Published online May 15, 2020. doi: 10.4251/wjgo.v12.i5.514
Peer-review started: December 17, 2019
First decision: February 20, 2020
Revised: March 18, 2020
Accepted: March 28, 2020
Article in press: March 28, 2020
Published online: May 15, 2020
Processing time: 149 Days and 0.3 Hours
Colorectal cancer (CRC) is a worldwide problem, which has been associated with changes in diet and lifestyle pattern. As a result of colonic fermentation of dietary fibres, short chain free fatty acids are generated which activate free fatty acid receptors (FFAR) 2 and 3. FFAR2 and FFAR3 genes are abundantly expressed in colonic epithelium and play an important role in the metabolic homeostasis of colonic epithelial cells. Earlier studies point to the involvement of FFAR2 in colorectal carcinogenesis.
To understand the role of short chain FFARs in CRC.
Transcriptome analysis console software was used to analyse microarray data from CRC patients and cell lines. We employed short-hairpin RNA mediated down regulation of FFAR2 and FFAR3 genes, which was validated using quantitative real time polymerase chain reaction. Assays for glucose uptake and cyclic adenosine monophosphate (cAMP) generation was done along with immunofluorescence studies to study the effects of FFAR2/FFAR3 knockdown. For measuring cell proliferation, we employed real time electrical impedance-based assay available from xCELLigence.
Microarray data analysis of CRC patient samples showed a significant down regulation of FFAR2 gene expression. This prompted us to study the FFAR2 in CRC. Since, FFAR3 shares significant structural and functional homology with FFAR2, we knocked down both these receptors in CRC cell line HCT 116. These modified cell lines exhibited higher proliferation rate and were found to have increased glucose uptake as well as increased level of glucose transporter 1. Since, FFAR2 and FFAR3 signal through G protein subunit (Gαi), knockdown of these receptors was associated with increased cAMP. Inhibition of protein kinase A (PKA) did not alter the growth and proliferation of these cells indicating a mechanism independent of cAMP/PKA pathway.
Our results suggest role of FFAR2/FFAR3 genes in increased proliferation of colon cancer cells via enhanced glucose uptake and exclude the role of PKA mediated cAMP signalling. Alternate pathways could be involved that would ultimately result in increased cell proliferation as a result of down regulated FFAR2/FFAR3 genes. This study paves the way to understand the mechanism of action of short chain FFARs in CRC.
Core tip: Free fatty acid receptors (FFAR) have been reported to be associated with colorectal cancer (CRC). In this report, we studied short chain FFAR2 and FFAR3 and have provided preliminary evidence about the possible involvement of increased glucose uptake in FFAR2 and FFAR3 knockdown clones of CRC cell line. We generated double knock down for FFAR2 and FFAR3 genes in a CRC cell line (HCT116) and studied possible mechanisms of increased cell proliferation in these cells.
- Citation: Al Mahri S, Al Ghamdi A, Akiel M, Al Aujan M, Mohammad S, Aziz MA. Free fatty acids receptors 2 and 3 control cell proliferation by regulating cellular glucose uptake. World J Gastrointest Oncol 2020; 12(5): 514-525
- URL: https://www.wjgnet.com/1948-5204/full/v12/i5/514.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v12.i5.514
Colorectal cancer (CRC) is a disease that is associated with the dietary patterns, metabolism and inflammation[1]. Type of diet significantly modifies the risk for development of CRC[2]. There is evidence linking high carbohydrate-low fibre diet to increased risk for CRC[3-6]. The site of this cancer is the location for processing of food aided by the gut microbiota. The effect of diet on CRC has been studied with different perspectives of associated factors. Metabolism of nutrients, role of gut microbiota and familial factors are being studied to better understand the causal factors in diet related initiation and progression of CRC[7]. Type of food intake, its digestion and metabolism are an upcoming area of research with potential to develop preventive and therapeutic strategies. Characterization of gut microbiota with newer technologies is allowing the possibility of customized probiotic treatment for the prevention of CRC[7,8]. Short Chain Fatty Acids (SCFAs) are produced in the distal gut by bacterial fermentation of macro-fibrous material that escapes digestion in the upper gastrointestinal tract and enters the colon[9,10]. SCFAs such as butyrate and propionate exert anticancer effect on colon as they have been shown to induce differentiation, growth arrest and apoptosis, mainly due to their intracellular actions, through inhibition of histone deacetylase[11,12]. This suggests that SCFAs produced in the gut could have protective properties against development of CRC. SCFAs are cognate ligands for a group of G-protein coupled receptors, free fatty acid receptor (FFAR) 2 and FFAR3 also known as GPR43 and GPR41, respectively[13,14]. FFAR2 and FFAR3 genes are abundantly expressed in human colon[15]. FFAR2 recognizes all three major SCFAs, but the affinities for FFAR3 are in the order of propionate > butyrate > acetate[16]. Activated FFAR2 initiate signalling through the Gαi pathway to suppress cyclic adenosine monophosphate (cAMP) levels, and through the Gαq pathway to enhance calcium mobilization[17]. Negative impact of FFAR2 on cAMP levels results in inhibition of protein kinase A (PKA) (primary downstream effector of cAMP) and its substrate, cAMP response element binding protein. Together, this leads to reduced expression of histone deacetylase[18]. Several studies on FFARs suggested their involvement in the onset and progression of colon cancer[15,19]. It has been suggested that FFAR2 plays a role in the cancer cell metabolism and growth. Loss of FFAR2 was observed using immunohistochemistry in malignant colon adenocarcinoma tissues by 80% compared to normal human colon mucosa tissue[15]. Transforming activity of loss of FFAR2 and its oncogenic potential was validated in fibroblasts[20]. However, there is no conclusive study to suggest the role of FFAR2 as well as FFAR3 in cancer cell growth and metabolism[21].
The aim of this study is to contribute towards our understanding the connection between gut microbiota and CRC. In this report, we focused on genes encoding for receptors for SCFAs in CRC patient samples. We analysed gene expression level of FFAR2 and FFAR3 in our patient cohort. Using microarray analysis of matched tumour-normal tissues, we provide evidence that gene expression of FFAR2 is significantly reduced in CRC patients. While some patients had reduced FFAR3 gene expression, we did not observe significant differences in gene expression levels in our patient cohort. To further characterize the mechanistic effect of reduced level of expression of these receptors, we generated knockdown (KD) clones of FFAR2 and FFAR3 genes in HCT116 CRC cell line. While FFAR2 KD exhibited increased cell proliferation, it was further increased with subsequent KD of FFAR3 gene. These KD clones were found to exhibit a significant increase in their glucose uptake as well as increased expression of glucose transporter 1 (GLUT1). Simultaneously, increased levels of cAMP were observed in these cells. These results provide evidence to suggest the role of FFAR2 as well as FFAR3 in progression of CRC via a previously unknown mechanism of increased glucose uptake.
CRC patient samples were analysed for gene expression changes using microarray analyses of the dataset we previously reported (GEO accession number: GSE 50421)[22]. For the analysis of differential gene expression between tumour and normal, we used Transcriptome Analysis Console software available from Affymetrix (Thermofisher Scientific, United States). Sample IDs and respective expression values for FFAR2 and FFAR3 genes in tumour and normal samples are given in Supplementary Table 1. Gene expression values were generated using Transcriptome Analysis Console and further analysed using GraphPad prism 7 software. Paired t-test was used to compare matched tumor-normal samples and detailed results are provided in Supplementary Table 2.
The study is approved by the Institutional Review Board at King Abdullah International Medical Research Center. Procedural and ethical consent forms were generated and approved by the Institutional Review Board office at at King Abdullah International Medical Research Center. Each patient prior to sample collection signed the procedural and ethical consent forms. The permissions and consents were obtained according to the journal's guidelines and standards.
HCT116 cell line (ATCC® CCL-247™) was obtained from American Type Culture Collection (Manassas, VA, United States). Cells were cultured in 5%CO2 at 37 °C. Cells were grown in advanced Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS, 100 IU/mL of penicillin, 100 μg/mL of streptomycin and 2 mmol/L L-glutamine (Gibco, Thermo Fisher Scientific, Waltham, MA, United States).
FFAR2 short-hairpin RNA (shRNA), targeting 4 different sequences of the FFAR2 gene, and a control scrambled shRNA plasmid were obtained from Origene Technologies, Inc. 1 × 105 HCT116 cells were plated in a 24-well plate and transfected with 0.5 μg of plasmid DNA using Lipofectamine3000 with a ratio of 1:2 (DNA:Lipo3000) according to the manufacturer’s protocol. Transfected HCT116 cells were selected in 0.8 μg/mL of puromycin (As per data available at http://cell-lines.toku-e.com/Cell-Lines_1422.html for 4 wk then isolated colonies were maintained in 0.8 μg/mL of puromycin antibiotic. Colonies were screened for FFAR2 KD using quantitative real time polymerase chain reaction (qRT-PCR) and one colony with most efficient KD was selected for subsequent experiments. One colony from scrambled shRNA construct was also selected to serve as control in all experiments.
We used pre-packaged Lentiviral particles harbouring 3 different shRNA sequences against the FFAR3 gene and a control-scrambled shRNA that were obtained from GeneTarget, Inc. To generate FFAR2/FFAR3 double KD cells. Earlier selected colony with most efficient KD of FFAR2 were transduced with the FFAR3 Lentiviral particles with a multiplicity of induction (MOI) = 5. Cells were selected in 5 μg/mL of blasticidin antibiotic (Dose was determined for HCT 116 cells using kill curve for blasticidin) for 4 wk then colonies were maintained in 5 μg/mL of blasticidin and 0.8 μg/mL of puromycin antibiotic. Colonies were screened for FFAR2 and FFAR3 KD using qRT-PCR and one colony with most efficient KD was selected for subsequent experiments.
Total RNA from five patients’ tumour-normal paired samples was extracted using the QIAGEN RNeasy Mini Kit (Qiagen; Cat# 74104). 2 μg of RNA was used for cDNA synthesis using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific). qRT-PCR was performed by using ABI 7900HT PCR system (Applied Biosystems, Foster City, CA, United States). GAPDH was used as housekeeping control. Sequence of primers used are GAPDH-F: 5’ ACAACTTTGGTATCGTGGAAGG 3’, GAPDH-R: 5’ GCCATCACGCCACAGTTTC 3’; FFAR2-F: 5’ TGCTACGAGAACTTCACCGAT 3’; FFAR2-R: 5’ GGAGAG-CATGATCCACACAAAAC 3’; FFAR3-F: 5’ GCGTGGAGGATCTACGTGAC 3’; FFAR3-R: 5’ TGTGAGTGTTCACTGGTCTTTC 3’. PowerUp SYBR Green Master Mix (Applied Biosystems) was used. All reactions performed in triplicate and the qRT-PCR data was analysed by using the RQ method (2-ΔΔCt) by SDS RQ manager and expression Suite version 1.1. Complete details with Ct values have been provided in Supplementary Tables 3 and 4.
One × 104 HCT116 cells from control-scrambled, FFAR2 KD and FFAR 2/3 double KD clones were plated in each well of a 96 well plate. Next day, cells were washed twice with phosphate buffer saline and incubated in glucose free DMEM. Glucose uptake was determined using Glucose uptake assay kit (Cayman Chemicals, MI, United States) following manufacturers protocol. Briefly, cells were incubated with 100 μg/ml fluorescent 2-N-7-Nitrobenz-2-oxa-1, 3-diazol-4-yl-Amino-2-Deoxyglucose (2-NBDG) in glucose free medium for 1h. Cells were washed with assay buffer 3 times and analysed immediately. 2-NBDG taken up by cells was detected on a Tecan Infinite 200 pro fluorimeter (Tecan Group Ltd. Männedorf, Switzerland) with fluorescent filters designed to detect fluorescein (excitation/emission = 485/535 nm). Cells without 2-NBDG incubation were used as blank control.
Three x 104 HCT116 cells from control-scrambled, FFAR2 KD and FFAR2/FFAR3 KD were plated on a 48 well plate overnight. Next day, cells were incubated with 500 μmol/L IBMX (3-isobutyl-1-methylxanthine) for 1h. cAMP in each well was measured using cAMP screen Immunoassay System (Thermo-fisher scientific) by following the manufactures instructions. Briefly, after IBMX incubation, cells were lysed in 100 μL lysis buffer (provided with the kit) by incubating at 37 °C for 30 min. 50 μL of cell lysates were added to the pre-coated 96 well plate followed by addition of cAMP-alkaline phosphatase conjugate and cAMP antibodies to each well. A cAMP standard was also prepared ranging 0.002-2000 pmol of cAMP. Plate was incubated at room temp for 2 h with constant shaking. Each well was washed 6 times with wash buffer followed by addition of CSPD/Sapphire-II™ RTU substrate/enhancer solution and incubation for 30 min. Measurements were made using a single-mode luminometer (Molecular Devices). Standard curve was made using the reading from cAMP standards and cAMP was measured in each well with reference to the standard curve. The experiment was repeated twice with 8 technical repeats for each condition.
Rate of cell proliferation was measured using xCELLigence real time cell analyser-dual purpose (RTCA-DP) available from ACEA biosciences (San Diego, United States). E-16 plates were used for monitoring cell adhesion and growth. This system works on the principle of electrical impedance. A unitless parameter termed Cell Index (CI) is used to measure the relative change in electrical impedance to represent cell status. CI is a relative and dimensionless value since it represents the impedance change divided by a background value. When there are no cells present in the medium, the sensor’s electronic property will not be affected and the impedance will be small. When there are more cells on the electrodes, the impedance will be larger. CI calculation is based on the following formula: CI = (Zi – Z0)/15 ς, where Zi is the impedance at an individual point of time during the experiment and Z0 is the impedance at the start of the experiment. CI is a self-calibrated value derived from the ratio of measured impedances. For cell proliferation experiments, we plated 40000 cells from scrambled control, FFAR2 KD and FFAR 2/FFAR3 KD HCT116 cells on E-16 plate in duplicate and cell proliferation was monitored for 60 h. The experiment was repeated 3 times and slope values of the growth curve were plotted using in-built software and later analysed on Graph pad prism. H89 (PKA inhibitor) was added at a concentration of 50 μmol/L after 24 h of plating and cell growth was monitored up to 60 h.
Fifty thousand HCT116 cells from control-scrambled, FFAR2 KD and FFAR2/FFAR3 KD were grown on cover slips were washed with PBS and fixed with 4% formaldehyde. Cells were incubated with anti-GLUT1 antibody (Catalogue #PA1-46152, Thermo Fisher Scientific, United States) in PBS containing 5% serum and 0.1% Triton for 30min at 37 °C. Cells were washed four times with PBS and incubated with fluorescein isothiocyanate (FITC) conjugated antibodies for 30 min. Cells were washed four times and immediately imaged using EVOS FL Auto imaging system (ThermoFisher Scientific). Images were quantified for GLUT1 expression using metamorph image processing software (Molecular devices).
One million HCT116 cells from control-scrambled, FFAR2 KD and FFAR2/FFAR3 KD cells were lysed with Immunoprecipitation buffer (Thermo Fisher Scientific) containing protease inhibitors. The scraped cells were kept on ice for 30 min and centrifuged at 10000 g for 5 min and the supernatant was collected. Protein concentrations were determined by Qubit protein assay kit (Thermo Fisher Scientific). Twenty μg of lysate was boiled at 95 °C for 5 min and loaded onto ready-made gel 4%–20% Mini-PROTEAN® TGX™ Precast Protein Gels (Bio-Rad), subjected to electrophoresis and transferred to PVDF membranes (Bio-Rad). The membranes were blocked using 5% BSA (MILLIPORE) in Tris buffer saline + Tween 20 (TBST) buffer for 1 h with shaking at room temperature. The following primary antibodies were used: PKA-PAS (Cell signalling) isotype anti-rabbit at recommended dilution of 1:1000 in 5% BSA with gentle shaking overnight at 4 °C. The blots were incubated with the appropriate Horse Radish Peroxidase-conjugated secondary antibody (Bio-Rad), and the signals were detected with Chemiluminiscent HRP Substrate (Bio-Rad). Images were captured and analysed using Chemidoc gel documentation system (Bio-Rad).
Data were presented as the mean ± SD and analysed for statistical significance using two tailed students t test available in the GraphPad prism 7 software version 7.03. Each bar represents an average of at least two independent experiments with multiple technical replicates in each experiment. Significance was set for a P value of < 0.05.
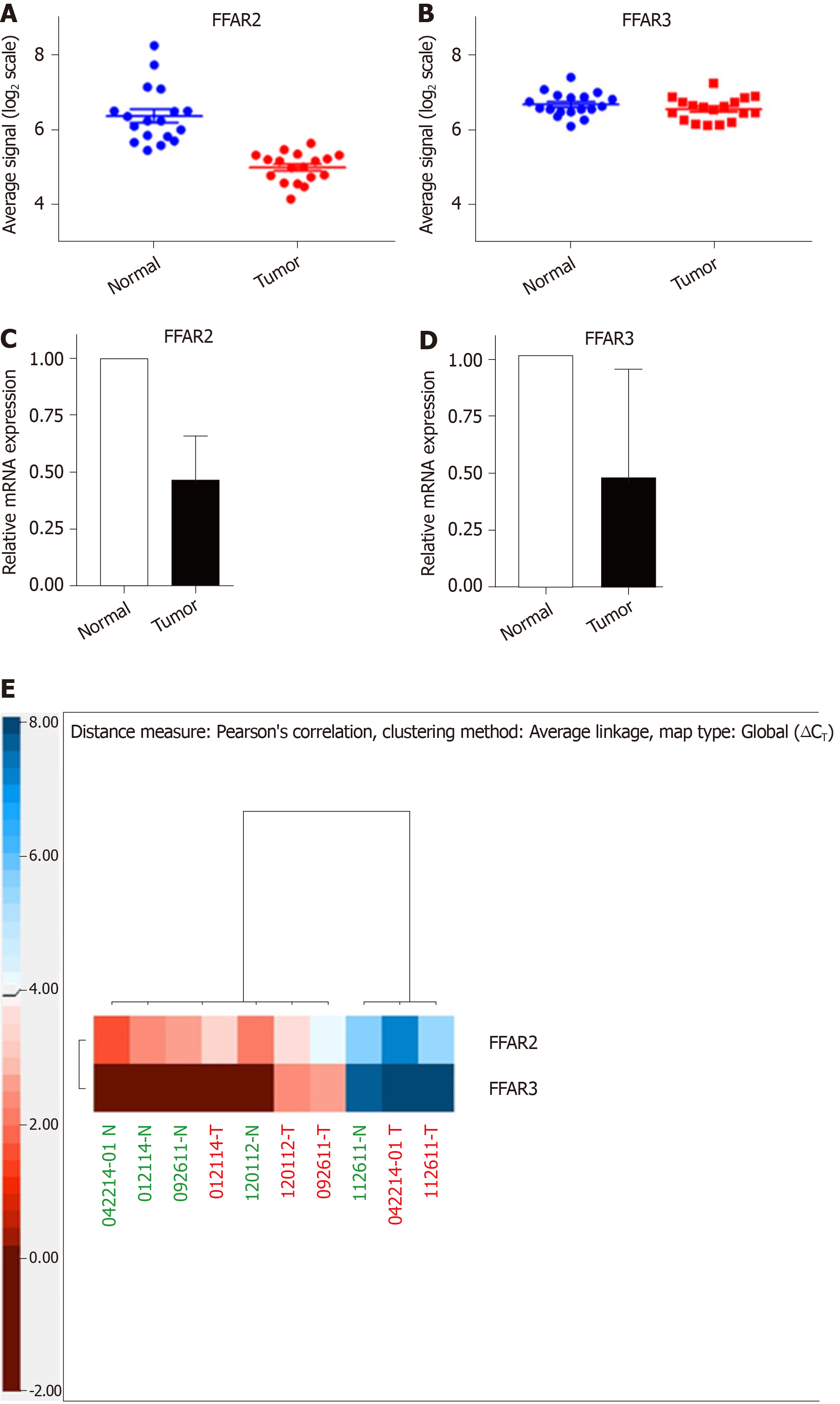
In a cohort of 18 CRC patients, we used microarray data to compare the expression levels of FFAR2 gene in matched tumour-normal tissue samples and found it to be significantly downregulated (Figure 1A). There was -1.388 ± 0.2065 times difference between means of normal tissue versus matched tumour for FFAR2 gene signals (P < 0.0001). But there was no significant difference in expression levels of FFAR3 gene (Figure 1B). We did qRT-PCR analysis for FFAR2 and FFAR3 genes on five CRC patient samples that were available from our patient cohort. In these five CRC patients, there was significant downregulation of both FFAR2 and FFAR3 genes. FFAR2 gene showed 5.388 times down regulation in these samples (P = 0.0066) and FFAR3 was also down regulated more than two times (Figure 1C and D). A heat map visualization of FFAR2 and FFAR3 expression in each of the patient tumour-normal matched tissues reflects the inter-patient heterogeneity (Figure 1E). We analysed other members of FFARs namely FFAR1 and FFAR4 in these patients and found no significant difference in their expression levels (Supplementary Figure 1 and Supplementary Table 5).
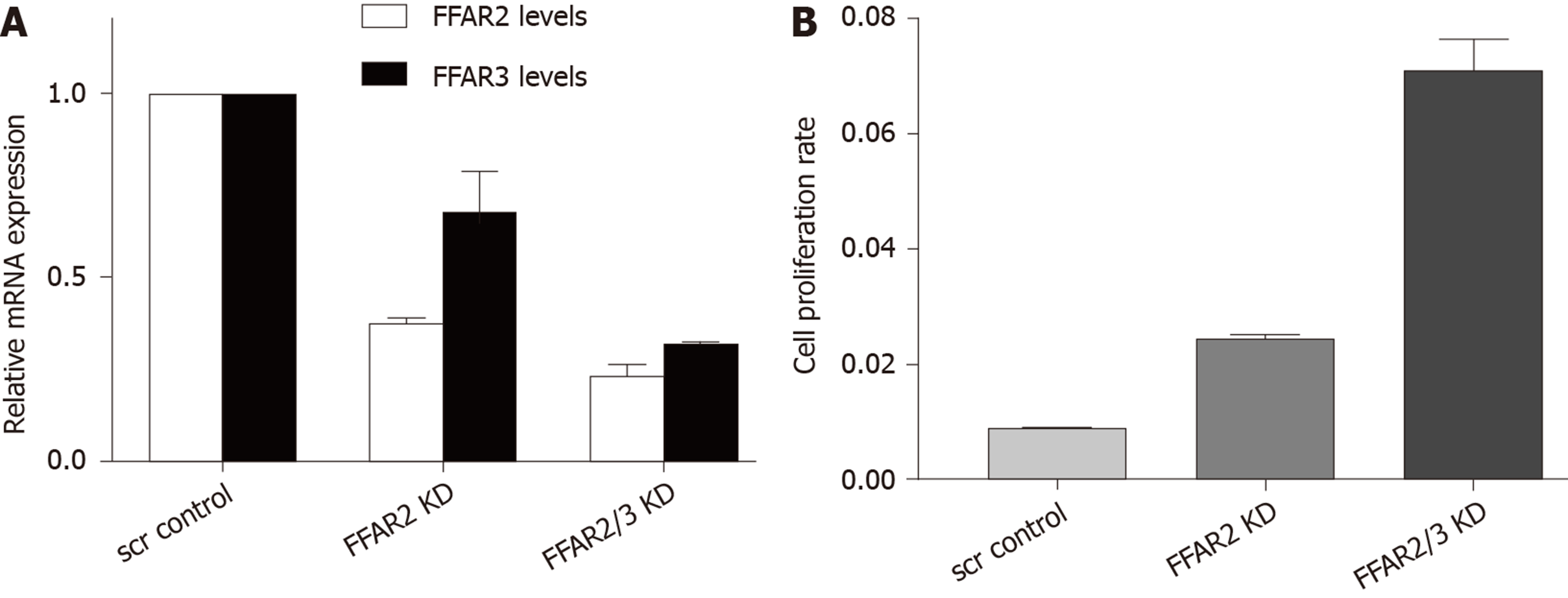
We engineered HCT116 cells to downregulate the expression of FFAR2 and FFAR3 genes. First, we knocked down FFAR2 gene and obtained 63% reduced stable expression. In this cell line FFAR3 levels were also found to be slightly affected with 32% reduction in expression. Next, we down regulated FFAR3 levels in this cell line. We found stable reduced expression of FFAR2 (77%) and FFAR3 (68%) in this cell clone. These two cell lines were chosen for all further experiments (Figure 2A). We measured the rate of proliferation of these HCT116 KD cells and found increased rate of proliferation as reflected by the cell index values in FFAR2 knock down cells (2.84 times compared to scrambled control. Double KD cells showed even higher increase in cell proliferation with 8.26 times cell index as compared to scrambled control (Figure 2B).
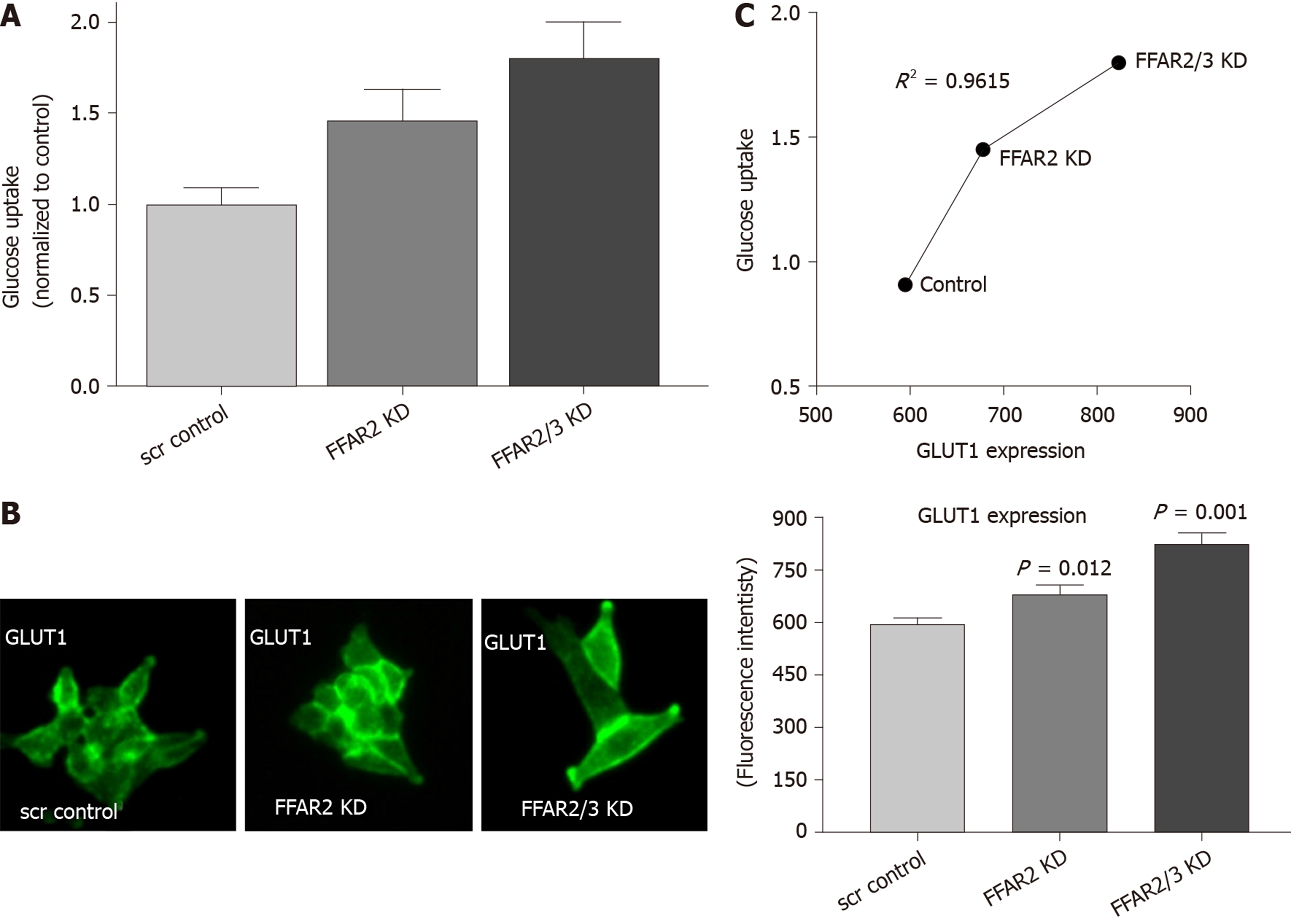
We measured the uptake of glucose in the HCT116 KD cells. HCT116 with FFAR2 KD showed about 1.5 times more glucose uptake whereas double KD exhibited even more with 1.8 times increase over the control-scrambled HCT116 cells (Figure 3A). We subsequently measured the levels of GLUT1 expression in these cells and found it to be significantly increased as higher fluorescence intensity was observed in modified cells (Figure 3B). GLUT1 expression pattern correlated with the glucose uptake in the modified cells as reflected in coefficient of determination value of 0.9615 (Figure 3C).
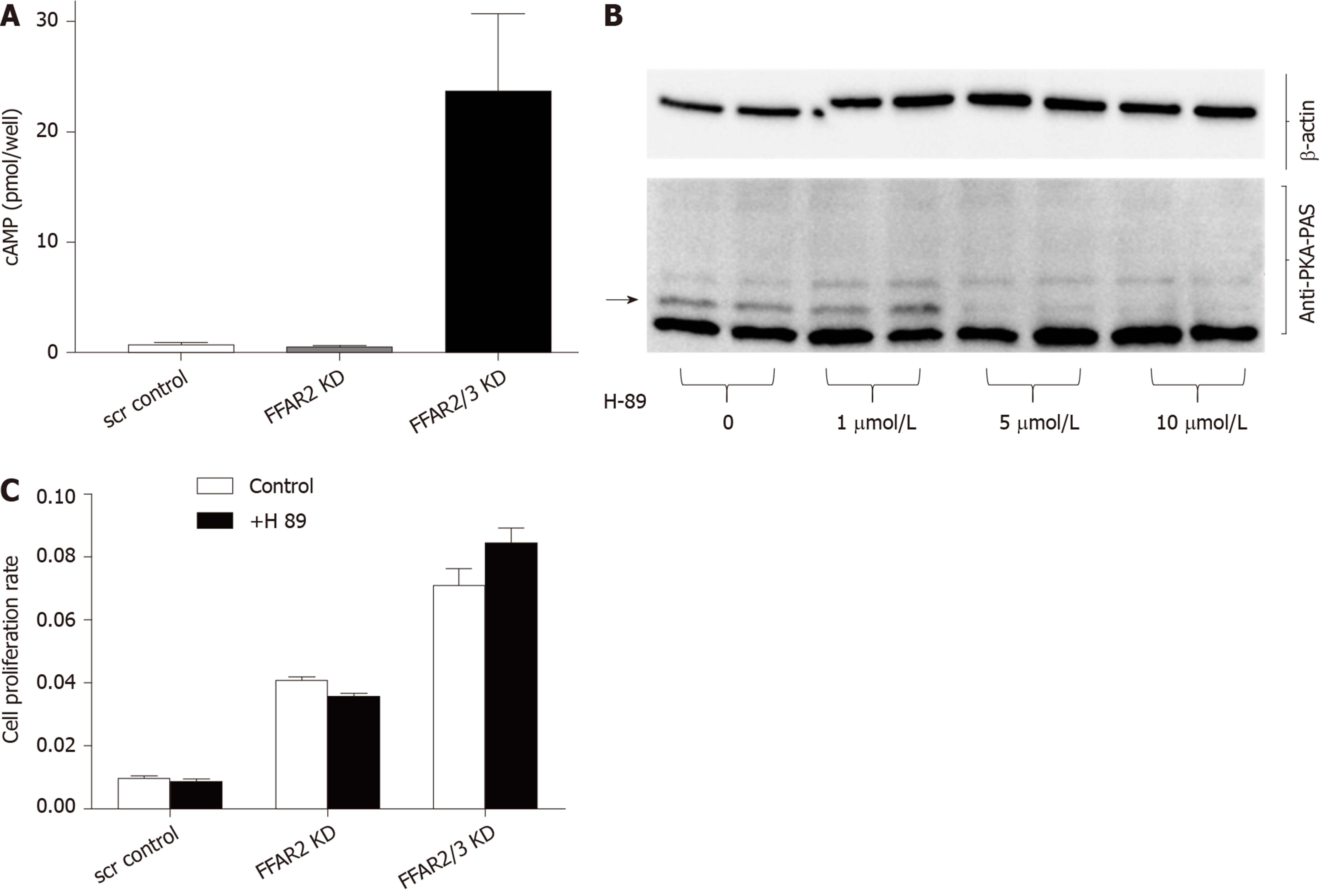
In order to understand the mechanistic role of FFARs in CRC, we attempted to interrogate the involvement of cAMP pathway. We found highly significant increase in cAMP levels in HCT116 double KD cells with more than 23 times increased cAMP levels as compared to FFAR2 KD and control-scrambled HCT116 cells (Figure 4A). Interestingly, the lack of significant change in cAMP level in FFAR2 KD HCT116 cells with a significant increase in rate of cell proliferation suggests a cAMP independent effect of FFAR2. To further validate this hypothesis, we blocked the effect of cAMP using the pharmacological inhibitor H89, which blocks the function of PKA, and tested the rate of cell proliferation in the presence or absence of H89. H89 showed a saturated inhibitory effect on HCT116 cells above 1 μmol/L concentration (Figure 4B). As expected, we did not observe any difference in rate of cell proliferation in the presence or absence of H89 in HCT116 KD clones confirming our hypothesis of an PKA independent mechanism of cAMP activity in HCT116 KD cells (Figure 4C). H89 also showed sustained inhibition up to 48h, which confirmed the efficacy during the entire duration of the cell proliferation experiment (Supplementary Figure 2).
As the evidence is supporting the strong connection between diet and CRC, there is an increased quest to understand the underlying molecular mechanism. Both long and short chain free fatty acids have been shown to be associated with cancer and metastasis[23-27]. This becomes relevant especially in CRC where high fat diet has been strongly correlated with the initiation and progression of disease. Receptors for these fatty acids would be good targets for designing prevention strategies. But the available evidence so far is not clear on establishing their role in different types of cancer. These receptors have shown to mediate increased cancerous activity as well as reduction in growth in different types of cancer cells. Also, these receptors belong to family of G-protein coupled receptors which are favourite molecules as drug targets[28]. In the present study, we focused on understanding the role of gut microbiota derived SCFA receptors in CRC. We used both patient samples and HCT116 cell lines in this study. FFAR2 and FFAR3 are well known receptors for SCFAs. FFAR2 has been implicated in CRC[29] but there is no known evidence for FFAR3 association. There are reports where the heterodimers of these receptors have been suggested to signal the short chain fatty acids. Earlier, we had reported cytogenetic and gene expression profile of CRC patients[22,30,31]. We used this data to analyse the expression profile of short chain FFARs in patient cohort. While we found significant down regulation of FFAR2 gene in few CRC patients, the variability of expression was high. This further supports the notion of inter-patient heterogeneity observed in cancer especially CRC[32,33] and strengthens argument in favour of personalized medicine[8]. We further validated our results using available CRC patient mRNA samples and carried out qRT-PCR based expression analysis. Both microarray and qRT-PCR analyses confirmed down regulation of FFAR2 gene in tumour samples. Our results thus confirm previous reports of down regulated FFAR2 gene in CRC[29]. However, our observations regarding FFAR3 are novel and this study underscores the importance of studying the two receptors together. While we observed no significant difference in expression level of FFAR3 gene in CRC patient cohort, there was significant down regulation observed in qRT-PCR data as reported earlier[29]. This could be due to the variability observed in these selected patient samples and/or due to higher sensitivity of qRT-PCR assay and possible differences in the two techniques[34].
In order to establish the role of short chain FFARs, we engineered HCT116, a colon cancer cell line to reduce the expression of FFAR2 and FFAR3 genes using shRNA technology. Our hypothesis suggested increased cell proliferation in HCT116 cells with reduced expression of short chain FFARs. Two cell line clones from HCT116 were generated – One with reduced expression of FFAR2 gene alone and another with reduced expression of both FFAR2 and FFAR3 genes. FFAR2 KD cells showed highly significant increase in cell proliferation whereas double KD showed comparatively enhanced effect in cell proliferation as well as glucose uptake and cAMP production. This is a clear evidence of FFAR2 and FFAR3 function as tumour suppressors via mechanism that needs to be fully understood. A recent report suggested epigenetic dysregulation of inflammation suppressors by FFAR2[19]. CRC cells are known to uptake glucose at a higher rate and Warburg effect is a hallmark of cancer cells[35]. HCT116 with reduced levels of FFAR2 and FFAR3 also displayed increased uptake of glucose in an additive manner, which could be responsible for increased cell proliferation. We suggest the involvement of FFAR2 and FFAR3 in glucose metabolism but this needs to be further studied to better understand the network affected by the reduced expression of these genes. Increased glucose uptake in the engineered cells was accompanied with an increased expression of GLUT1 – a well-known glucose transporter. Overexpression of GLUT1 has been suggested as a negative prognostic biomarker in CRC and indicator of clinically aggressive disease[36]. Our results thus suggest a previously unknown important connection between FFAR2/FFAR3 and glucose metabolism. Increased glucose metabolism has been known to be induced by short chain fatty acids[37].
To further understand the effect of FFAR2/FFAR3 on the signalling pathways, we measured the cAMP levels in engineered cells. Intracellular cAMP level has been shown to regulate cellular motility[38]. cAMP has also been shown to suppress apoptosis in CRC cells[39]. There was a huge increase in cAMP levels in double KD cells which was correlated with increased cell proliferation. However, FFAR2 single KD cells showed an increased cell proliferation and glucose uptake without any changes in cAMP levels. These results suggest that impact of FFAR2/FFAR3 on cell proliferation and glucose uptake are independent of cAMP signalling pathway. To further evaluate the role cAMP pathway, we inhibited PKA, a known downstream target of cAMP signalling by H89 molecule which is a known PKA inhibitor[40]. Inhibition of PKA had no impact on the rate of cell proliferation. Some studies have shown FFAR2/FFAR3 to signal through other pathways like p38 and JNK signalling[41] and Hippo-Yap pathway[42]. These pathways may be involved in mediating FFAR2/FFAR3 effect in our study and are an interesting area of research for future projects.
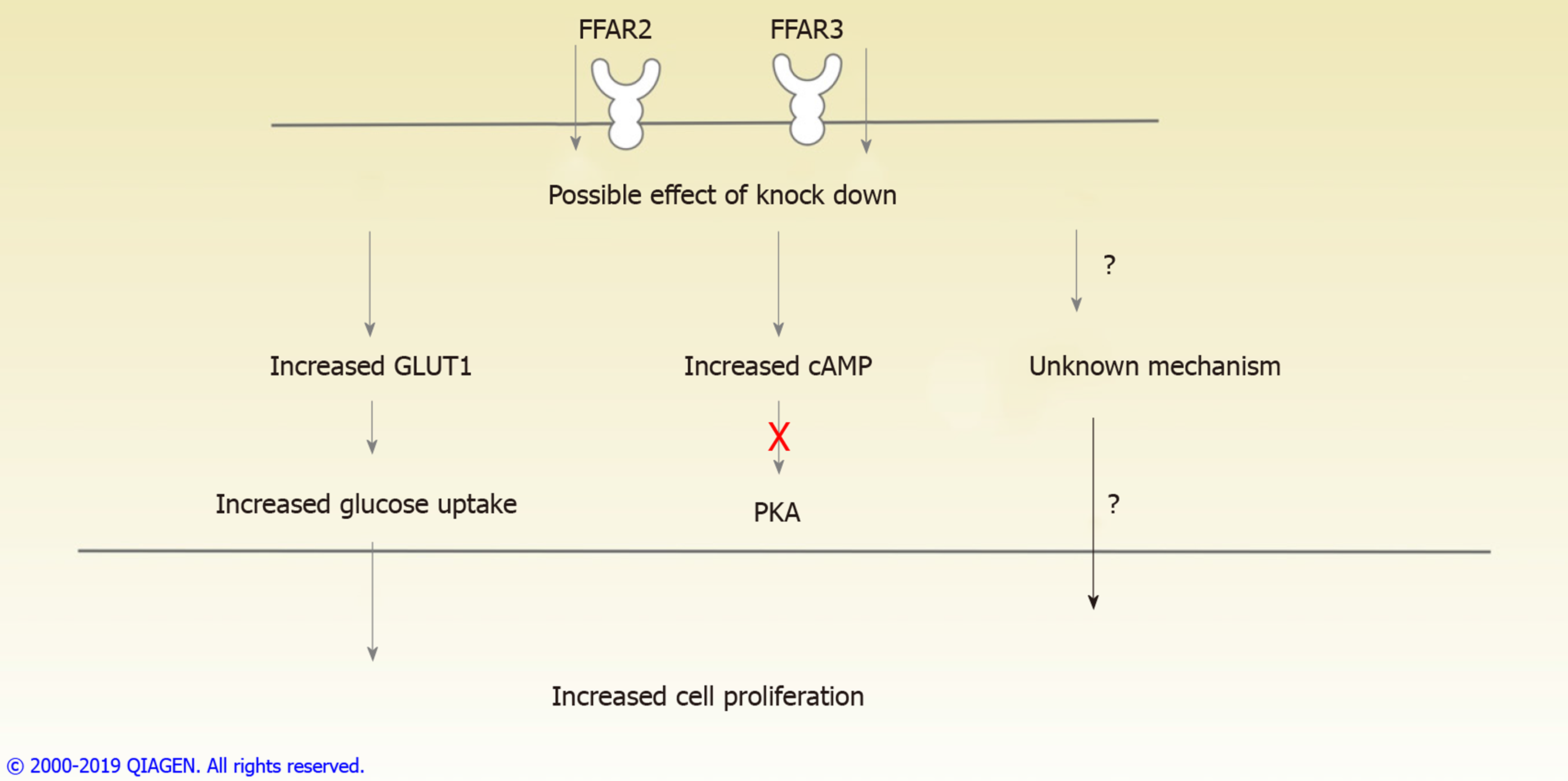
As illustrated in (Figure 5), our results conclusively establish the role of FFAR2 and FFAR3 in increased proliferation of CRC cells. This study also provides evidence to suggest the involvement of GLUT1 and PKA independent cAMP signalling pathway which needs to be further studied for identifying therapeutic targets and biomarkers for CRC progression.
Colorectal cancer (CRC) has been linked with free fatty acid receptors (FFARs). However, the mechanism of action of FFARs in CRC needs to be better studied.
To generate evidence that can better explain the role of diet in CRC and its association with gut microbiome.
To understand how FFAR2 and FFAR3 contribute to CRC cell growth and metabolism.
Cell culture, RNA interference (RNAi), Transfection, quantitative real time PCR, Western blot, Glucose uptake assay, cAMP assay, real time cell proliferation assay, Immunofluorescence, statistical and computational analyses.
FFAR2 is downregulated in CRC patient samples. CRC cells with reduced levels of FFAR2 and FFAR3 genes expression show increased rate of proliferation. Increased levels of glucose transporter and subsequent increase in glucose uptake is observed alongside increased cAMP levels in cells with reduced expression of FFAR2 and FFAR3.
Short chain FFARs FFAR2 and FFAR3 may contribute in increased cell proliferation by increased glucose uptake.
Processing of food by gut microbiota could be associated with initiation, progression and severity of CRC. Modifying dietary profiles for high-risk individuals may be a preventive measure for CRC.
Manuscript source: Unsolicited Manuscript
Corresponding Author's Membership in Professional Societies: American Association for Cancer research.
Specialty type: Oncology
Country/Territory of origin: Saudi Arabia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Handra-Luca A S-Editor: Wang YQ L-Editor: A E-Editor: Liu JH
| 1. | Moug SJ, Bryce A, Mutrie N, Anderson AS. Lifestyle interventions are feasible in patients with colorectal cancer with potential short-term health benefits: a systematic review. Int J Colorectal Dis. 2017;32:765-775. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 2. | Key TJ, Spencer EA. Carbohydrates and cancer: an overview of the epidemiological evidence. Eur J Clin Nutr. 2007;61 Suppl 1:S112-S121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 3. | Rossi M, Mirbagheri SEYEDS, Keshavarzian A, Bishehsari F. Nutraceuticals in colorectal cancer: A mechanistic approach. Eur J Pharmacol. 2018;833:396-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 4. | Yang J, Yu J. The association of diet, gut microbiota and colorectal cancer: what we eat may imply what we get. Protein Cell. 2018;9:474-487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 207] [Cited by in RCA: 205] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 5. | Li R, Grimm SA, Mav D, Gu H, Djukovic D, Shah R, Merrick BA, Raftery D, Wade PA. Transcriptome and DNA Methylome Analysis in a Mouse Model of Diet-Induced Obesity Predicts Increased Risk of Colorectal Cancer. Cell Rep. 2018;22:624-637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 6. | Trock B, Lanza E, Greenwald P. Dietary fiber, vegetables, and colon cancer: critical review and meta-analyses of the epidemiologic evidence. J Natl Cancer Inst. 1990;82:650-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 314] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 7. | Azcárate-Peril MA, Sikes M, Bruno-Bárcena JM. The intestinal microbiota, gastrointestinal environment and colorectal cancer: a putative role for probiotics in prevention of colorectal cancer? Am J Physiol Gastrointest Liver Physiol. 2011;301:G401-G424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 173] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 8. | Aziz MA, Yousef Z, Saleh AM, Mohammad S, Al Knawy B. Towards personalized medicine of colorectal cancer. Crit Rev Oncol Hematol. 2017;118:70-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 9. | Cummings JH, Macfarlane GT. Role of intestinal bacteria in nutrient metabolism. JPEN J Parenter Enteral Nutr. 1997;21:357-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 328] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 10. | Russell WR, Hoyles L, Flint HJ, Dumas ME. Colonic bacterial metabolites and human health. Curr Opin Microbiol. 2013;16:246-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 269] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 11. | Hague A, Elder DJ, Hicks DJ, Paraskeva C. Apoptosis in colorectal tumour cells: induction by the short chain fatty acids butyrate, propionate and acetate and by the bile salt deoxycholate. Int J Cancer. 1995;60:400-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 277] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 12. | Medina V, Edmonds B, Young GP, James R, Appleton S, Zalewski PD. Induction of caspase-3 protease activity and apoptosis by butyrate and trichostatin A (inhibitors of histone deacetylase): dependence on protein synthesis and synergy with a mitochondrial/cytochrome c-dependent pathway. Cancer Res. 1997;57:3697-3707. [PubMed] |
| 13. | Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, Pike NB, Strum JC, Steplewski KM, Murdock PR, Holder JC, Marshall FH, Szekeres PG, Wilson S, Ignar DM, Foord SM, Wise A, Dowell SJ. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312-11319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1583] [Cited by in RCA: 1740] [Article Influence: 79.1] [Reference Citation Analysis (0)] |
| 14. | Layden BT, Angueira AR, Brodsky M, Durai V, Lowe WL. Short chain fatty acids and their receptors: new metabolic targets. Transl Res. 2013;161:131-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 252] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 15. | Tang Y, Chen Y, Jiang H, Robbins GT, Nie D. G-protein-coupled receptor for short-chain fatty acids suppresses colon cancer. Int J Cancer. 2011;128:847-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 210] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 16. | Ichimura A, Hasegawa S, Kasubuchi M, Kimura I. Free fatty acid receptors as therapeutic targets for the treatment of diabetes. Front Pharmacol. 2014;5:236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 131] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 17. | Lee T, Schwandner R, Swaminath G, Weiszmann J, Cardozo M, Greenberg J, Jaeckel P, Ge H, Wang Y, Jiao X, Liu J, Kayser F, Tian H, Li Y. Identification and functional characterization of allosteric agonists for the G protein-coupled receptor FFA2. Mol Pharmacol. 2008;74:1599-1609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 125] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 18. | Shao RH, Tian X, Gorgun G, Urbano AG, Foss FM. Arginine butyrate increases the cytotoxicity of DAB(389)IL-2 in leukemia and lymphoma cells by upregulation of IL-2Rbeta gene. Leuk Res. 2002;26:1077-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 19. | Pan P, Oshima K, Huang YW, Agle KA, Drobyski WR, Chen X, Zhang J, Yearsley MM, Yu J, Wang LS. Loss of FFAR2 promotes colon cancer by epigenetic dysregulation of inflammation suppressors. Int J Cancer. 2018;143:886-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 20. | Hatanaka H, Tsukui M, Takada S, Kurashina K, Choi YL, Soda M, Yamashita Y, Haruta H, Hamada T, Ueno T, Tamada K, Hosoya Y, Sata N, Yasuda Y, Nagai H, Sugano K, Mano H. Identification of transforming activity of free fatty acid receptor 2 by retroviral expression screening. Cancer Sci. 2010;101:54-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 21. | Ulven T. Short-chain free fatty acid receptors FFA2/GPR43 and FFA3/GPR41 as new potential therapeutic targets. Front Endocrinol (Lausanne). 2012;3:111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 202] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 22. | Aziz MA, Periyasamy S, Al Yousef Z, AlAbdulkarim I, Al Otaibi M, Alfahed A, Alasiri G. Integrated exon level expression analysis of driver genes explain their role in colorectal cancer. PLoS One. 2014;9:e110134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 23. | Shimomoto T, Luo Y, Ohmori H, Chihara Y, Fujii K, Sasahira T, Denda A, Kuniyasu H. Advanced glycation end products (AGE) induce the receptor for AGE in the colonic mucosa of azoxymethane-injected Fischer 344 rats fed with a high-linoleic acid and high-glucose diet. J Gastroenterol. 2012;47:1073-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Ohmori H, Luo Y, Fujii K, Sasahira T, Shimomoto T, Denda A, Kuniyasu H. Dietary linoleic acid and glucose enhances azoxymethane-induced colon cancer and metastases via the expression of high-mobility group box 1. Pathobiology. 2010;77:210-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 25. | Ohmori H, Fujii K, Kadochi Y, Mori S, Nishiguchi Y, Fujiwara R, Kishi S, Sasaki T, Kuniyasu H. Elaidic Acid, a Trans-Fatty Acid, Enhances the Metastasis of Colorectal Cancer Cells. Pathobiology. 2017;84:144-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 26. | Fujii K, Luo Y, Fujiwara-Tani R, Kishi S, He S, Yang S, Sasaki T, Ohmori H, Kuniyasu H. Pro-metastatic intracellular signaling of the elaidic trans fatty acid. Int J Oncol. 2017;50:85-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 27. | Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002;132:1012-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 371] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 28. | Hopkins MM, Meier KE. Free Fatty Acid Receptors and Cancer: From Nutrition to Pharmacology. Handb Exp Pharmacol. 2017;236:233-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 29. | Sivaprakasam S, Gurav A, Paschall AV, Coe GL, Chaudhary K, Cai Y, Kolhe R, Martin P, Browning D, Huang L, Shi H, Sifuentes H, Vijay-Kumar M, Thompson SA, Munn DH, Mellor A, McGaha TL, Shiao P, Cutler CW, Liu K, Ganapathy V, Li H, Singh N. An essential role of Ffar2 (Gpr43) in dietary fibre-mediated promotion of healthy composition of gut microbiota and suppression of intestinal carcinogenesis. Oncogenesis. 2016;5:e238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 102] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 30. | Eldai H, Periyasamy S, Al Qarni S, Al Rodayyan M, Muhammed Mustafa S, Deeb A, Al Sheikh E, Afzal M, Johani M, Yousef Z, Aziz MA. Novel genes associated with colorectal cancer are revealed by high resolution cytogenetic analysis in a patient specific manner. PLoS One. 2013;8:e76251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 31. | Aziz MA, Periyasamy S, Yousef Z, Deeb A, AlOtaibi M. Colorectal cancer driver genes identified by patient specific comparison of cytogenetic microarray. Genom Data. 2014;2:29-31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 32. | Miyauchi T, Yaguchi T, Kawakami Y. Inter-patient and Intra-tumor Heterogeneity in the Sensitivity to Tumor-targeted Immunity in Colorectal Cancer. Nihon Rinsho Meneki Gakkai Kaishi. 2017;40:54-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 33. | Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational implications of tumor heterogeneity. Clin Cancer Res. 2015;21:1258-1266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 415] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 34. | Sinicropi D, Cronin M, Liu ML, Ferrari M, Ozkan M, Heller MJ. Gene Expression Profiling Utilizing Microarray Technology and RT-PCR. In: Ferrari M, Ozkan M, Heller MJ. BioMEMS and Biomedical Nanotechnology: Volume II: Micro/Nano Technologies for Genomics and Proteomics. Ferrari M, Ozkan M, Heller MJ. Boston: Springer, 2007: 23-46. |
| 35. | Fang S, Fang X. Advances in glucose metabolism research in colorectal cancer. Biomed Rep. 2016;5:289-295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 36. | Yang J, Wen J, Tian T, Lu Z, Wang Y, Wang Z, Wang X, Yang Y. GLUT-1 overexpression as an unfavorable prognostic biomarker in patients with colorectal cancer. Oncotarget. 2017;8:11788-11796. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 37. | Anderson JW, Bridges SR. Short-chain fatty acid fermentation products of plant fiber affect glucose metabolism of isolated rat hepatocytes. Proc Soc Exp Biol Med. 1984;177:372-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 98] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 38. | Mokashi S, Delikatny SJ, Orr FW. Relationships between chemotaxis, chemotactic modulators, and cyclic nucleotide levels in tumor cells. Cancer Res. 1983;43:1980-1983. [PubMed] |
| 39. | Nishihara H, Hwang M, Kizaka-Kondoh S, Eckmann L, Insel PA. Cyclic AMP promotes cAMP-responsive element-binding protein-dependent induction of cellular inhibitor of apoptosis protein-2 and suppresses apoptosis of colon cancer cells through ERK1/2 and p38 MAPK. J Biol Chem. 2004;279:26176-26183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 87] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 40. | Lochner A, Moolman JA. The many faces of H89: a review. Cardiovasc Drug Rev. 2006;24:261-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 262] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 41. | Kobayashi M, Mikami D, Kimura H, Kamiyama K, Morikawa Y, Yokoi S, Kasuno K, Takahashi N, Taniguchi T, Iwano M. Short-chain fatty acids, GPR41 and GPR43 ligands, inhibit TNF-α-induced MCP-1 expression by modulating p38 and JNK signaling pathways in human renal cortical epithelial cells. Biochem Biophys Res Commun. 2017;486:499-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 149] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 42. | Thirunavukkarasan M, Wang C, Rao A, Hind T, Teo YR, Siddiquee AA, Goghari MAI, Kumar AP, Herr DR. Short-chain fatty acid receptors inhibit invasive phenotypes in breast cancer cells. PLoS One. 2017;12:e0186334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |