INTRODUCTION
Gastrointestinal (GI) cancers, including malignancies in the gastrointestinal tract and accessory organs of digestion, represent the leading causes of death worldwide as most of them exhibit poor prognosis[1]. According to the American Cancer Society's estimation, GI cancers will be responsible for approximately 338090 new cases and 169280 deaths in 2021 in the United States[2]. Although accumulating research has enriched the pathogenesis and provided therapeutic strategies for GI cancers, many of them are hard to diagnose and treat. The prognosis of GI cancers remains poor. Some patients even miss the chance of being cured once diagnosed[3-5]. As a result, it is necessary to investigate the potential molecular target of GI cancers.
Proliferating cell nuclear antigen (PCNA) clamp associated factor (PCLAF), a 15-KD protein-containing PCNA-binding motif [PIP box, Qxx(L/I/M)xx(F/Y)(F/Y)], is a potential molecular target of GI cancers. PCLAF is associated with several types of GI tract cancers, such as esophageal cancer[6,7], gastric cancer (GC)[8-10], colorectal cancer (CRC)[11], and accessory organs of digestion cancers, including liver cancer[12-14], and pancreatic cancer (PC)[15].
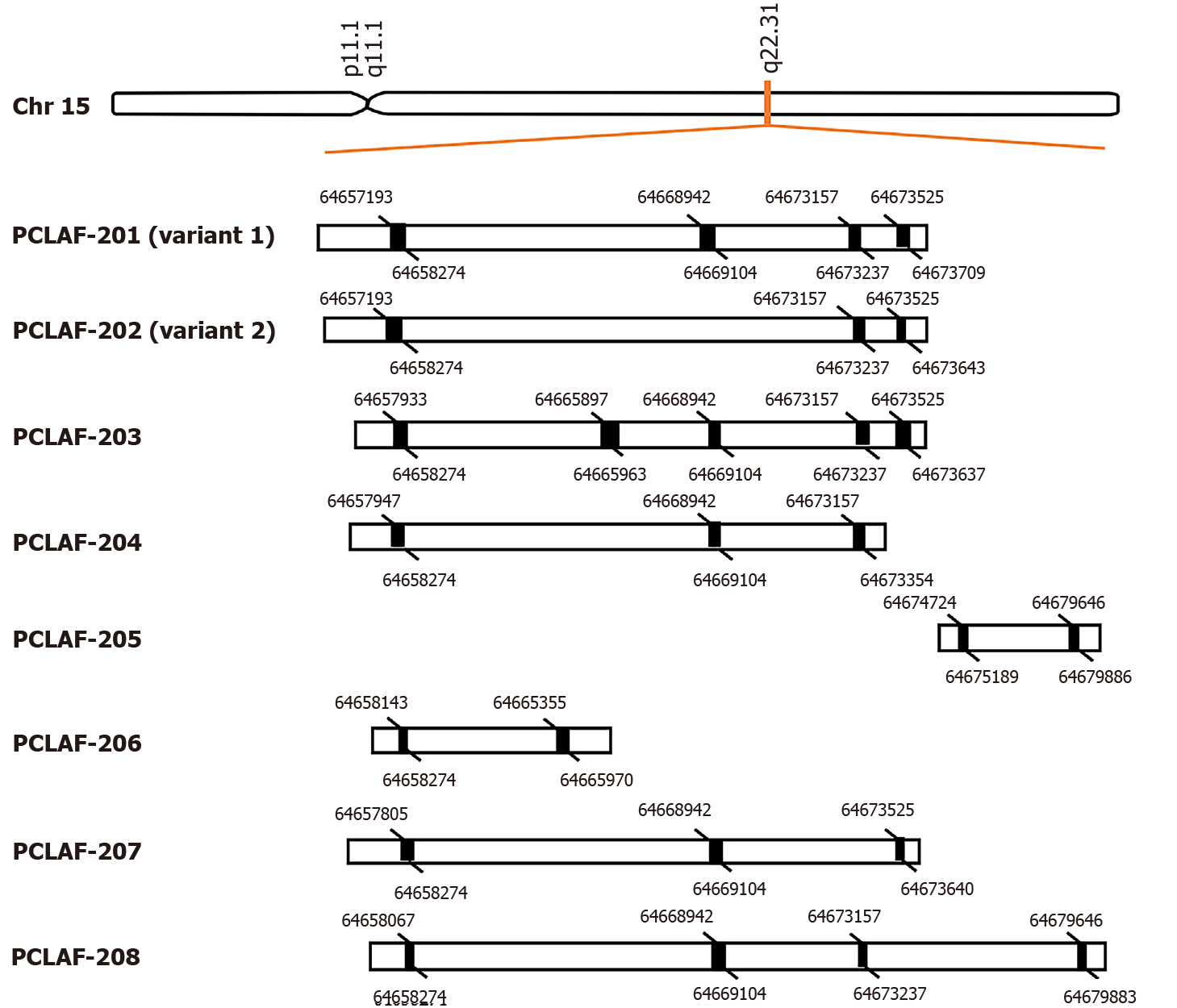
PCLAF was initially identified in 2001 as a PCNA associated protein[16]. It is also named KIAA0101, PCNA associated factor (p15), overexpressed in anaplastic thyroid carcinoma-1, or hepatitis C virus (HCV) nonstructural protein 5A-transactivated protein 9[16-19]. The gene has eight transcript variants, including two known protein-coding transcripts, two putative protein-coding transcripts, and four processed transcripts (Ensemble: ENSG00000166803) (Figure 1). Most studies focused on the known protein-coding variant 1 of PCLAF.
Figure 1 Alternative pre-mRNA splicing of proliferating cell nuclear antigen clamp associated factor.
Proliferating cell nuclear antigen clamp associated factor (PCLAF), maps to human chromosome 15q22.31, can be spliced into eight different transcripts, including two known protein-coding transcripts (PCLAF-201; 202), two putative protein-coding transcripts (PCLAF-203; 207), and four known processed transcripts (PCLAF-204; 205; 206; 208). The black rectangles represent exons. The figure indicates the spliced bases of each exon.
The capacity of PCLAF for biological functions is attributed to its interaction with PCNA[20]. It plays a transcription factor in regulating several targets to modulate cellular processes, such as DNA replication, DNA repair, apoptosis, and cell cycle regulation[18,21-24]. Based on the biological function of PCLAF, there is no doubt that dysregulation of PCLAF in human cells might lead to malignant tumors. In addition to GI tumors, PCLAF mRNA and protein are highly expressed in various other tumors, including lung cancer[25,26], adrenal cancer[27], breast cancer (BC)[28], and ovarian cancer[29]. The extensive involvement in oncogenesis extends far beyond a simple role. Various studies have documented that PCLAF activates some signal pathways that lead to tumorigenesis[26,29]. It also suppresses the anti-tumor genes in cancers[14,28]. Fortunately, several articles report that individual microRNAs can directly target PCLAF in different types of cancers[30-32]. In this review, we will mainly summarize the biological function of PCLAF in GI cancers and propose molecular mechanisms to examine the relationship between PCLAF and human malignancies.
BIOLOGICAL ROLE OF PCLAF
According to the data files in the National Center for Biotechnology Information, the PCLAF gene maps to human chromosome 15q22.31, a region harboring a tumor suppressor gene for CRC[33]. Variant 1 consists of 2132 bp with four exons, which encodes a protein of 111 amino acids. It has the PIP box, and overcomes the p21-mediated inhibitory function in cell cycle progression[16]. PCLAF variant 2 consists of 1969 bp with three exons. Its protein lacks the PIP box domain as exon 3 is skipped during splicing. There is little information on the biological function of variant 2. In our early study, we found that PCLAF variant 2 is markedly expressed in normal liver tissues, and it plays a competition role with PCLAF variant 1 on binding and regulating the function of p53[34].
Current studies suggest that PCLAF is expressed in some normal human tissues; however, the expression level is weak, and there are individual differences and specificity. Northern blot shows that PCLAF is highly expressed in the colon and thymus but low in spleen, ovary, testis, small intestine, and kidney[35]. Yu et al[16] reported that PCLAF mRNA is expressed in the liver, pancreas, and placenta at high levels but not in the heart or whole adult brain. From semi-quantitative polymerase chain reaction (PCR) results, Mizutani and his colleague determined that PCLAF is expressed ubiquitously, including in the placenta, kidney, spleen, thymus, small intestine, and thyroid tissue. However, the gene expression is weak[19]. Immunohistochemical (IHC) staining shows nuclear staining of PCLAF in the endometrium, lymph node, epidermis, and small and large intestines, but not in the brain, pancreas, lung, liver, heart, and myometrium[21]. Another study also found no PCLAF expression in normal pancreatic ductal or acinar cells, or any of the vital normal organs, including lung, heart, liver, and kidney[15]. In conclusion, PCLAF is positively expressed in some lymphoid organs, including the kidney, spleen, thymus, lymph node, and some rapidly dividing tissues, such as the placenta, endometrium, epidermis, and so on.
There is also scope for dispute about the subcellular localization. The IHC analysis of PCs and PC cells shows positive staining of PCLAF in the nuclei[15]. Similar results are also available in primary invasive breast tumors[36], and colon cancer cell lines[11]. In HeLa cells, PCLAF was observed in nuclei and perinuclear compartments by immunofluorescence microscopy. Additionally, in dual-labeling experiments of HeLa and Hs578T cells, a bright focus of PCLAF stain colocalized with a single site consistent with the centrosome, indicating its localization in the region of centrosomes[37]. In addition to detection in nuclei, PCLAF protein was found in the cytoplasm in adrenal cancer samples[27]. Moreover, PCLAF immunofluorescence coincided with the image of cytochrome oxidase subunit I, an inner mitochondrial membrane protein, which suggests that PCLAF is localized in mitochondria[38]. PCLAF is also a secreted protein as it can be detected in plasma or peripheral blood mononuclear cells[31,39]. As a result, PCLAF protein in the nucleus might predominate. However, it can also translocate to the cytoplasm or be secreted into blood.
Current studies reveal that PCLAF may play vital roles in DNA replication and DNA repair, mainly in S phase. As variant 1 of PCLAF contains the PIP box, a domain can interact with PCNA, an essential molecule for DNA replication and DNA repair through interaction with several DNA replication proteins, such as DNA ligase, endonuclease, and DNA polymerases[40]. Turchi et al[24] showed that prevention of the interaction between PCLAF and PCNA significantly alters DNA repair mechanisms, suggesting that PCLAF is responsible for the correct function of PCNA during DNA repair. Mechanism studies have demonstrated that UHRF1 (ubiquitin-like PHD and RING finger domains 1), an E3 ubiquitin ligase, ubiquitinates PCLAF at Lys 15 and Lys 24 and promotes its binding to PCNA during unperturbed late S-phase[20]. While DNA replication is challenged, rapid, proteasome-dependent removal of ubiquitylated PCLAF from PCNA is triggered, which facilitates bypass of replication-fork-blocking lesions by allowing recruitment of translesion DNA synthesis polymerase to mono-ubiquitylated PCNA[22]. Moreover, the PCLAF promoter contains 3 E2F-binding motifs. E2F4 and E2F6 binding to its promoter caused transcriptional repression of PCLAF, which leads to inhibition of cell proliferation, S-phase progression, and DNA synthesis[21]. PCLAF is also the substrate of Cdh1 from the multi-subunit E3 ubiquitin ligase anaphase-promoting complex or cyclosome (APC/C) through binding to the KEN box of PCLAF, which also contributes to the degradation of PCLAF[23]. Studies have also determined that PCLAF is involved in the DNA damage response, and controls cell cycle progression[18,23], implying dysregulation of PCLAF might contribute to diseases.
Recent studies have further expanded our knowledge on PCLAF. Paiva et al[41] found novel menstruation-associated genes, including PCLAF, through genome-wide expression arrays of endometrial biopsies. However, the role of PCLAF in menstruation is still not clear. Sang et al[42] explored the effects of TGF-beta3 and BMP2 on chondrogenesis in mesenchymal stem cells (MSCs) by performing gene expression profiles of MSCs treated with TGF-beta3 and BMP2. They reported that PCLAF and another two genes were important in chondrogenesis by PPI network analysis. Combined with the specific expression of PCLAF in mouse embryogenesis[35], this strongly indicates that PCLAF may play notable roles in growth and development in humans.
PCLAF CONTRIBUTES TO THE DEVELOPMENT OF GI TUMORS
Studies in recent years have revealed that PCLAF is overexpressed in several types of GI tract tumors, including esophageal cancer[6,7], GC[8-10], and CRC[39,43]. Some studies have also shown a strong association between PCLAF and tumors in accessory organs of digestion, including PC[15] and liver cancer[12-14,44-46]. This review describes current studies on the role of PCLAF in GI tumors in detail.
Esophageal cancer
Esophageal cancer is a significant malignant tumor worldwide[1]. There are two main histological types with different biological characteristics, geographical distributions, and risk factors, namely esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma[47]. ESCC accounts for approximately 90% of esophageal cancer in Asia[48]. PCLAF mRNA level is significantly increased in various malignancies, including esophageal, breast, uterine, cervix, brain, kidney, and lung tumors. However, elevated PCLAF is especially marked in esophageal tumors, and is increased over 10-fold compared with normal esophageal tissues. This result implies that PCLAF might predict esophageal cancer progression[16]. Cheng et al[7] showed that PCLAF protein expression was higher in esophageal cancer tissues than in corresponding normal tissues using IHC analysis, suggesting that PCLAF is associated with esophageal cancer. Further study has indicated that PCLAF is associated with higher stage, tumor recurrence, and poor survival[7]. PCLAF overexpression promoted cell viability in the esophageal carcinoma Eca-109 cell line and ESCC TE1 cell line using the MTT assay. Knockdown of PCLAF increases cell numbers in the G1 phase by flow cytometry analysis, and downregulates the protein levels of cyclin A and B, indicating that PCLAF might control esophageal cancer cell viability via induction of cyclin A and B[7]. PCLAF also promoted cell migration and invasion in ESCC EC9706 and TE1 cell lines using the transwell assay[6]. Moreover, elevated PCLAF expression predicts a worse response to chemotherapy. Clinical data has proved these findings. An in vitro experiment has also confirmed that overexpression of PCLAF enhances Eca-109 cells' resistance to cisplatin-induced cell growth inhibition[7]. These results suggest that PCLAF plays a positive role in the development of esophageal cancer. However, the mechanism still needs further study.
GC
GC is the third most lethal malignancy worldwide[1]. Helicobacter pylori infection is the leading risk factor[49]. PCLAF was found to be highly expressed in three gastric carcinoma cell lines, including MKN-28, SGC-7901, and MKN-45, especially in MKN-45[10]. Clinical sample analysis confirmed that PCLAF mRNA and protein were upregulated in GC tissues compared to corresponding non-tumorous gastric tissues[8,9]. Moreover, GC patients with highly expressed PCLAF exhibit high recurrence and low 2-year survival rates. These results suggest that PCLAF might be a marker of recurrence in GC[9]. PCLAF mRNA and protein levels are also significantly upregulated in peripheral blood mononuclear cells from GC patients[50]. In addition, PCLAF expression correlates with the 3-year survival rate. Combined with high diagnostic sensitivity and specificity, PCLAF can be a prognostic target in GC[50]. Knockdown of PCLAF expression using siRNA in GC cell lines significantly inhibits cell viability, cell cycle progression, and cell migration[9,10], indicating that PCLAF promotes GC development in vitro. However, the molecular mechanism remains unknown. Wang et al[8] analyzed PCLAF associated genes using the cBioPortal database in The Cancer Genome Atlas, followed by Gene Ontology (GO) analysis. They reported that PCLAF associated genes were involved in RNA processing, cell cycle process, DNA metabolic process, and so on. Involved pathways of differential expressed genes were cell cycle, spliceosome, DNA replication, etc. An in-silico analysis provided a potential mechanism of PCLAF in GC for further investigation.
CRC
CRC is responsible for the second leading cause of cancer death worldwide, according to the global cancer statistics 2018[1]. High risk factors include dietary patterns characterized by high intakes of red meat, processed meat, sugar-sweetened beverages, refined grains, desserts, and potatoes[47]. Immunostaining of colon cancer tissue microarray confirmed that PCLAF is strongly expressed in colon cancer tissues compared to normal intestine[11]. Consistently, PCLAF is overexpressed and mainly localized in the nucleus of colon cancer cell lines. Knockdown of PCLAF using shRNA inhibits cell proliferation. Furthermore, this process is not dependent on the interaction between PCLAF and PCNA[11]. Genomic profiling of plasma RNA was performed by cDNA microarray hybridization from CRC patients and healthy donors. The authors selected three potential markers, including PCLAF, for further confirmation using quantitative RT-PCR. It was shown that PCLAF, which was combined with the other two elevated plasma genes EPAS1 and UBE2D3, can classify CRC samples before and after surgery[39,43]. These results indicate that PCLAF may be an early plasma marker in CRC patients. However, its specificity and sensitivity in the prediction of CRC remain to be investigated further. A recent study by Wang et al[51] also showed that PCLAF may be a potential therapeutic target of CRC following bioinformatics analysis of the Gene Expression Omnibus database. However, there is no available research on PCLAF function in CRC. Further research of the underlying role of PCLAF in CRC and its mechanisms is still needed.
PC
PC is the seventh most lethal cancer worldwide. Smoking, obesity, and longstanding type 2 diabetes are known risk factors of PC[47]. Few studies have determined the role of PCLAF in PC, a common malignancy with a low 5-year survival rate[1]. In the study by Hosokawa et al[15], increased expression of PCLAF was found in PC tissues and cell lines. Knockdown of PCLAF using siRNA significantly reduced cell viability and colony formation. Overexpression of PCLAF promoted cell proliferation and tumor formation in nude mice. These results suggest that PCLAF is involved in the development of PC. Inhibiting the interaction between PCLAF and PCNA by the cell-permeable 20-amino-acid PIP box dominant-negative peptide resulted in significant growth suppression of the PC cell line KLM-1[15], implying that the interaction between PCLAF and PCNA is essential in PC tumorigenesis.
Liver cancer
From 1990 to 2015, there was a 75% increase in global liver cancer cases. Liver cancer, one of the most successful killers, can be classified into two types, either primary or metastatic (secondary). Based on the type of cells that becomes cancerous, primary liver cancer has several types, including hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma, mucinous cystic neoplasms, angiosarcoma and hemangiosarcoma, and hepatoblastoma. HCC is the most common form of primary liver cancer in adults. Hepatitis B virus infection was responsible for 33% of deaths from HCC, alcohol consumption for 30%, HCV infection for 21%, and other causes for 16% of incidents of HCC worldwide[47]. The relationship between PCLAF and HCC has been studied but the results are inconsistent[47]. Guo et al[38] reported that PCLAF is a down-regulated and growth-inhibitory gene in HCC. The PCLAF protein level is lower in HCC tissues than in normal tissues, and there is no significant correlation between the clinicopathological stages of HCC with PCLAF expression. However, contrary to the study by Guo et al[38], our study found that both the mRNA and protein levels of PCLAF variant 1 were overexpressed in HCC tissues compared with those in normal tissues, especially in higher stage (3-4) tumors[14]. This observation is similar to that in another report where PCLAF protein was highly expressed in HCC tissues and was associated with serum HBsAg positive, high AFP, and high-grade (2-4) tumors[46]. We speculate that the discrepancy may be related to the differential expression of the PCLAF variants. The antibody used in Guo’s experiment was prepared by themselves, which may bind to both variants 1 and 2 of PCLAF, while our group used the antibody specific to PCLAF variant 1. As a result, it is speculated that variant 2 of PCLAF may have a different expression pattern from variant 1 in HCC. Our further study has shown that the protein level of PCLAF variant 2 was markedly expressed in nontumorous tissues compared to HCC tissues[34], while Yuan et al[46] reported that PCLAF variant 2 mRNA was highly expressed in HCC tissues. The discrepancy may be due to Yuan and his colleague using a pair of primers to detect both variants 1 and 2 simultaneously, while we assessed the protein of PCLAF tv2 instead of the mRNA of PCLAF tv2. Interestingly, we also found that the overexpression of PCLAF variant 1 prevents doxorubicin-induced apoptosis[14].
Gene integrated bioinformatics analysis indicates that PCLAF is one of the core genes associated with HCC[52]. PCLAF mRNA levels are increased in HCC[53]. A recent study has shown that PCLAF is involved in the microvascular invasion in HCC by activating epithelial-mesenchymal transition[13]. PCLAF is highly expressed in HCC stem cells-enriched hepatoma spheres compared to monolayer-cultured cells. Furthermore, knockdown of PCLAF reduces the percentage of CD133 or EpCAM positive cells, which represented HCC stem cells, indicating that PCLAF can enhance the self-renewal of HCC stem cells[54]. In addition, analysis of 760 samples in the ONCOMINE database showed that high PCLAF expression was associated with worse overall survival in HCC[12]. A twenty gene-based gene set (including PCLAF) is associated with the pathological progression from cirrhosis to HCC. This set is also detectable in peripheral blood and serves as an independent prognostic factor for recurrence-free survival and overall survival of HCC[55]. These results suggest that PCLAF may be a non-invasive prediction target in HCC. Increased PCLAF mRNA level in whole blood and peripheral blood mononuclear cells of HCC patients also correlates with a higher stage and lower survival rate[44,45]. Furthermore, both sensitivity and specificity of whole blood PCLAF mRNA in the diagnosis and prognosis of HCC are higher than those of AFP and CEA[44]. In conclusion, PCLAF may be an ideal diagnostic and prognostic marker in HCC.
PCLAF PARTICIPATES IN OTHER DISEASES
PCLAF in other cancers
In addition to GI cancers, PCLAF is also overexpressed in other types of malignancies, such as lung cancer[26,56], fibrosarcoma[30,32], ovarian cancer[29,57], BC[28,37], renal cell carcinoma[58], adrenal cancer[27], chronic lymphocytic leukemia[59], and nasopharyngeal carcinoma[60].
Several in-silico analyses have demonstrated that PCLAF overexpression is crucial in the process of non-small cell lung cancer (NSCLC) and predicts poor progression and low survival in patients with NSCLC[26,61-63]. IHC staining of clinical samples also showed a high level of PCLAF expression in NSCLC tissues, which correlated with male gender, tumor progression, lymph node metastasis, non-adenocarcinoma histological classification, and smoking history[64]. Another study revealed a similar result where PCLAF was an independent prognostic factor for overall survival in NSCLC cases. Furthermore, depletion of PCLAF induces G1 phase cell cycle arrest, inhibits NSCLC cell proliferation and migration, and decreases tumor volume in nude mice[26,56], suggesting that PCLAF may contribute to the development of NSCLC. Mechanism research has found that PCLAF mediates the premature degradation of spindle assembly checkpoint (SAC) components by coordination with a SAC regulator-UbcH10 to cause further dysfunction of SAC and neoplastic proliferation[25]. A recent study showed that PCLAF is also correlated with poor prognosis of lung adenocarcinoma. Upregulated PCLAF modulates cell cycle genes via the DREAM complex, which leads to the proliferation of lung cancer cells[65].
PCLAF also contributes to the most common and lethal cancers - BC and ovarian cancer - in women. PCLAF protein is overexpressed in BC tumor samples and is directly implicated in increased disease severity[37]. Knockdown of PCLAF markedly inhibits breast cell growth, cell cycle progression, and colony formation. The expression of three cell cycle-related genes – CCNE2, CDK6, and CDKN1A – is also suppressed by PCLAF silencing in BC cell lines[28]. However, in another study, depletion of PCLAF failed to affect the cell cycle. PCLAF binds to BRCA1 in centrosomes, and extreme abundance of PCLAF may result in centrosome number defects, which are involved in BC[37]. In addition to BC, PCLAF can be transcriptionally activated in ovarian cancer tissues. High expression of PCLAF predicts poor prognosis and drives proliferation and metastasis of ovarian cancer cells. In vitro experiments also indicate that PCLAF resulted in cisplatin resistance through inhibition of cisplatin-induced apoptosis and autophagy in ovarian cancer cells[29,57]. These results imply that PCLAF is an underlying predictive and therapeutic target in ovarian cancer.
Two miRNAs have been reported to suppress metastatic properties, such as migration, invasion, and anchorage-independent growth of fibrosarcoma, possibly through targeting PCLAF[30,32], suggesting that PCLAF also participates in sarcoma genesis. Furthermore, PCLAF also promotes cell proliferation and metastasis in cell lines of renal cell carcinoma[58], and adrenal cancer[27]. Bioinformatics assays to identify differentially expressed genes in brain cancer (including oligodendroglioma, glioblastoma, and pituitary cancer)[66-68], and malignant pleural mesothelioma[69] also show that PCLAF is a potentially associated gene. However, the function of PCLAF in these types of cancer requires further investigation.
PCLAF may also be involved in chronic lymphocytic leukemia, a type of B cell malignancy. PCLAF is highly expressed in CD19+ B cells isolated from the peripheral blood of chronic lymphocytic leukemia patients. Knockdown of PCLAF inhibits cell proliferation and induces cell cycle arrest and cell apoptosis in chronic lymphocytic leukemia cells, indicating the positive role of PCLAF in chronic lymphocytic leukemia progression[59].
PCLAF in immune and inflammation-associated diseases
Multiple sclerosis (MS) and rheumatoid arthritis (RA) are the most frequent autoimmune diseases[70,71]. A study demonstrated that 27 differentially expressed genes, including PCLAF, were overexpressed in peripheral blood mononuclear cells from MS patients when the PARK7 interactome data from the GDS3920 profile were analyzed, which revealed underlying novel targets of MS[72]. In addition to MS, PCLAF is also associated with RA. Increased CD4+ T cells are the primary regulators in the damaging inflammatory process leading to RA[73]. Aterido et al[74] selected the most influential genes in the CD4+ T cell-specific networks in RA using a new dimensionality reduction approach. They found that genes associated with the expression of PCLAF and BIRC5 were highly probably related to the CD4+ T cell pathophysiology in RA. PCLAF might increase proliferation and reduce apoptosis of CD4+ T cells in the synovial membrane in RA. A study evaluated the immune system-related genes in acute/chronic antibody-mediated and T-cell mediated rejections in patients with renal allografts[75]. It was found that a series of genes, including PCLAF, are upregulated in failed grafts caused by chronic rejection, suggesting that PCLAF may be involved in chronic inflammation. In conclusion, PCLAF may participate in the proliferation of immune cells, and regulate inflammatory pathways in diseases.
SIGNALING PATHWAYS OF PCLAF INVOLVED IN CANCERS
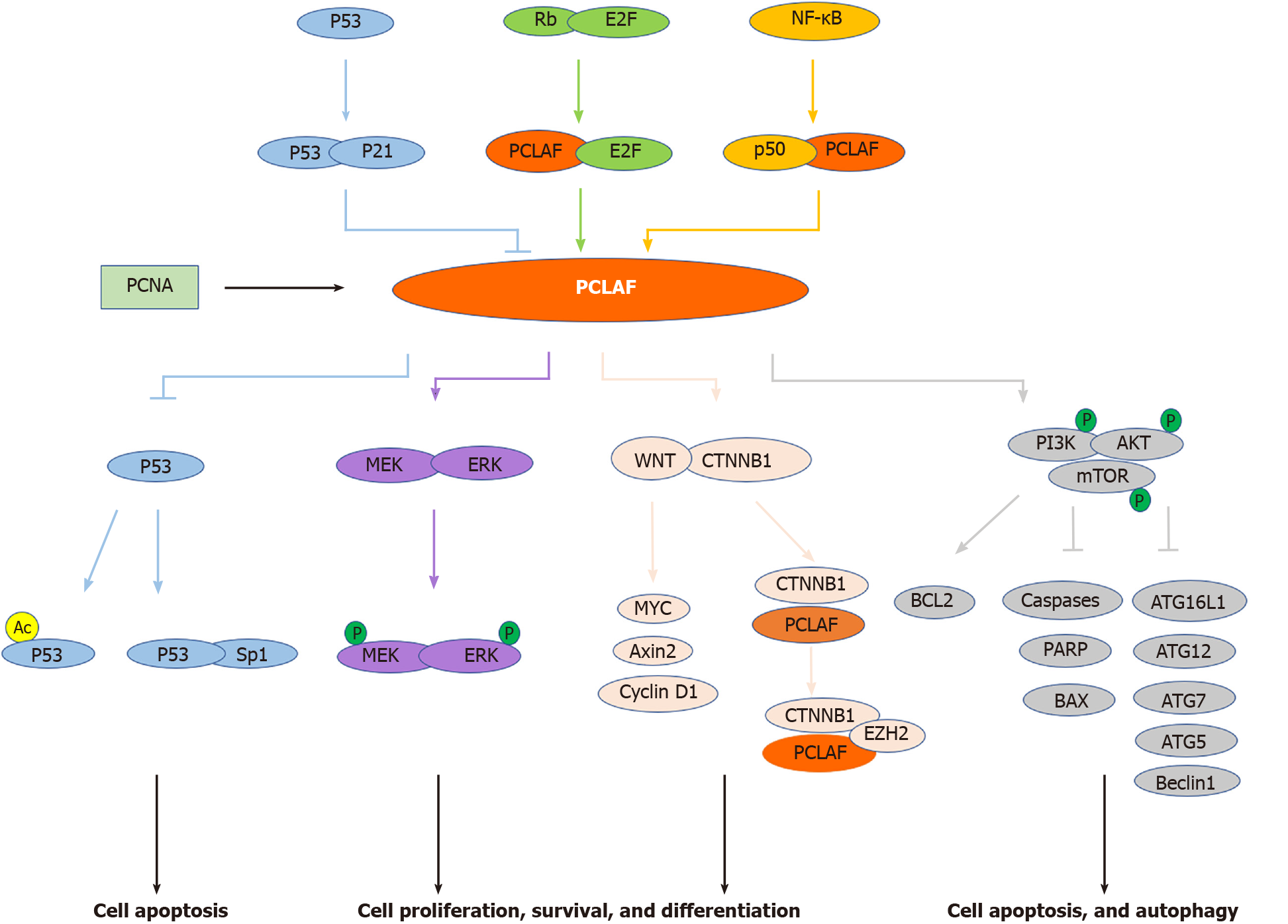
Although many studies have reported the relationship between PCLAF and tumors, whether PCLAF participates in tumor development and occurrence is still unclear. The potential mechanism may be that PCLAF is involved in the signaling pathway leading to cellular response and tumor development and occurrence, including pathways targeting PCLAF and PCLAF-mediated cascades (Figure 2).
Figure 2 Signal pathways involved in proliferating cell nuclear antigen clamp associated factor-mediated cancers.
Proliferating cell nuclear antigen (PCNA) clamp associated factor (PCLAF) is a PCNA associated protein. Several pivotal pathways, including the Rb/E2F pathway, NF-κB pathway, and p53-p21 cascade, are implicated in PCLAF-mediated diseases. In addition, PCLAF contributes to some conditions through dysregulation of the p53 pathway, WNT signal pathway, MEK/ERK pathway, and PI3K/AKT/mTOR signal cascade (see text). Lines with an arrowhead indicate promotion, and lines with a bar indicate inhibition in steady-state or stress conditions. The distinct line colors indicate the different pathways implicated with PCLAF.
Rb/E2F signaling pathway
The Rb/E2F signaling pathway is the central regulatory mechanism mediating S-phase gene transcription[76]. Moreover, this pathway is involved in many tumors[77,78]. A study showed that the Rb/E2F pathway inhibits the PCLAF promoter. Knockdown of Rb by siRNA enhances the expression of PCLAF in the BC cell line MCF7. There are three potential E2F binding motifs, located at -94 to -87, -29 to -22, and 17 to 25 bp of the transcription start site, in the PCLAF promoter. Site-directed mutagenesis indicated that all three sites are functional[21]. These results indicate that PCLAF is a target of the Rb/E2F signaling pathway.
NF-κB signaling pathway
The activation of NF-κB plays a key role in the pathogenesis of some inflammatory diseases. Recent studies have shown that NF-κB plays a key role in cell-cycle regulation, apoptosis, and tumorigenesis[79]. Li et al[17] constructed three PCLAF promoter sequence fragments: p656 (-529 to +127), p288 (-161 to +127), and p132 (-5 to +127). NF-κB subunit p50 interacted with p656 and p288, but not with p132, suggesting that the binding region is localized at the fragment from nucleotides -5 to -161 bp upstream of the transcription start site. We suggest that PCLAF is a target gene of NF-κB and may play a role in NF-κB-mediated oncogenesis. Ma’s group also indicated that Bcl3, p50, and RelB bind to the promoter of PCLAF and positively regulate the expression of PCLAF, suggesting that the NF-κB p50/RelB complex can regulate the expression of PCLAF by binding to its promoter[60].
P53-p21 signaling pathway
The p53-p21 pathway is the most-studied molecular mechanism of PCLAF. Many studies have revealed that the expression of several DNA replication factors, including PCNA, POLD1, and FEN1, are regulated by the p53 pathway[80,81]. Studies have also investigated the regulation of p53 on PCLAF. Extensive-expression analysis of p53-regulated genes suggests that PCLAF is also downregulated by the adenovirus-mediated introduction of p53[15]. Hosokawa et al[15] disclosed that the mRNA and protein levels of endogenous PCLAF were significantly decreased in HCT116 p53+/+ cells, while no change was observed in HCT116 p53-/- cells after treatment with doxorubicin. HCT 116 p21-/- cells showed a similar result. As both p21 and PCLAF have the same PIP box domain, and PCLAF can compete with p21 for binding to PCNA[16], we propose that regulation of PCLAF expression is dependent on the p53-p21 pathway.
In reverse, PCLAF can also regulate the activity of p53. GO analysis identified the effect of PCLAF on the GC-related pathways and showed several implicated pathways, including the p53 signal transduction pathway[8]. In our previous study, we discovered that PCLAF can inhibit p53 transcriptional activity and decrease the expression levels of p53 targeting genes. We also found that PCLAF prevents doxorubicin-induced apoptosis in HepG2 cells by inhibiting the acetylation of p53 at lys382[14]. Additionally, PCLAF knockdown promotes the interaction between p53 and Sp1. Silencing p53 counteracts the inhibitory effect of PCLAF knockdown on cell proliferation and cell cycle progression, suggesting that PCLAF plays a role in cancer via the regulation of p53[28].
WNT/CTNNB1 signaling pathway
The WNT/CTNNB1 signaling pathway plays a critical role in animal development and tissue homeostasis. Numerous studies have indicated that its deregulation is involved in some human diseases, including cancer[82]. PCLAF can aberrantly activate the WNT/CTNNB1 pathway in several types of cancers, including colon cancer, NSCLC, and ovarian cancer[11,26,57]. PCLAF silencing by siRNA or shRNA decreases the protein level of total CTNNB1 in the NSCLC cell line A549, HCC cell lines Huh7 and HCCLM3[26,54]. Following X-ray irradiation treatment, knockdown of PCLAF further inhibited CTNNB1 expression, suggesting that PCLAF can regulate the expression of CTNNB1 in NSCLC[26]. Another study further indicated that PCLAF activates the WNT/CTNNB1 pathway in ovarian cancer cells using the TOP FLASH reporter assay. Depletion of PCLAF inhibits nuclear translocation of CTNNB1 and the expression of WNT/CTNNB1 downstream genes, including Axin2 and MYC. These results indicate that PCLAF is essential in activating the WNT/CTNNB1 signal in ovarian cancer cells[57]. A similar result in colon cancer showed that PCLAF depletion decreases CTNNB1 reporter activation, downregulates the expression of Axin2, MYC, and cyclin D1, and inhibits WNT3A-induced transcriptional activation of Axin2. A further protein binding assay showed that PCLAF interacts with CTNNB1 and activates CTNNB1 target genes by recruiting EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit), a specific CTNNB1 coactivator, to promoters. PCLAF knockdown decreases the binding of EZH2 to the promoters[11]. These results suggest that PCLAF is essential in WNT/CTNNB1 activation.
PCLAF overexpression can also hyperactivate the WNT/CTNNB1 signal pathway in vivo. Whole-mount immunostaining of mouse embryos showed that PCLAF was explicitly expressed in the apical ectodermal ridge of the limb bud, where WNT signaling is activated, implying that PCLAF modulates the WNT signaling pathway in the mouse embryo. In Xenopus laevis embryo, axis-duplication assays showed that secondary axis formation was only seen in the group injected with both CTNNB1 and PCLAF mRNA. Groups injected with CTNNB1 or PCLAF alone failed to induce secondary axes[11]. These results suggest that PCLAF is involved in WNT/CTNNB1 signaling in vivo.
MEK/ ERK signaling pathway
ERK is an essential downstream component of the mitogen-activated protein kinase cascade, which is a vital signal pathway participating in the regulation of normal cell proliferation, survival, and differentiation. MEK is the crucial kinase that phosphorylates ERK[83,84]. The MEK/ERK signaling pathway plays a role in tumor development. Knockdown of PCLAF represses MEK/ERK expression and decreases their phosphorylation levels in the normal and irradiated NSCLC cell line A549[26]. Nevertheless, in another study, the protein levels of MEK and ERK showed no significant difference in HCV NS5A expression and PCLAF knockdown, while the phosphorylation levels of MEK and ERK were significantly elevated. These results show that PCLAF depletion activates the MEK/ERK pathway under HCV NS5A protein expression[85]. As a result, PCLAF may regulate cell behavior by regulating MEK/ERK molecules.
PI3K/AKT/mTOR pathway
The PI3K/AKT/mTOR pathway is one of the most common deregulated signal pathways and therapeutic targets in many human malignancies[86]. PCLAF overexpression significantly elevates the phosphorylation levels of PI3K [p-PI3K(Tyr607)], AKT [p-AKT(Ser473)], and mTOR [p-mTOR (Ser2448)], while knockdown of PCLAF results in downregulation of these proteins in ovarian cancer cells. A further study demonstrated that PCLAF increases the anti-apoptotic protein BCL2 and decreases pro-apoptotic proteins caspase 9, caspase 7, poly (ADP-ribose) polymerase 1, and BAX. Also, PCLAF downregulates autophagy pathway genes, including ATG16L1, ATG12, ATG7, ATG5, LC3 II/LC3 I, and Beclin1, and upregulates the expression of p62. These observations imply that PCLAF may activate the PI3K/AKT/mTOR pathway and further inhibit apoptosis and autophagy processes[29].
POTENTIAL THERAPEUTIC STRATEGY BY TARGETING PCLAF
PCLAF participates in tumor progression by promoting cell proliferation and cell cycle progression by binding to PCNA through the PIP box. As a result, disturbing the interaction between PCLAF and PCNA may be a strategy for cancer therapy, such as the PIP box dominant-negative peptide used in the study by Hosokawa et al[15], which competes with the PIP box of PCLAF for binding with PCNA.
Another way to inhibit the oncogenic role of PCLAF is to suppress the mRNA or protein level of PCLAF. Current studies have shown that some anti-tumor drugs, including doxorubicin[14,15], and cisplatin[7,29], can suppress the expression level of PCLAF. A putative anti-metastatic agent, silibinin, can also suppress the transcriptional level of PCLAF[87]. However, these drugs are multitargeting and not specific to PCLAF.
A study demonstrated that ultraviolet irradiation strongly decreases the ubiquitylated forms of PCLAF, but total PCLAF expression levels were not markedly altered[22]. However, overexpression of PCLAF decreases ultraviolet radiation-induced cell death[35]. In contrast, knockdown of PCLAF inhibits cell proliferation after X-ray irradiation and ultraviolet radiation treatment in vitro, demonstrating that reduced expression of PCLAF enhances radiosensitivity[22,26]. Therefore, knockdown of PCLAF expression may be helpful for therapy.
Furthermore, PCLAF is the target of miRNAs, including miR-1[88], miR-34a[51], miR-139-3p[31], miR-183[66,89], miR-197-5p[30], miR-216a-5p[6], and miR-429[32]. A study of prostate cancer showed that PCLAF was negatively correlated with miR-1 and was identified as a crucial miR1 target gene[88]. The bioinformatics analysis tool TargetScan predicts that PCLAF, as one of miR-34a and miR-139-3p potential target genes, is negatively correlated with miR-139-3p expression and related to poor prognosis in CRC and HCC, respectively[31,48]. PCLAF lacking the 3’-untranslated region (3’-UTR) prevents the inhibitory effects of miR-183 on cell proliferation and reverses UV-induced DNA damage in human trabecular meshwork cells, suggesting that PCLAF is the target of miR-183[89]. A similar result was found in pituitary tumors. Furthermore, the expression of miR-183 and PCLAF was significantly correlated with Ki-67 and p53, markers of the aggressiveness of pituitary tumors[66]. MiR-197-5p and miR-429, as tumor suppressors, suppress cell migration, invasion, and anchorage-independent growth of fibrosarcoma cells possibly by targeting PCLAF during sarcoma genesis[30,32], suggesting that PCLAF is also an underlying target of miR-197-5p and miR-429. A recent study has also shown that miR216a5p suppresses the proliferation, migration, and invasion of ESCC cell lines (EC9706 and TE1) and negatively regulates PCLAF expression by directly targeting the 3’-UTR of PCLAF mRNA[6]. In the above studies, PCLAF serves as a therapeutic target in different tumors. Downregulation of PCLAF expression suppresses the development of tumors in vitro.
Additionally, researchers have investigated two types of potential specific inhibitors of PCLAF. Firstly, the shRNA or siRNA specific to PCLAF have already been proved to be effective in the knockdown of PCLAF expression and inhibited the malignant properties of cancer cells in many types of cancers, including esophageal cancer[7], HCC[34], lung cancer[26], BC[28], renal cell carcinoma[58], etc. Secondly, variant 2 of PCLAF can inhibit the expression of variant 1. A further study also showed that PCLAF variant 2 acts as an endogenous competitor of variant 1 by competing for binding of P53[34]. As a result, suppressing PCLAF variant 1 using variant 2 is a possible biological therapeutic strategy for tumors. However, the expression pattern and role of variant 2 on variant 1 of PCLAF in different types of tumors still require further investigation.
CONCLUSION
The research on PCLAF is still in its infancy. Although many studies indicate that PCLAF may be a potential gene in the medical diagnosis of several cancers and a promising candidate target for several cancer therapeutic drugs, many questions are still unanswered. What are the functions of PCLAF variants 1 and 2 in normal tissues and cancers? Can PCLAF level be used in the early diagnosis of cancer? What is the molecular mechanism of PCLAF in oncogenesis? Future experiments are needed to determine the novel signaling pathway of PCLAF and the function of the other PCLAF variants. Deciphering the mechanism of PCLAF will help improve cancer prognosis and treatment in the future.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Molinari C S-Editor: Zhang H L-Editor: Webster JR P-Editor: Li X