Published online Dec 15, 2019. doi: 10.4251/wjgo.v11.i12.1218
Peer-review started: March 4, 2019
First decision: June 5, 2019
Revised: July 9, 2019
Accepted: October 3, 2019
Article in press: October 3, 2019
Published online: December 15, 2019
Processing time: 284 Days and 4 Hours
Artificial intelligence, such as convolutional neural networks (CNNs), has been used in the interpretation of images and the diagnosis of hepatocellular cancer (HCC) and liver masses. CNN, a machine-learning algorithm similar to deep learning, has demonstrated its capability to recognise specific features that can detect pathological lesions.
To assess the use of CNNs in examining HCC and liver masses images in the diagnosis of cancer and evaluating the accuracy level of CNNs and their performance.
The databases PubMed, EMBASE, and the Web of Science and research books were systematically searched using related keywords. Studies analysing pathological anatomy, cellular, and radiological images on HCC or liver masses using CNNs were identified according to the study protocol to detect cancer, differentiating cancer from other lesions, or staging the lesion. The data were extracted as per a predefined extraction. The accuracy level and performance of the CNNs in detecting cancer or early stages of cancer were analysed. The primary outcomes of the study were analysing the type of cancer or liver mass and identifying the type of images that showed optimum accuracy in cancer detection.
A total of 11 studies that met the selection criteria and were consistent with the aims of the study were identified. The studies demonstrated the ability to differentiate liver masses or differentiate HCC from other lesions (n = 6), HCC from cirrhosis or development of new tumours (n = 3), and HCC nuclei grading or segmentation (n = 2). The CNNs showed satisfactory levels of accuracy. The studies aimed at detecting lesions (n = 4), classification (n = 5), and segmentation (n = 2). Several methods were used to assess the accuracy of CNN models used.
The role of CNNs in analysing images and as tools in early detection of HCC or liver masses has been demonstrated in these studies. While a few limitations have been identified in these studies, overall there was an optimal level of accuracy of the CNNs used in segmentation and classification of liver cancers images.
Core tip: Artificial intelligence, such as convolutional neural networks (CNNs), have been used in the interpretation of images, including pathology and radiology images with potential application in the diagnosis of hepatocellular cancer (HCC) and liver masses. CNN, a machine-learning algorithm similar to deep learning, has demonstrated its capability to recognise specific features that can detect pathological lesions. The primary aim of this review is to assess the use of CNNs in examining HCC and liver masses images in the diagnosis of cancer. The second aim is to evaluate the accuracy level of CNNs and their clinical performance.
- Citation: Azer SA. Deep learning with convolutional neural networks for identification of liver masses and hepatocellular carcinoma: A systematic review. World J Gastrointest Oncol 2019; 11(12): 1218-1230
- URL: https://www.wjgnet.com/1948-5204/full/v11/i12/1218.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i12.1218
Significant progress has been made in image recognition primarily due to the recent revival of deep learning, particularly the convolutional neural network (CNN), a class of artificial neural networks that have been widely used in biomedical and clinical research[1]. For example, the potential use of CNNs has been shown in the detection of gastrointestinal bleeding in wireless capsule endoscopy images using handcrafted and CNN features[2], diagnosis of Helicobacter pylori infection based on endoscopy images[3,4], and detection of gastrointestinal polyps using endoscopy images[5,6]. There is also a surge of interest in the potential of CNNs in radiology research[1,7] and in cellular and histopathological examinations[8]. Several studies have shown the ability of the CNN algorithms in (1) Lesion detection, a prevalent task for endoscopists, radiologists, and pathologists to detect abnormalities with medical images. These include the detection of colonic polyps, the detection of lesions on radiological images, and the detection of histopathological malignant changes on biopsy images[1,5-9]. CNN algorithms are also useful for (2) Classification, the CNNs utilise target lesions depicted in medical images, and these lesions are classified into classes. One of these is classifying lesions into particular categories (lesions or normal; malignant or benign). Other examples may include classification of precancerous gastric disease using the CNNs[10] or classification of skin cancer[11]. Therefore, the task is to determine “optimal” boundaries for separating classes in the multi-dimensional feature space that is formed by input features. At least three significant techniques have been described that use CNNs for medical image classifications, including training the CNNs from scratch[12], using “off-the-shelf CNN” features as complementary information channels to existing hand-crafted image features[13], and performing unsupervised pre-training on natural or medical images and fine-tuning using deep learning models[14]. (3) Segmentation of organs or anatomical structures is a functional image processing technique for the analysis of medical images such as quantitative evaluation of clinical parameters and computer-aided diagnosis system[15]; and (4) Image reconstruction, which may include obtaining a noiseless computed tomography (CT) image reconstructed from a subsampled sonogram[16]. With the above information in mind, this study aims at reviewing and identifying the applications and uses of CNNs in the interpretation of liver cancers, including hepatocellular carcinoma (HCC), liver metastasis (secondaries), and other liver masses.
Primary liver cancer, mainly HCC, is the fifth most common cancer in men and the seventh most common in women and is the third leading cause of cancer-related death worldwide[17]. In general, the disease is less common in females, and in most areas in the world, the male to female liver cancer rates are two- to three-fold higher, possibly due to the higher prevalence of risk factors in males and differences in sex steroid hormones, and perhaps epigenetic factors[18]. Studies showed that there is an increasing rate of HCC worldwide, which may be related partly to hepatitis B virus and hepatitis C virus infections, obesity, diabetes, metabolic syndrome, and non-alcoholic fatty infiltration of the liver[19].
However, the burden of HCC varies depending on geographical location. For example, in the Asia-Pacific region, it is a significant public health problem[20]. Because the liver is a common site of metastasis from cancers of other organs, mainly colorectal, gastric, pancreatic, breast, and lung cancers, secondaries to the liver add to the burden of liver cancer[21].
Currently and as per the guidelines of the American and European liver societies and World Gastroenterology Organisations, ultrasound is widely used in surveillance of HCCs, and CT and magnetic resonance images (MRI) are indicated to characterise a focal lesion suspected in the liver[22-24]. The diagnosis of HCC relies on either MR images or contrast-enhanced CT-scan, which enable the identification of up to 65% of small cell nodules < 2 mm in size[22-25]. However, the detection of small nodules is dependent on vascular dynamic enhancement pattern throughout the different phases of the study[26]. Also, there is inter-operator variability induced by visual qualitative assessment[27]. Therefore, the use of computer-aided diagnosis framework may enable us to resolve these limitations and enhance the diagnosing outcomes of these radiological modalities.
Based on these findings, this study aims to evaluate the use of CNN in HCC, liver metastasis (secondaries), or images of other liver masses. The rationales for the study were to assess the current status of convolutional neural networks and their applications in liver oncology images, identify gaps and deficiencies in ongoing research in this new field, particularly in relation to liver oncology images, and discuss future directions and research priorities that may maximise the applications of research in hepatic oncology. Therefore, our research questions are: (1) What is the current status of research output in the use of CNN in assessing HCC, liver metastases (secondaries), or images of other liver masses? and (2) What is the accuracy of CNN, deep learning systems, in lesion detection, classification, or segmentation of these images?
This manuscript was reported in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analysis guidelines[28].
The databases PubMed, EMBASE, and the Web of Science were searched for studies on CNNs in liver cancer and liver masses images. Also, research books that published full papers from conferences and scientific meetings were searched. The search covered studies up to January 2019. Only studies in the English language and conducted on humans were included. Studies on animals or animal models were not included. We searched for articles with contributions of the subject headings and the following key words: “Cancer”, “Liver”, “Hepatocellular carcinoma”, “HCC”, “Liver mass”, “Metastasis”, “Hepatic”, “Secondaries in liver”, “Radiology”, “Pathology”, “Histology”, “Histopathology”, “Malignancy”, “Primary liver cancer”, “Ultrasound”, “Computed tomography”, and “Magnetic resonance images”. To maximize the yield of the search, another search was performed manually by searching the list of references of the primary articles and reviews to identify studies not found by the database search[29].
We also searched the journals listed by the Journal Citation Reports-2017 of the Web of Science under the categories Gastroenterology and Hepatology (n = 32 journals), Oncology (n = 41 Journals), Radiology (n = 6 journals), Pathology (n = 14 journals), Computer Sciences and Engineering (n = 18 journals), and Medical Informatics (n = 7).
To identify targeted studies, we created a PICOS framework (Population, Intervention, Comparison, Outcome, Studies) for the inclusion and exclusion. Table 1 summarises the PICOS framework used. The following inclusion and exclusion criteria were used in selecting studies. Studies that reported data on the use of the CNNs in liver cancers images (HCC or liver metastasis/other liver masses) were included. Full research papers of conferences and scientific meetings were also considered if they fulfil the research purpose. The search was limited to studies in the English language and conducted on humans. Studies on animals or animal models were not included. Reviews, editorials, commentaries, letters to the Editors, abstracts published in conference proceedings, were not included.
| Population | Worldwide, datasets, patients, males and females, humans |
| Intervention | Use of CNN-diagnostic proposals and computer models |
| Comparison | Normal population, use of manual detection by radiologists, hepatologists, or anatomical pathologists examining images/slides. |
| Outcome | Lesion detection, classification, segmentation, or image reconstruction. |
| Studies | Controlled, or comparison to manual (routine) assessment, or comparison to benchmark or other artificial intelligence models. |
Two researchers (the author and a research assistant) independently reviewed the titles and abstracts of all citations identified by the literature search. Relevant studies were retrieved and reviewed in detail. Any disagreement was discussed by the two evaluators. The full texts of potentially relevant articles were sought, and the selection criteria were applied. Reviewers were not blinded to authors’ names or institutions. Studies were selected if they match the selection criteria.
Data were extracted independently by the two researchers using a predefined extraction form. The following data were abstracted in the form: (1) First author’s name; (2) Year of publication; (3) Objectives/research question; (4) Method used; (5) Liver cancer/liver masses investigated; (6) Main results; (7) Accuracy, sensitivity, and specificity of method used; and (8) Institute, university, city, and country where the study was conducted. Details on reported statistical associations and comparison of the results obtained with those obtained by using other methods were also evaluated. The agreement between evaluators measured by the degree of inter-rater agreement using Cohen kappa coefficient was also carried out using SPSS software (Armonk, NY, United States)[30].
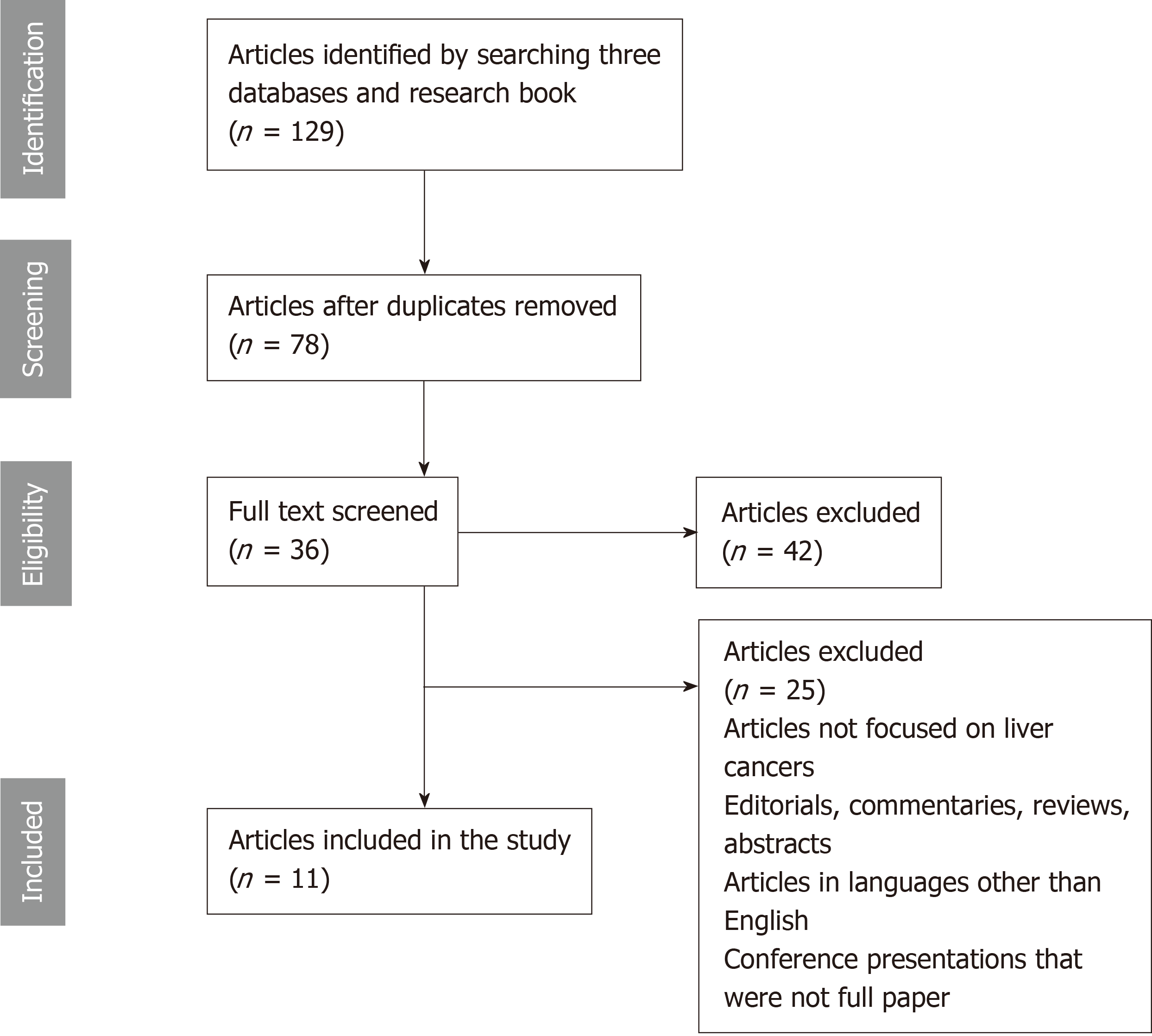
Figure 1 is a flow diagram summarizing the search results and selection process of articles. One hundred and twenty-nine potentially relevant publications were identified through the search of the three databases and research books. After removal of duplicates, 78 articles remained. Of these, 42 were not relevant to the inclusion criteria. Thirty-six full-text articles were assessed for eligibility. Finally, we identified 11 articles that met our selection criteria and were consistent with the aims of the systematic review[31-41].
Table 2 summarises details of the 11 studies included[31-41]. The studies demonstrated the ability of CNN models in analysed images of liver cancers as follows: Classification of liver masses into five categories: category A: Classic HCC; category B: Malignant liver tumour other than HCC; category C: intermediate masses (early HCC, dysplastic nodules, or benign liver masses; category D: Haemangiomas; category E: Cysts[31], detection of small metastasis in the liver[32], discrimination between primary liver cancer (HCC) and secondaries in the liver[33], differentiation between chronic liver diseases such as cirrhosis and the presence of HCC on top of cirrhosis[34], classification of grade of HCC nuclei and segmentation of HCC nuclei on pathology images[35,36], classification of liver lesions[31,33,34,37,41], and detection of liver tumour or liver masses and identification of their types and phases[38-40]. While these studies examined liver CT images[31,32,37-40], ultrasound images[34], and 3D multi-parameter MRI scan images[33,41], other images such as cellular and histopathological images were also included[35,36].
| Ref. | Research question/Purpose | Method used | Key findings | Conclusions |
| Yasaka et al[31], 2018 | Can deep learning techniques enable the classification of liver masses into categories? | A retrospective study. CNN together with dynamic contrast enhanced CT image set of liver masses were used over 3 phases (55 536 images obtained from 460 patients used in training and 100 liver mass image sets were used in testing). | The CNN model enabled classification of liver masses into 5 categories. The median area under the receiver operating characteristic curve to differentiate between categories was 0.92. | Deep learning with CNN enabled classification of liver masses and differentiate between 5 categories at dynamic CT with high accuracy. |
| Ben-Cohen et al[32], 2018 | Can a proposed CNN enable the detection of liver metastasis in CT examinations of the liver? | The method involved both a global context and a fully convolutional network and a local patch level analysis with superpixel sparse based classification. A total of 20 patients with a total of 68 lesions and a testing set with CT examinations from 14 patients with overall 55 lesions were included. | The true positive rate was 94.6% with 2.9 false positive per case. | The system enhanced the detection of small metastasis in the liver and is clinically promising. |
| Trivizakis et al[33], 2019 | Evaluate the use of a novel 3D CNN model in tissue classification of medical images and application for discriminating between primary and metastatic liver tumours. | The proposed model consisted of 4 consecutive strided 3D convolutional layers with 3 × 3 × 3 Kernel size and ReLU for activation functions. | The classification performance was 83% for 3D vs 69.6% and 65.2% for two 2D CNNs models; demonstrating significant tissue classification accuracy improvement compared to two 2D CNNs. | The proposed 3D CNN architecture can differentiate between primary and secondary liver tumours. |
| Bharti et al[34], 2018 | Test a proposed model to differentiate between chronic liver and cirrhosis and presence of hepatocellular carcinoma (HCC). | The system is based on higher order features, a hierarchical organisation and multi-resolution, which enabled the characterisation of echotexture and roughness of liver surface. | The proposed CNN feature was able to differentiate four liver stages acquired from ultrasound images. The classification accuracy of the model in differentiating between the four groups was 96.6%. | The model is able to differentiate between normal liver, chronic liver disease, cirrhosis and HCC evolved on top of cirrhosis. |
| Li et al[35], 2017 | Can a joint multiple fully connected CNN with extreme learning machine (MFC-CNN-ELM) architecture be used in hepatocellular carcinoma (HCC) nuclei grading? | Centre-proliferation segmentation (CPS) method was used and labels of grayscale image patch were marked under the guidance of three pathologists. A multiple fully convolutional neural network (MFC-CNN) was designed to extract the multi-form feature vectors of each input image automatically. Finally, the CNN-ELM model was used to grade HCC nuclei. | External validation using ICPR 2014 HEP-2 cell shows the good generalisation of MFC-CNN-EL architecture. | The proposed MFC-CNN-ELM has superior performance in grading HCC nuclei compared with related works. |
| Li et al[36], 2018 | Can a proposed structured convolutional extreme learning machine (SC-ELM) and case-based shape template (CBST) method used in HCC nucleus segmentation? | The model SC-ELM was developed for global segmentation of pathology images and is used for coarse segmentation. | The model was used for experimentation with 12.7 liver pathology images. | The proposed model demonstrated higher performance compared with related and published work. |
| Frid-Adar et al[37], 2018 | Can generated medical images be used for synthetic data augmentation and improving the performance of CNN medical image classification? | Using a limited data of CT images of 182 liver lesions (53 cysts, 64 metastasis and 65 haemangiomas), and generating synthetic medical images using recently presented deep learning Generative Adversarial Networks (GANs), the authors then used CNN to compare classification. Performance was compared with synthetic data augmentation and classic data augmentation. | The classification performance on using classic data augmentation yielded 78.6% sensitivity and 88.4% specificity. The synthetic data augmentation resulted in an increase to 85.7% for sensitivity and 92.4% for specificity. | The approach of synthetic data augmentation can be generalized to other classification applications. |
| Vivanti et al[38], 2018 | Can CNN-based method enable robust automatic liver tumour delineation in longitudinal CT studies? | The inputs are baseline scans and the tumour delineation, a follow-up scan, and a liver tumour global CNN. | Results from 222 tumours from 31 patients yielded an average overlap error of 17% (SD = 11.2) and surface distance of 2.1 mm (SD = 1.8) which is far better than stand-alone segmentation. | The method shows that follow-up framework yields accurate tumour tracking with a small training dataset. |
| Vivanti et al[39], 2017 | Automatic detection and segmentation of new tumours in longitudinal CT scan studies and for liver tumours burden quantification. | Inputs were the baseline and follow-up CT scans. The outputs were the new tumours segmentations in the follow-up scans, and measurement of tumour burden quantification and tumour burden change. | 246 tumours of which 97 were new tumours from 37 longitudinal liver CT studies, yielded positive new tumours detection rate of 86% vs 72% with stand-alone detection, and a tumour burden volume overlap error of 16%. | Compared to existing methods, this method enables accurate and reliable detection of known tumours and detection of new tumours in the follow-up scans. |
| Todoroki et al[40], 2018 | Can a deep CNN method detect liver masses regardless to their types and phases (stages) from CT images? | The proposed method is based on deep learning which used multi-layered CNN. The Tumour detection methods comprised two steps: (1) Segmentation of the liver from CT liver images; and (2) Calculation of probability of each pixel in the segmented liver. | 3D multi-phase contrast-enhanced CT liver images of 75 cases were used in the study. The cases comprised 5 types of lesions-cysts, focal nodular hyperplasia, hepatocellular carcinoma, haemangioma, and metastases, each type was represented in 15 cases. The detection results for each type was superior to detected boundaries by doctors and outperformed other methods. | The proposed deep CNN tumour detection method demonstrated an ability to discriminate between the 5 different types, and the performance outperformed that of other convolutional methods. |
| Zhang et al[41], 2018 | Can a proposed 3D multi-parameter magnetic resonance images and a novel deep CNN method classify different types of liver tissue in patients with hepatocellular carcinoma? | A novel deep CNN incorporating auto-context elements with U-net-like architecture was developed. The model uses multi-level hierarchical architecture and multi-phase training procedures. | Liver magnetic resonance images from 20 patients yielded promising results in classifying liver tissues. The method was compared with a benchmark method, multi-resolution input single-phase training and single-resolution-input, single-phase training and demonstrated higher discriminatory performance. | The multi-resolution input, the auto-context design, and the multi-phase training procedure have helped in improving the performance of the model. |
Geographically, these studies were performed in Japan (2), China (3), the United States (2), India (1), Greece (1), and Israel (4). Some papers were written by authors from two countries. Top universities, hospitals, and research institutes that led such research were: Department of Radiology, the University of Tokyo Hospital, Tokyo, Japan; Faculty of Engineering, Department of Biomedical Engineering Medical Image Processing Laboratory, University of Tel Aviv, Israel; Department of Informatics Engineering Technology Education Institute of Crete, Greece; Thapar Institute of Engineering and Technology, Patiala, India; Manipal Hospital, Bangalore, India; Software College, Northeastern University, Shenyang, China; Electric and Computer Engineering Stevens Institute of Technology, NJ, United States; Department of Diagnostic Images, the Chaim Sheba Medical Centre; School of Computer Science and Engineering, The Hebrew University of Jerusalem, Jerusalem, Israel; Information Science and Engineering, Ritsumeikan University, Shiga, Japan; Medical School, Zhejiang University, Hangzhou, China; Department of Biomedical Engineering, Yale University, CT, United States; and Department of Electrical Engineering, Yale University, CT, United States.
Description of reported methods: The description of the method used in these studies and the content described provided significant detail about the dataset, the generation of the CNN algorithm, the architecture used, the experiments carried out to assess system performance, system evaluation, and accuracy of automatic liver segmentation or classification[14,31-41]. However, in most studies identified no details were given regarding clinical information, such as number of patients included, sources and number of images used, and clinical procedures carried out. This imbalance in the methods described may be related to the background of the authors of these studies and the journals that published these studies. The 11 studies were published by 58 authors; of these, five were from radiology departments, one was from the pathology department, and two were possibly with medical background. The remaining 50 authors were non-medical and were from engineering, computer science, and medical image processing laboratory. Except for Radiology[31], and Ultrasonic Imaging[34], the majority of these articles were published in journals specialised in computer science and Biomedical informatics, such as Neurocomputing[32,36,37], IEEE Journal of Biomedical Health Informatics[33], Computers in Biology and Medicine[35], Medical and Biological Engineering and Computing[38], and International Journal of Computer Assisted Radiology and Surgery[39]. These two factors may explain the dominance of technical information regarding computer science information in the study methods.
Datasets and number of patients: The number of patients and images used in the studies varied significantly. Ben-Cohen et al[32] reported that their study involved 20 patients with 68 lesions in total and that they were tested on a testing set that included CT examinations for 14 patients with a total of 55 lesions[32]. Another study that used CT imaging was Frid-Adar et al[37], who reported a limited dataset of 182 liver lesions (53 cysts, 64 metastases, and 65 haemangiomas). The authors highlighted difficulties in obtaining datasets, and although there are medical datasets available online, they felt that they are still limited and only applicable to specific medical problems[37]. Also, the study by Todoroki et al[40] used 3D multi-phase contrast-enhanced liver CT images from 75 patients. The cases comprised five different lesions, namely-cysts, focal nodular hyperplasia, HCC, haemangioma, and metastases[40]. The study by Yasaka et al[31] is the fourth study that used contrast agent-enhanced CT images of the liver. The study was a retrospective study and used images of liver masses over three phases (non-contrast-agent enhanced, arterial, and delayed). The masses were grouped under five categories, as stated earlier[31]. On the other hand, the study by Trivizakis et al[33] examined upper abdominal MRI scans of 134 patients (37.7% primary liver mass and 62.3% metastatic lesion). The study by Zhang et al[41] also used MRI scan images of 20 patients, with HCC generating 1700 non-overlapping patches. Only one study used liver ultrasound images acquired from 94 patients. The images datasets comprised 48 normal liver, 50 chronic liver, 50 cirrhosis, and 41 HCC evolved over cirrhosis. Other datasets comprised experimental data involving 127 liver pathology images[35,36].
Methods and study design: Methods using CNN models to study HCC and liver masses (secondaries or metastases or other liver masses) in these 11 studies varied significantly. While the study by Ben-Cohn et al[32] used a global context with a fully convoluted network and a local patch level analysis with superpixel sparse based classification, the study by Trivizakis et al[33] proposed a CNN model comprising four consecutive strided 3D convolutional layers with 3 × 3 × 3 kernel size and ReLU as activation functions. The design has a fully connected layer with 2048 neurons and a soft max layer for binary classification. For training and validation, a dataset of 130 Diffusion Weighted MR images scans was used[33]. The method used by Bharti et al[34] aimed at examining ultrasound images through visual interpretation of liver images.
The accuracy measurements used in these studies varied and can be summarised as follows: (1) Assessing system performance and system evaluation[32,34]; (2) Assessing accuracy of automatic liver segmentation and lesion detection[32,34]; (3) Generating operating characteristics curves and precision recall[33]; (4) Comparing the method used outcomes with the results obtained from other CNNs[35,37,41]; (5) Measuring sensitivity, specificity, and accuracy parameters[37,38]; (6) Comparing accuracy of the deep CNN method with those measured by Bayesian model and the benchmark method[40,41]; (7) Comparing the outcomes with visual inspection by expert radiologists (precision and recall rates)[37]; and (8) Comparing the performance of the CNN-based system to non-network state-of-the-art methods for liver lesion classification[37]. Some studies did not measure sensitivity or specificity[34,40,41].
The studies demonstrated a medium accuracy of differential diagnosis of liver masses of 0.84 for test data and the under receiver operating characteristic curve was 0.92[31]. Ben-Cohen et al[32] reported satisfactory results and demonstrated that on using 3-fold cross validation, experiments resulted in a true positive rate of 94.6% with 2.9 false positive result per case. The classification performance of the work conducted by Trivizakis et al[33] was 83% for 3D vs 69.6% and 65.2% for 2D, which indicates significance of tissue classification accuracy compared to two 2D CNNs. Another classification CNN system of four liver stages by Bharti et al[34] had an accuracy of 96.6%. Other studies showed that the proposed method had superior performance when compared with related works by other techniques[35,36,40,41]. Li et al[35] showed that external validation of the proposed method multiple fully connected CNN with extreme learning machine model they created by using Hep-2 cells indicates that their method can be generalised in grading HCC nuclei. The work of Vivanti et al[39] showed an overlap error of 17% (SD = 11.2) and surface distance of 2.1 mm (SD = 1.8), which was far better than stand-alone segmentation. The proposed system did not require large annotated training datasets, which is an advantage compared to other systems[38]. Another study by Vivanti et al[39] showed that the experiments resulted in a yield of true positive new tumours detection rate of 86% versus 72% with stand-alone detection. The tumour burden volume overlap error was 16%, which means that the follow-up CT scans enable not only the detection of new tumours but also the estimation of tumour burden volumetric; both are required in the diagnosis and management of liver tumours[39].
The inter-rater agreement between evaluators had overall κ scores in the range of 0.779-0.894.
During the last 5-6 years a significant shift in research trends in image processing has taken place, and the use of artificial intelligence in the field has moved from hand crafted algorithms to deep learning architectures[14]. In this study, our aims were to assess the use of the CNNs in deep learning of liver cancer (HCC and secondaries in the liver or liver masses) images. In this review, 11 studies were identified. Because of the heterogeneous data, gaps in the reported results, and the variability in the design of methods, it was not possible to conduct a meta-analysis. Nevertheless, certain parameters were evident from these studies. First, deep learning architectures and particularly the CNNs have been usefully implemented into medical imaging domain. The studies have shown the accuracy of the CNNs in analysing and offering the diagnosis (segmentation, and classification) of radiology images (CT scans, ultrasound, and MRI scans) and histopathology and cellular images of liver cancers. Although the number of full original articles on each cancer was small, the 11 studies were published in the years 2017 and 2018; reflecting the fact that deep learning as outlined in the CNNs and their applications in medical sciences is a recently developed discipline[42]. Second, although the technical information and the description of the test data preparation are important, the training and evaluation algorithm development are vital, and information about the sources of images, the patients involved in the study, and the clinical information collected is equally important. In this review, it was noted that the methods used varied and were not standardised. While these differences might be related to differences in the focus of the journals that published these studies and the background of the authors (medical vs engineering), there is a need for more studies that equally represent authors from medical, computer science, and engineering and address both aspects in the methods and results to maximise the research readability and applications of publications across disciplines involved. Third, it would be of interest to assess the performance of different research articles that were validated on the same datasets; however, most studies did not provide enough information about the exact sources of their datasets and it was not possible to trace which studies used the same testing protocol[43]. Such information is vital for comparison and assessment. We hope that journals interested in publishing such studies on deep learning, the CNNs, and artificial intelligence develop standardized guidelines that require authors to state such information, including details about the sources of datasets and protocols used in testing.
This study, however, is not free from limitations. Considering this diversity and lesions included, we must interpret the findings with caution. There could be publication bias that precluded the publication of negative studies. Studies included were only those published in the English language, and there could be good articles related to this topic that were published in other languages. However, this study presents what has been accomplished in this area in the English language literature. Studies varied in terms of patient type, study design, design of the CNN models, pathology of diseases included, and details of methods used[44].
This study highlights a number of future directions for research that uses CNNs to interpret liver cancer images. These can be summarised as follows: First, while the improvement in the design of the CNNs has required relatively small training sets of images, as it is the case with the models presented by Vivanti et al[38,39], there is a need for multi-institute and multicentre collaborations in studies including a large number of patients with cirrhosis due to different pathological causes and patients with HCC on top of cirrhosis, liver secondaries, or liver masses. Such collaborations could resolve concerns about the insufficient amount of training data in the medical image domain and enable measurement of accuracy and performance of the CNNs algorithms. Several researchers reported difficulties in obtaining images, which make the direct application of machine learning algorithms inappropriate for medical datasets and hence affect the capacity to conduct image classification or image segmentation with high accuracy[42,44]. Second, differentiation between primary and secondary (metastases in the liver) or other liver masses is difficult on the basis of radiological imaging. The studies included in this review showed that CT-based deep learning methods can enable the categorisation of liver metastases[32,33]. Although such differentiation between primary and secondary liver tumours may be clinically useful, other priorities that we need to explore further are: (1) Examination of HCC on top of liver cirrhosis to observe the characteristics of HCC on cirrhosis and non-cirrhotic liver tissue[34]; and (2) Conducting longitudinal liver CT scan studies and comparing outcomes with existing stand-alone and follow-up methods. We hypothesise that longitudinal studies could enable researchers to compare changes with the baseline scan and thus could offer a better detection of new small tumours[38,39]. Third, we need case control studies where the use of CNN could be compared with manual assessment of images by experts, radiologists, hepatologists, and pathologists. One of the major challenges we face with the use of CNN is the difficulty in choosing discriminant features to represent the clinical characteristics and using them as the key features in the CNN algorithm in segmentation and in classification functions. Again, this goal cannot be achieved without the collaboration of medical experts, radiologists, pathologists, computer engineering programmers, and experts designing these systems. Fourth, future studies should give more attention to the assessment of accuracy and sensitivity of the CNNs in evaluating the performance of systems and calculating the positive predictive values. Ideally, a study should use two or three different methods and compare accuracy parameters for the same set of images using these different methods. Currently, we are lacking such studies in the literature, and so any comparison of accuracy is not optimum because of several variables interfering with the methods/models reported[45].
With increasing research applying CNNs in liver cancer images there is a demand to evaluate carefully their accuracy and define future research directions. While current studies have covered major liver cancers, the number of studies conducted so far is small and limited, and more research is needed to answer questions about the accuracy and sensitivity of the CNN algorithms. The CNNs have demonstrated abilities in segmentation, classification, and lesion detection of radiological and anatomical pathology images of common cancers. However, several deficiencies in current studies were observed. In most studies there was no balance in the content of methods among the description of patients involved, the medical component, and the technical computer related component. Furthermore, comparing the use of the CNNs with other models is needed particularly in regard to accuracy and sensitivity of each model on the same set of images. Therefore, future studies that focus on these areas should be multi-institute and the outcome of multicentre collaborations should include with a large number of patients. This is particularly important in view of the growing demand of CNNs in liver oncology.
This study highlighted several aspects related to convolutional neural networks (CNNs). First, CNNs have potential use in identifying HCC and differentiating HCC from other liver masses with high accuracy. Second, CNNs can offer several functions concerning liver cancer, including lesion detection, classification, and segmentation. Third, the use of CNN in liver cancer is not limited to radiological images, but it is of value in pathological and cellular studies. However, the study identified several limitations in the literature in this area, mainly the smaller number of studies on the topic and the lack of studies from multi-centres as well as the lack of longitudinal liver computed tomography (CT) scan studies that can enable comparing outcomes with existing stand-alone and follow-up methods. These longitudinal studies could allow researchers to compare changes with the baseline scan and thus could offer better detection of new small liver tumours.
Recently, an increasing interest in the use of deep learning has emerged in research, particularly CNNs, a class of artificial intelligence that has been widely used in biomedical research. This study reviews the current literature on the use of CNNs in assessing hepatocellular carcinoma (HCC) and liver masses and how such advanced technology can help improve clinical diagnosis.
While the study focuses on an evolving field in gastroenterology and oncology with promising outcomes, several researchers reported difficulties in obtaining images, which make the direct application of machine learning algorithms inappropriate for medical datasets and hence affect the capacity to conduct image classification with high accuracy. Therefore, improvement in the design of CNNs and multi-institute and multi-centre collaborations with a large number of patients with cirrhosis due to different pathological causes and patients with HCC on top of cirrhosis or liver secondaries is needed.
The study aimed at assessing the use of CNNs in examining HCC and liver masses images in the diagnosis of cancer and evaluating the accuracy level of the CNNs and their performance.
Several databases, including PubMed, EMBASE, and Web of Science, were systematically searched for studies that covered pathological anatomy, cellular, and radiological images on HCC or liver masses using the CNNs. The data were extracted as per a predefined extraction protocol, and the accuracy level and performance of the CNNs in detecting cancer or early stages of cancer were analysed. The primary outcomes of the study were investigating the type of cancer or liver mass and identifying the type of images that showed optimum accuracy in cancer detection.
A small number of studies were identified. The studies demonstrated the ability to differentiate liver masses, differentiate HCC from other liver lesions, and differentiate HCC from cirrhosis or development of new tumours. Two studies focused on HCC nuclei grading or segmentation. In these studies, the CNNs showed satisfactory levels of accuracy. The studies aimed at detecting lesions, classification, and segmentation. Several methods were used to assess the accuracy of CNN models used.
While the current studies have covered liver cancers, the number of studies conducted so far is small and limited, and more research is needed to answer questions about the accuracy and sensitivity of the CNN algorithms. The CNNs demonstrated abilities in segmentation, classification, and lesion detection in radiological and anatomical pathology images of common cancers. However, several deficiencies in current studies were observed.
A large multi-centre trial is needed to evaluate carefully the use of CNNs and their clinical applications in HCC and liver masses. Differentiation between primary and secondary (metastases in the liver) or other liver masses is hard based on radiological imaging. The studies included in this review showed that CT-based deep learning methods could enable the categorisation of liver metastases from primary liver cancers. Future studies should give more attention to the assessment of accuracy and sensitivity of the CNNs in evaluating the performance of systems and calculating the positive predictive values.
The author would like to thank Dr Sarah Azer of St Vincent Hospital, University of Melbourne, for her help during writing this research article.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country of origin: Saudi Arabia
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Aykan NF S-Editor: Dou Y L-Editor: Filipodia E-Editor: Liu MY
| 1. | Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, Kadoury S, Tang A. Deep Learning: A Primer for Radiologists. Radiographics. 2017;37:2113-2131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 511] [Cited by in RCA: 689] [Article Influence: 98.4] [Reference Citation Analysis (0)] |
| 2. | Xiao Jia, Meng MQ. Gastrointestinal bleeding detection in wireless capsule endoscopy images using handcrafted and CNN features. Conf Proc IEEE Eng Med Biol Soc. 2017;2017:3154-3157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 3. | Shichijo S, Nomura S, Aoyama K, Nishikawa Y, Miura M, Shinagawa T, Takiyama H, Tanimoto T, Ishihara S, Matsuo K, Tada T. Application of Convolutional Neural Networks in the Diagnosis of Helicobacter pylori Infection Based on Endoscopic Images. EBioMedicine. 2017;25:106-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 4. | Itoh T, Kawahira H, Nakashima H, Yata N. Deep learning analyzes Helicobacter pylori infection by upper gastrointestinal endoscopy images. Endosc Int Open. 2018;6:E139-E144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 130] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 5. | Billah M, Waheed S. Gastrointestinal polyp detection in endoscopic images using an improved feature extraction method. Biomed Eng Lett. 2017;8:69-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 6. | Billah M, Waheed S, Rahman MM. An Automatic Gastrointestinal Polyp Detection System in Video Endoscopy Using Fusion of Color Wavelet and Convolutional Neural Network Features. Int J Biomed Imaging. 2017;2017:9545920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 7. | Fu Y, Mazur TR, Wu X, Liu S, Chang X, Lu Y, Li HH, Kim H, Roach MC, Henke L, Yang D. A novel MRI segmentation method using CNN-based correction network for MRI-guided adaptive radiotherapy. Med Phys. 2018;45:5129-5137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 8. | Sirinukunwattana K, Ahmed Raza SE, Yee-Wah Tsang, Saltz JH, Snead DR, Cree IA, Rajpoot NM. Locality Sensitive Deep Learning for Detection and Classification of Nuclei in Routine Colon Cancer Histology Images. IEEE Trans Med Imaging. 2016;35:1196-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 784] [Cited by in RCA: 510] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 9. | Wen S, Kurc TM, Hou L, Saltz JH, Gupta RR, Batiste R, Zhao T, Nguyen V, Samaras D, Zhu W. Comparison of Different Classifiers with Active Learning to Support Quality Control in Nucleus Segmentation in Pathology Images. AMIA Jt Summits Transl Sci Proc. 2018;2017:227-236. [PubMed] |
| 10. | Zhang X, Hu W, Chen F, Liu J, Yang Y, Wang L, Duan H, Si J. Gastric precancerous diseases classification using CNN with a concise model. PLoS One. 2017;12:e0185508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5683] [Cited by in RCA: 5353] [Article Influence: 669.1] [Reference Citation Analysis (0)] |
| 12. | Menze BH, Jakab A, Bauer Sl, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J. Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp Ç, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SM, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Van Leemput K. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging. 2015;34:1993-2024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3112] [Cited by in RCA: 2001] [Article Influence: 200.1] [Reference Citation Analysis (0)] |
| 13. | Ciompi F, de Hoop B, van Riel SJ, Chung K, Scholten ET, Oudkerk M, de Jong PA, Prokop M, van Ginneken B. Automatic classification of pulmonary peri-fissural nodules in computed tomography using an ensemble of 2D views and a convolutional neural network out-of-the-box. Med Image Anal. 2015;26:195-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 151] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 14. | Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging. 2016;35:1285-1298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3594] [Cited by in RCA: 1921] [Article Influence: 213.4] [Reference Citation Analysis (0)] |
| 15. | Zhang Y, Chandler DM, Mou X. Quality Assessment of Screen Content Images via Convolutional-Neural-Network-Based Synthetic/Natural Segmentation. IEEE Trans Image Process. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Mahmood F, Durr NJ. Deep learning and conditional random fields-based depth estimation and topographical reconstruction from conventional endoscopy. Med Image Anal. 2018;48:230-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 17. | Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 400] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 18. | Zheng B, Zhu YJ, Wang HY, Chen L. Gender disparity in hepatocellular carcinoma (HCC): multiple underlying mechanisms. Sci China Life Sci. 2017;60:575-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 89] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 19. | Margini C, Dufour JF. The story of HCC in NAFLD: from epidemiology, across pathogenesis, to prevention and treatment. Liver Int. 2016;36:317-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 183] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 20. | Omata M, Cheng AL, Kokudo N, Kudo M, Lee JM, Jia J, Tateishi R, Han KH, Chawla YK, Shiina S, Jafri W, Payawal DA, Ohki T, Ogasawara S, Chen PJ, Lesmana CRA, Lesmana LA, Gani RA, Obi S, Dokmeci AK, Sarin SK. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int. 2017;11:317-370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1628] [Cited by in RCA: 1639] [Article Influence: 204.9] [Reference Citation Analysis (0)] |
| 21. | Lubienski A, Leibecke T, Lubienski K, Helmberger T. Liver metastases. Recent Results Cancer Res. 2006;167:79-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 22. | Mitchell DG, Bruix J, Sherman M, Sirlin CB. LI-RADS (Liver Imaging Reporting and Data System): summary, discussion, and consensus of the LI-RADS Management Working Group and future directions. Hepatology. 2015;61:1056-1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 362] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 23. | Tang A, Cruite I, Sirlin CB. Toward a standardized system for hepatocellular carcinoma diagnosis using computed tomography and MRI. Expert Rev Gastroenterol Hepatol. 2013;7:269-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Ferenci P, Fried M, Labrecque D, Bruix J, Sherman M, Omata M, Heathcote J, Piratsivuth T, Kew M, Otegbayo JA, Zheng SS, Sarin S, Hamid S, Modawi SB, Fleig W, Fedail S, Thomson A, Khan A, Malfertheiner P, Lau G, Carillo FJ, Krabshuis J, Le Mair A; World Gastroenterology Organisation Guidelines and Publications Committee. World Gastroenterology Organisation Guideline. Hepatocellular carcinoma (HCC): a global perspective. J Gastrointestin Liver Dis. 2010;19:311-317. [PubMed] |
| 25. | Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5972] [Cited by in RCA: 6572] [Article Influence: 469.4] [Reference Citation Analysis (1)] |
| 26. | Iavarone M, Piscaglia F, Vavassori S, Galassi M, Sangiovanni A, Venerandi L, Forzenigo LV, Golfieri R, Bolondi L, Colombo M. Contrast enhanced CT-scan to diagnose intrahepatic cholangiocarcinoma in patients with cirrhosis. J Hepatol. 2013;58:1188-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 27. | Dubus L, Gayet M, Zappa M, Abaleo L, De Cooman A, Orieux G, Vilgrain V. Comparison of semi-automated and manual methods to measure the volume of liver tumours on MDCT images. Eur Radiol. 2011;21:996-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 28. | Miao ZF, Liu XY, Wang ZN, Zhao TT, Xu YY, Song YX, Huang JY, Xu H, Xu HM. Effect of neoadjuvant chemotherapy in patients with gastric cancer: a PRISMA-compliant systematic review and meta-analysis. BMC Cancer. 2018;18:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 29. | Azer SA, Azer D. Group interaction in problem-based learning tutorials: a systematic review. Eur J Dent Educ. 2015;19:194-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 30. | Wagner WE. Using Ibm® Spss® statistics for research methods and social science statistics. 6th Edition. California. SAGE Publication, 2017. . |
| 31. | Yasaka K, Akai H, Abe O, Kiryu S. Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology. 2018;286:887-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 393] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 32. | Ben-Cohen A, Klang E, Kerpel A, Konen E, Amitai M, Greenspan H. Fully convolutional network and sparsity-based dictionary learning for liver lesion detection in CT examinations. Neurocomputing. 2018;275, 1585-1594. [RCA] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 33. | Trivizakis E, Manikis GC, Nikiforaki K, Drevelegas K, Constantinides M, Drevelegas A, Marias K. Extending 2-D Convolutional Neural Networks to 3-D for Advancing Deep Learning Cancer Classification With Application to MRI Liver Tumor Differentiation. IEEE J Biomed Health Inform. 2019;23:923-930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 34. | Bharti P, Mittal D, Ananthasivan R. Preliminary Study of Chronic Liver Classification on Ultrasound Images Using an Ensemble Model. Ultrason Imaging. 2018;40:357-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 35. | Li S, Jiang H, Pang W. Joint multiple fully connected convolutional neural network with extreme learning machine for hepatocellular carcinoma nuclei grading. Comput Biol Med. 2017;84:156-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 36. | Li S, Jiang H, Yao Y-D, Pang W, Sun Q, Kuang L. Structure convolutional extreme learning machine and case-based shape template for HCC nucleus segmentation. Neurocomputing. 2018;312:9-26. [RCA] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Frid-Adar M, Diamant I, Klang E, Amitai M, Goldberger J, Greenspan H, Member, IEEE. GAN-based Synthetic Medical Image Augmentation for increased CNN Performance in Liver Lesion Classification. Neurocomputing. 2018;321:321-331. [RCA] [DOI] [Full Text] [Cited by in Crossref: 703] [Cited by in RCA: 554] [Article Influence: 79.1] [Reference Citation Analysis (0)] |
| 38. | Vivanti R, Joskowicz L, Lev-Cohain N, Ephrat A, Sosna J. Patient-specific and global convolutional neural networks for robust automatic liver tumor delineation in follow-up CT studies. Med Biol Eng Comput. 2018;56:1699-1713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 39. | Vivanti R, Szeskin A, Lev-Cohain N, Sosna J, Joskowicz L. Automatic detection of new tumors and tumor burden evaluation in longitudinal liver CT scan studies. Int J Comput Assist Radiol Surg. 2017;12:1945-1957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 40. | Todoroki Y, Han X-H, Iwamoto Y, Lin L, Hu H, Chen Y-W. Detection of liver tumor candidates from CT images using deep convolutional neural networks. In: Chen YW, Tanaka S, Howlett R, Jain L. (eds) Innovation in Medicine and Healthcare 2017. KES-InMed 2017; 2017. Smart Innovation, Systems and Technologies, vol 71. Springer, Cham. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 41. | Zhang F, Yang J, Nezami N, Laage-Gaupp F, Chapiro J, De Lin M, Duncan J. Liver tissue classification using an auto-context-based deep neural network with a multi-phase training framework. In: Bai W, Sanroma G, Wu G, Munsell B, Zhan Y, Coupé P. (eds) Patch-Based Techniques in Medical Imaging. Patch-MI 2018. Lecture Notes in Computer Science, vol 11075. Springer, Cham. [RCA] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 42. | Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol. 2017;10:257-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 420] [Article Influence: 52.5] [Reference Citation Analysis (0)] |
| 43. | Feng M, Wang Y, Liu J, Zhang L, Zaki HFM, Mian A. Benchmark Data Set and Method for Depth Estimation from Light Field Images. IEEE Trans Image Process. 2018;27:3586-3598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 44. | Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9:611-629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1202] [Cited by in RCA: 1186] [Article Influence: 169.4] [Reference Citation Analysis (0)] |
| 45. | Shi W, Gong Y, Tao X, Wang J, Zheng N. Improving CNN Performance Accuracies With Min-Max Objective. IEEE Trans Neural Netw Learn Syst. 2018;29:2872-2885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |