INTRODUCTION
Malignancies that occur in the sites from the esophagus to the rectum can be roughly classified as gastrointestinal (GI) cancers. These include the tumors rooting in the solid digestive organs and those occurring in the digestive tract. Some of them can develop from the neuroendocrine cells in the digestive system. It was estimated that about 333680 digestive cancer cases were diagnosed in the United States in 2020[1]. Many of the tumors, such as pancreatic carcinoma and hepatocellular carcinoma (HCC), have a poor prognosis even with intensive treatment. As a multifactorial process, both the individual’s genetic and the relevant environmental factors contribute to oncogenesis[2]. As there is no effective therapy for cancerous diseases, early diagnosis and timely intervention play key roles in reducing mortality. Varied imaging modalities are available in cancer clinics. Because of their lower cost and easier availabilities, blood biomarkers are highly recommended by many guidelines for tumor screening, diagnosis, and therapeutic effect evaluation[3,4].
Most of the approved biomarkers for GI cancer diagnosis are proteins. With the achievement of oncogenesis research and the advances of modern analytical techniques, many other macromolecules have been explored as new types of biomarkers. For example, a panel consisting of seven plasma micro ribonucleic acids was reported to be efficient for HCC diagnosis, especially for early-stage HCC[5]. Cell-free deoxyribonucleic acid (cfDNA) was readily detected in liquid biopsy samples[6]. With some traditional protein biomarkers, cfDNA could also be used in early-stage HCC screening[7]. These newly explored biomarkers contribute to GI cancer diagnosis and management to a varied extent.
Besides macromolecules, small molecular metabolites are also indispensable for an organism. Metabolites are the direct executors of metabolism. The entity of the whole metabolites in an organism constitutes its unique metabolome. A given metabolite profile is phenotype-specific, and phenotype is substantially modulated by metabolites[8,9]. Most of the inborn metabolic diseases (IMDs) exhibit metabolite concentration abnormalities[10]. Treatment of many IMDs involves limiting intake of certain kinds of chemicals[11]. Mass spectrometry (MS) is the earliest technology that was introduced into clinical laboratories for IMD diagnosis purposes[12].
Except for acting as the substrates and the products of enzymatic reactions, metabolites can also be the biomarkers for cancer diagnosis and treatment. This review would focus on the advances in using metabolites for GI cancer study and clinical practice.
METABOLOMICS
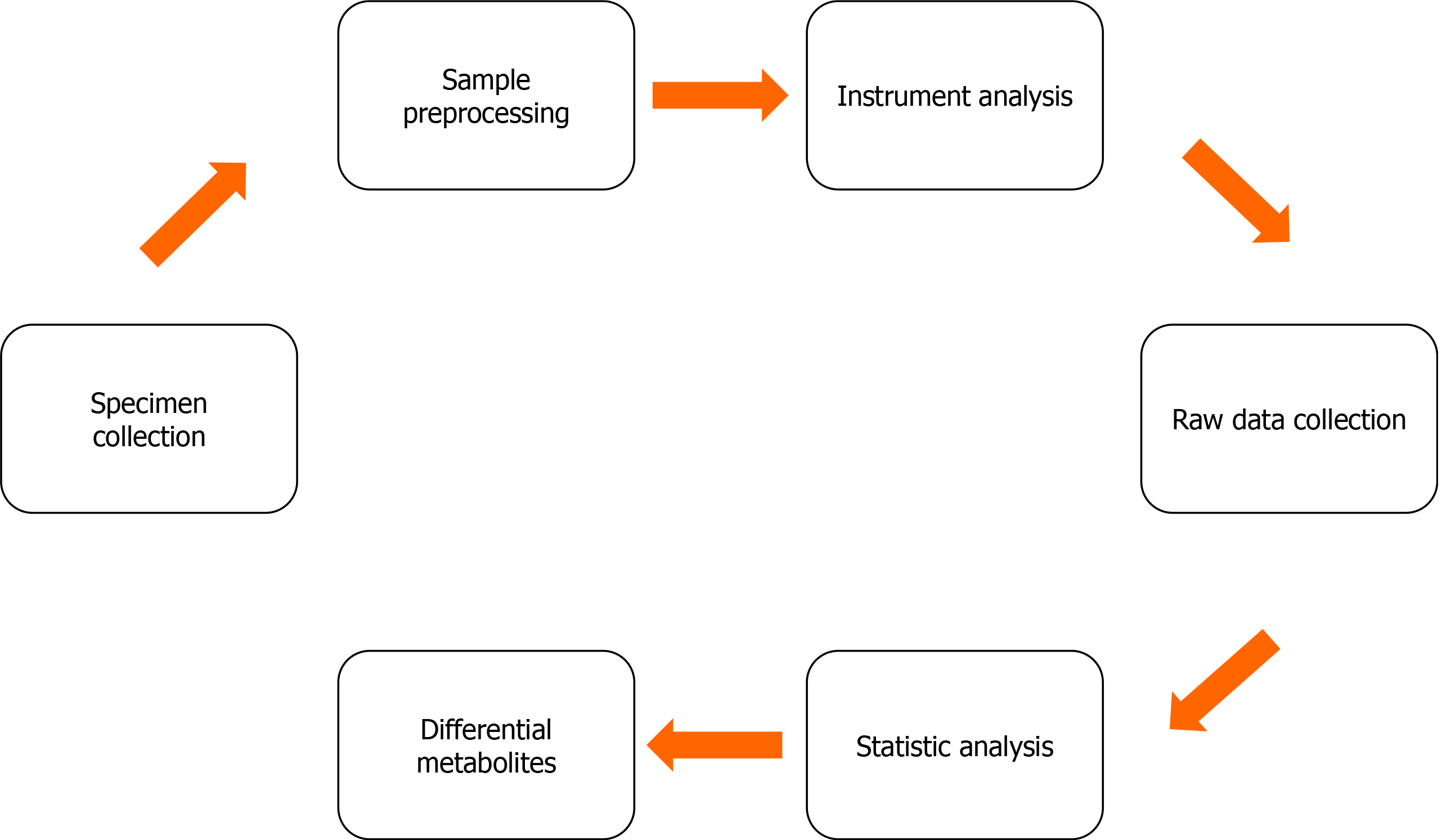
Genomics, transcriptomics, and proteomics are the high-throughput analysis of specific molecules in biological samples. Compared with the other omics, metabolomics is a newly coined conception. It aims at quantifying/qualifying as many metabolites as possible in a metabolome[8,13] (Figure 1). Since the advent of modern analytical technologies, high-throughput analyzing a metabolome has become possible. Nearly all the clinical specimens are compatible with metabolomic analysis[14]. Metabolomics aims at the compounds with molecular weights less than 1500 Dalton[15]. The leading techniques in this arena are MS and nuclear magnetic resonance (NMR) spectroscopy[16]. Both tactics have their inherent advantages in different analytical aspects[17]. For example, NMR is superior to MS in its analysis speed and noninvasive features[18]. MS is characterized by its high sensitivity and resolution[19]. Coupled with some separation technologies, MS or NMR can provide improved analytical abilities. This gave birth to the hyphened metabolomic analytical measures, such as liquid chromatography-MS (LC-MS), gas chromatography-MS (GC-MS), and capillary electrophoresis-MS. So far, most of the metabolomics studies were finished by employing the hyphened techniques. Many scientific groups tried to integrate NMR and MS. This approach provides distinctive advantages, especially for the analysis using isotopes[18].
Figure 1 The basic workflow of metabolomics.
Samples aiming at different purposes are first collected. The applicable specimen types include blood, biopsy, biofluid, cell, and urine samples. Some specimens must be preprocessed before they are analyzed with various equipment. The manipulations include metabolite extraction, condensation, or derivatization as possible. The metabolomics data are usually collected with the corresponding software equipped with the instruments. Some software also provides data pre-procession (e.g., to remove noise signals) and statistical analysis functions. The differential metabolites are first screened out by statistical methods. These selected metabolites should be verified using another set of samples if possible. It is better to ascertain the concentration changes of each metabolite using a robust quantitation method.
Metabolites have different polarities, volatilities, and hydrophilic properties owning to their elementary compositions. These physical aspects provide analysts with the opportunity to develop varied analytical methods to meet different needs. Therefore, there have been many derivative omics conceptions from metabolomics. For example, lipidomics is the metabolomic analysis of lipids exclusively. Metabolomic analysis focusing on carbohydrates can be called glycometabolomics[20]. Nucleosides include limited members. The concentration changes of modified nucleosides are frequently encountered in different diseases. Several metabolomics groups have paid more attention to the modified nucleoside detection[21].
According to whether the potential analytes were predefined, the metabolomic analysis could be divided into targeted and untargeted analysis[22]. The former is to detect the metabolites with definite identities, and the latter is to analyze all the measurable metabolites that are compatible with the adopted methods. The targeted analysis is frequently applied to studies with definite purposes, such as for verification or accurate quantitation. The untargeted strategy is suitable for global screening or catching a glimpse of the samples. Additionally, there is an analysis called pseudotargeted metabolomics[23]. This tactic is based on the principle that certain precursor molecules can produce definite daughter ions under a specific ionized circumstance. The ion fragmentation features are compound-specific. These structurally correlated ions could be monitored in parallel by some types of MS[24]. The pseudotargeted metabolomic analysis is independent of any identity knowledge of the analytes.
For biomarker exploration, a metabolomic study should consider untargeted analysis first. This analysis helps to lock the potential valuable metabolites. Then, a targeted metabolomic analysis is carried out. It is better to employ the quantitative analytical method that is most suitable for the targeted analytes. For quantitation accuracy, any untargeted analysis method is only compatible with limited types of metabolites. The following targeted analysis with robust quantitation capacities helps to corroborate whether the untargeted analysis findings are reliable and reproducible. Ideally, the targeted analysis should use another set of samples.
A great challenge in metabolomics is metabolite identification. It is better to build a database in which all the analytical features of the metabolites are recorded. Unfortunately, it is unknown how many metabolites might exist in different biological samples. Some groups have tried to set up a database according to their routine needs. Many of the databases are free to non-commercial use[25,26]. To simplify metabolite identification, many software programs have been developed. Some of them could directly use the data collected with the analytical equipment[27]. Statistical and bioinformatic analysis is necessary for biomarker selection and annotation. Many software programs provide various online analysis tools[28].
GI CANCER PREDICTION
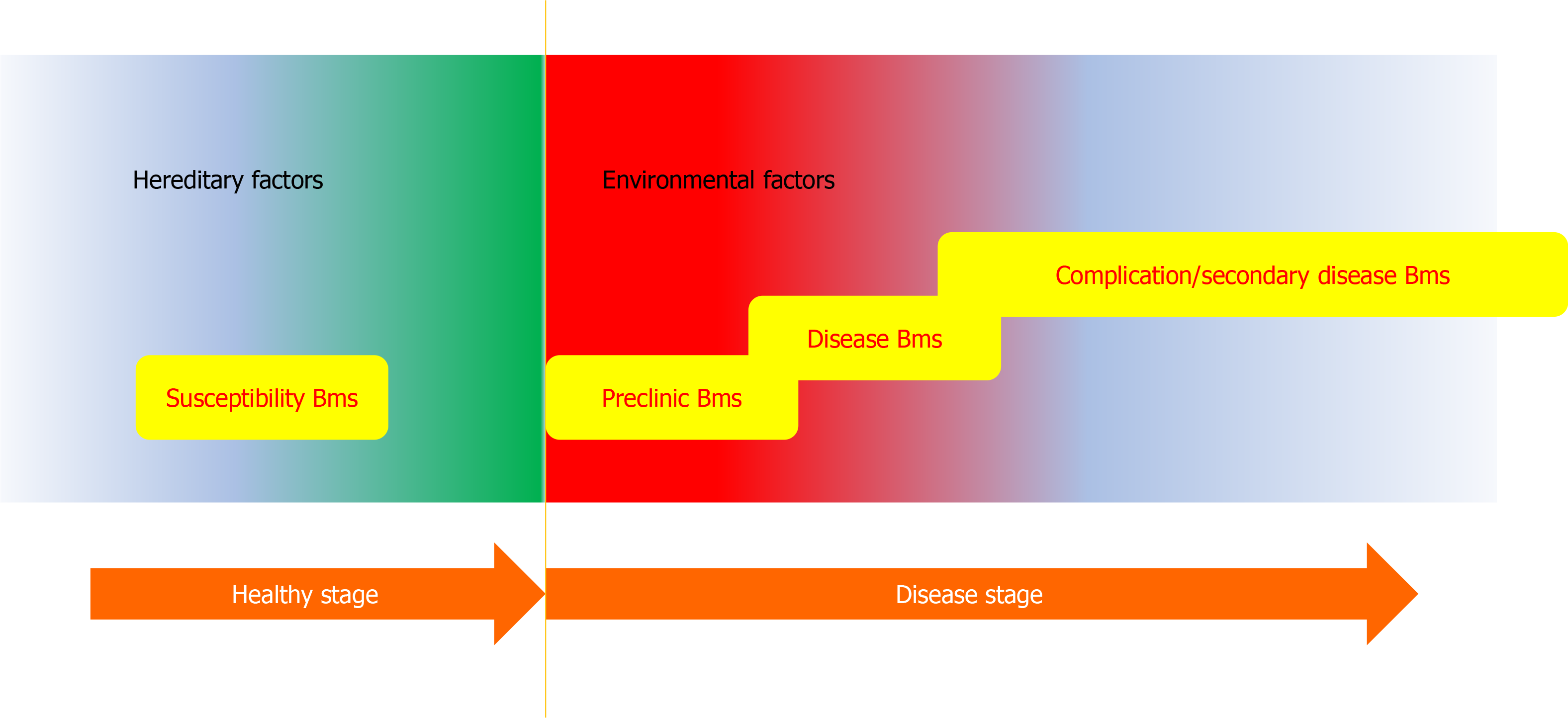
Any disease, including cancerous diseases, obeys their regular development progression. There must be some clues existing in the preclinical stages (Figure 2). This provides opportunities to predict diseases. In a prospective study based on LC-MS, plasma valine, leucine, and isoleucine were reported to be valuable for pancreatic ductal adenocarcinoma (PDAC) prediction especially for the onset within 2-5 years[29]. Subjects with these amino acid changes had two times higher risks than the control ones. The three branched-chain amino acids (BCAAs) belong to necessary amino acids. Whereas, the authors demonstrated that the raised plasma concentrations of these BCAAs were not the results of excessive ingestion. They were linked to early-stage tissue protein breakdown driven by the K-ras gene. Interestingly, if the three BCAAs were combined with tyrosine and phenylalanine, they could be used to predict future diabetes onset. A 12-year follow-up study indicated that individuals with elevated blood concentrations of the five amino acids were at higher risks to develop type 2 diabetes (T2DM)[30]. T2DM and PDAC had a reciprocal relationship[31]. Thus, it is better to introduce other metabolites to improve the prediction accuracy when the metabolite panels are overlapped. To enlarge the metabolite coverage, a study simultaneously employed LC-MS and GC-MS to analyze the blood samples. The study included 226 pairs of case and control subjects. The plasma phosphatidylcholine [PC (15:0/18:2)], coumarin, and picolinic acid levels were found to be positively related to pancreatic cancer. Six glycerophospholipids were inversely associated with pancreatic cancer incidence. After excluding the interference factors including T2DM, the PC (18:1/18:4), instead of PC (15:0/18:2), was found to be most valuable especially for predicting the onset within 5 years[32]. From the perspective of epidemiology, factors that are inversely correlated to diseases are protective. Although both studies utilized LC-MS and selected the subjects of similar backgrounds[29,32], the potential prediction markers were not identical. One reason is that tumorigenesis is a complex process. It can be triggered by different combinations of driver factors. The other reason might be that lifestyles, food appetite, and genetic backgrounds vary greatly amid different races and populations. For instance, African Americans have a higher colorectal cancer (CRC) rate than rural South Africans. Epidemic investigation proved that the former consumed more animal protein and fat in their daily life[33]. On the contrary, the latter ingested more fibers. If the food styles were exchanged between them, fecal water and urine metabolomes changed accordingly. If they ingested more protein and fat-rich food, both the Americans and the Africans were characterized with abundant fecal choline and urine trimethylamine-N-oxide[33].
Figure 2 Schematic representation of the fluctuation of biomarker in the whole period of a disease.
Disease susceptibility is usually defined by the individual’s genetic background. The susceptibility biomarker (Bms) can be detected by genetic analysis most possibly. The onset of the disease would be triggered by many environmental factors. At the very beginning (preclinical stage), some prediction Bms appears. When a disease progresses to the clinical stage (with clear symptoms) the diagnosis Bms could be detected. If the disease advances further, some complications and secondary hurts would emerge. These end events give birth to the opportunities to develop the relevant Bms. Metabolomics could be applied to the whole disease period. Besides, prognosis and treatment efficacy Bms could also be explored by metabolomic analysis.
Diet affects not only cancer risks but also the prognosis [34-36]. A follow-up study enrolled 463 postmenopausal CRC women. The researchers found that diet and food with anti-inflammatory potential could improve overall survival[37]. The relationship between dietary exposures and diseases was the key theme of nutritional metabolomics[36]. Unfortunately, up to now, large-scale meta-analysis data for GI cancer prediction using metabolite markers are rare. Fortunately, metabolomics analyses have identified many candidate biomarkers about specific food exposures. For example, meat and/or seafood consumption resulted in elevated plasma essential amino acids, polyunsaturated fatty acids, and D-glucose[38]. Shellfish consumption affected plasma phosphatidylethanolamine (p36: 4). Plasma 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid was related to fish intakes in the Asian population[38]. What should be mentioned is that if the fish ingestion study is carried out in European people, the candidate marker should be trimethylamine-N-oxide instead of 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid[39].
Besides tumor onset prediction, metabolites can be used to predict prognosis. Redalen et al[40] reported that tumor glycine was an adverse prognostic factor for locally advanced rectal cancer. Cancers with rapid growth rates were demonstrated to consume glycine excessively[41]. Too many reasons can affect the concentrations of a single amino acid. As the lessons from protein biomarker applications, a biomarker panel including several (kinds of) metabolites might be more valuable and reliable than a single metabolite.
GI CANCER DIAGNOSIS AND EARLY DIAGNOSIS
CRC poses a great challenge to public health, especially in developed countries. Early diagnosis is important to reduce mortality. To early detect CRC, plasma samples collected from stage 0/I/II patients and the controls were subjected to GC/triple-quadrupole MS (TMS) analysis[42]. A regression model consisting of eight metabolites [pyruvic acid-meto-TMS, glycolic acid-2TMS, tryptophan-3TMS (/SI), palmitoleic acid-TMS, fumaric acid-2TMS (/SI), ornithine-4TMS (/SI), lysine-4TMS, and 3-hydroxyisovaleric acid-2TMS] could realize satisfying CRC diagnosis with a sensitivity of 99.3% and specificity of 93.8%. In that study, the traditional protein markers carcinoma embryonic antigen and carbohydrate antigen19-9 showed good specificities, but their sensitivities were low (< 20%). The authors also pointed out that this model could not be applied for aggressive CRC (e.g., stage III/IV). When invasive CRC metastasizes, it might affect and spread to many organs. It can be expected that the systemic metabolic changes caused by local and metastasis tumors are different.
Another notorious GI cancer is HCC. Alpha-fetoprotein (AFP) has been used for HCC surveillance and diagnosis for decades. Its limited specificity is obvious. The rapid advances of imaging modalities have excluded the utilization of AFP according to the recently approved guidelines[43]. Unfortunately, imaging examination could miss many solid neoplasms with a diameter less than 3 cm. Thus, the early diagnosis needs some alternative solutions. In this light, a large-scale metabolomic study was conducted. To pursue the robustness of the diagnosis, many diseases that might interfere with HCC were included as possible[44]. It was found that serum phenylalanyl-tryptophan and glycocholate showed good performance in HCC diagnosis and differential diagnosis. Even for small HCC, the combined use of the two metabolites could achieve an area under the receiver-operating characteristic curve (AUC) of 0.866. According to the results, elevated glycocholate was positively correlated to HCC. Phenylalanyl-tryptophan was negatively correlated to HCC. An appropriate tumor biomarker should be in high concentrations in the blood because of its excessive release or production. Pathologically, the decreased phenylalanyl-tryptophan might be the result of tumor-related overconsumption. Technically, biomarkers with decreased concentrations causes the quantitation difficulty.
Except for the blood samples, feces sample is also a valuable specimen for metabolomics. In theory, components in the feces reflect the intestinal physiological and pathological status. A pilot metabolomic study detected 527 reproducible metabolites in the feces samples from CRC patients. Three fecal heme-related molecules, 18 peptides/amino acids, palmitoyl-sphingomyelin, mandelate, p-hydroxy-benzaldehyde, acetaminophen metabolites, tocopherols, sitostanol, 3-dehydrocarnitine, pterin, conjugated-linoleate-18-2N7, N-2-furoyl-glycine, and p-aminobenzoate were found to be valuable for CRC diagnosis[45]. However, metabolites in the feces varied greatly due to diet styles and gut microflora. Many metabolites contributing to the CRC diagnosis in the above-mentioned study were bacterial metabolites or co-metabolites of human beings and the gut microbes. This resulted in the observation that not all the biomarkers were elevated in the CRC feces. For the stabilities, feces were not comparable to blood samples[45]. The markers indicating the storage stabilities of blood samples have been explored and identified[46,47]. Similar studies about feces were seldom conducted. Gut microbiota affects the intestinal microenvironment. Unhealthy microbiota contributes to many diseases including CRC. In this light, the fecal metabolomic analysis might be more valuable for prediction use[48,49].
Small-intestine neuroendocrine tumors (SINETs) are a common GI cancer stemming from the neuroendocrine cells in the small bowel. Many of these tumors have features of metastasis. By performing NMR-based metabolomic analysis, Imperiale et al[50] found that succinate, glutathion, taurine, myoinositol, and glycerophosphocholine were elevated in the tumor samples. The normal small intestine tissues were rich in alanine, creatine, ethanolamine, and aspartate. When the hepatic metastasis lesions were compared with the normal liver, acetate, succinate, choline, phosphocholine, taurine, lactate, and aspartate were found to be rich in the lesions. The primary SINETs were characterized with increased succinate, valine, and myoinositol when they were compared with the metastases. This study demonstrated that identical tumors found in different microenvironments could exhibit distinctive phenotypes[50].
Cholangiocarcinoma was thought to be related to bile acid metabolism[51]. Zhang et al[52] analyzed 329 plasma samples collected from the controls, benign biliary diseases, cholangiocarcinoma, gallbladder cancer, and HCC populations. Taurochenodeoxycholic acid and chenodeoxycholic acid played key roles in separating cholangiocarcinoma both from the healthy controls and from the HCC patients. The diagnostic performance was even superior to the commonly used carbohydrate antigen 19-9.
Recurrence is a key theme in the tumor research field. From three independent cohorts, Qiu et al[53] found 14 upregulated and 1 downregulated metabolite biomarkers to predict CRC relapse. The authors also pointed out the inconsistency of these metabolite changes amid different cohorts. No matter what potential uses, to validate biomarkers must need more effort.
PATHOLOGICAL DIAGNOSIS OF GI CANCER
Traditional pathological diagnosis is dependent on slice samples. Preparing a satisfying slide sample is a time-consuming and labor-intensive task. The intraoperative histological examination costs only half an hour but is expensive. Also, the diagnosis accuracy is affected by the expertise of both the technologists and the pathologists. What makes the matter worse is that the traditional pathological slides only afford limited tissues or cells. It brings about inevitable sampling bias. When it comes to metabolomics, most or all the resected tissues can be used to extract the metabolites. Additionally, the extracts can be subjected to various preprocessing such as condensation, dilution, or derivatization to meet different analytical needs.
Endoscopic examination is widely used in CRC screening. The morphological characteristics of advanced adenomas and CRC tissues are inadequate for differentiation purposes. In a study, an untargeted MS-based metabolomic technique was first employed to analyze CRC and matched paracancerous tissues. This profiling strategy narrows the cancer-related metabolic changes to amino acid metabolism. Then, another MS-based targeted amino acid analysis was performed. The results showed that combined use of methionine, tyrosine, valine, and isoleucine was enough to distinguish CRC from advanced adenoma[54]. The notable advantages of metabolomics are its simplicity and rapidness.
As widely admitted, MS analysis is characterized by its high specificity and rich chemical information. The traditional pathological tactic has a distinguished resolution. If the advantages of both are combined, pathologists will gain more deep insight into the slice samples[55]. Fortunately, scientists have developed applicable strategies to integrate the two techniques and applied the so-called MS imaging (MSI) strategy to cancer pathological studies. Desorption electrospray ionization mass spectrometry (DESI-MS) can give chemical information from the surfaces of an intact or processed tissue specimen under ambient conditions[56]. Nagai et al[57] first performed an untargeted analysis of HCC and benign tissue samples by MS. They found that TG 16:0/18:1 (9Z)/20:1 (11Z) (m/z 904.83) and TG 16:0/18:1 (9Z)/18:2 (9Z, 12Z) (m/z 874.79) played roles in separating the two kinds of samples. Then, they employed MSI to explore the tissue distribution of the two TGs. Despite the overlap at the boundary regions, condensed TG 16:0/18:1 (9Z)/20:1 (11Z) distribution in the tumor regions and abundant TG 16:0/18:1 (9Z)/18:2 (9Z, 12Z) (m/z 874.79) in the nontumor regions was obvious. The results were consistent with the previous reports about the saturated and unsaturated fatty acid distribution in the tumor and nontumor tissues. These fusion images integrated traditional hematoxylin and eosin staining and MS ion imaging. The strategy provided high-quality pathological pictures at 10 μm-resolution[55]. The most valuable use of MSI might be to explore extremely small local and metastasis lesions.
MSI can not only be used to help pathological diagnosis, but it can also be used to aid tumor-related enzyme exploration. Sun et al[58] first employed airflow-assisted DESI-MSI to profile region-specific metabolites in esophageal squamous cell carcinoma (ESCC) and corresponding normal samples. Then, they performed metabolic pathway matching analysis based on the selected differential metabolites to lock potential tumor-associated metabolic enzymes. Subsequently, immunohistochemical staining was performed to validate the enzyme expression changes. Finally, they found that proline biosynthesis, glutamine metabolism, uridine metabolism, histidine metabolism, fatty acid biosynthesis, and polyamine biosynthesis pathways were altered in ESCC. Pyrroline-5-carboxylate reductase 2 and uridine phosphorylase 1 was upregulated in ESCC tissues. This high-coverage-based MSI analysis provided valuable information on new drug development and therapeutic target identification.
Direct, real-time, and non-invasive examination of intact tissues is highly appreciated in surgical rooms. It is affordable that partial normal tissues are damaged in some surgical operations. However, in neurosurgical resections, damaging normal brain tissues has always been avoided. Traditional DESI-MS can work under ambient conditions, but it suffers from technical incompatibilities in many facets such as the use of organic solvents, high-pressure nebulizing gas, and high voltages[59]. Zhang et al[59] developed a device called MasSpec Pen based on the DESI-MS. The MS was equipped with a handheld probe that could squeeze a discrete water droplet under control. The droplet was delivered on the surface of the target tissue. Metabolites in the tissue could be extracted into the droplet and transferred to the analysis system-an Orbitrap mass spectrometer. The authors employed the MasSpec Pen to analyze several kinds of benign and malignant solid tissue samples. The results demonstrated that this device could realize a diagnostic sensitivity of 96.4% and specificity of 96.2%. The overall accuracy was 96.3%. Furthermore, MasSpec Pen has ever been introduced into the porcine upper GI tracts in a study. The accuracy of distinguishing the liver from the stomach tissues in vivo was 98%[60]. In fact, utilizing MasSpec Pen for any cancer diagnosis was solely dependent on the availability of the corresponding tissue-specific database[59].
Like MasSpec Pen, iKnife is another rapid evaporative ionization mass spectrometry (REIMS)-based metabolomic diagnosis device. It can not only realize real-time pathological analysis but also act as an “electric lancet". iKnife does not rely on the liquid media to dissolve the metabolites. It directly analyzes the gas components released from the burned tissues. Electrosurgical devices are prevailing in the operation rooms because of their simultaneous dissection and hemostasis functions. The burned tissues would release smoke containing many oxidized metabolites. This previously discarded smoke is collected with a specifically designed device and then transferred to REIMS to be analyzed. The chemical information in the smoke can be used to identify the properties of the tissues releasing the smoke[61]. Balog et al[61] analyzed 1624 cancerous, 1231 healthy, and 78 inflammatory bowel disease samples. They found a different distribution of lipid species across the specimens. Alexander et al[62] applied iKnife to diagnose CRC. The overall accuracy was 94.4%. Phosphatidylserines and bacterial phosphatidylglycerols were rich in the cancer samples. Ceramides were condensed in the adenomas. The normal tissues were characterized by elevated plasmalogens and triacylglycerols[62]. iKnife can be used to identify the origins of the metastatic tumors. When differentiating healthy liver parenchyma from metastasis colonic adenocarcinomas, the iKnife could give a diagnostic accuracy of 96% (73/76).
PERSONALIZED GI CANCER TREATMENT
Chemotherapy is necessary for GI cancer treatment. Chemotherapeutical drug administration brings about several side or toxic effects. Even if the physicians can correctly make their chemotherapy decisions, the one-size-fits-all approaches do not guarantee a good prognosis for all the patients. Precise prediction of the chemosensitivities would benefit both the patients and the physicians. Pharmacometabolomics is the science utilizing metabolomics to predict patient responses to drug treatments. A pilot study based on serum metabolomics indicated that elevated serum deoxyribose 1-phosphate and decreased S-lactoylglutathione correlated to chemotherapy sensitivities[63]. Capecitabine is an antimetabolic agent that could be metabolized to 5-fluorouracil-the active form for CRC treatment. Side effects of capecitabine are largely originated from its intermediate metabolite 5′-deoxy-5-fluorouridine (5′-DFUR). By performing 1H NMR spectrometer-based metabolomic analysis of 52 CRC serum samples, Backshall et al[64] found that patients with higher LDL-like lipid particles and choline phospholipid were prone to suffering from 5′-DFUR toxicity. Also helped by NMR metabolomics, Bertini et al[65] analyzed 153 serum samples from metastasis CRC patients before cetuximab and irinotecan administration. They found that the patients with long and short overall survival (OS) time could be identified with an accuracy of 78.5%. The patients with OS > 24 mo and < 3 mo showed different serum metabolite profiles. They also pointed out that the potential differential metabolites contributing to separation of the two groups were also affected by some other factors such as obesity.
Postoperation chemoradiotherapies are indispensable, even if surgical resection is performed in the early stage of esophageal cancer. However, not all the cases benefit from the adjuvant strategies. A metabolomics study found that decreased serum arabitol, glycine, L-serine, and L-arginine indicated a positive response to chemoradiotherapies[66]. For predicting the chemoradiotherapy responses, the combined use of the four metabolites generated an AUC > 0.7.
Chemoresistance is frequently encountered clinically. The resistance could be acquired or innate. Many chemotherapy drugs are antimetabolites and affect cell metabolism. The built-in metabolic plasticity and the robustness of the metabolic networks render the cells with conspicuous capacities to resist perturbations from the environment. Cells can reprogram their metabolism to resist the perturbations from the chemotherapy drugs. Those cells that can not adapt to the drug stimuli will be killed. Intracellular metabolite pools are dynamic in size. The pool sizes were affected by the metabolic flux rates of the relevant metabolic pathways[67]. Cells can keep hemostasis by redirecting the metabolic fluxes of the relevant metabolic pathways. The flux rates can be calculated. The most widely used metabolic flux analysis (MFA) is 13C MFA. The analysis uses the 13C-labeled substrate (usually the 13C-labeled glucose or amino acids) to feed the cells. After proper incubation, intracellular metabolites are quantified by metabolomic analysis. The detected metabolites are then used to calculate the metabolic fluxes through chemometrics according to the labeled element distribution in the metabolic pools[68]. Mathematically, a metabolic network is a set of stoichiometric equations. Each equation is defined by a real enzymatic reaction that can be easily retrieved from biochemical textbooks or public databases. Because the metabolic networks contain hundreds to thousands of pathways, the calculation is a tough job. Most of the tasks are finished by software models run on computers. Combined with computational and mathematical modeling tactics, MFA could shed light on cellular phenotypes from another angle[69].
Highly expressed hexokinase 2 (HK2) is frequently found in HCC cells. An MFA using (1,2-13C) glucose and (U-13C) glutamine as tracers exhibited that glucose uptake and lactate secretion rates dropped by 40% in Huh7 cells with HK2 silencing. Glutamine and branched-chain amino acid uptakes, secretion of alanine and glutamate, and the tricarboxylic acid cycle-related fluxes were not affected. The HK2 silencing cells were more sensitive to one-carbon unit depletion. There was a 2-fold increase in serine uptake and glycine secretion. There was no obvious change in the intracellular glucose to serine flux. The study also found that silencing HK2 synergized sorafenib, which provided a clue to treat HCC by manipulating HK2[70].
Flux balance analysis is another type of MFA. It sets rational constraints on a metabolic network and presumes that the network is in its steady-state. Nikmanesh et al[71] constructed a model integrating expression data from Gene Expression Omnibus and metabolomics data. The metabolic model included 3748 reactions and 2766 metabolites. Using this model, the authors compared the metabolic flux difference of 56 normal and 67 CRC cells. Compared to the normal cells, cancer cells exhibited 503 upregulated and 560 downregulated fluxes. Reactions catalyzed by retinol dehydrogenase, bicarbonate transporter, cytosine deaminase, glutathione peroxidase, and mitochondrial adenosine diphosphate/adenosine triphosphate (ATP) transporter were the notably downregulated ones. The other pathways with decreased metabolic flux rates included pathways involving palmitoyl-CoA desaturase, glutamine synthetase, ATP synthase, and uridine triphosphate-glucose-1-phosphate uridylyltransferase. The nucleotide metabolism (catalyzed by nucleoside-diphosphate kinase) and pyruvate metabolism (catalyzed by L-lactate dehydrogenase) pathways had increased flux rates. Some reactions involved in purine catabolism, glycolysis/gluconeogenesis, and hyaluronan metabolism showed increased flux rates. In that model, the authors also included the point mutation information. This coupling strategy helped to discover the driver regulatory modules. Thus, with the help of data mining and integrating tools, metabolomics could potentially be used to uncover potential therapeutic targets and new tumor driver mechanisms. This would be good at formulating personalized therapeutic strategies.
Traditionally, the enzyme catalyzing the slowest step in a metabolic pathway is deemed as the rate-limiting enzyme. The relevant step is regarded as the rate-limiting step. At the very beginning, metabolic engineering aims at manipulating these enzymes. Unfortunately, overexpressing the relevant enzymes fails frequently. Metabolic control analysis (MCA) introduces a new conception to determine the real rate-limiting step by considering how a given enzyme exerts its influence on the fluxes and the concentrations of the involved metabolites[72]. As hemostasis is maintained by metabolism, some key metabolite changes might be lethal. The enzymes catalyzing the relevant reactions could be drug targets potentially. One of the prominent pilot studies using MCA to identify therapeutic targets was reported in the practice of treating trypanosomiasis. Scientists found that the glucose transporter, aldolase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, and glycerol-3-phosphate dehydrogenase were the Archil's heels of parasites instead of red blood cells[73,74]. Thus, relational treatment strategies could be developed by circumventing the targets that might damage the hosts.
Using MCA, Koit et al[75] found that HCC tissues showed suppressed respiratory chain complexes I functions. But, it was not the case for breast cancer tissues. Mitochondrial membrane permeabilities were different between the two types of tumor cells. These clues were valuable on how to select effective anti-tumor drugs. Many tumor therapies share the same drugs or drugs with similar mechanisms. Physicians could make more personalized therapeutic decisions with the MCA results.
Although the variability of a single person’s metabolome is universal, every individual has his/her relatively stable metabolic phenotype. It dominates the specific responses to specific stimuli. Assfalg et al[76] collected 40 urine specimens from 22 healthy persons across 3 mo. According to the 1H NMR urine metabolomic data, the interindividual difference was larger than the intraindividual difference. Fifteen metabolites were enough to confirm an unknown sample origin with 100% confidence. The individual-specific phenotypes contained subject-specific nutrition tolerance, drug efficacy and toxicity, disease risk, and much physical and pathological response information[76]. The authors also implied that to define an individual’s phenotype needs specimens collected in a long period. This could exclude the casual influence. Thus, metabolomics could be a valuable tool for personalized medicine.
CONCLUSION
Genomics, transcriptomics, and proteomics studies have been applied in tumor fields for many decades. The findings from a single omic analysis are prone to being misinterpreted due to the tumor heterogeneities. Many analytical skills and tools could be selected to perform metabolomic analysis. Compared to the other omics, metabolomics is still in its infancy. New methods of metabolite identification, bioinformatic analysis of the data, noise signal removal for the spectroscopic data, and analytical speed improvement are still under development. It should be noticed that all the above-mentioned GI cancer metabolite biomarkers are not “new” metabolites. All of them could be found in physiological conditions. Also, their concentration changes can be found in non-cancerous diseases. Nearly all the mentioned GI cancer metabolite biomarkers can be found in other cancers. Identical metabolite markers can be found in different GI cancers and even can be used for different purposes. Unlike the protein and the mutant gene biomarkers, metabolite biomarker concentrations are severely affected by diet styles and circadian rhythms. To use metabolite biomarkers should follow an intensive verification procession and must consider the backgrounds against which the metabolite markers are identified. Compared to the other omics, metabolomics had many advantages[77]: (1) Changes taking place at the gene or protein levels can be amplified at the metabolome level; (2) Metabolomic analysis does not need the complete gene sequence information; (3) The members of a metabolome are smaller than those of a genome or proteome; and (4) Performing a metabolomic analysis is cheaper than performing a transcriptome or a proteome analysis. Besides the above-mentioned applications, metabolomics has been used to explore gene functions[78], drug mechanisms[79], enzyme functions[80], and tumor driver metabolites (oncometabolites)[81]. Although the applications are scattered in different bioscience fields, it can be concluded that metabolomics is undoubtedly a valuable complement to the other techniques in prompting GI cancer research.
Manuscript source: Invited manuscript
Specialty type: Medical laboratory technology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sridharan G S-Editor: Zhang L L-Editor: Wang TQ P-Editor: Li JH