Published online Sep 15, 2019. doi: 10.4251/wjgo.v11.i9.705
Peer-review started: March 18, 2019
First decision: March 28, 2019
Revised: April 10, 2019
Accepted: July 16, 2019
Article in press: July 17, 2019
Published online: September 15, 2019
Processing time: 181 Days and 1.5 Hours
The molecular mechanisms involved in microRNAs (miRNAs) have been extensively investigated in gastric cancer (GC). However, how miR-331 regulates GC pathogenesis remains unknown.
To illuminate the effect of miR-331 on cell metastasis and tumor growth in GC.
The qRT-PCR, CCK8, Transwell, cell adhesion, Western blot, luciferase reporter and xenograft tumor formation assays were applied to explore the regulatory mechanism of miR-331 in GC.
Downregulation of miR-331 associated with poor prognosis was detected in GC. Functionally, miR-331 suppressed cell proliferation, metastasis and tumor growth in GC. Further, miR-331 was verified to directly target musashi1 (MSI1). In addition, miR-331 inversely regulated MSI1 expression in GC tissues. Furthermore, upregulation of MSI1 weakened the inhibitory effect of miR-331 in GC.
miR-331 inhibited development of GC through targeting MSI1, which may be used as an indicator for the prediction and prognosis of GC.
Core tip: Gastric cancer (GC) has become one of the main threats to human life and health. MicroRNAs (miRNAs) have been reported to act as promoters and inhibitors in GC. In this study, the effect of miR-331 on cell metastasis and tumor growth was illuminated in GC. The results showed that miR-331 was downregulated in GC, which predicted poor prognosis in GC patients. Moreover, overexpression of miR-331 inhibited cell metastasis and tumor growth in GC cells. Further, musashi1 (MSI1) is a direct target gene of miR-331. MiR-331 inhibited GC progression through the suppression of MSI1.
- Citation: Yang LY, Song GL, Zhai XQ, Wang L, Liu QL, Zhou MS. MicroRNA-331 inhibits development of gastric cancer through targeting musashi1. World J Gastrointest Oncol 2019; 11(9): 705-716
- URL: https://www.wjgnet.com/1948-5204/full/v11/i9/705.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i9.705
In recent years, gastric cancer (GC) has become one of the main threats to human life and health, of which more than 50% of cases occur in East Asia[1]. In China, the incidence rate of GC ranks first among all kinds of tumors. About 170000 people die of GC every year, almost one-fourth of the total number of malignant tumor deaths. Furthermore, more than 20000 new patients with GC are found every year[2]. The pathogenesis of GC is complex and still unclear, involving many etiological factors and genetic changes. Although great progress has been made in understanding the pathogenesis of malignant tumors, the treatment of GC still has limitations[3]. Moreover, low survival rate is found in GC patients, and cell metastasis is the main causes of poor prognosis in GC patients[4]. Despite the treatment strategy for GC has been improved, it is necessary to explore high-sensitivity and low-cost treatments.
It has been demonstrated that microRNAs (miRNAs) can act as promoters and inhibitors in various cancers including GC by inhibiting gene expression[5]. For example, miR-133a was downregulated in GC and inhibited tumor growth, migration, and EMT process by targeting PSEN1[6]. Conversely, upregulation of miR-592 was identified in GC and promoted cell proliferation, invasion and migration through the PI3K/AKT and MAPK/ERK signaling pathways[7]. Recently, the dysregulation of miR-331 in different cancers has aroused our concern. It has been demonstrated that miR-331 can function as a marker for diagnosis and prognosis of hepatocellular carcinoma patients[8]. It had been reported that miR-331 was overexpressed in malignant breast tumors[9]. Moreover, miR-331 promoted hepatocellular carcinoma cell metastasis and proliferation through regulating ING5[10]. In addition, miR-331 was found to enhance Epithelial-to-Mesenchymal Transition (EMT) in prostate cancer[11]. However, downregulation of miR-331 had been identified in esophageal adenocarcinoma and predicted tumor recurrence[12]. In addition, miR-331-3p regulated expression of NRP-2 to inhibit development and progression of glioblastoma[13]. In particularly, Guo et al[14] had shown that miR-331 directly targeted E2F1 and induced the inhibition of GC tumor growth. However, how miR-331 regulates GC cell viability and metastasis remains blurry and need to be investigated.
Musashi1 (MSI1) is a neural RNA-binding protein required for Drosophila adult external sensory organ development[15]. Moreover, the function of MSI1 has been identified in normal and cancer stem cells[16]. Recently, MSI1 has been found to regulate tumor growth and proliferation in several human cancers. MSI1 had been identified as carcinogenesis, progression, and poor prognosis related biomarker for gallbladder adenocarcinoma[17]. Furthermore, MSI1 can regulate breast cancer proliferation and is an indicator for poor survival[18]. More importantly, upregulation of MSI1 has been identified in human colorectal adenomas and GC[19,20]. Moreover, knockdown of MSI1 resulted in tumor regression in colon cancer[21]. However, the role of MSI1 and its relationship with miR-331 have not been reported in previous studies.
Therefore, the regulatory mechanism of miR-331 with MSI1 was investigated in GC. We mainly focused on how miR-331 regulates GC tumor growth and metastasis. The findings will contribute to better understanding the pathogenesis of GC.
The GC tissues used in this experiment and their clinical and follow-up information were provided by Affiliated Hospital Taishan Medical University. All 78 patients involved in the experiment did not receive radiotherapy or chemotherapy prior to surgery. Participants provided written informed consent before designing the research, and Human Ethics Committee of Affiliated Hospital Taishan Medical University approved the experiment.
A normal gastric cell GES1 and SGC-7901, BGC-803, MKN-45 GC cell lines were purchased from Beijing Zhongke Quality Inspection Biotechnology Co., Ltd. (Beijing, China). The cells then were seeded in RPMI-1640 medium with 10% fetal bovine serum (FBS) and incubated in an atmosphere with 5% CO2 at 37 °C.
MiR-331 mimics, miR-331 inhibitors, and MSI1 plasmid (RiboBio Co, Ltd, Guangzhou, China) were transferred into MKN-45 cells respectively with Lipofectamine 2000 (Invitrogen, CA, United States) based on experimental needs. Untreated MKN-45 cells were set as the control. The nucleotide sequences were: miR-331 mimics, 5’-GCC CCU GGG CCU AUC CUA GAA-3’, antisense: 5’-CUA GGA UAG GCC CAG GGG CUU-3’; miR-331 inhibitor, 5’-UUC UAG GAU AGG CCC AGG GGC-3’.
We extracted total RNA from MKN-45 cell by using TRIZOL reagent (TaKaRa Bio, United States). The First-Strand cDNA Synthesis Kit (Promega, United States) was added to obtain cDNA. The mixture of the qRT-PCR standard reaction system was then added to SYBR Green PCR Master Mix (Applied Biosystems, CA, United States). The prepared PCR reaction solution was placed on ABI7300 real-time PCR machine (Applied Bio-systems) for PCR amplification reaction. U6 and GAPDH were respectively used as the controls for miR-331 and MSI1, which were quantified with the 2−△△ct method. The forward and reverse primers of qRT-PCR are given in Table 1.
| Gene | Primers sequence (5-3’) |
| MiR-331 | F: GCCCCTGGGCCTATCCTAGAA |
| R: ACGCGTCGACTTTTAGGGCTAAGTTGCTTC | |
| MSI1 | F: CTCCAAAACAATTGACCCTA |
| R: GCTCAAAATATTGCTTCACG | |
| U6 | F: AGAGCCTGTGGTGTCCG |
| R: CATCTTCAAAGCACTTCCC | |
| GAPDH | F: ATGGGGAAGGTGAAG GTCG |
| R: GGGGTCATTGATGGCAACAAT A |
MKN-45 cells were pre-incubated in a 96-well plate for 24 h (at 37 °C, 5% CO 2). MKN-45 (2 × 104) cells were incubated for 24, 48, 72 and 96 h. 10 mL of CCK-8 (Dojindo, Kumamoto, Japan) solution was used to incubate the cells for 4 h. The absorbance at 450 nm was observed with a microplate reader (Molecular Devices).
The xenograft study was approved by the Animal Care and Use Committee of Affiliated Hospital Taishan Medical University. We purchased nude mice (4 weeks old) from the Shanghai Lab Animal Research Center (Shanghai, China). Then 3 × 106 MKN-45 cells with stably overexpressing miR-331 or miR-NC were injected into mice subcutaneous. The tumor volume was observed every one week. After 4 wk, the mice were sacrificed and tumors were used for further study.
Ki67 Cell Proliferation Kit was purchased from Sangon Biotech Co., Ltd. (Shanghai, China). The experiment was performed based on their protocol. The section of gastric cancer tissues were dewaxed, hydrated, and washed twice with PBS for 5 min. After blocking with Blocking Buffer for 30-60 min at room temperature, Anti-ki67 Rabbit antibody (1:50) was added to incubate overnight at 4 °C in a humidified atmosphere. Then, secondary antibody HRP-conjugated Donkey anti-Rabbit IgG (1:500) was added and incubated for 60 min at room temperature. After washing 3 times with PBS, DAB mixture was added to each slide, and incubated for 5-10 min at room temperature protected from light. The section was washed, counterstained, dehydrated, transparentized and mounted. Images were captured using microscope.
The upper chamber surface of the bottom membrane of the Transwell chamber (8-μm pore size membranes) was coated with Matrigel (BD), and the Matrigel was polymerized into a gel at 37 °C for 30 min. Moreover, the transfected MKN-45 cells were starved prior to invasion assay. 5 × 104 mL MKN-45 cell suspension (100 μL) was added to the Transwell chamber, and then a medium containing 20% FBS (600 μL) was added to the lower chamber with the 24-well plate. After routine incubation for 24 h, the Transwell chamber was fixed and stained. The number of invading cells observed under a microscope of 400 times. The Transwell cell migration assay performed without Matrigel, other process is basically same as the invasion assay.
A pGL3 luciferase vector (Invitrogen) containing the wild or mutant type of 3’-UTR of MSI1 gene was prepared. The luciferase vector and miR-331 mimics were co-transfected into MKN-45 cells. After 48 h, Dual-Luciferase Reporter Assay System (Promega) was used to examine the luciferase activity. MSI1 gene information was wild type (Ensembl number ENSG00000135097) and mutated sequence (5’-UGGCGAGGGCAGACCGGUCCCCA-3’).
Protein samples were lysed using RIPA buffer (Beyotime, Shanghai, China). 10% concentrated SDS-PAGE protein loading buffer was added to the collected protein samples. After denaturing the protein, the protein sample was directly loaded into the SDS-PAGE gel and then transferred into PVDF membrane. PVDF membrane was incubated with the corresponding primary antibodies overnight at 4 °C, including rabbit polyclonal antibody to MSI1 (1:1000, ab21628, Abcam, Shanghai, China) and rabbit monoclonal antibody to GAPDH (1:1000, ab181602, Abcam, Shanghai, China). The washing solution was added for 5-10 min, and the diluted Goat anti-Rabbit. Goat anti-Rabbit IgG(H+L) HRP secondary antibody (1:500, ab205718, Abcam, Shanghai, China) was added and incubated at room temperature for 1 h. Finally, ECL reagent (Millipore, MA, United States) was used to detect proteins. In addition, E-cadherin, N-cadherin and Vimentin antibodies were all obtained from Abcam (1:1000, Shanghai, China).
Data were analyzed using SPSS 13.0 and Graphpad Prism 6. The difference between the groups was calculated through Chi-squared Test or Tukey’s one-way ANOVA. The log-rank test Kaplan-Meier analysis was used to compare the survival differences. The data was shown as mean ± SD. When P < 0.05, the data is considered statistically significant.
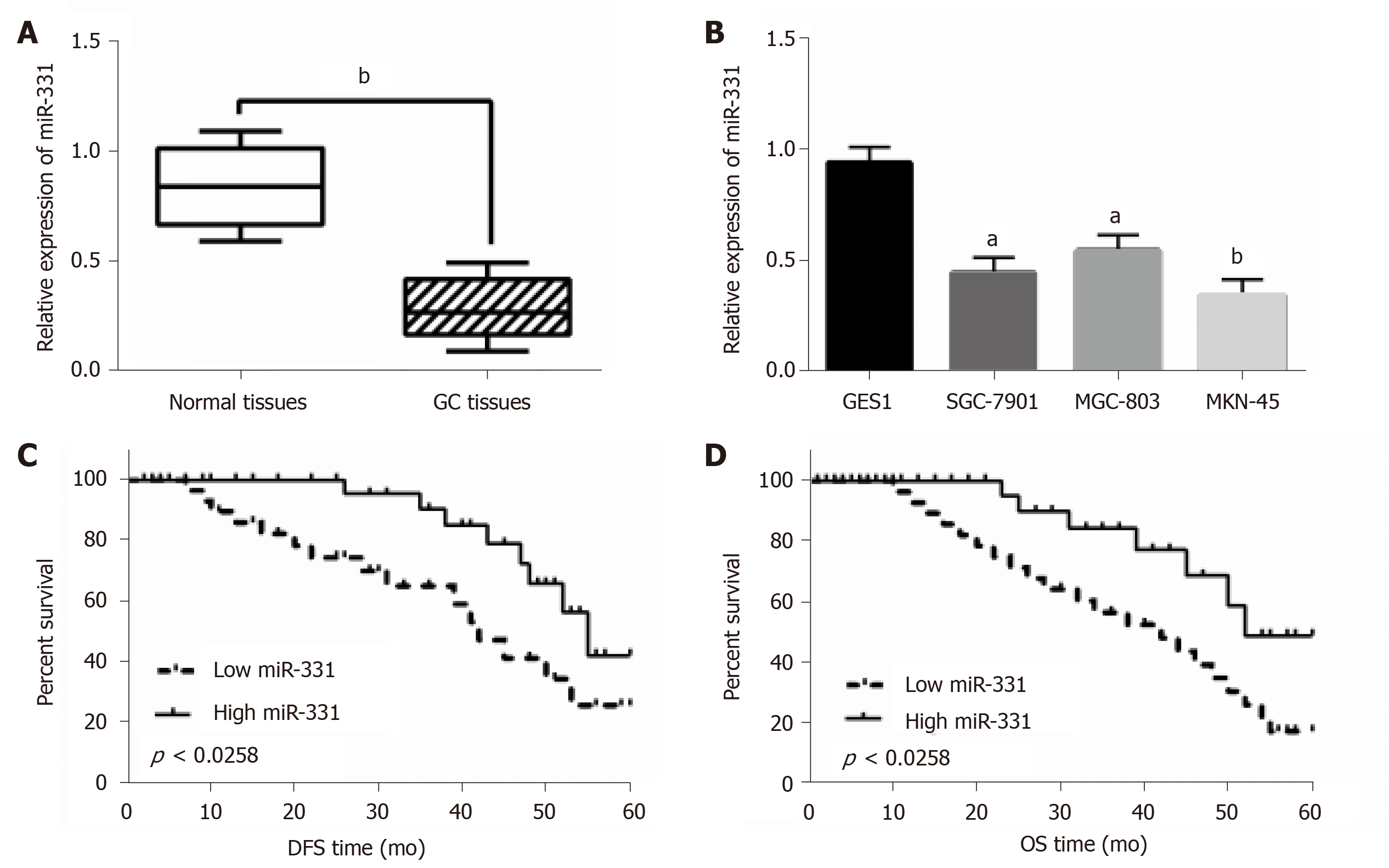
In GC tissues and cell lines, miR-331 expression was observed by qRT-PCR assay. First, low miR-331 expression was identified in GC tissues contrast to normal tissues (Figure 1A). Meanwhile, the reduction of miR-331 expression was found in SGC-7901, MGC-803, MKN-45 cells contrasted to GES1 cells (Figure 1B). MKN-45 cell line was selected for subsequent experiments because of the significant differences in expression of miR-331. Based on the expression of miR-331, these cases were divided into a high miR-331 expression group and a low expression group based on its median value in GC patients as a cutoff point (cutoff point = 0.75). Furthermore, abnormal miR-331 expression was correlated with lymph nodes metastasis and TNM stage in GC patients (P < 0.05, Table 2). In addition, shorter disease free survival (DFS) and overall survival (OS) was correlated with low miR-331 expression in GC patients (Figure 1C and 1D). Therefore, miR-331 expression was reduced, which predicted poor prognosis of GC patients.
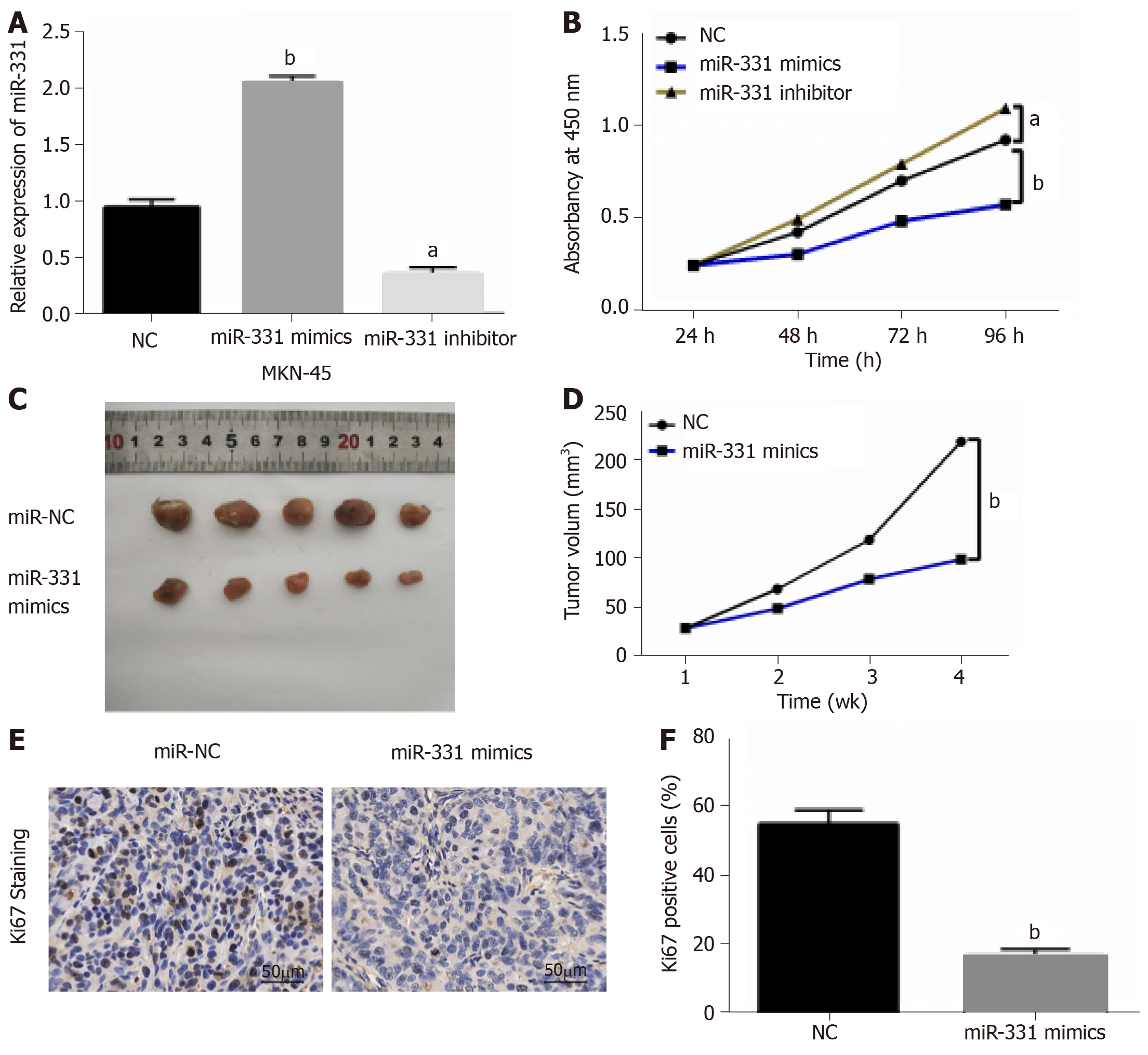
MiR-331 mimic or inhibitor was transfected into MKN-45 cells to perform gain-loss experiment. MiR-331 expression was obviously increased by its mimics, but decreased by its inhibitor (Figure 2A). Next, we found that miR-331 mimics inhibited the proliferation of MKN-45 cells. And miR-331 inhibitor showed the opposite results (Figure 2B). Moreover, the effect of miR-331 on tumor growth was analyzed in GC. MiR-331 mimics was found to decline the tumor volume and suppressed tumor growth compared to control group (Figure 2C and 2D). Additionally, miR-331 overexpression led to a significant decrease in the number of hyperproliferative Ki-67+ tumor cells (Figure 2E). Collectively, miR-331 inhibited cell proliferation and tumor growth in GC.
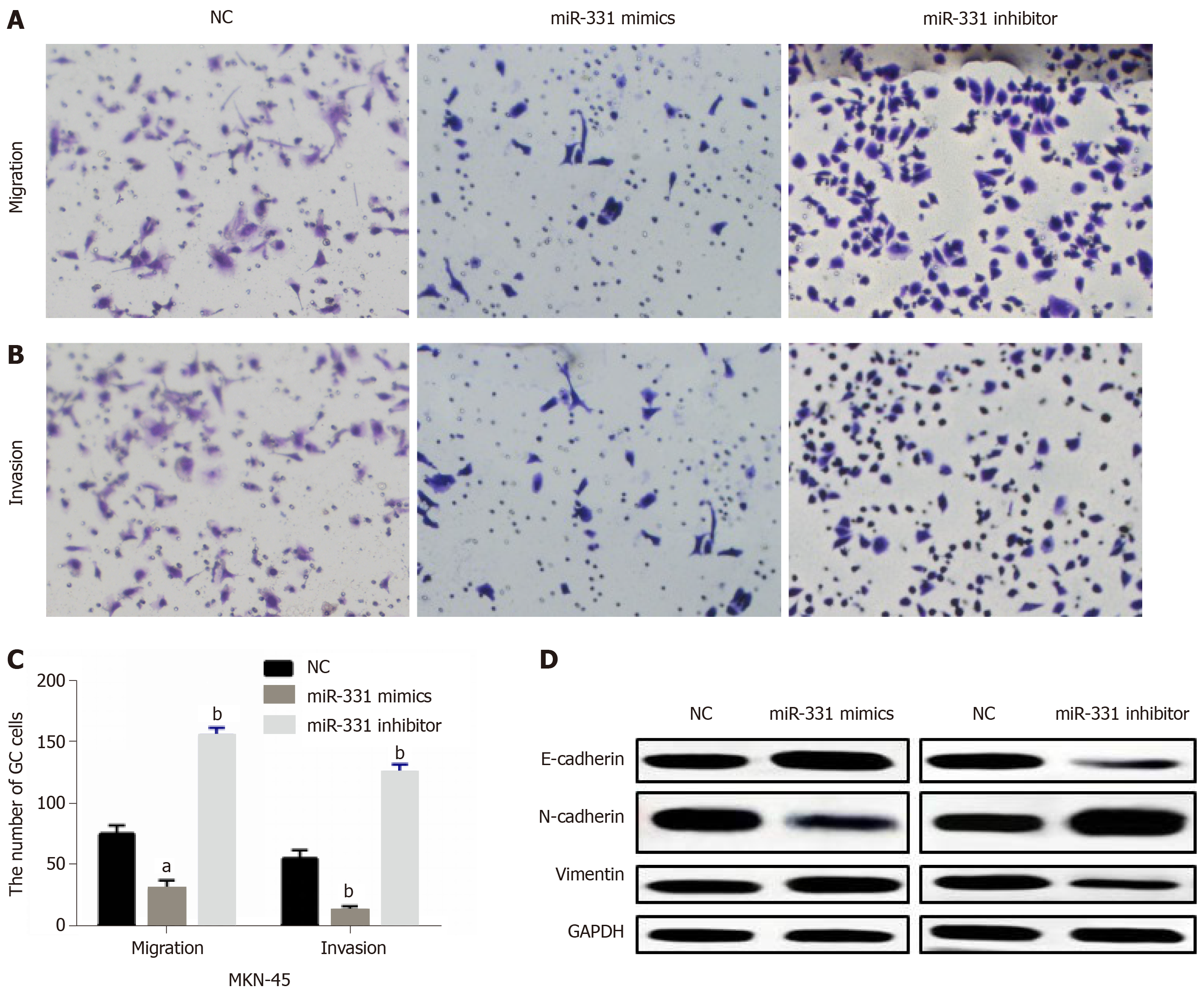
Then, how miR-331 regulates cell metastasis was investigated in MKN-45 cells. Transwell assay suggested that miR-331 overexpression suppressed cell migration, while miR-331 knockdown promoted MKN-45 cell migration (Figure 3A and 3C). For cell invasion in GC, the same effect of miR-331 overexpression and knockdown was also identified (Figure 3B and 3C). Next, the effect of miR-331 on EMT was explored in GC cells. Overexpression of miR-331 facilitated E-cadherin expression and hindered expressions of N-cadherin and Vimentin. In contrast, knockdown of miR-331 blocked E-cadherin expression and promoted expression levels of N-cadherin and Vimentin (Figure 3D), indicating that miR-331 blocked EMT in GC. Briefly, miR-331 inhibited cell metastasis in GC.
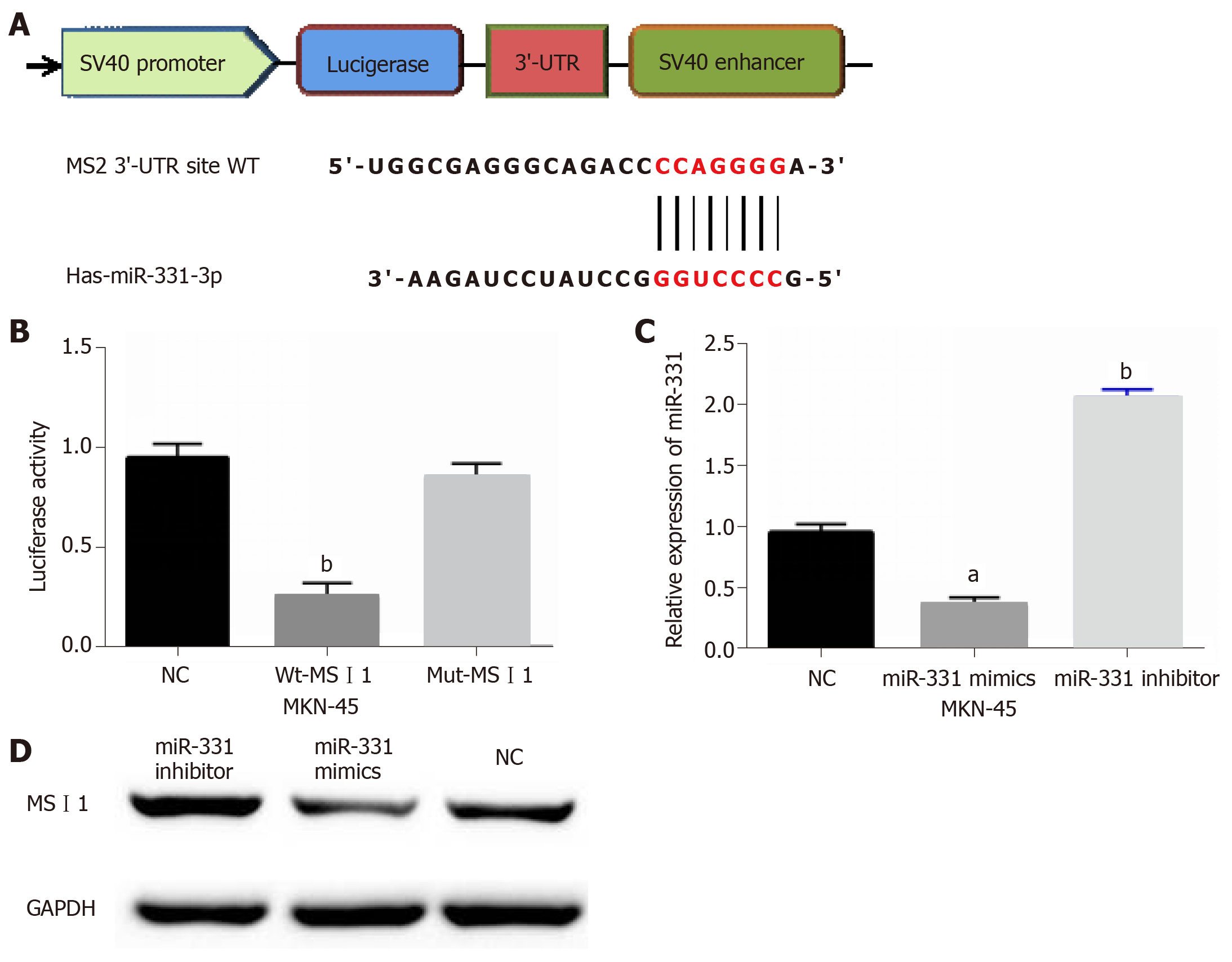
The target genes of miR-331 were searched in TargetScan databases to disclose how miR-331 suppresses GC progression. The binding sites between miR-331 and MSI1 were showed in Figure 4A. Next, luciferase reporter assay was designed to confirm that prediction. As predicted, miR-331 mimics inhibited the luciferase activity of Wt-MSI1, but had no effect on Mut-MSI1 luciferase activity (Figure 4B). Then, MSI1 expression regulated by miR-331 mimics or inhibitor was assessed in MKN-45 cells. The mRNA and protein MSI1 expression was inhibited by miR-331 mimics, but promoted by miR-331 inhibitor (Figure 4C and 4D). Thus, we believe that miR-331 directly targets MSI1 and regulates MSI1 expression.
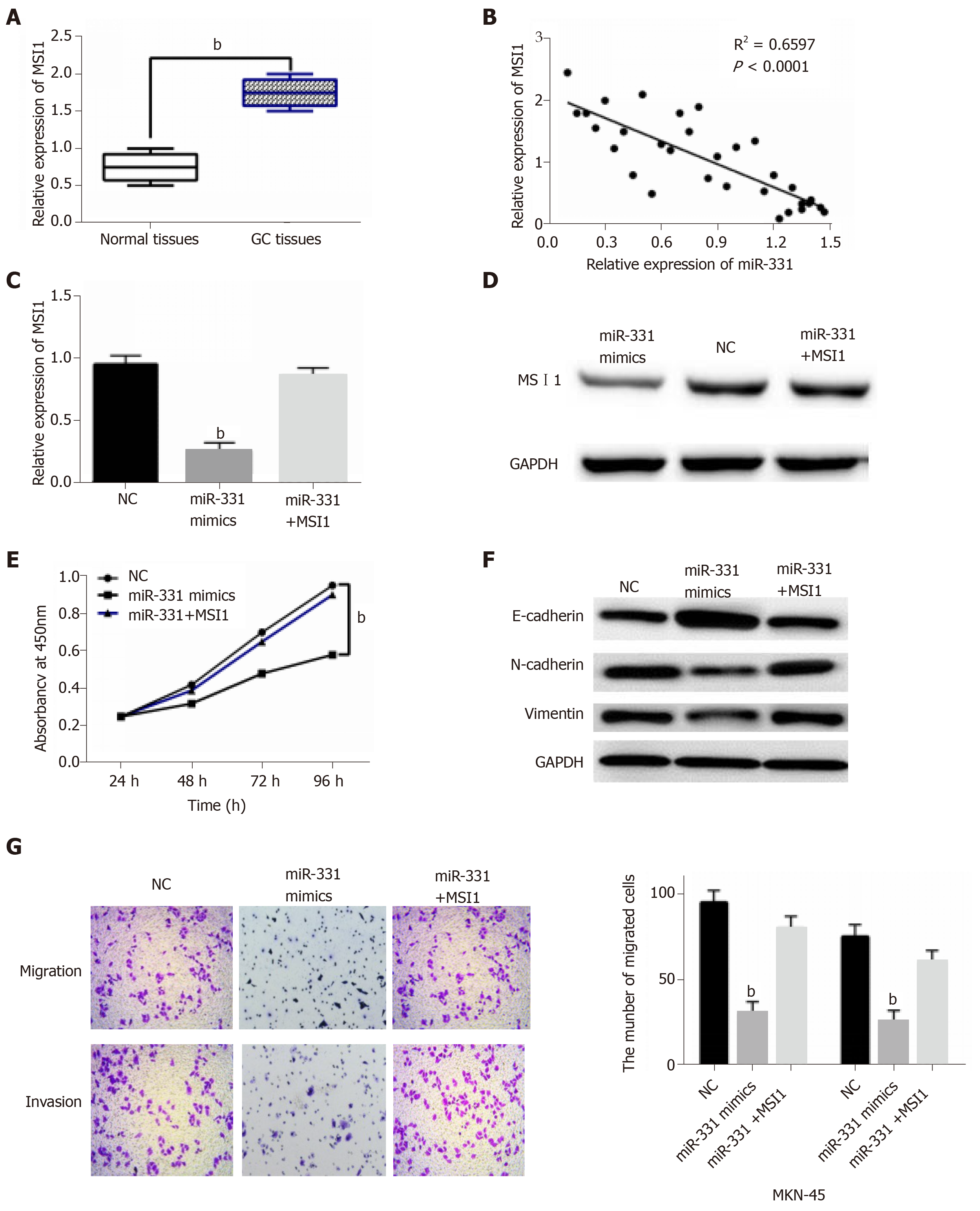
In GC tissues, the abnormal MSI1 expression was then observed. MSI1 expression was dramatically increased in GC tissues in comparison with normal tissues (Figure 5A). Moreover, miR-331 expression was identified to negatively regulate MSI1 expression in GC tissues (Figure 5B). It indicates that the interaction between miR-331 and MSI1 may exist in GC. MiR-331 mimics and MSI1 vector were co-transfected into MKN-45 cells to verify the above conjecture. As we suspected, the reduction of MSI1 expression induced by miR-331 mimics was recovered by MSI1 vector in MKN-45 cells (Figure 5C and 5D). Functionally, MSI1 vector impaired the inhibitory effect of miR-331 on cell proliferation (Figure 5E). Upregulation of MSI1 also restored the inhibitory effect of miR-331 on EMT in MKN-45 cells (Figure 5F). For cell migration and invasion, the suppressive effect of miR-331 was impaired by upregulation of MSI1 (Figure 5G). Taken together, upregulation of MSI1 weakened the inhibitory effect of miR-331 on GC progression.
As a new research hotspot, the regulatory mechanism of miRNAs in GC has been widely reported, indicating the strong potential of miRNA in the future treatment of GC. In our research, miR-331 was downregulated in GC, which predicted poor prognosis of GC patients. Functionally, miR-331 acted as an inhibitor in GC through suppressing cell proliferation, metastasis and tumor growth. Further, miR-331 directly targets MSI1 and inversely regulates its expression. Furthermore, upregulation of MSI1 impaired the inhibitory effect of miR-331 in GC. Briefly, miR-331 inhibited GC progression through targeting MSI1.
In many human cancers, the deregulation of miR-331 is involved in their tumorigenesis. Decreased miR-331 expression has been identified in colorectal cancer, prostate cancer and glioblastoma[13,22,23]. The biological function of miR-331 was also investigated in previous studies. For example, the inhibition of cell proliferation induced by miR-331 had been observed in urothelial carcinoma[24]. Chen et al[25] proposed that miR-331 inhibited melanoma cell proliferation and invasion through regulating AEG-1. Moreover, miR-331 overexpression reduced PCa cell migration and invasion, as well as xenograft tumor initiation[23]. Consistent with above previous studies, the inhibitory effect of miR-331 was also confirmed in GC. In addition, we also found that overexpression of miR-331 blocked EMT in GC cells, which has not been reported in other studies. These findings indicate that miR-331 can be used as a suppressor in GC progression.
Further, many target genes of miR-331 were confirmed in different cancers, such as ST7L and ERBB-2[26,27]. However, as far as we know, there is no research about the relationship between miR-331 and MSI1. Here, MSI1 was verified as a direct target of miR-331. And MSI1 expression was increased in GC tissues. The upregulation of MSI1 had also been found in GC tissues in previous report, which further confirm our results[28]. Moreover, overexpression of MSI1 was related to GC progression and poor prognosis in GC patients[29], indicating that MSI1 was involved in the development of GC. Additionally, upregulation of MSI1 has also been detected in cervical cancer, which promoted cell proliferation and tumor growth[30]. It is reasonable to know that miR-331 inhibits cell viability and metastasis via targeting MSI1 in GC. In line with previous studies, we testified above results in current research. Upregulation of MSI1 was found to weaken the inhibitory effect of miR-331 in GC.
In summary, it is firstly proposed the downregulation of miR-331 that associated with poor prognosis in GC patients in current research. Further, miR-331 directly targets MSI1 and inversely regulates its expression. Moreover, miR-331 inhibited cell viability and metastasis through targeting MSI1 in GC. Therefore, miR-331 may be a potential therapeutic target for GC.
As one of the most frequent cancers, gastric cancer (GC) caused more than 700000 deaths in just 2012 worldwide. Although the molecular mechanisms involved in microRNAs (miRNAs) have been extensively investigated in GC, how miR-331 regulates GC pathogenesis remains unknown.
To find more molecular mechanism or biomarker for diagnosis and treatment of GC
This study aims to explore the anti-cancer effect of miR-331 in GC and investigate its molecular mechanism against GC cells.
MiR-331 expression was observed by qRT-PCR assay in GC tissues and cell lines. MiR-331 mimic or inhibitor was transfected into MKN-45 cells to perform gain-loss experiment to observe effect of miR-331 on GC cell viability and migration. Bioinformatics analysis is used to predict the target gene of miR-331. The antagonistic effect between and MSI1 was confirmed by gain-loss experiment and detection of proliferation and migration. The expression of crucial proteins was measured by western blotting.
We found that downregulation of miR-331 was associated with poor prognosis in GC. In addition, miR-331 significantly inhibited GC cell growth, migration and invasion. Further, MSI1 was verified to directly target miR-331 and can effectively be regulated in GC tissues. Furthermore, upregulation of MSI1 weakened the inhibitory effect of miR-331 in GC. Western blotting analysis showed that E-cadherin, N-cadherin and Vimentin expression markedly affected by miR-331 and MSI1 in GC cell line, suggesting that EMT is a very direct regulated target of miR-331 and MSI1 in GC.
Our study demonstrated that miR-331 can significantly inhibit GC cell growth, migration and invasion. Furthermore, it can work through MSI1. Therefore, our study provides some molecular mechanism and two new biomarkers for GC.
In the future, research may reveal the important role of miR-331 that enhances the sensitivity of GC detection and further develop for its application in anti-cancer treatments. The identification of the miR-331/MSI1 molecular axis may further explain the underlying mechanism.
Thank Professor Ya-Ling Liu (Department of Pathology, Taishan Medical College) and Professor Ji-Xue Shi (Emergency Department, Affiliated Hospital of Taishan Medical College) for their selfless help. From topic selection, experiment design, result analysis to article writing and paper revision, it provides careful guidance and help. I would like to express my deep gratitude.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chivu-Economescu M, de Melo FF, Matsuo Y S-Editor: Gong ZM L-Editor: Filipodia E-Editor: Qi LL
| 1. | Zheng Y, Xu D, Bu Z. Chinese version of NCCN Clinical Practice Guidelines in Oncology officially authorized by NCCN. Chin J Cancer Res. 2016;28:144-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 2. | Zong L, Abe M, Seto Y, Ji J. The challenge of screening for early gastric cancer in China. Lancet. 2016;388:2606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 271] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 3. | Qi X, Liu Y, Wang W, Cai D, Li W, Hui J, Liu C, Zhao Y, Li G. Management of advanced gastric cancer: An overview of major findings from meta-analysis. Oncotarget. 2016;7:78180-78205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 4. | Kishi K, Fujiwara Y, Yano M, Motoori M, Sugimura K, Takahashi H, Ohue M, Sakon M. Usefulness of diagnostic laparoscopy with 5-aminolevulinic acid (ALA)-mediated photodynamic diagnosis for the detection of peritoneal micrometastasis in advanced gastric cancer after chemotherapy. Surg Today. 2016;46:1427-1434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 5. | Xu Z, Li C, Qu H, Li H, Gu Q, Xu J. MicroRNA-195 inhibits the proliferation and invasion of pancreatic cancer cells by targeting the fatty acid synthase/Wnt signaling pathway. Tumor Biol. 2017;39:101042831771132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Chen XB, Li W, Chu AX. MicroRNA-133a inhibits gastric cancer cells growth, migration, and epithelial-mesenchymal transition process by targeting presenilin 1. J Cell Biochem. 2019;120:470-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 7. | He Y, Ge Y, Jiang M, Zhou J, Luo D, Fan H, Shi L, Lin L, Yang L. MiR-592 Promotes Gastric Cancer Proliferation, Migration, and Invasion Through the PI3K/AKT and MAPK/ERK Signaling Pathways by Targeting Spry2. Cell Physiol Biochem. 2018;47:1465-1481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 8. | Chen L, Chu F, Cao Y, Shao J, Wang F. Serum miR-182 and miR-331-3p as diagnostic and prognostic markers in patients with hepatocellular carcinoma. Tumour Biol. 2015;36:7439-7447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 9. | Papadopoulos EI, Papachristopoulou G, Ardavanis A, Scorilas A. A comprehensive clinicopathological evaluation of the differential expression of microRNA-331 in breast tumors and its diagnostic significance. Clin Biochem. 2018;60:24-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | Cao Y, Chen J, Wang D, Peng H, Tan X, Xiong D, Huang A, Tang H. Upregulated in Hepatitis B virus-associated hepatocellular carcinoma cells, miR-331-3p promotes proliferation of hepatocellular carcinoma cells by targeting ING5. Oncotarget. 2015;6:38093-38106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 11. | Fujii T, Shimada K, Tatsumi Y, Tanaka N, Fujimoto K, Konishi N. Syndecan-1 up-regulates microRNA-331-3p and mediates epithelial-to-mesenchymal transition in prostate cancer. Mol Carcinog. 2016;55:1378-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 12. | Gu J, Zhang J, Zheng L, Ajani JA, Wu X, Ye Y. Serum miR-331-3p predicts tumor recurrence in esophageal adenocarcinoma. Sci Rep. 2018;8:14006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 13. | Epis MR, Giles KM, Candy PA, Webster RJ, Leedman PJ. miR-331-3p regulates expression of neuropilin-2 in glioblastoma. J Neurooncol. 2014;116:67-75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 14. | Guo X, Guo L, Ji J, Zhang J, Zhang J, Chen X, Cai Q, Li J, Gu Q, Liu B, Zhu Z, Yu Y. miRNA-331-3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochem Biophys Res Commun. 2010;398:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 92] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 15. | Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67-81. [PubMed] |
| 16. | Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 284] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 17. | Liu DC, Yang ZL, Jiang S. Identification of musashi-1 and ALDH1 as carcinogenesis, progression, and poor-prognosis related biomarkers for gallbladder adenocarcinoma. Cancer Biomark. 2010;8:113-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Wang XY, Penalva LO, Yuan H, Linnoila RI, Lu J, Okano H, Glazer RI. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol Cancer. 2010;9:221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 115] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 19. | Fan LF, Dong WG, Jiang CQ, Xia D, Liao F, Yu QF. Expression of putative stem cell genes Musashi-1 and beta1-integrin in human colorectal adenomas and adenocarcinomas. Int J Colorectal Dis. 2010;25:17-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Wang T, Ong CW, Shi J, Srivastava S, Yan B, Cheng CL, Yong WP, Chan SL, Yeoh KG, Iacopetta B, Salto-Tellez M. Sequential expression of putative stem cell markers in gastric carcinogenesis. Br J Cancer. 2011;105:658-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 21. | Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, Bishnupuri KS, Natarajan G, Anant S, Houchen CW. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448-1458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 138] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 22. | Zhao D, Sui Y, Zheng X. MiR-331-3p inhibits proliferation and promotes apoptosis by targeting HER2 through the PI3K/Akt and ERK1/2 pathways in colorectal cancer. Oncol Rep. 2016;35:1075-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 23. | Epis MR, Giles KM, Beveridge DJ, Richardson KL, Candy PA, Stuart LM, Bentel J, Cohen RJ, Leedman PJ. miR-331-3p and Aurora Kinase inhibitor II co-treatment suppresses prostate cancer tumorigenesis and progression. Oncotarget. 2017;8:55116-55134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 24. | Morita K, Fujii T, Itami H, Uchiyama T, Nakai T, Hatakeyama K, Sugimoto A, Miyake M, Nakai Y, Tanaka N, Shimada K, Yamazaki M, Fujimoto K, Ohbayashi C. NACC1, as a Target of MicroRNA-331-3p, Regulates Cell Proliferation in Urothelial Carcinoma Cells. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 25. | Chen L, Ma G, Cao X, An X, Liu X. MicroRNA-331 inhibits proliferation and invasion of melanoma cells by targeting astrocyte-elevated gene-1. Oncol Res. 2018; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 26. | Chen X, Luo H, Li X, Tian X, Peng B, Liu S, Zhan T, Wan Y, Chen W, Li Y, Lu Z, Huang X. miR-331-3p Functions as an Oncogene by Targeting ST7L in Pancreatic Cancer. Carcinogenesis. 2018; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 27. | Epis MR, Giles KM, Barker A, Kendrick TS, Leedman PJ. miR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J Biol Chem. 2009;284:24696-24704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 131] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 28. | Kuang RG, Kuang Y, Luo QF, Zhou CJ, Ji R, Wang JW. Expression and significance of Musashi-1 in gastric cancer and precancerous lesions. World J Gastroenterol. 2013;19:6637-6644. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 14] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 29. | Shou Z, Jin X, He X, Zhao Z, Chen Y, Ye M, Yao J. Overexpression of Musashi-1 protein is associated with progression and poor prognosis of gastric cancer. Oncol Lett. 2017;13:3556-3566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 30. | Liu X, Yang WT, Zheng PS. Msi1 promotes tumor growth and cell proliferation by targeting cell cycle checkpoint proteins p21, p27 and p53 in cervical carcinomas. Oncotarget. 2014;5:10870-10885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |