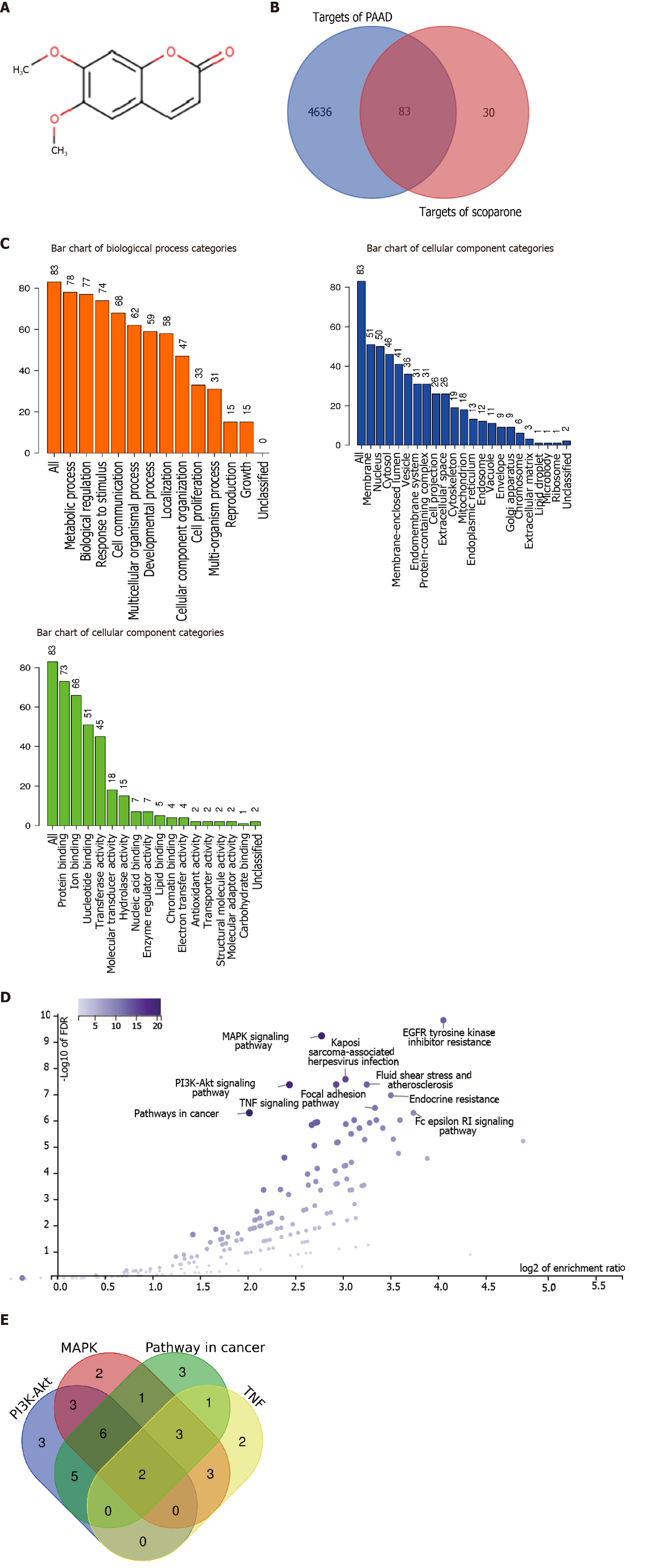
Scoparone, also known as 6,7-dimethoxycoumarin, belongs to the coumarin class of natural organic compounds. It is primarily extracted from the dry aerial part of Artemisia capillaris Thunb., which is widely used in the prevention and treatment of neonatal jaundice in Asia. Scoparone also has anticoagulant, anti-inflammatory, antifibrosis, and antioxidation activities[12-15], and can act as a vasodilator, reduce blood lipid levels, and exert an antiatherosclerosis effect on the cardiovascular system[16-18]. Jang et al[19] found that scoparone may suppress the expression of chemokines (interleukin 8 and monocyte chemotactic protein-1) by inhibiting the nuclear factor-κappa B signaling pathway in leukemia cells activated by phorbol 12-myristate 13-acetate. In another study, scoparone could reduce the viability and migration of laryngeal cancer cells in a dose-dependent manner by inducing apoptosis without affecting the activity of ERK1/2 and Akt kinase[20]. Scoparone may also inhibit the proliferation of prostate cancer cells by directly interfering with the transcription of signal transducer and activator of transcription 3[21]. In our study, we evaluated the antitumor activity of scoparone on pancreatic cancer cells and investigated the molecular mechanism of action, providing evidence of a potential drug or adjuvant for pancreatic cancer treatment.
Prediction of drug targets: The following two databases were used to predict targets of scoparone activity: SwissTargetPrediction (http://www.swisstargetprediction.ch/), a database for predicting drug targets based on the similarity of two- or three-dimensional structures with known compounds[22]; and STITCH (http://stitch.embl.de/, Version:5.0), a platform for searching known and predicted interactions between compounds and proteins, containing 9643763 proteins from 2031 species[23].
Prediction of pancreatic cancer targets: The following two databases were used to predict pancreatic cancer-related targets: GeneCards (https://www.genecards.org, Version:5.0), a searchable comprehensive database that integrates data from over 150 sources[24]; and Comparative Taxonomic Database (CTD, http://ctdbase.org, updated 10/28/2020), which provides information on interactions or relationships between compounds and genes, compounds and proteins, compounds and diseases, or genes and diseases[25]. Targets with a score greater than the median were screened out, and the duplicates were obtained as pancreatic cancer-related targets. In addition, the overlaps of drug and disease targets were the effective targets of scoparone to treat pancreatic cancer.
Enrichment analysis of potential targets: WebGestalt (http://www.webgestalt.org, updated 01/14/2019) is an online website focusing on functional enrichment analysis[26]. Biological processes, cellular composition, and molecular function were selected for Gene Ontology (GO) analysis, and Kyoto Encyclopedia of Genes and Genomes (KEGG)[27] was selected for pathway analysis for the predicted pancreatic cancer-related targets of scoparone.
Protein-protein interaction network: STRING (https://string-db.org/cgi/input.pl/, Version 11.0) is an online database for predicting protein-protein interactions (PPIs)[28]. The predicted targets were uploaded into the STRING database to obtain the PPI network, which was then imported into Cytoscape 3.7.2, an open software platform for constructing data analysis, integration, and visualization networks[29]. The software plugins Cytohubba and Network Analyzer[30,31] were used to analyze the network characteristics, in which the degree of the nodes represents the importance of the proteins in the network. The higher the degree value, the darker the node color.
Expression patterns of candidate hub genes: Gepia (http://gepia.cancer-pku.cn/) is a database that can be used for differential expression analyses between tumor and normal tissues as well as survival and correlation analyses[32]. The expression patterns of candidate hub genes identified by pathway and PPI network analyses were evaluated using Gepia.
Cell culture: Pancreatic cancer cell lines Capan-2 and SW1990 were respectively purchased from American Type Culture Collection (ATCC, Manassas, VA, United States) and the cell resource center of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). Capan-2 cells were cultivated in RPMI-1640 medium (Biological Industries, Beit HaEmek, Israel) and SW1990 cells in L15 medium (Biological Industries) mixed with 10% fetal bovine serum (FBS, Biological Industries) in a 5% CO2 incubator at 37 °C.
Cell viability assay: Capan-2 and SW1990 cells were seeded onto 96-well plates at a density of 4000 cells/well. After 12 h, the medium was replaced with different scoparone (Sigma-Aldrich, St. Louis, MO, United States) concentrations (0, 0.5, 1, 5, 10, 50, 80, 100, 200, 300, 400, 600, and 800 μmol/L) for 48 h. Cell viability was measured with the CCK-8 assay kit (ApexBio Technology, Houston, TX, United States) according to the instructions. Next, the optical density (OD) value at 450 nm was measured, and the IC50 value was calculated. Cells were then inoculated again with 100, 200, and 400 μmol/L scoparone (1/2 IC50, IC50, and 2 IC50) for 24, 48, and 72 h, and the OD value was measured as compared to the control group (0 μmol/L scoparone with the same amount of DMSO solution of 400 μmol/L scoparone).
Wound healing assay: Capan-2 and SW1990 cells were pretreated with 0 (Con), 100, 200, and 400 μmol/L scoparone for 24 h. A vertical wound was drawn in the cells with a 200-μL sterile pipette tip. After washing with phosphate buffer saline (PBS) (Biological Industries), serum-free culture medium was added, and images of the wound were taken at 0, 24, 48, and 72 h using an Eclipse microscope (Nikon Corporation, Tokyo, Japan).
Transwell assay: Capan-2 and SW1990 cells were pretreated with 0, 100, 200, and 400 μmol/L scoparone for 24 h, following which the cells were resuspended in serum-free medium. A 200-μL aliquot of cell suspension (2.5 × 105 cells/mL) was added to the upper layer of the cell culture chamber, while 600-μL medium containing 10% FBS was added to the lower level and then incubated in a 5% CO2 incubator at 37 °C for 16-24 h. The membrane of the upper level was fixed with 4% paraformaldehyde for 2 min, permeated with 100% methanol for 20 min, and then stained with 0.2% crystal violet at 25 °C for 15 min. Under an Eclipse microscope (Nikon Corporation), five fields (200 × magnification) were randomly observed and imaged.
The cell invasion assay was conducted by adding 100 μL of Matrigel (Corning Inc., New York, NY, United States) diluted at a 1:8 ratio in a serum-free medium to the upper level, followed by incubation overnight at 37 °C. The other steps were perfor
Apoptosis by flow cytometry: Following the instructions, apoptosis was measured using the PE Annexin V Apoptosis Detection Kit (BD Biosciences, San Jose, CA, United States) by flow cytometry. Capan-2 and SW1990 cells were pretreated with 0, 100, 200, and 400 μmol/L scoparone for 24 h. Cells were rinsed twice with cold PBS after digestion with trypsin without EDTA, followed by resuspension and dilution to 1 × 106 cells/mL with 1 × Annexin V Binding Buffer; 5 μL of PE Annexin V and 5 μL of 7-AAD reagent were added and then incubated in dark place for 15 min. The FACSCalibur flow cytometer (BD Biosciences) was used to measure the apoptosis rate.
Cell cycle by flow cytometry: Following the instructions, cell cycle was measured using PI/RNase Staining Buffer (BD Biosciences) by flow cytometry. Capan-2 and SW1990 cells were pretreated with 0, 100, 200, and 400 μmol/L scoparone for 24 h. Cells were resuspended with cold PBS after digestion and fixed with 70% ethanol overnight at -20 °C and then 500 μL of PI/RNase staining buffer was added to each flow tube and stained in dark place for 15 min. The FACSCalibur flow cytometer (BD Biosciences) was used to measure cell cycle.
Quantitative reverse transcription polymerase chain reaction: TRIzol reagent (Invitro
Western blot analysis: Capan-2 and SW1990 cells were pretreated with 0, 100, 200, and 400 μmol/L scoparone for 24 h. RIPA lysis buffer (Beyotime Biotech, Jiangsu, China) was used to lyse the cells and then proteins were separated and transferred to polyvinylidene fluoride (PVDF) membranes (GE Healthcare, Chicago, IL, United States). After blocking with 5% skimmed milk or 5% bovine serum albumin (Beyotime) for 2 h, the membranes were incubated with primary antibodies overnight at 4 °C and then incubated with secondary antibodies at room temperature for 2 h. Finally, an ECL Western blotting substrate (Tanon Science & Technology Co., Ltd., Shanghai, China) was used to visualize the bands with the Amersham imager 680 chemiluminescence imaging system (Amersham imager 680; GE Healthcare, United States). GAPDH (1:10000; Proteintech Group, Wuhan, China) served as a control for normalization, and primary antibodies against the following antigens were employed: PI3K (1:1000; Cell Signaling Technology, Inc., Danvers, MA, United States), Akt (1:2000; Proteintech Group), p-Akt (1:2000; Cell Signaling Technology), MMP9 (1:1000; Proteintech Group), Bcl-2 (1:2000; Proteintech Group), Bax (1:10000; Proteintech Group), and cleaved caspase-3 (1:1000; Cell Signaling Technology).
Xenograft tumor model in nude mice: The animal experiment was approved by the Animal Ethics Committee of the Institute (Ethical code number: 2020PS766K). Twelve four-week-old BALB/C nude mice were purchased from Beijing Huafukang Bioscience (Beijing, China) and were acclimatized to laboratory conditions (23 °C, 12 h/12 h light/dark, 50% humidity, ad libitum access to food and water) for 2 wk prior to experimentation following the regulations of the guidelines of the Animal Care and Ethics Committee of Shengjing Hospital of China Medical University (Shenyang, Liaoning, China). Nude mice received subcutaneous injections of 100-μL Capan-2 cells (2 × 106). Tumor length (L) and width (W) and mouse weight were measured every second day. Tumor volume was calculated using the following formula: Tumor volume (V) = (L × W2)/2. When V reached approximately 100 mm3 after 7-10 d, 12 nude mice were randomly and equally divided into either a control group or an experimental group. The control group was intraperitoneally injected with 50 μL of normal saline solution, while the experimental group was injected with 50 μL of 200 μmol/L scoparone every 2 d for 3 wk. The nude mice were euthanized by sodium pentobarbital overdose (150 mg/kg) by intraperitoneal injection, following which the tumors were excised, photographed, measured, and weighed.
Immunohistochemistry: Tumor samples were fixed with 4% paraformaldehyde for 48 h and then dehydrated and paraffin-embedded. Tissue sections (3 μm) were deparaffinized, rehydrated, and antigen-repaired. After blocking, sections were incubated with primary antibodies anti-Ki67 (1:200; Abcam, Cambridge, United Kingdom) and anti-PCNA (1:500; Proteintech Group) for 12-16 h at 4 °C, then incubated with the secondary antibody goat anti-rabbit immunoglobulin G at room temperature for 30 min. Sections were stained with 3,3'-diaminobenzidine (DAB), counter-stained with hematoxylin, dehydrated with gradient ethanol, cleared with xylene, sealed with neutral gum, observed, and photographed under an Eclipse microscope (Nikon Corporation).
Data are represented as the mean ± SD, and t-test (between two groups) or one-way ANOVA (multiple groups) was used for statistical analyses with IBM SPSS Statistics v. 25.0 software (IBM Corp., Armonk, NY, United States) and GraphPad Prism 8.0 software (GraphPad Software, La Jolla, CA, United States). A P value < 0.05 indicated statistical significance. All experiments were repeated at least three times.
One hundred and thirteen predicted scoparone targets were identified by searching the SwissTargetPrediction database and STITCH platform. Meanwhile, 11691 and 27717 pancreatic cancer-related targets were identified by searching the GeneCards and CTD databases, respectively. Among them, 5806 disease targets with scores > the median 2.1 and 13855 targets with scores > the median 10.98 were screened out; moreover, 4719 duplicates were obtained as pancreatic cancer-related targets. Finally, a total of 83 therapeutic pancreatic cancer targets of scoparone were obtained by overlapping disease and drug targets (Figure 1B).
Eighty-three predicted pancreatic cancer targets of scoparone were imported into WebGastalt for functional enrichment analysis, including GO analysis (Figure 1C) and KEGG analysis (Figure 1D). The top ten enrichment pathways among the tumor-related signaling pathways in which scoparone might be involved were mapped into a volcanic plot. These included the MAPK signaling pathway, the PI3K/Akt signaling pathway, pathways in cancer, and the tumor necrosis factor (TNF) signaling pathway. These four pathways involved 34 targets, among which two targets, IKBKB and Akt1, participated in the four pathways, whereas nine targets, including FLT4, EGFR, IGF1R, MET, PDGFRb, ERBB2, MAPK8, MAPK10, and RPS6KA5, were involved in three of the four pathways (Figure 1E).
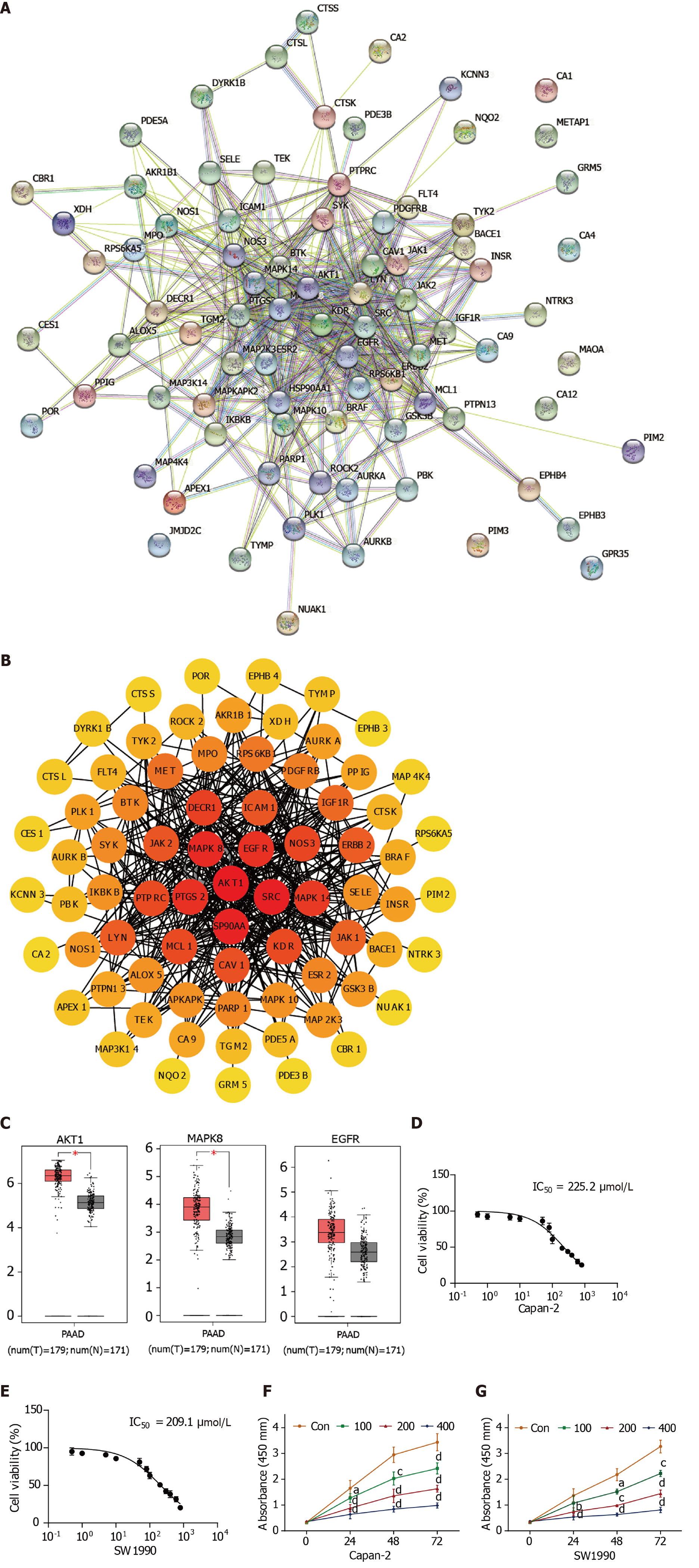
The PPI network between 83 scoparone and pancreatic cancer targets was identified using the STRING database, in which eight proteins, including CA1, CA4, CA12, GPR35, KDM4C, MAOA, METAP1, and PIM3, were not involved in the interaction network (Figure 2A). Then, the PPI network was visualized by Cytoscape (Figure 2B). The top five targets were Akt1, SRC, HSP90AA1, MAPK8, EGFR, and PTGS2 (EGFR and PTGS were both in the fifth place), which overlapped with targets related to the signaling pathways. AKT1 was the most likely candidate hub gene to be influenced by scoparone in pancreatic cancer, followed by MAPK8 and EGFR.
The Gepia database was used to analyze AKT1, MAPK8, and EGFR expression differences (Figure 2C). AKT1 and MAPK8 expression differed significantly between pancreatic cancer and normal pancreatic tissues, in which the expression of AKT1 was significantly higher than that of MAPK8. From this analysis, it can be inferred that scoparone may suppress pancreatic cancer by inhibiting the expression of AKT1, followed by that of MAPK8.
The viability of Capan-2 and SW1990 cells was significantly reduced by scoparone treatment in a dose-dependent manner, exhibiting IC50 values of 225.2 μmol/L and 209.1 μmol/L, respectively (Figure 2D and E). Compared with the control group, scoparone inhibited the proliferation of Capan-2 and SW1990 cells in a dose- and time-dependent manner (P < 0.0001, Figure 2F and G).
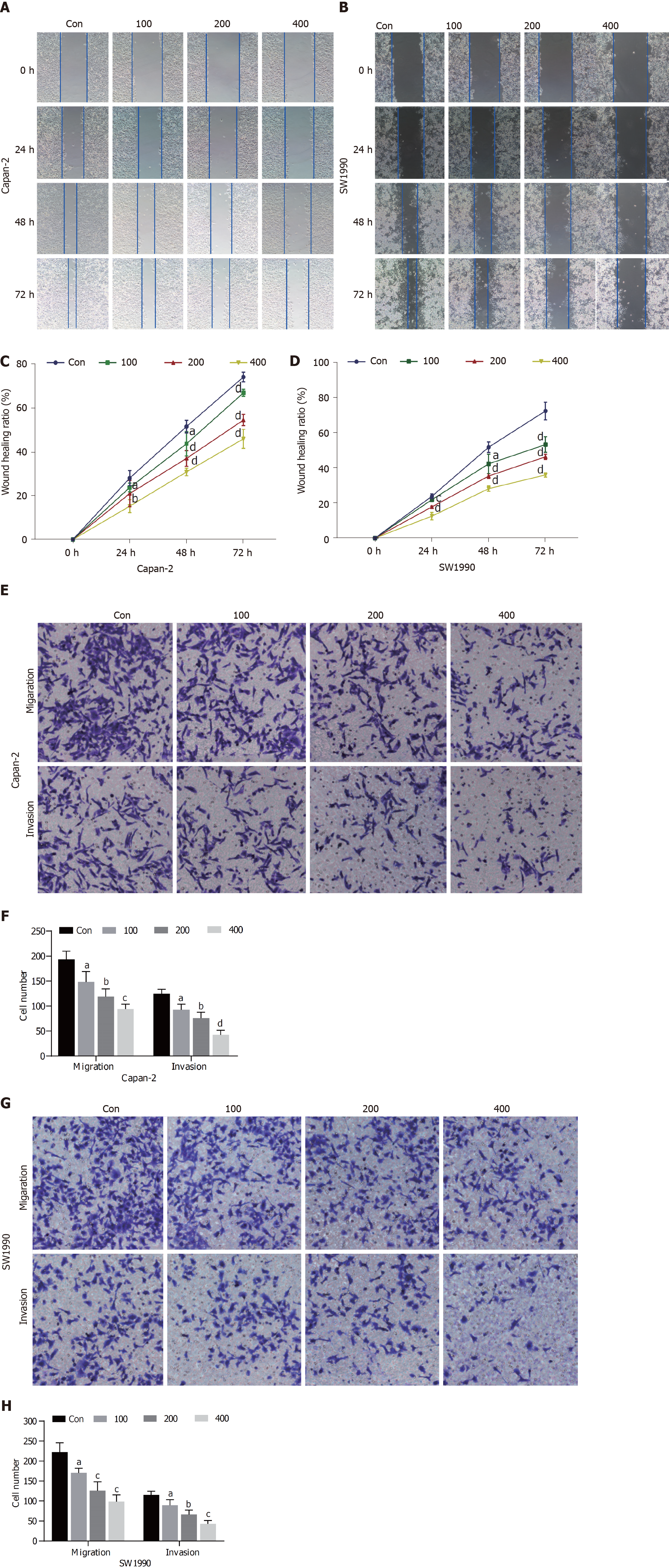
The results of the wound healing assay demonstrated that the migration ability of Capan-2 and SW1990 cells was reduced in a dose-dependent manner compared with the control group after treatment with scoparone (P < 0.05; Figure 3A-D). Further, the results of the transwell assay revealed that the migration and invasion abilities of Capan-2 and SW1990 cells were significantly decreased by scoparone treatment compared with the control group (P < 0.05; Figure 3E-H).
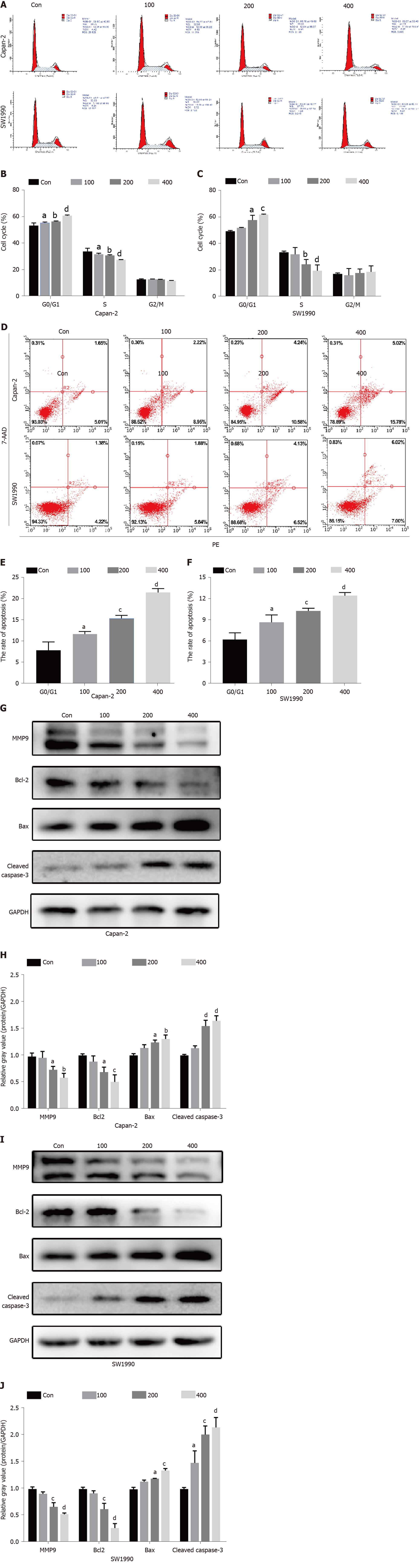
Flow cytometry results indicated that the abundance of Capan-2 and SW1990 cells increased in the G0/G1 phase compared with the control group, whereas that decreased in the S phase, and no noticeable change was observed in the abundance of cells in the G2/M phase (Figure 4A-C). Meanwhile, scoparone treatment could induce the apoptosis of Capan-2 and SW1990 cells, as the 100, 200, and 400 μmol/L scoparone groups had significantly higher apoptosis rates than the control group (Figure 4D-F). Flow cytometry verified that scoparone could inhibit cell proliferation by causing cell cycle arrest in the G0/G1 phase and inducing apoptosis.
To further explore the antitumor molecular mechanism of scoparone, quantitative reverse transcription PCR (qRT-PCR) and Western blot were performed. First, qRT-PCR confirmed that AKT1 expression was decreased in the 200 μmol/L scoparone group compared with the control group, whereas MAPK8 expression was increased (Figure 5E and F). The PI3K/Akt signaling pathway, the key pathway most likely related to Akt1 expression and the antitumor effect, was analyzed using Western blot. Scoparone treatment did not decrease the total expression of PI3K and Akt, but it inhibited p-Akt expression, which may have led to significantly decreased MMP9 and Bcl-2 expression and increased Bax and cleaved caspase-3 expression (Figure 5G-J). These results suggested that scoparone could reduce the migration and invasion of pancreatic cancer cells and induce apoptosis through PI3K/Akt signaling pathway.
To investigate the suppression of pancreatic cancer cell proliferation in vivo, xenograft tumors were derived from Capan-2 cells in nude mice. Xenograft tumors in the 200 μmol/L scoparone group were smaller in volume and lighter in weight than those in the control group (Figure 5G-I). Immunohistochemistry (IHC) revealed that the percentages of Ki67- and PCNA-positive cells in the 200 μmol/L scoparone group were significantly decreased compared with those of the control group (Figure 5J-L). It is suggested that scoparone could suppress the growth of pancreatic cancer in vivo.
Pancreatic cancer, known as “the king of cancers”, has the lowest survival rate among all malignant tumors. At present, chemotherapy can improve the survival of patients with pancreatic cancer; however, drug resistance, toxicity, adverse effects, and the high cost of chemotherapy limit its effectiveness. Therefore, exploring new therapeutic targets and drugs remains an important strategy for the treatment of pancreatic cancer. In recent years, natural products have attracted considerable attention due to their reliable efficacy and relatively few side effects[34]. Scoparone, a phenylpropanoid monomer used in traditional Chinese medicine, has a wide range of pharmacological properties, and its antitumor activity has been confirmed in leukemia, laryngeal cancer, and prostate cancer. However, to date, no study has demonstrated its effect on other tumors or determined its underlying molecular mechanism.
The inhibitory effect of scoparone in vitro and in vivo on pancreatic cancer and its associated molecular mechanism were investigated in this study. The current study results concur with those of previous studies in terms of inhibiting cell proliferation; scoparone inhibited pancreatic cell proliferation by causing cell cycle arrest in the G0/G1 phase and inducing apoptosis. Meanwhile, Western blot analysis demonstrated that the expression of the anti-apoptotic protein Bcl-2 was reduced, whereas that of the proapoptotic proteins, Bax, and cleaved caspase-3 was increased after scoparone treatment. Downregulation of anti-apoptotic protein and upregulation of pro-apoptotic protein can lead to increased apoptosis. In addition to the wound healing and transwell assay results, the expression of MMP9, which is associated with cancer cell invasion and metastasis, was decreased after scoparone treatment, further indicating that scoparone inhibited the migration and invasion of pancreatic cancer cells. Finally, xenograft tumor and IHC results confirmed the inhibitory effect of scoparone on pancreatic cancer cell proliferation in vivo, which is an advantage over previous studies.
In the present study, the hub genes involved in the suppression of pancreatic cancer by scoparone were obtained by network bioinformatics analyses using public databases and platforms, including SwissTargetPrediction, STITCH, GeneCards, CTD, STRING, WebGestalt, Cytoscape, and Gepia. Ultimately, AKT1 was confirmed, using qRT-PCR, to be the hub gene involved in pancreatic cancer suppression by scoparone. Serine/threonine kinase Akt, also known as protein kinase B, consists of isomers Akt1/PKB α, Akt2/PKB β, and Akt3/PKB γ, which are the products of three different genes, rather than alternative splicing of one gene[35]. Although the genes encoding these isomers are different, their amino acid homology reaches 80%[36,37], of which Akt1 is the most important and characteristic subtype that participates in the occurrence and development of various tumors through numerous signaling pathways. Among these, the PI3K/Akt signaling pathway is one of the most frequently activated pathways in tumors. Increasing studies have confirmed that the PI3K/Akt signaling pathway is involved in tumor cell survival, proliferation, metabolism, growth, metastasis, and drug resistance[38,39]. In pancreatic cancer, the PI3K/Akt signaling pathway also functions as an antitumor drug target. Xu et al[40] found that canagliflozin exerted antitumor effects by inhibiting glycolysis through the PI3K/Akt/mTOR signaling pathway, which was improved when combined with gemcitabine, revealing the potential of this pathway in the clinical treatment of pancreatic cancer. However, previous studies investigating the antitumor mechanism of scoparone were insufficient and ambiguous. The findings of this study showed that scoparone inhibits proliferation, metastasis, and invasion of pancreatic cancer cells by cell cycle arrest and apoptosis through the PI3K/Akt signaling pathway.
In summary, the present study confirmed that scoparone plays an antitumor role in pancreatic cancer through the PI3K/Akt signaling pathway. However, there are some limitations to this study. First, the targets predicted using the publicly available databases may not be integrated. Second, other tumor-related signaling pathways, such as MAPK and TNF, may also participate in the antitumor effect of scoparone. Finally, pharmacokinetic analyses of scoparone were not conducted in the present study, thus the clinical applications warrant further investigations. Nevertheless, the results of the present study suggest that scoparone is likely to be an effective drug or adjuvant for pancreatic cancer treatment and provide directions for exploring new antitumor drugs.
Pancreatic cancer is a highly malignant tumor whose treatment is not effective, so it is necessary to develop new anti-pancreatic cancer treatments. Scoparone has attracted considerable attention for its antitumor activity.
To explore a new method to develop anti-tumor drugs.
To explore the potential antitumor effect of scoparone on pancreatic cancer and the possible molecular mechanism of action.
The target genes of scoparone were determined using the bioinformatics analyses. The effect of scoparone on pancreatic cancer cell proliferation, migration, invasion, cell cycle, and apoptosis was detected in vitro. The expression of hub genes was tested using quantitative reverse transcription polymerase chain reaction, and the molecular mechanism was analyzed using Western blot. The in vivo effect of scoparone on pancreatic cancer cell proliferation was detected using a Xenograft tumor model in nude mice and immunohistochemistry.
Our findings indicated that scoparone inhibited pancreatic cancer cell proliferation in vitro and in vivo, inhibited migration and invasion, and induced cycle arrest and apoptosis in vitro through the PI3K/Akt signaling pathway.
Scoparone inhibits pancreatic cancer cell proliferation, migration, and invasion, and induces cycle arrest and apoptosis through the PI3K/Akt signaling pathway.
Pharmacokinetic analyses of scoparone were not conducted in the present study, thus the clinical applications warrant further investigations.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Oberhardt V, Trivedi P S-Editor: Gao CC L-Editor: Wang TQ P-Editor: Ma YJ
| 1. | Sãftoiu A, Bhutani MS, Itoi T, Arcidiacono PG, Bories E, Cazacu IM, Constantin A, Coronel E, Dietrich CF, Duda DG, Garcia JI, Hocke M, Ignee A, Jenssen C, Jinga M, Khor C, Oppong KW, Pereira S, Petrone MC, Santo E, Seicean A, Seo DW, Siyu S, Vilmann P, Waxman I, Yeaton P. Changes in tumor vascularity depicted by contrast-enhanced EUS as a predictor of prognosis and treatment efficacy in patients with unresectable pancreatic cancer (PEACE): A study protocol. Endosc Ultrasound. 2019;8:235-240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 2. | Bhutani MS, Klapman JB, Tuli R, El-Haddad G, Hoffe S, Wong FCL, Chasen B, Fogelman DR, Lo SK, Nissen NN, Hendifar AE, Varadhachary G, Katz MHG, Erwin WD, Koay EJ, Tamm EP, Singh BS, Mehta R, Wolff RA, Soman A, Cazacu IM, Herman JM. An open-label, single-arm pilot study of EUS-guided brachytherapy with phosphorus-32 microparticles in combination with gemcitabine +/- nab-paclitaxel in unresectable locally advanced pancreatic cancer (OncoPaC-1): Technical details and study protocol. Endosc Ultrasound. 2020;9:24-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 3. | Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, Xia C, Sun K, Yang Z, Li H, Wang N, Han R, Liu S, Mu H, He Y, Xu Y, Fu Z, Zhou Y, Jiang J, Yang Y, Chen J, Wei K, Fan D, Wang J, Fu F, Zhao D, Song G, Jiang C, Zhou X, Gu X, Jin F, Li Q, Li Y, Wu T, Yan C, Dong J, Hua Z, Baade P, Bray F, Jemal A, Yu XQ, He J. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6:e555-e567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 795] [Cited by in RCA: 977] [Article Influence: 139.6] [Reference Citation Analysis (2)] |
| 4. | Zhao C, Gao F, Li Q, Liu Q, Lin X. The Distributional Characteristic and Growing Trend of Pancreatic Cancer in China. Pancreas. 2019;48:309-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 5. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55843] [Article Influence: 7977.6] [Reference Citation Analysis (132)] |
| 6. | Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913-2921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5379] [Cited by in RCA: 5140] [Article Influence: 467.3] [Reference Citation Analysis (0)] |
| 7. | Esposito I, Konukiewitz B, Schlitter AM, Klöppel G. Pathology of pancreatic ductal adenocarcinoma: facts, challenges and future developments. World J Gastroenterol. 2014;20:13833-13841. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 63] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (1)] |
| 8. | Arya N, Wyse JM, Jayaraman S, Ball CG, Lam E, Paquin SC, Lightfoot P, Sahai AV. A proposal for the ideal algorithm for the diagnosis, staging, and treatment of pancreas masses suspicious for pancreatic adenocarcinoma: Results of a working group of the Canadian Society for Endoscopic Ultrasound. Endosc Ultrasound. 2020;9:154-161. [PubMed] |
| 9. | Costache MI, Cazacu IM, Dietrich CF, Petrone MC, Arcidiacono PG, Giovannini M, Bories E, Garcia JI, Siyu S, Santo E, Popescu CF, Constantin A, Bhutani MS, Saftoiu A. Clinical impact of strain histogram EUS elastography and contrast-enhanced EUS for the differential diagnosis of focal pancreatic masses: A prospective multicentric study. Endosc Ultrasound. 2020;9:116-121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 10. | Ge N, Brugge WR, Saxena P, Sahai A, Adler DG, Giovannini M, Pausawasdi N, Santo E, Mishra G, Tam W, Kida M, de la Mora-Levy JG, Sharma M, Umar M, Katanuma A, Lee L, Garg PK, Eloubeidi MA, Yu HK, Raijman I, Arturo Arias BL, Bhutani M, Carrara S, Rai P, Mukai S, Palazzo L, Dietrich CF, Nguyen NQ, El-Nady M, Poley JW, Guaraldi S, Kalaitzakis E, Sabbagh LC, Lariño-Noia J, Gress FG, Lee YT, Rana SS, Fusaroli P, Hocke M, Dhir V, Lakhtakia S, Ratanachu-Ek T, Chalapathi Rao AS, Vilmann P, Okasha HH, Irisawa A, Ponnudurai R, Leong AT, Artifon E, Iglesias-Garcia J, Saftoiu A, Larghi A, Robles-Medranda C, Sun S. An international, multi-institution survey of the use of EUS in the diagnosis of pancreatic cystic lesions. Endosc Ultrasound. 2019;8:418-427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Zeng S, Pöttler M, Lan B, Grützmann R, Pilarsky C, Yang H. Chemoresistance in Pancreatic Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 414] [Article Influence: 69.0] [Reference Citation Analysis (0)] |
| 12. | Cho DY, Ko HM, Kim J, Kim BW, Yun YS, Park JI, Ganesan P, Lee JT, Choi DK. Scoparone Inhibits LPS-Simulated Inflammatory Response by Suppressing IRF3 and ERK in BV-2 Microglial Cells. Molecules. 2016;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 13. | Liu B, Deng X, Jiang Q, Li G, Zhang J, Zhang N, Xin S, Xu K. Scoparone improves hepatic inflammation and autophagy in mice with nonalcoholic steatohepatitis by regulating the ROS/P38/Nrf2 axis and PI3K/AKT/mTOR pathway in macrophages. Biomed Pharmacother. 2020;125:109895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 132] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 14. | Wang Y, Wang M, Chen B, Shi J. Scoparone attenuates high glucose-induced extracellular matrix accumulation in rat mesangial cells. Eur J Pharmacol. 2017;815:376-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 15. | Xu M, Cai J, Wei H, Zhou M, Xu P, Huang H, Peng W, Du F, Gong A, Zhang Y. Scoparone Protects Against Pancreatic Fibrosis via TGF-β/Smad Signaling in Rats. Cell Physiol Biochem. 2016;40:277-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 16. | Chen YL, Huang HC, Weng YI, Yu YJ, Lee YT. Morphological evidence for the antiatherogenic effect of scoparone in hyperlipidaemic diabetic rabbits. Cardiovasc Res. 1994;28:1679-1685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Jung SH, Lee GB, Ryu Y, Cui L, Lee HM, Kim J, Kim B, Won KJ. Inhibitory effects of scoparone from chestnut inner shell on platelet-derived growth factor-BB-induced vascular smooth muscle cell migration and vascular neointima hyperplasia. J Sci Food Agric. 2019;99:4397-4406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Park S, Kim JK, Oh CJ, Choi SH, Jeon JH, Lee IK. Scoparone interferes with STAT3-induced proliferation of vascular smooth muscle cells. Exp Mol Med. 2015;47:e145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Jang SI, Kim YJ, Kim HJ, Lee JC, Kim HY, Kim YC, Yun YG, Yu HH, You YO. Scoparone inhibits PMA-induced IL-8 and MCP-1 production through suppression of NF-kappaB activation in U937 cells. Life Sci. 2006;78:2937-2943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 20. | Kielbus M, Skalicka-Wozniak K, Grabarska A, Jeleniewicz W, Dmoszynska-Graniczka M, Marston A, Polberg K, Gawda P, Klatka J, Stepulak A. 7-substituted coumarins inhibit proliferation and migration of laryngeal cancer cells in vitro. Anticancer Res. 2013;33:4347-4356. [PubMed] |
| 21. | Kim JK, Kim JY, Kim HJ, Park KG, Harris RA, Cho WJ, Lee JT, Lee IK. Scoparone exerts anti-tumor activity against DU145 prostate cancer cells via inhibition of STAT3 activity. PLoS One. 2013;8:e80391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 22. | Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42:W32-W38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 596] [Cited by in RCA: 1005] [Article Influence: 91.4] [Reference Citation Analysis (0)] |
| 23. | Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44:D380-D384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 712] [Cited by in RCA: 1059] [Article Influence: 105.9] [Reference Citation Analysis (0)] |
| 24. | Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1-1.30.33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1289] [Cited by in RCA: 2902] [Article Influence: 322.4] [Reference Citation Analysis (0)] |
| 25. | Davis AP, Grondin CJ, Johnson RJ, Sciaky D, Wiegers J, Wiegers TC, Mattingly CJ. Comparative Toxicogenomics Database (CTD): update 2021. Nucleic Acids Res. 2021;49:D1138-D1143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 619] [Cited by in RCA: 693] [Article Influence: 173.3] [Reference Citation Analysis (0)] |
| 26. | Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199-W205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1238] [Cited by in RCA: 2207] [Article Influence: 441.4] [Reference Citation Analysis (0)] |
| 27. | Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18868] [Cited by in RCA: 24761] [Article Influence: 990.4] [Reference Citation Analysis (0)] |
| 28. | Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10161] [Cited by in RCA: 11750] [Article Influence: 1958.3] [Reference Citation Analysis (1)] |
| 29. | Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-2504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24663] [Cited by in RCA: 33594] [Article Influence: 1599.7] [Reference Citation Analysis (0)] |
| 30. | Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1127] [Cited by in RCA: 1253] [Article Influence: 69.6] [Reference Citation Analysis (0)] |
| 31. | Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4:S11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1658] [Cited by in RCA: 3766] [Article Influence: 342.4] [Reference Citation Analysis (0)] |
| 32. | Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5550] [Cited by in RCA: 7089] [Article Influence: 886.1] [Reference Citation Analysis (0)] |
| 33. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 133995] [Article Influence: 5583.1] [Reference Citation Analysis (1)] |
| 34. | Shen B. A New Golden Age of Natural Products Drug Discovery. Cell. 2015;163:1297-1300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 449] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 35. | Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, González-Barón M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30:193-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1407] [Cited by in RCA: 1788] [Article Influence: 85.1] [Reference Citation Analysis (0)] |
| 36. | Lee RS, House CM, Cristiano BE, Hannan RD, Pearson RB, Hannan KM. Relative Expression Levels Rather Than Specific Activity Plays the Major Role in Determining In Vivo AKT Isoform Substrate Specificity. Enzyme Res. 2011;2011:720985. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 37. | Zhang X, Zhang S, Yamane H, Wahl R, Ali A, Lofgren JA, Kendall RL. Kinetic mechanism of AKT/PKB enzyme family. J Biol Chem. 2006;281:13949-13956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 38. | Yang H, Liu JX, Shang HX, Lin S, Zhao JY, Lin JM. Qingjie Fuzheng granules inhibit colorectal cancer cell growth by the PI3K/AKT and ERK pathways. World J Gastrointest Oncol. 2019;11:377-392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 39. | Gu JF, Fu W, Qian HX, Gu WX, Zong Y, Chen Q, Lu L. TBL1XR1 induces cell proliferation and inhibit cell apoptosis by the PI3K/AKT pathway in pancreatic ductal adenocarcinoma. World J Gastroenterol. 2020;26:3586-3602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 5] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (1)] |
| 40. | Xu D, Zhou Y, Xie X, He L, Ding J, Pang S, Shen B, Zhou C. Inhibitory effects of canagliflozin on pancreatic cancer are mediated via the downregulation of glucose transporter1 and lactate dehydrogenase A. Int J Oncol. 2020;57:1223-1233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |