Published online Aug 15, 2020. doi: 10.4251/wjgo.v12.i8.857
Peer-review started: February 10, 2020
First decision: March 24, 2020
Revised: April 6, 2020
Accepted: June 17, 2020
Article in press: June 17, 2020
Published online: August 15, 2020
Processing time: 184 Days and 3.9 Hours
Gastric cancer (GC) is the most commonly diagnosed malignancy worldwide. Increasing evidence suggests that it is necessary to further explore genetic and immunological characteristics of GC.
To construct an immune-related gene (IRG) signature for accurately predicting the prognosis of patients with GC.
Differentially expressed genes (DEGs) between 375 gastric cancer tissues and 32 normal adjacent tissues were obtained from The Cancer Genome Atlas (TCGA) GDC data portal. Then, differentially expressed IRGs from the ImmPort database were identified for GC. Cox univariate survival analysis was used to screen survival-related IRGs. Differentially expressed survival-related IRGs were considered as hub IRGs. Genetic mutations of hub IRGs were analyzed. Then, hub IRGs were selected to conduct a prognostic signature. Receiver operating characteristic (ROC) curve analysis was used to evaluate the prognostic performance of the signature. The correlation of the signature with clinical features and tumor-infiltrating immune cells was analyzed.
Among all DEGs, 70 hub IRGs were obtained for GC. The deletions and amplifications were the two most common types of genetic mutations of hub IRGs. A prognostic signature was identified, consisting of ten hub IRGs (including S100A12, DEFB126, KAL1, APOH, CGB5, GRP, GLP2R, LGR6, PTGER3, and CTLA4). This prognostic signature could accurately distinguish patients into high- and low- risk groups, and overall survival analysis showed that high risk patients had shortened survival time than low risk patients (P < 0.0001). The area under curve of the ROC of the signature was 0.761, suggesting that the prognostic signature had a high sensitivity and accuracy. Multivariate regression analysis demonstrated that the prognostic signature could become an independent prognostic predictor for GC after adjustment for other clinical features. Furthermore, we found that the prognostic signature was significantly correlated with macrophage infiltration.
Our study proposed an immune-related prognostic signature for GC, which could help develop treatment strategies for patients with GC in the future.
Core tip: Gastric cancer (GC) is the most commonly diagnosed malignancy worldwide. Increasing evidence suggests that it is necessary to further explore genetic and immunological characteristics of GC. Our study identified an immune-related prognostic signature for GC, which could accurately distinguish patients into high- and low- risk groups. High risk patients had a poorer prognosis. Multivariate Cox regression analysis demonstrated that the prognostic signature could independently predict GC prognosis. Furthermore, it was significantly associated with immune cell infiltration (especially macrophages). Therefore, the signature may possess prognostic value as a prediction tool for identification of patients who will benefit from immunotherapy.
- Citation: Qiu XT, Song YC, Liu J, Wang ZM, Niu X, He J. Identification of an immune-related gene-based signature to predict prognosis of patients with gastric cancer. World J Gastrointest Oncol 2020; 12(8): 857-876
- URL: https://www.wjgnet.com/1948-5204/full/v12/i8/857.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v12.i8.857
Gastric cancer (GC) is the fourth most commonly diagnosed malignancy and the second leading cause of cancer-related death worldwide[1]. Although the incidence of GC has decreased year by year, the prognosis of GC patients is still not optimistic, especially in China[2]. Yet, its pathogenesis remains unclear, therefore, identification of effective biomarkers and therapeutic targets needs to be addressed.
The tumor microenvironment (TME), especially the immune system, plays a pivotal role in the occurrence and development of GC[3]. Dysfunction of the immune system assists tumor cells to avoid immune surveillance. Immunotherapy such as programmed death-1 (PD-1) blockade has become a promising strategy for GC treatment[4]. However, the clinical outcomes of GC patients are still unsatisfactory, and most of novel immunotherapies are still in the early stages of clinical research[5,6]. The underlying mechanisms of the immune checkpoint blockade response are complex. Thus, deeper genetic and immunological characterization of GC is required to guide clinicians in selecting and determining the best treatment options. The prognosis of GC is closely related to crosstalk between immune cells and tumor cells[7]. Nevertheless, the role of immune-related genes (IRGs) in predicting GC patients’ prognosis has not yet been elucidated.
Considering the prognostic potential of IRGs in GC, in this study, we studied immune-related molecular features. We analyzed IRGs using a large amount of transcription data of GC and explored their potential molecular mechanisms. Based on differentially expressed IRGs, we developed an immune-related prognostic model. Our results will help develop treatment strategies for patients with GC.
Transcriptome RNA-seq data and corresponding clinical information of GC were retrieved from the Genomic Data Commons (GDC) data portal (https://gdc.http://xenahubs.net), including 375 GC tissues and 32 normal adjacent tissues[8]. A list of IRGs were downloaded from the Immunology Database and Analysis Portal (ImmPort) database[9]. Furthermore, we derived a list of transcription factors (TFs) from Cistrome database (http://cistrome.org/CistromeCancer/)[10]. Mutation data were obtained from the Broad GDAC Firehose.
Differential expression analysis was performed using the edgeR package (http://bioconductor.org/packages/edgeR/) in R[11]. The raw data were normalized by Trimmed mean of M values (TMM). The genes with |log fold change (FC)| > 1 and false discovery rate (FDR) < 0.01 were considered as differentially expressed genes (DEGs). Differentially expressed IRGs and TFs were extracted from these DEGs.
The clusterProfiler package was used to annotate DEGs, including gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)[12]. GO terms include biological process, molecular function, and cellular component. P < 0.05 was considered to be significantly enriched.
Cox univariate survival analysis of IRGs was performed using the survival package in R. P < 0.05 was set as the screening criterion. Survival-related IRGs were identified. Furthermore, differentially expressed survival-related IRGs were considered as hub IRGs.
Hub IRGs were analyzed based on the STRING database (https://string-db.org/). Protein-protein interaction (PPI) network was analyzed using Cytoscape. The interactions among proteins could be involved in the progression of diseases. The nodes with high degree could possess potential as hub genes/proteins.
Mutation data of GC samples from Broad GDAC Firehose website were used to analyze genetic alterations of hub IRGs. The results were visualized into waterfall maps using maftools[13].
Hub IRGs were analyzed by multivariate regression analysis. Then, hub IRGs were selected to conduct a prognostic signature. The risk score was calculated by the expression levels of hub IRGs and Cox regression coefficient. All patients with GC were divided into a high risk group and low risk group according to the median value of risk score. To verify the prognostic potential, the area under the time-dependent receiver operating characteristic (ROC) curve (AUC) was calculated with the survival ROC package[14]. The correlation between clinical features and the prognostic signature was evaluated. Immune infiltrate levels of GC were obtained from the TIMER database[15]. The tumor-infiltrating immune cells included macrophages, B cells, CD4+ T cells, CD8+ T cells, dendritic cells, and neutrophils. The relationships between immune cells and the prognostic signature were calculated.
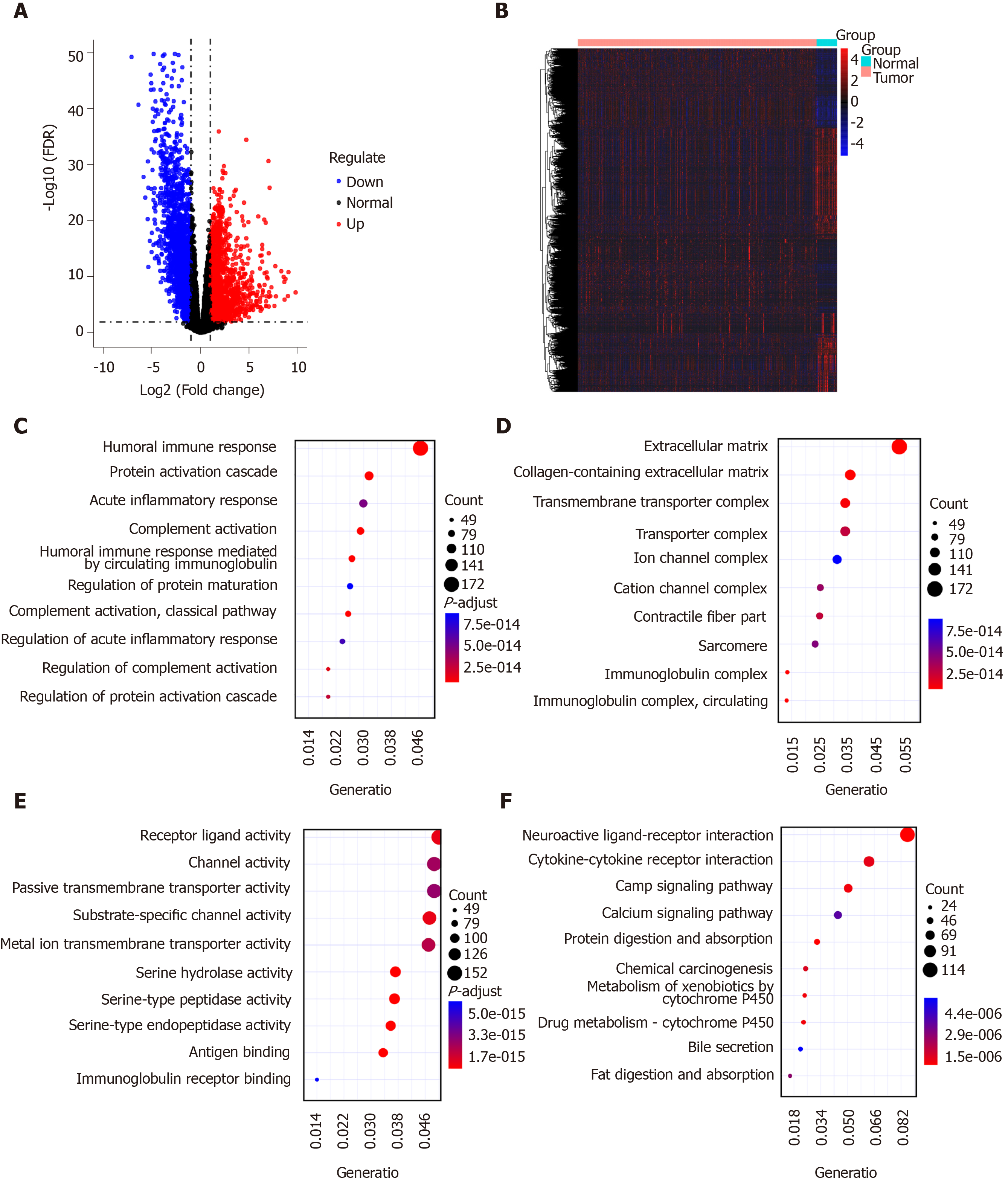
Four thousand two hundred and fifty-nine DEGs with |logFC| > 1 and FDR < 0.01 were identified in 375 GC tissues compared to 32 normal adjacent tissues, including 1951 up-regulated and 2356 down-regulated genes, as depicted in volcano plot (Figure 1A). Hierarchical clustering analysis showed that these DEGs can obviously distinguish GC tissue samples from normal adjacent tissue samples (Figure 1B). These DEGs were significantly associated with immune-related biological processes like humoral immune response and acute inflammatory response (Figure 1C). The top ten cellular components and molecular functions enriched by DEGs are depicted in Figure 1D and 1E, respectively. As shown in Figure 1F, the DEGs were mainly enriched in several pathways related with GC, such as neuroactive ligand-receptor interaction, cytokine-cytokine receptor interaction, chemical carcinogenesis and so on. These results suggested that these DEGs might be involved in the pathogenesis of GC.
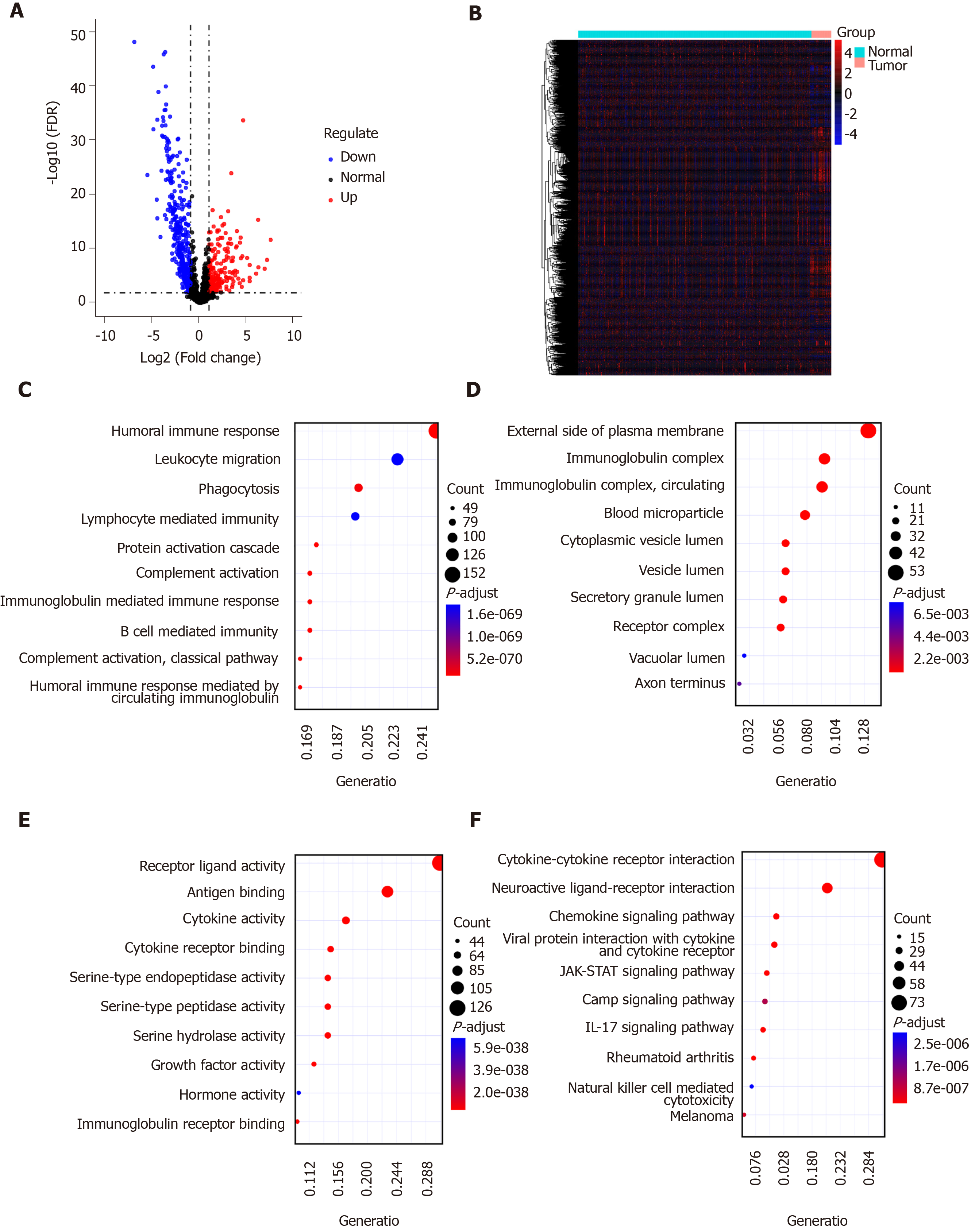
A list of IRGs were downloaded from the ImmPort database. Differentially expressed IRGs were screened from all DEGs, including 181 up-regulated and 354 down-regulated IRGs. The results are visualized into volcano plot (Figure 2A) and heatmap (Figure 2B). In the biological process results, the differentially expressed IRGs were mainly enriched in immune-related processes such as humoral immune response, phagocytosis, and B cell mediated immunity (Figure 2C). As for CC, the genes were in association with immunoglobulin complex, receptor binding, and circulating immunoglobulin complex (Figure 2D). Intriguingly, these IRGs have the molecular immune-related functions like receptor ligand activity, antigen binding, and cytokine activity (Figure 2E). As expected, these differentially expressed IRGs were mainly enriched in immune-related pathways such as cytokine-cytokine receptor interaction, neuroactive ligand-receptor interaction, and viral protein interaction with cytokine and cytokine receptor (Figure 2F).
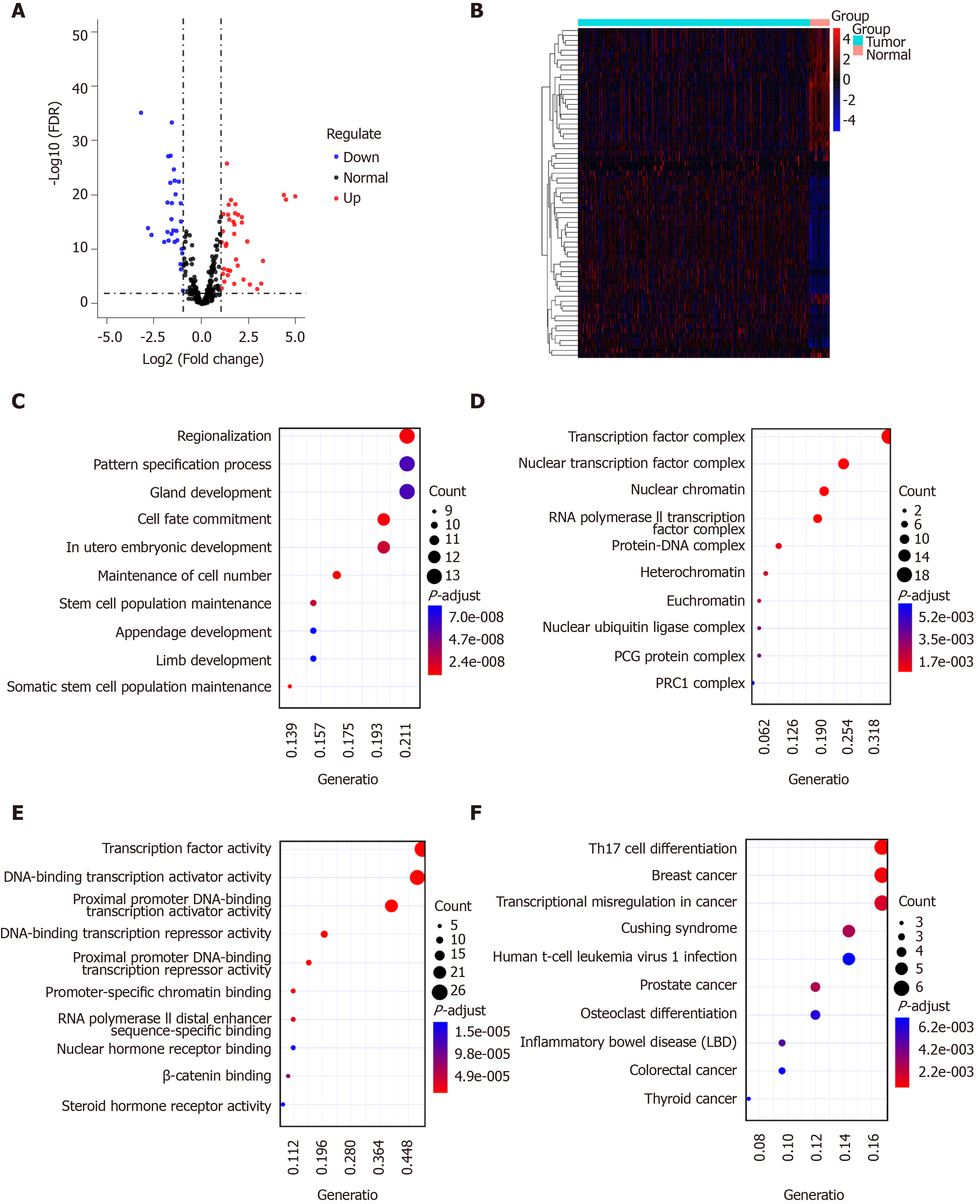
Sixty-seven TFs were differentially expressed between GC tissues and normal tissues, including 37 up-regulated and 30 down-regulated TFs, as shown in volcano plot and heatmap (Figure 3A and B). We further explored their potential functions. The results showed that these TFs are mainly involved in regionalization, pattern specification process, and gland development (Figure 3C). As expected, these TFs could regulate TF complex, nuclear TF complex, and nuclear chromatin (Figure 3D). Similarly, they have the function of TF activity (Figure 3E). According to KEGG pathway enrichment results, these TFs are involved in several cancers (Figure 3F).
Cox univariate survival analysis of all IRGs was performed using the survival package in R. The results showed that 183 IRGs were significantly related to overall survival of patients with GC (P < 0.05).
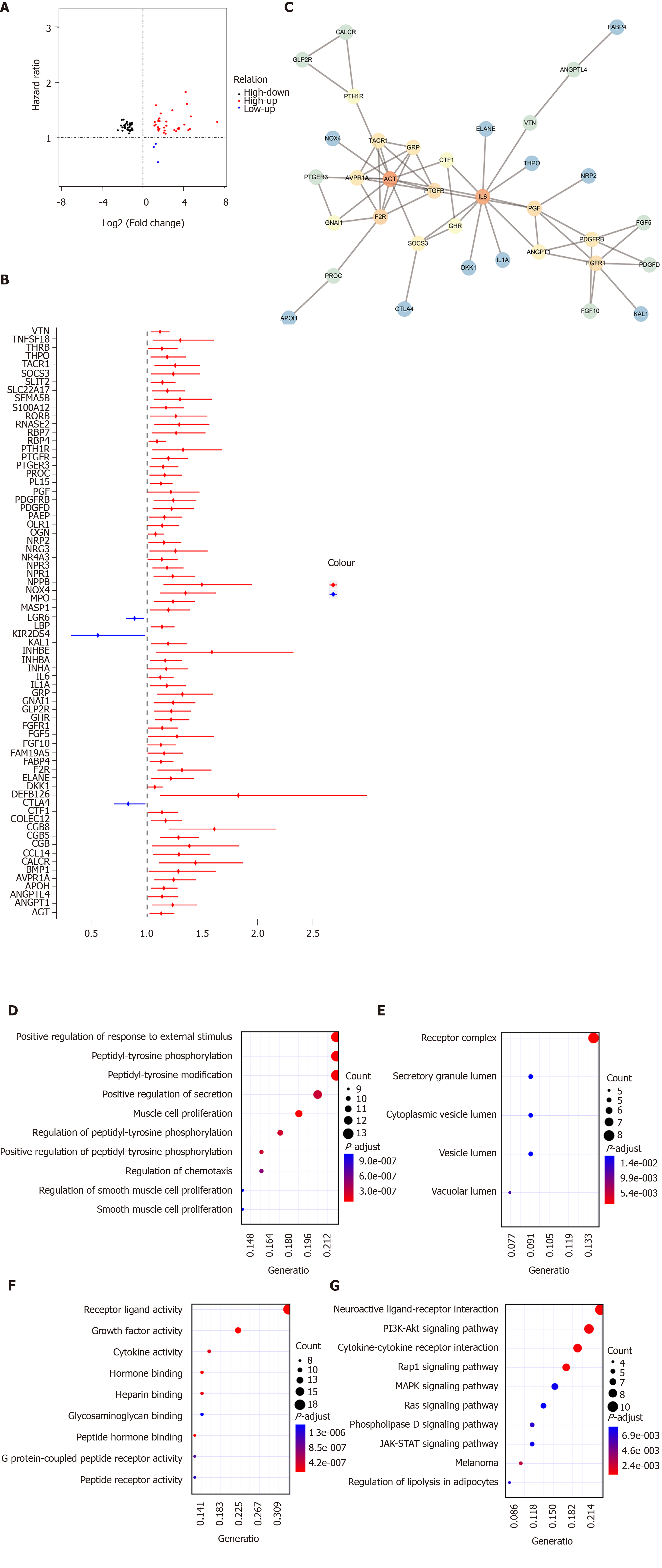
By intersection of differentially expressed IRGs and survival-related IRGs, 70 hub IRGs were obtained for GC (Figure 4A). The forest plot of hazard ratios (HRs) suggested that except CTLA4, LGR6, and KIR2DS4, other hub IRGs were risk factors for GC (Figure 4B). As shown in PPI network, IL6, F2R, and AGT were the top three hub genes (Figure 4C). GO enrichment analysis results showed that these hub IRGs were enriched in many biological processes like positive regulation of response to external stimulus, muscle cell proliferation, and peptidyl-tyrosine phosphorylation (Figure 4D). In the cellular component results, receptor complex, vacuolar lumen, and secretory granule lumen were mainly enriched (Figure 4E). As for molecular function, the hub IRGs were significantly associated with receptor ligand activity, growth factor activity, and peptide hormone binding (Figure 4F). As shown in Figure 4G, hub IGRs are mainly involved in GC-related pathways such as neuroactive ligand-receptor interaction, Rap1 signaling pathway, PI3K-Akt signaling pathway, and cytokine-cytokine receptor interaction.
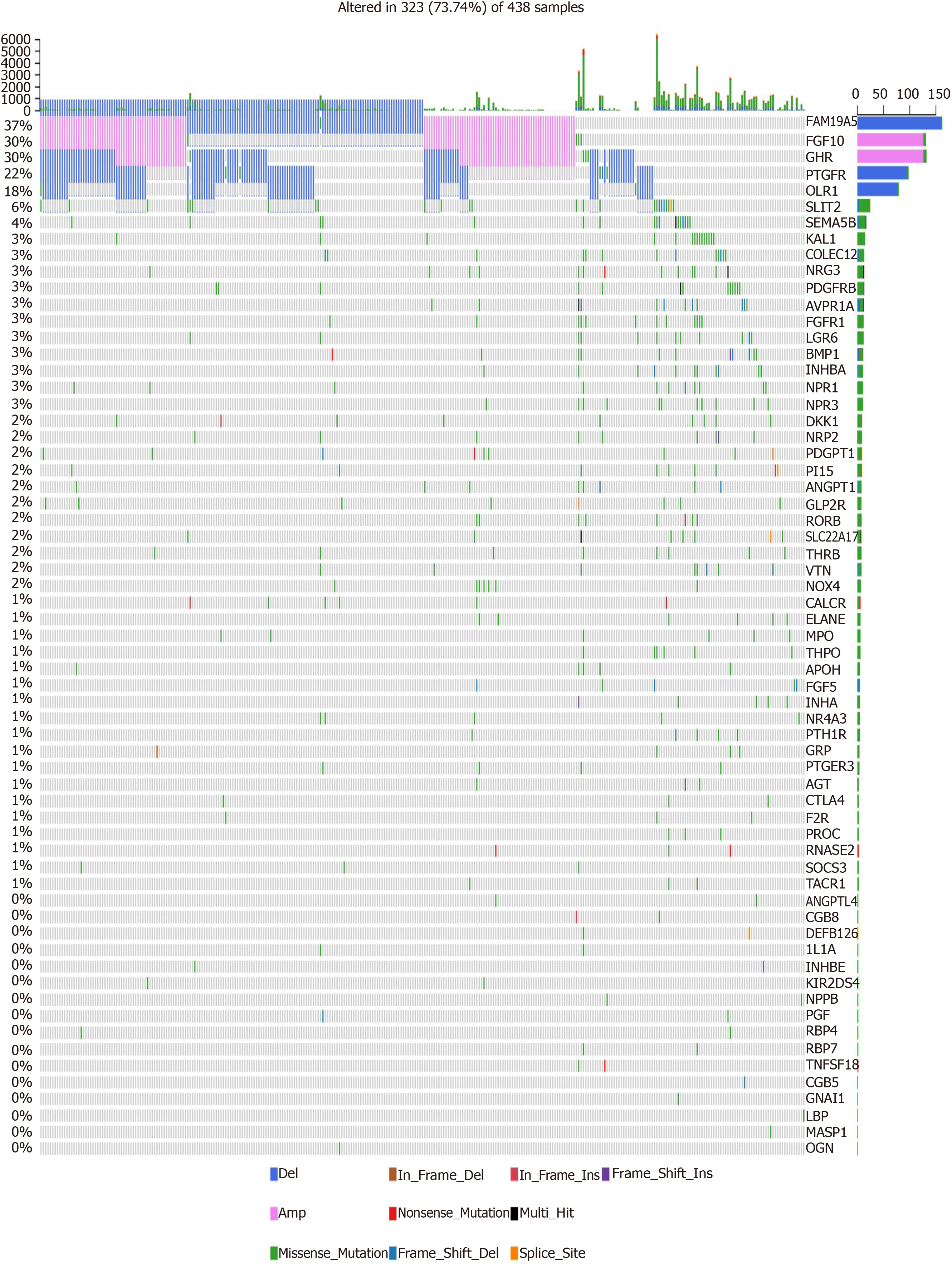
We further analyzed the molecular features of hub IRGs for GC. Their genetic alterations were detected. Among 438 samples, 323 had genetic alterations. The results showed that deletions and amplifications were the two most common types of genetic mutations (Figure 5).
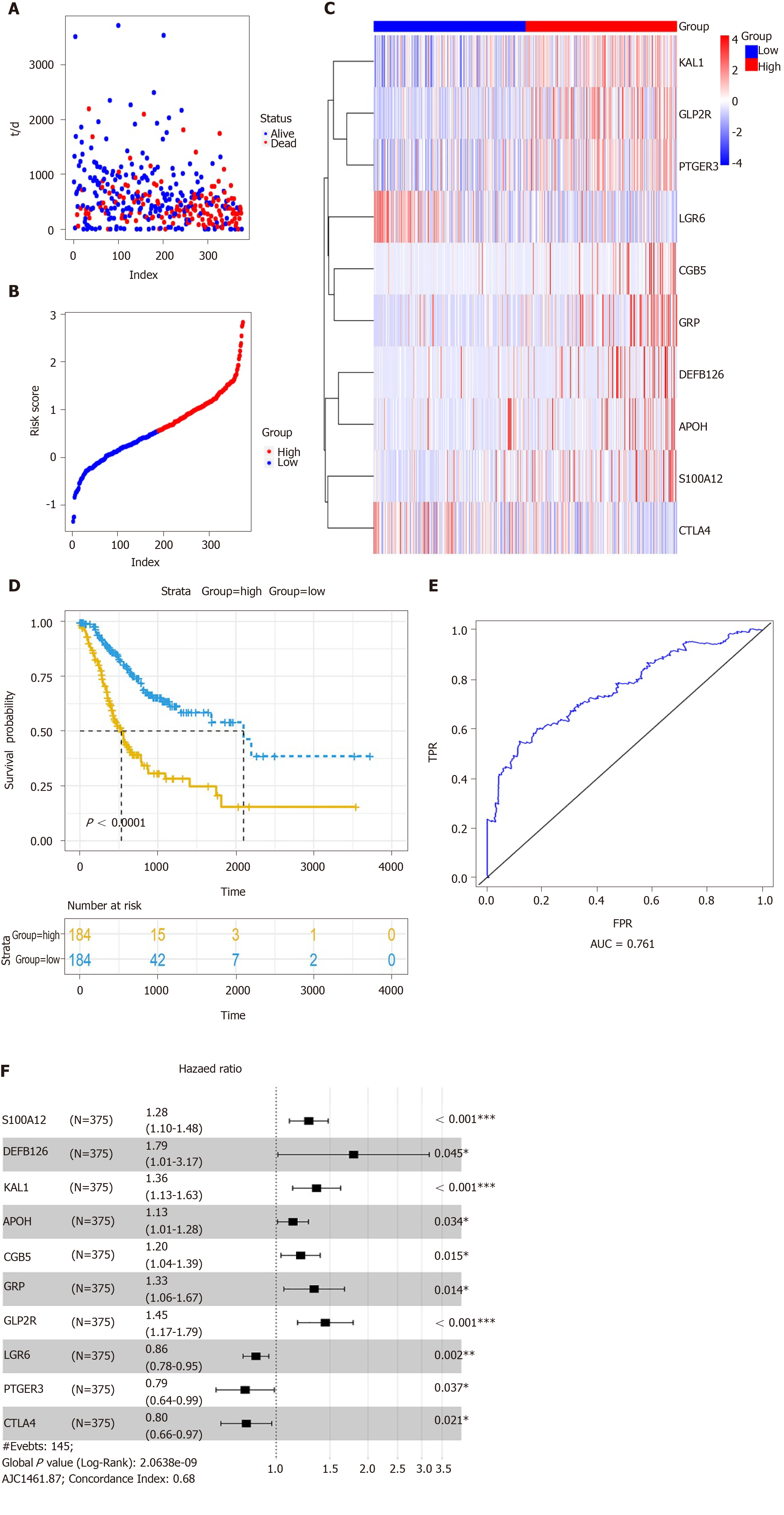
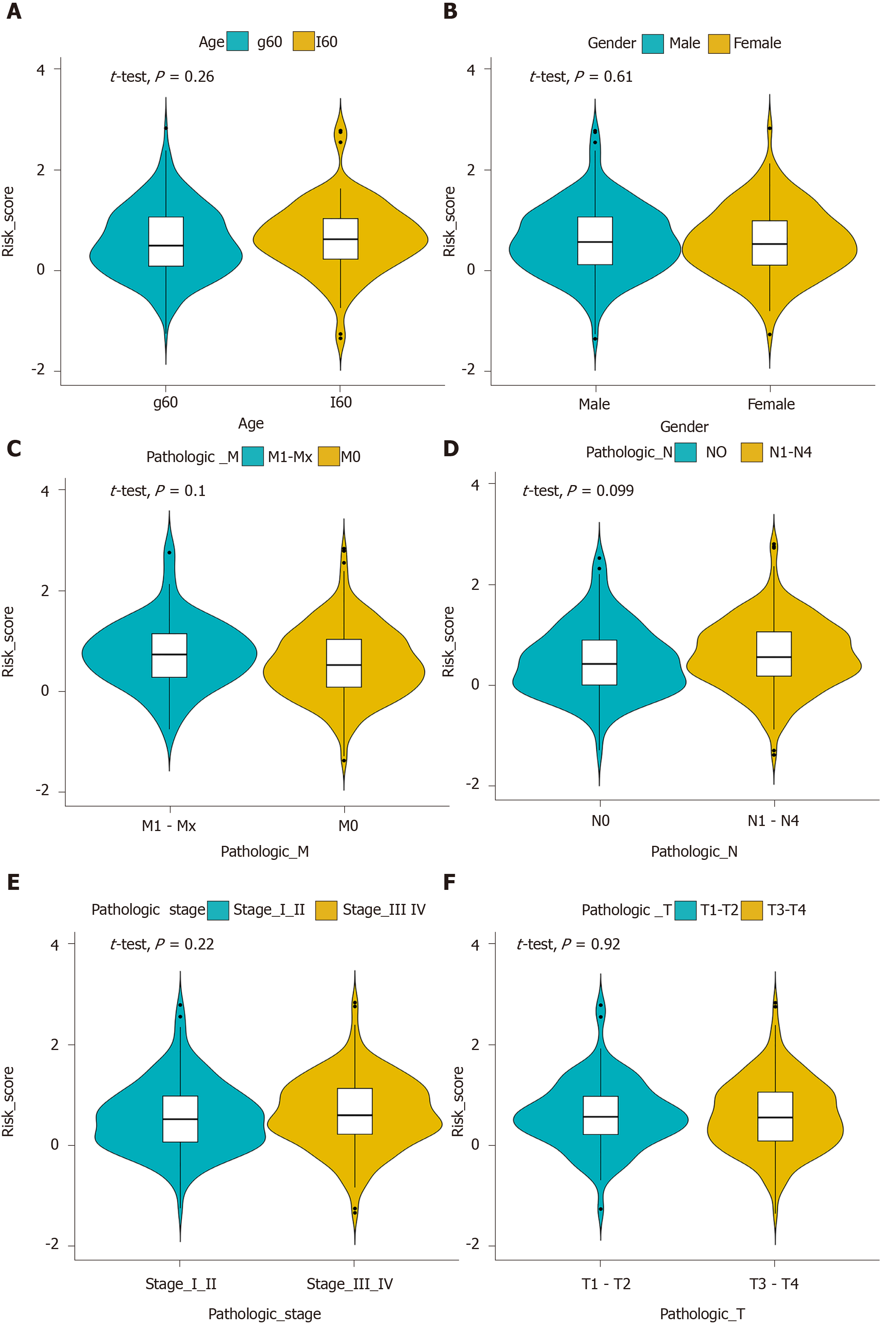
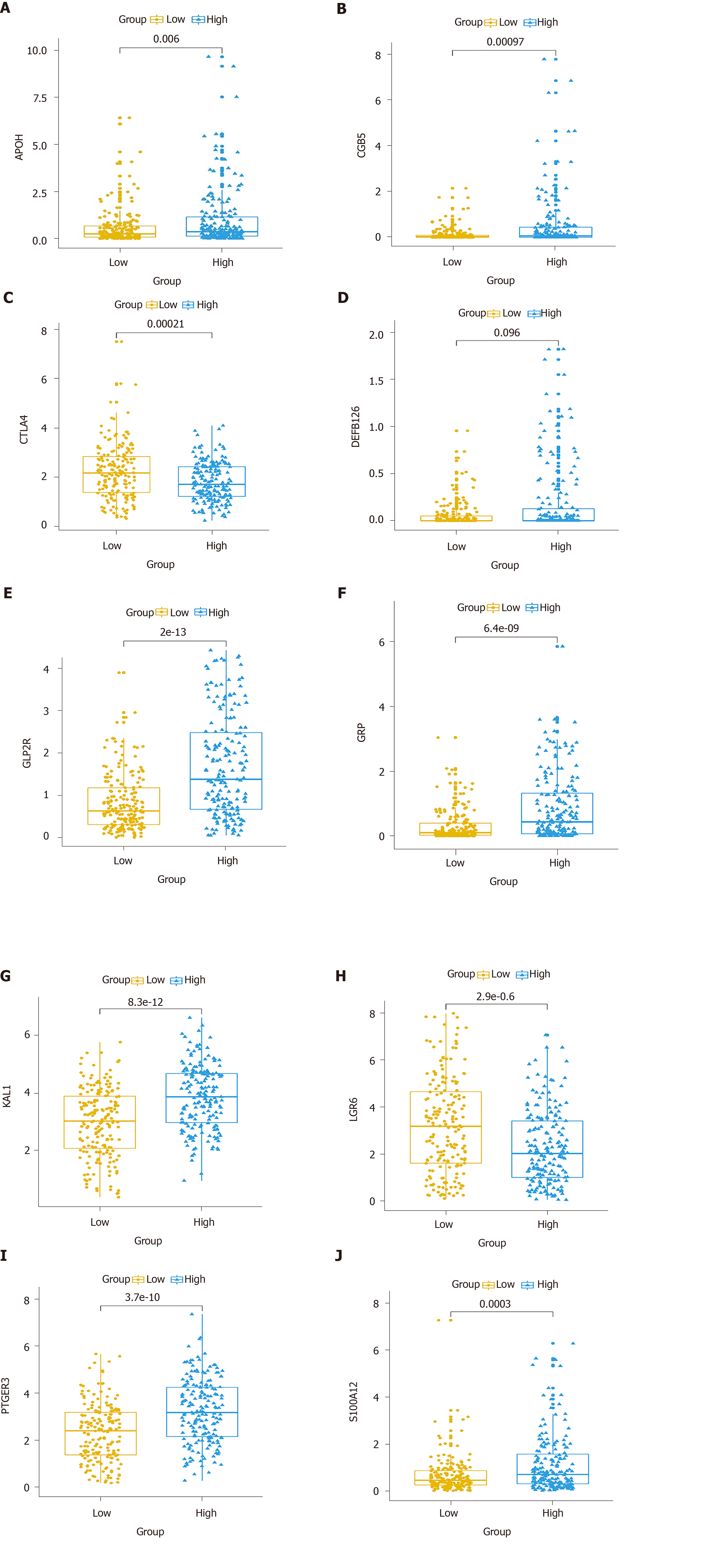
Multivariate Cox regression analysis was performed based on the hub IRGs using the survival package in R. Ten hub IRGs were selected to construct a prognostic signature for GC. As depicted in the forest plot, each hub IRG in the prognostic signature can accurately predict GC patients’ prognosis (Figure 6A). Among them, S100A12 (HR = 1.28; 95% confidence interval [CI]: 1.1-1.48; P = 0.001), DEFB126 (HR = 1.79; 95%CI: 1.01-3.17; P = 0.045), KAL1 (HR = 1.36; 95%CI: 1.13-1.63; P = 0.001), APOH (HR = 1.13; 95%CI: 1.01-1.28; P = 0.034), CGB5 (HR = 1.2; 95%CI: 1.04-1.39; P = 0.015), GRP (HR = 1.33; 95%CI: 1.06-1.67; P = 0.014), and GLP2R (HR = 1.45; 95%CI: 1.17-1.79; P = 0.001) were risk factors for GC, while LGR6 (HR = 0.86; 95%CI: 0.78-0.95; P = 0.002), PTGER3 (HR = 0.79; 95%CI: 0.64-0.99; P = 0.037), and CTLA4 (HR = 0.8; 95%CI: 0.66-0.97; P = 0.021) were protective factors for GC. Risk score was calculated and the patients were divided into a high risk group and low risk group based on the median value of risk score (Figure 6B, C). Heatmap depicts the expression patterns of the ten hub IRGs between the high risk group and low risk group (Figure 6D). Overall survival analysis showed that high risk patients had shortened survival time than t low risk patients (Figure 6E; P < 0.0001). The AUC was 0.761, suggesting that the prognostic signature had a high sensitivity and accuracy (Figure 6F). Correlation analysis showed that the prognostic signature was not significantly associated with clinical features including age (Figure 7A), gender (Figure 7B), pathologic T (Figure 7C), pathologic N (Figure 7D), pathologic M (Figure 7E), and pathologic stage (Figure 7F). As shown in multivariate regression analysis results, the prognostic signature could become an independent prognostic predictor after adjustment for other factors including age, gender, pathologic T, pathologic N, pathologic M, and pathologic stage (Table 1). Furthermore, we also found a difference in expression patterns of the ten hub IRGs in the prognostic signature between GC tissues and normal tissues, including APOH (Figure 8A; P = 0.006), CGB5 (Figure 8B; P = 0.00097), CTLA4 (Figure 8C; P = 0.00021), DEFB126 (Figure 8D; P = 0.096), GLP2R (Figure 8E; P = 2e-13), GRP (Figure 8F; P = 6.4e-09), KAL1 (Figure 8G; P = 8.3e-12), LGR6 (Figure 8H; P = 2.9e-06), PTGER3 (Figure 8I; P = 3.7e-10), and S100A12 (Figure 8J; P = 0.0003).
| Variable | Univariate analysis | Multivariate analysis | ||
| Hazard ratio (95%CI) | P value | Hazard ratio (95%CI) | P value | |
| Age | 1.554 (1.064- 2.268) | 0.022424 | 2 (1.36-2.95) | 4.15E-04 |
| Gender | 1.274 (0.896- 1.812) | 0.177496 | 1.12 (0.78-1.6) | 0.537783 |
| Pathologic T | 1.753 (1.156- 2.659) | 0.00829 | 1.39 (0.87-2.21) | 0.170758 |
| Pathologic N | 1.877 (1.242- 2.835) | 0.002784 | 1.15 (0.66-2.01) | 0.625769 |
| Pathologic M | 1.939 (1.208- 3.112) | 0.006063 | 1.61 (0.99-2.61) | 0.054747 |
| Pathologic stage | 1.988 (1.398- 2.827) | 1.30E-04 | 1.49 (0.89-2.47) | 0.126572 |
| Risk score | 2.718 (2.13-3.47) | < 0.0001 | 2.74 (2.13-3.52) | < 0.0001 |
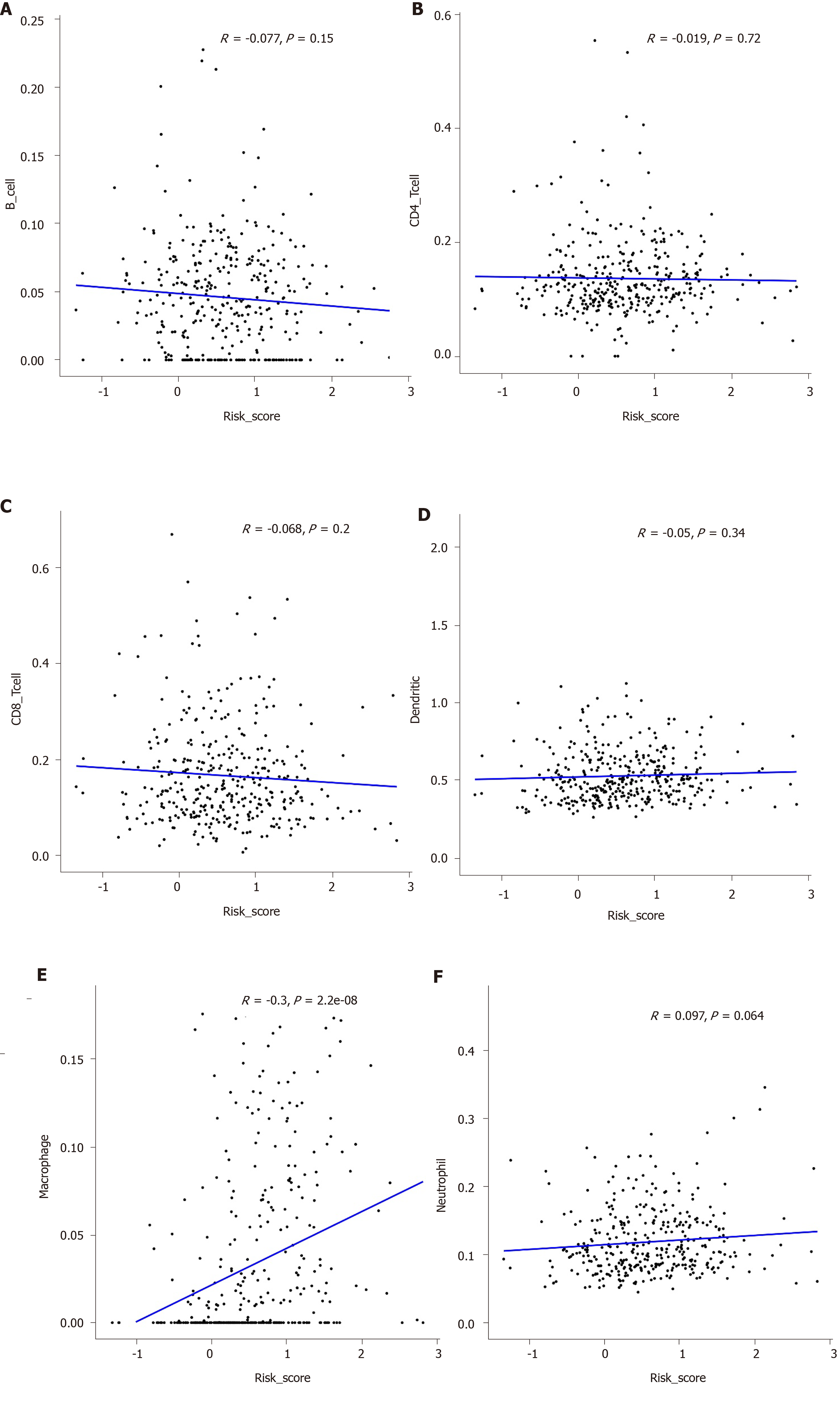
To explore whether the hub IRGs in the prognostic signature are involved in the tumor immune microenvironment, the correlation between immune cell infiltration and prognostic signature was analyzed. We found that the prognostic signature was significantly correlated with macrophage infiltration (Figure 9A). However, there was no significant correlation between the prognostic signature and infiltration of other immune cells including B cells (Figure 9B), CD4+ T cells (Figure 9C), CD8+ T cells (Figure 9D), dendritic cells (Figure 9E), and neutrophils (Figure 9F).
In this study, we comprehensively analyzed the transcriptome RNA-seq data of GC and identified 4259 DEGs. Among all DEGs, 181 up-regulated and 354 down-regulated DEGs were IRGs. These differentially expressed IRGs were mainly enriched in immune-related processes and pathways such as humoral immune response, phagocytosis, B cell mediated immunity, and cytokine-cytokine receptor interaction. Humoral immune response is in association with the progression of GC[16]. B cells play a role in regulating the immune response in GC. B cell depletion may be a useful strategy to enhance anti-tumor immune response[17,18]. To explore the potential molecular mechanisms of GC, we also analyzed TFs among all DEGs. The results showed that 67 TFs were differentially expressed in GC. Functional enrichment analysis confirmed that these TFs have the function of TF activity. Thus, it is necessary to further explore the potential mechanisms of TFs we identified in the pathogenesis of GC.
One hundred and eighty-three survival-related IRGs were identified for GC by Cox univariate survival analysis. Among them, 70 hub IRGs were differentially expressed and associated with overall survival of GC. PPI network analysis indicated that IL6, F2R, and AGT were the top three hub genes. For example, a previous study has found that IL-6 could become a target to overcome chemotherapy resistance in GC[19]. These hub IGRs are mainly involved in GC-related pathways such as Rap1 signaling pathway[20], PI3K-Akt signaling pathway[21], and cytokine-cytokine receptor interaction[22]. Among 438 samples, 323 had genetic alterations. Deletions and amplifications frequently occurred for all hub IRGs. Genetic alteration could become a promising target for the therapy of GC[23]. In this study, we constructed an immune-related prognostic signature consisting of ten hub IRGs. Each hub IRG could separately predict the overall survival of patients with GC. Among them, a member of the S100 family, S100A12, as a component of ubiquitinylation complex, could be involved in β-catenin degradation[24]. A previous study found that S100A12 could be significantly associated with invasion and metastasis of GC. It has been considered as an independent prognostic factor for GC, which is consistent with our results[25]. Furthermore, CGB5 is correlated with a poor prognosis in patients with advanced GC[26,27]. It has been reported that LGR6 might be involved in the development of GC via the PI3K/AKT/mTOR axis[28]. However, the functions of other hub IRGs in GC remain unclear.
Our findings suggested that high risk patients had shortened overall survival time than low risk patients based on the median value of risk score. ROC curve analysis confirmed that the prognostic signature had an excellent performance. Our further analysis showed that the prognostic signature could become an independent prognostic factor after adjusting for other prognostic factors including age, gender, pathologic T, pathologic N, pathologic M, and pathologic stage. Moreover, we found that the prognostic signature was positively correlated with the level of macrophage infiltration. M2 macrophages have been shown to be associated with a poor prognosis in a variety of cancers, including GC[29-31].
Our research has the following limitations. On the one hand, our prognostic markers were based on gene expression profiles. Due to their shortcomings such as high price, long translation cycle, and high requirements for bioinformatics, it is difficult to popularize them in routine clinical applications. However, some alternative methods may be worthy of further exploration, such as screening for optimized signatures from prognostic characteristics through immunohistochemistry. On the other hand, the training cohort of the immune signature model we constructed was from retrospective studies. Therefore, the model requires to be validated by more datasets.
In summary, we have identified differentially expressed IRGs in this study, which may provide a promising perspective for the treatment of GC. We also found that the signature is positively correlated with immune cell infiltration (especially macrophages) and inflammatory responses. The immune gene signature could effectively predict GC patients’ survival, which may be a useful prediction tool to identify patients who will benefit from immunotherapy.
Gastric cancer (GC) is the most commonly diagnosed malignancy worldwide. Increasing evidence suggests that it is necessary to further explore genetic and immunological characteristics of GC.
The prognosis of GC is closely related to the crosstalk between immune cells and tumor cells. Nevertheless, the role of immune-related genes in predicting GC patients’ prognosis has not yet been elucidated.
In this study, we aimed to construct an immune-related gene signature for accurately predicting the prognosis of patients with GC.
Cox univariate survival analysis was performed to screen survival-related immune-related genes (IRGs). Differentially expressed survival-related IRGs were considered as hub IRGs. Hub IRGs were selected to conduct a prognostic signature. Receiver operating characteristic (ROC) curve analysis was performed to evaluate its prognostic performance. The correlation of the signature with clinical features and tumor-infiltrating immune cells was analyzed.
Our study constructed a prognostic signature consisting of ten hub IRGs (including S100A12, DEFB126, KAL1, APOH, CGB5, GRP, GLP2R, LGR6, PTGER3, and CTLA4), and it could be an independent prognostic predictor for GC. Furthermore, it was significantly associated with immune cell infiltration (especially macrophages).
We have proposed an immune-related prognostic signature for GC, which may possess prognostic value as a prediction tool for identification of patients who will benefit from immunotherapy.
The prognostic signature could help develop treatment strategies for patients with GC in the future.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Aurello P, Lazăr DC S-Editor: Ma YJ L-Editor: Wang TQ P-Editor: Wang LL
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11573] [Cited by in RCA: 13151] [Article Influence: 1878.7] [Reference Citation Analysis (4)] |
| 2. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13202] [Article Influence: 1466.9] [Reference Citation Analysis (3)] |
| 3. | Chen D, Chen G, Jiang W, Fu M, Liu W, Sui J, Xu S, Liu Z, Zheng X, Chi L, Lin D, Li K, Chen W, Zuo N, Lu J, Chen J, Li G, Zhuo S, Yan J. Association of the Collagen Signature in the Tumor Microenvironment With Lymph Node Metastasis in Early Gastric Cancer. JAMA Surg. 2019;154:e185249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 4. | Wu X, Gu Z, Chen Y, Chen B, Chen W, Weng L, Liu X. Application of PD-1 Blockade in Cancer Immunotherapy. Comput Struct Biotechnol J. 2019;17:661-674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 356] [Article Influence: 59.3] [Reference Citation Analysis (0)] |
| 5. | Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, Pitot HC, Hamid O, Bhatia S, Martins R, Eaton K, Chen S, Salay TM, Alaparthy S, Grosso JF, Korman AJ, Parker SM, Agrawal S, Goldberg SM, Pardoll DM, Gupta A, Wigginton JM. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455-2465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5599] [Cited by in RCA: 6275] [Article Influence: 482.7] [Reference Citation Analysis (0)] |
| 6. | Muro K, Chung HC, Shankaran V, Geva R, Catenacci D, Gupta S, Eder JP, Golan T, Le DT, Burtness B, McRee AJ, Lin CC, Pathiraja K, Lunceford J, Emancipator K, Juco J, Koshiji M, Bang YJ. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): a multicentre, open-label, phase 1b trial. Lancet Oncol. 2016;17:717-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 878] [Cited by in RCA: 909] [Article Influence: 101.0] [Reference Citation Analysis (1)] |
| 7. | Lazăr DC, Avram MF, Romoșan I, Cornianu M, Tăban S, Goldiș A. Prognostic significance of tumor immune microenvironment and immunotherapy: Novel insights and future perspectives in gastric cancer. World J Gastroenterol. 2018;24:3583-3616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 114] [Cited by in RCA: 111] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 8. | Rau A, Flister M, Rui H, Auer PL. Exploring drivers of gene expression in the Cancer Genome Atlas. Bioinformatics. 2019;35:62-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 9. | Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, Berger P, Desborough V, Smith T, Campbell J, Thomson E, Monteiro R, Guimaraes P, Walters B, Wiser J, Butte AJ. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58:234-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 577] [Article Influence: 57.7] [Reference Citation Analysis (0)] |
| 10. | Mei S, Meyer CA, Zheng R, Qin Q, Wu Q, Jiang P, Li B, Shi X, Wang B, Fan J, Shih C, Brown M, Zang C, Liu XS. Cistrome Cancer: A Web Resource for Integrative Gene Regulation Modeling in Cancer. Cancer Res. 2017;77:e19-e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 11. | Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22632] [Cited by in RCA: 28994] [Article Influence: 1812.1] [Reference Citation Analysis (0)] |
| 12. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11591] [Cited by in RCA: 22038] [Article Influence: 1695.2] [Reference Citation Analysis (0)] |
| 13. | Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747-1756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1228] [Cited by in RCA: 3080] [Article Influence: 440.0] [Reference Citation Analysis (0)] |
| 14. | Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1735] [Cited by in RCA: 2028] [Article Influence: 81.1] [Reference Citation Analysis (0)] |
| 15. | Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108-e110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2728] [Cited by in RCA: 4061] [Article Influence: 507.6] [Reference Citation Analysis (0)] |
| 16. | Yolanda LV, Sergio PD, Hugo ES, Isabel AF, Rafael BZ, Aldo TD, Gonzalo CR. Gastric cancer progression associated with local humoral immune responses. BMC Cancer. 2015;15:924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 17. | Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, Maeda T, Nagata Y, Kitanaka A, Mizuno S, Tanaka H, Chiba K, Ito S, Watatani Y, Kakiuchi N, Suzuki H, Yoshizato T, Yoshida K, Sanada M, Itonaga H, Imaizumi Y, Totoki Y, Munakata W, Nakamura H, Hama N, Shide K, Kubuki Y, Hidaka T, Kameda T, Masuda K, Minato N, Kashiwase K, Izutsu K, Takaori-Kondo A, Miyazaki Y, Takahashi S, Shibata T, Kawamoto H, Akatsuka Y, Shimoda K, Takeuchi K, Seya T, Miyano S, Ogawa S. Aberrant PD-L1 expression through 3'-UTR disruption in multiple cancers. Nature. 2016;534:402-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 512] [Article Influence: 56.9] [Reference Citation Analysis (0)] |
| 18. | Schwartz M, Zhang Y, Rosenblatt JD. B cell regulation of the anti-tumor response and role in carcinogenesis. J Immunother Cancer. 2016;4:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 190] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 19. | Ham IH, Oh HJ, Jin H, Bae CA, Jeon SM, Choi KS, Son SY, Han SU, Brekken RA, Lee D, Hur H. Targeting interleukin-6 as a strategy to overcome stroma-induced resistance to chemotherapy in gastric cancer. Mol Cancer. 2019;18:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 187] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 20. | Li X, Liu W, Wang H, Yang L, Li Y, Wen H, Ning H, Wang J, Zhang L, Li J, Fan D. Rap1 is indispensable for TRF2 function in etoposide-induced DNA damage response in gastric cancer cell line. Oncogenesis. 2015;4:e144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Zhang F, Li K, Yao X, Wang H, Li W, Wu J, Li M, Zhou R, Xu L, Zhao L. A miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop regulates tumour growth and chemoresistance in gastric cancer. EBioMedicine. 2019;44:311-321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 22. | Raja UM, Gopal G, Shirley S, Ramakrishnan AS, Rajkumar T. Immunohistochemical expression and localization of cytokines/chemokines/growth factors in gastric cancer. Cytokine. 2017;89:82-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 23. | Liang L, Fang JY, Xu J. Gastric cancer and gene copy number variation: emerging cancer drivers for targeted therapy. Oncogene. 2016;35:1475-1482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 113] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 24. | Ning X, Sun S, Hong L, Liang J, Liu L, Han S, Liu Z, Shi Y, Li Y, Gong W, Zhang S, Chen Y, Guo X, Cheng Y, Wu K, Fan D. Calcyclin-binding protein inhibits proliferation, tumorigenicity, and invasion of gastric cancer. Mol Cancer Res. 2007;5:1254-1262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Peng XC, Zeng Z, Huang YN, Deng YC, Fu GH. Clinical significance of TM4SF1 as a tumor suppressor gene in gastric cancer. Cancer Med. 2018;7:2592-2600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Nie K, Shi L, Wen Y, Pan J, Li P, Zheng Z, Liu F. Identification of hub genes correlated with the pathogenesis and prognosis of gastric cancer via bioinformatics methods. Minerva Med. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 27. | Yang Y, Shi Y, Hou Y, Lu Y, Yang J. CGB5 expression is independently associated with poor overall survival and recurrence-free survival in patients with advanced gastric cancer. Cancer Med. 2018;7:716-725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | Ke J, Ma P, Chen J, Qin J, Qian H. LGR6 promotes the progression of gastric cancer through PI3K/AKT/mTOR pathway. Onco Targets Ther. 2018;11:3025-3033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 29. | Chen Y, Zhang S, Wang Q, Zhang X. Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein. J Hematol Oncol. 2017;10:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 339] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 30. | Yamaguchi T, Fushida S, Yamamoto Y, Tsukada T, Kinoshita J, Oyama K, Miyashita T, Tajima H, Ninomiya I, Munesue S, Harashima A, Harada S, Yamamoto H, Ohta T. Tumor-associated macrophages of the M2 phenotype contribute to progression in gastric cancer with peritoneal dissemination. Gastric Cancer. 2016;19:1052-1065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 208] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 31. | Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie G, Ma Y, Shen L. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. 2017;36:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 326] [Cited by in RCA: 487] [Article Influence: 60.9] [Reference Citation Analysis (0)] |