Published online Oct 15, 2018. doi: 10.4251/wjgo.v10.i10.344
Peer-review started: July 5, 2018
First decision: July 24, 2018
Revised: August 5, 2018
Accepted: August 30, 2018
Article in press: August 31, 2018
Published online: October 15, 2018
Processing time: 103 Days and 9.6 Hours
To investigate the clinical utility of alpha-fetoprotein (AFP)-producing gastric cancer (AFPGC)-specific microRNA (miRNA) for monitoring and prognostic prediction of patients.
We performed a comprehensive miRNA array-based approach to compare miRNA expression levels between AFP-positive and AFP-negative cells in three patients with primary AFPGC. We next examined the expression levels of the selected miRNAs in five AFPGC and ten non-AFPGC tissue samples by quantitative reverse transcription-polymerase chain reaction to validate their utility. We also investigated the expression levels of the selected miRNA not only in tissue but also in plasma samples. Moreover, we investigated the relationship between plasma AFP levels and plasma selected miRNA expression levels, and also investigated the correlation of the selected miRNA expression levels and malignant potential.
Among the five miRNAs selected from the miRNA array results, the expression levels of miR-122-5p were significantly higher in the AFPGC patients than in the non-AFPGC patients (P < 0.05). In tissue samples, miR-122-5p expression level tended to be lower in the non-AFPGC tissue than the normal gastric mucosa. Conversely, in the AFPGC tissue, miR-122-5p expression level was significantly higher in the AFPGC tissue than both the normal gastric mucosa and the non-AFPGC tissue samples (P < 0.05). Plasma miR-122-5p expression levels were also significantly higher in the AFPGC patients than the health volunteers and the non-AFPGC patients (P < 0.05) and were strongly correlated with plasma AFP levels (r = 0.7975, P < 0.0001). Moreover, the correlation of miR-122-5p expression in tissue samples with malignant potential was stronger than that of plasma AFP level in the AFPGC patients. In contrast, no correlation was found between miR-122-5p expression levels and liver metastasis in the non-AFPGC patients.
miR-122-5p might be a useful biomarker for early detection and disease monitoring in AFPGC.
Core tip: We examined the microRNAs (miRNA) expression in alpha-fetoprotein (AFP)-producing gastric cancer (AFPGC) tissue samples using a comprehensive miRNA array-based approach, and also investigated the clinical utility of the identified AFPGC-specific miRNAs. We found the expression of miR-122-5p was significantly higher in the AFPGC tissues and plasma samples. Moreover, tissue miR-122-5p expression levels exhibited a stronger correlation with malignant potential than plasma AFP level in AFPGC patients. miR-122-5p might be a useful biomarker for early detection and disease monitoring in AFPGC.
- Citation: Maruyama S, Furuya S, Shiraishi K, Shimizu H, Akaike H, Hosomura N, Kawaguchi Y, Amemiya H, Kawaida H, Sudo M, Inoue S, Kono H, Ichikawa D. miR-122-5p as a novel biomarker for alpha-fetoprotein-producing gastric cancer. World J Gastrointest Oncol 2018; 10(10): 344-350
- URL: https://www.wjgnet.com/1948-5204/full/v10/i10/344.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v10.i10.344
Gastric cancer (GC) is one of the most common solid tumors and is the third leading cause of cancer-related deaths worldwide[1,2]. Despite improvements in treatment approaches, prognosis of patients with advanced GC remains poor even after curative resection.
Among various tumor subtypes, alpha-fetoprotein (AFP)-producing GC (AFPGC) is recognized as one of the most aggressive tumors, with a high propensity for liver metastasis and subsequent poor prognosis compared with other GC subtypes[3-7]. The incidence of AFPGC is low, ranging from 1.3% to 15% of all GCs[8-12]. Therefore, recent comprehensive molecular analyses have not yet referred to this minor subtype.
MicroRNAs (miRNAs) are endogenous, small, non-coding, single-stranded RNAs of 20-25 nucleotides that regulate the expression of target genes at post-transcriptional level by binding to complementary sequences[13]. Various miRNAs were shown to play crucial roles in cancer as well as normal cells were reported to act as tumor suppressors or oncogenes in a cell type-dependent manner in various cancers[14]. In addition, certain miRNAs have been used for cancer detection, monitoring of tumor dynamics, and predicting prognosis and chemoresistance[15-20].
In the present study, we examined the miRNAs expression in AFPGC tissue samples using a comprehensive miRNA array-based approach. We also investigated the clinical utility of the identified AFPGC-specific miRNAs in monitoring and prognostic prediction of patients with AFPGC and evaluated their potential as universal biomarker for liver metastases.
A total of 492 patients underwent gastrectomy for GC at the University of Yamanashi Hospital between 2012 and 2018. Tumor specimens and resected lymph nodes obtained at the time of surgery were immediately fixed in 10% neutral-buffered formalin and embedded in paraffin after fixation. None of the patients underwent preoperative chemotherapy or radiotherapy. Tissue samples of all five patients with primary AFPGC and those from ten patients with primary non-AFPGC at various stages as controls from the same cohort were selected. The selected AFPGC samples contained all AFPGC patients who were operated in our hospital.
Pre-operative plasma samples were also obtained from four AFPGC patients and twenty non-AFPGC patients with GC who underwent surgical resection at the University of Yamanashi Hospital between 2017 and 2018. Control plasma samples were collected from 12 healthy adult volunteers. A total of 5 mL blood samples were collected into ethylenediaminetetraacetic acid-coated tubes and immediately spun at 3000 rpm at 4°C for 10 min to separate serum, which was stored at -80°C for further processing. AFPGC was defined based on a plasma AFP level above 10 ng/mL or positive AFP immunoreactivity in tissue samples. This study was approved by the Ethics Committee of Yamanashi University and performed in accordance with the ethical standards of the Declaration of Helsinki and its amendments.
Formalin-fixed, paraffin-embedded tissue samples were cut into 10-µm-thick sections, and total RNA was extracted from tumor and normal gastric mucosa in each patient using RNeasy FFPE kit (Qiagen, Valencia, CA), according to the manufacturer’s protocol. In plasma samples, total RNA was extracted from 100 µL plasma using RNeasy Serum/Plasma kit (Qiagen), according to the manufacturer’s protocol.
Microarray analyses of the GC tissue samples were performed using 3D-Gene miRNA oligo chips (Toray Industries, Kamakura, Japan), with 2565 genes mounted onto each DNA chip. Results were compared between the AFP-positive and AFP-negative cells among AFPGC patient samples using macro-dissection. Tissue samples from the three AFPGC patients who underwent curative surgery were mixed equally. RNAs were labeled with the 3D-Gene miRNA labeling kit (Toray Industries). Fluorescent signals were scanned using a 3D-Gene scanner 3000 (Toray Industries) and analyzed with the 3D-Gene Extraction software (Toray Industries). In the current study, expression level of each miRNA was normalized using the median signal intensity of the all genes in each chip, and median signal intensity was adjusted to 25.
Levels of miRNAs were quantified by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) using a Human TaqMan MicroRNA Assay kit (Applied Biosystems, Foster City, CA), according to standard procedures. Reverse transcription was conducted with a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems). Tissue miRNA levels were normalized to the endogenous control RNU6B, and plasma miRNA levels were normalized to a synthetic RNA oligonucleotide, cel-miR-39-3p (Qiagen), by spiking the samples with the oligonucleotide which does not exist in human genome. The following primers were used for the Taqman assay (Thermo Fisher Scientific, CA, United States): human hsa-miR-122-5p (cat #002245), hsa-miR-144-5p (cat #002148), hsa-miR-20a-5p (cat #000580), hsa-miR-20b-5p (cat #001014), hsa-miR-106a-5p (cat #000578), RNU6B (cat #001093), and cel-miR-39-3p (cat #000200). ΔCt values for all miRNAs relative to the control gene RNU6B and cel-miR-39-3p were determined. ΔΔCt values were calculated using mean ΔCt values in non-AFPGC tissue, normal gastric mucosa, or healthy volunteer plasma samples. Plasma miR-122-5p expression was calculated using log10(2-ΔCt).
Statistical significance was determined using GraphPad Prism® version 5 (San Diego, CA). Quantitative values were expressed as means ± SD unless noted otherwise. Statistical significance was evaluated using the Student’s t test and one-way analysis of variance for each time point, followed by Tukey’s post hoc test. Pearson’s correlation coefficient was determined to assess the correlation between plasma AFP and plasma miR-122-5p levels. P values < 0.05 were considered to indicate statistical significance.
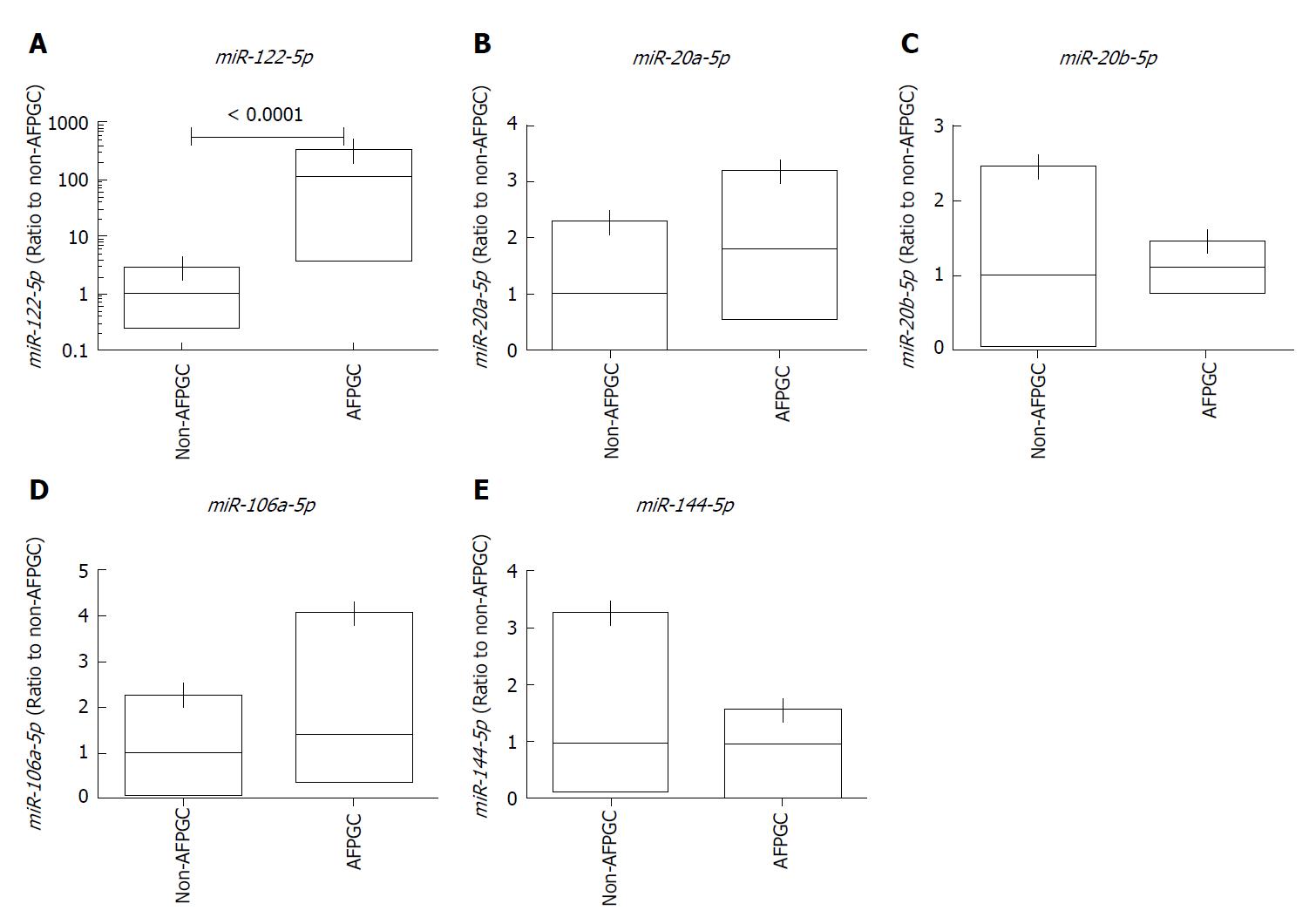
We selected miRNA candidates using a miRNA array-based approach. We compared the expression levels of each miRNA between the AFP-positive and AFP-negative cells in AFPGC patients. Of the 2565 candidates analyzed, we selected the following five miRNAs: miR-122-5p, miR-20a-5p, miR-20b-5p, miR-106a-5p, and miR-144-5p. The expression levels of these selected miRNAs were significantly different in AFP-positive cells compared with the AFP-negative cells, and the signal intensity of each miRNA was sufficient (Table 1).
| Signal intensity | Fold change | ||
| Gene ID | AFPGC | Non-AFPGC | AFPGC/non-AFPGC |
| hsa-miR-122-5p | 492 | 105 | 4.7 |
| hsa-miR-20a-5p | 245 | 113 | 2.2 |
| hsa-miR-20b-5p | 198 | 94 | 2.1 |
| hsa-miR-106a-5p | 304 | 195 | 1.6 |
| hsa-miR-145-5p | 712 | 1367 | 0.5 |
We examined the expression levels of the five selected miRNAs in five AFPGC and ten non-AFPGC tissue samples by qRT-PCR to validate their utility (Figure 1). Among these five miRNAs, the expression of miR-122-5p was significantly higher in the AFPGC patients than the non-AFPGC patients. Therefore, we selected miR-122-5p for further analyses in this study.
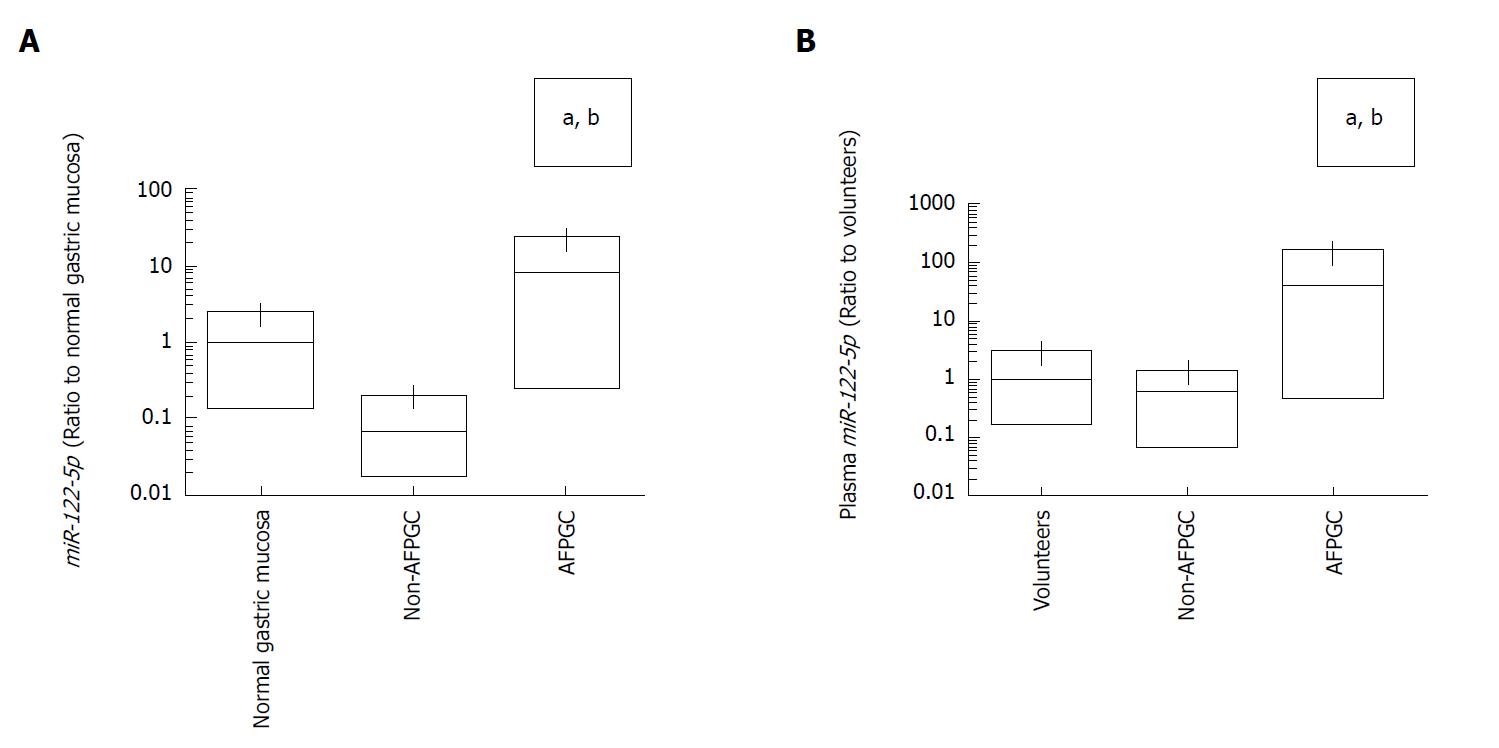
Next, we investigated the expression levels of miR-122-5p not only in tissue but also in plasma samples. In tissue samples, miR-122-5p expression levels tended to be lower in the non-AFPGC tissue samples than in the normal gastric mucosa samples. Conversely, miR-122-5p expression levels were significantly higher in the AFPGC tissue samples compared with the normal gastric mucosa and the non-AFPGC tissue samples (Figure 2A). The plasma expression levels of miR-122-5p were also significantly higher in the AFPGC patient samples than the samples from health volunteers and the non-AFPGC patients (Figure 2B).
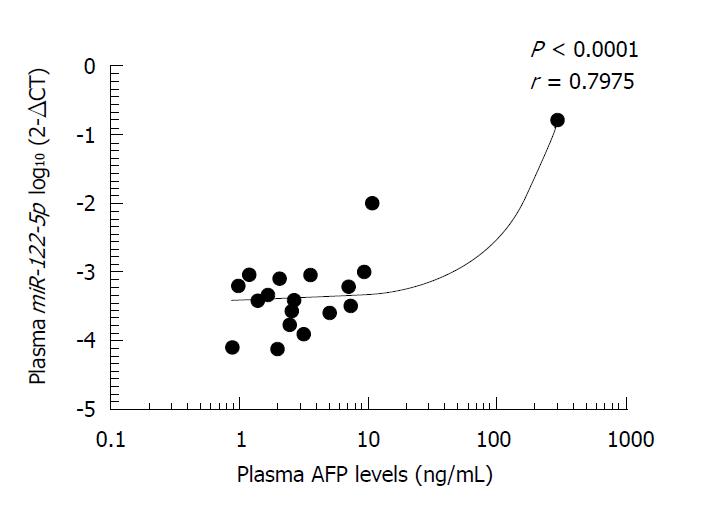
We next investigated the relationship between plasma AFP levels and plasma miR-122-5p expression levels in the AFPGC and non-AFPGC patients and found that miR-122-5p expression level in plasma was strongly correlated with plasma AFP level (r = 0.7975, P < 0.0001; Figure 3).
Figure 4 shows the correlation between malignant potential, all biomarkers, tissue miR-122-5p expression, and plasma AFP level in the AFPGC patients. We found that the expression level of miR-122-5p in tissue exhibited a stronger correlation with malignant potential (i.e., liver metastasis) than plasma AFP level in the AFPGC patients. Two patients with malignant potential were diagnosed morphologically as poorly differentiated adenocarcinoma and mucinous adenocarcinoma, and the other current alive patients were diagnosed as poorly differentiated adenocarcinoma and hepatoid adenocarcinoma.
AFPGC has been reported to be more likely to metastasize to liver and is therefore associated with extremely poor prognosis[8,11]. However, no genomic analyses have been conducted for AFPGC due to its rarity. Therefore, AFPGC-specific genomic and/or epigenomic alterations are not well known, which have urged us to examine the molecular characteristics specific to AFPGC. The current study investigated the molecular characteristics of AFPGC with a comprehensive analysis, with particular focus on miRNA expression.
The findings of the present study clearly demonstrated that the expression of miR-122-5p was significantly higher in the AFPGC tissues than the normal and non-AFPGC tissues. The expression levels of this miRNA were also higher in the plasma samples of patients with AFPGC compared with those of healthy volunteers and non-AFPGC patients and correlated with plasma AFP levels to a certain extent. Interestingly, the tissue expression level of miR-122-5p exhibited a stronger correlation with malignant potential than plasma AFP level in AFPGC patients, suggesting that miR-122-5p might have utility as a prognostic biomarker especially for liver metastasis in this small GC subgroup.
miR-122-5p expression has been increasingly examined in various normal and cancer tissue types. Several studies reported that miR-122-5p was specifically expressed in human liver and that hepatocyte-specific miR-122-5p regulated hepatocyte differentiation and metabolism[21-23]. Taken together, AFPGC might show characteristics of hepatocytes not only morphologically but also in its miRNA expression patterns. In fact, AFPGC was not necessarily hepatoid adenocarcinoma in this series, and two patients with aggressive development of liver metastasis were diagnosed morphologically as poorly differentiated adenocarcinoma and mucinous adenocarcinoma. Conversely, miR-122-5p was previously shown to function as a tumor suppressor and was reported to be downregulated in several cancer types such as hepatocellular carcinoma[24], non-small-cell lung cancer[25], gallbladder carcinoma[26], bladder cancer[27], and breast cancer[28]. In GC, the expression of miR-122-5p was reported to be lower in tumor tissue than the adjacent non-cancerous tissue. Furthermore, several studies reported that miR-122-5p inhibited proliferation, migration, and invasion in GC[15,29,30]. It’s not known exactly why miR-122-5p, which is known as suppressor gene, is higher in AFPGC. Some miRNA was reported that decreased in early cases and elevated again in staged-advanced cases[31]. Therefore, miR-122-5p decreased in carcinogenesis might be elevated during tumor evolution to AFPGC. However, the exact mechanism is unknown at the present time.
In the current study, we did not see a correlation between miR-122-5p in patients with non-AFPGC and development of liver metastasis, suggesting that the mechanism underlying liver metastasis might be distinct between AFPGC and non-AFPGC. We assume that AFPGC is completely different from non-AFPGC, and the mechanism of liver metastasis between AFPGC and non-AFPGC is also distinct.
Several reports demonstrated that the clinical behavior of AFPGC was distinct from that of non-AFPGC[32]. Recently, Lu et al[33] demonstrated that AFP contributed to invasion and metastasis directly. We speculate AFPGC has specific ability of liver metastasis, and correlated with miR-122-5p. Therefore, miR-122-5p might directly facilitate tumor proliferation, migration, and invasion, which raises the possibility of miR-122-5p as a potential therapeutic target in AFPGC. However, future studies are warranted to demonstrate the biological function underlying altered expression of miR-122-5p in AFPGC. The current study revealed miR-122-5p as a potentially useful biomarker for early detection, disease monitoring, and prognostic prediction in patients with AFPGC, which warrant further investigation.
Alpha-fetoprotein (AFP)-producing gastric cancer (AFPGC) is recognized as one of the most aggressive tumors, with a high propensity for liver metastasis and subsequent poor prognosis compared with other GC subtypes. Recent comprehensive molecular analyses have not yet referred to this minor subtype because of its rareness.
To discover universal biomarkers for liver metastasis by researching AFPGC-specific microRNAs (miRNAs).
To investigate the clinical utility of AFPGC-specific miRNA for monitoring and prognostic prediction of patients.
We performed a comprehensive miRNA array-based approach to compare miRNA expression levels between AFP-positive and AFP-negative cells, and also investigated the clinical utility of the identified AFPGC-specific miRNAs.
We found the expression of miR-122-5p was significantly higher in the AFPGC tissues than the normal and non-AFPGC tissues. The expression levels of this miRNA were also higher in the plasma samples of patients with AFPGC compared with those of healthy volunteers and non-AFPGC patients and correlated with plasma AFP levels. Moreover, the tissue expression level of miR-122-5p exhibited a stronger correlation with malignant potential than plasma AFP level in AFPGC patients.
miR-122-5p as a potentially useful biomarker for early detection and disease monitoring in patients with AFPGC.
We identified miR-122-5p as AFPGC-specific miRNA. miR-122-5p might be a clinical useful biomarker in AFPGC. Although studies are warranted to demonstrate the biological function underlying altered expression of miR-122-5p in AFPGC, the miR-122-5p might be a potential therapeutic target for liver metastasis in AFPGC.
The Authors are grateful to Motoko Inui and Makiko Mishina for expert technical assistance.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Jung Y, Takemura N S- Editor: Ji FF L- Editor: A E- Editor: Tan WW
| 1. | Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137-2150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2591] [Cited by in RCA: 2644] [Article Influence: 139.2] [Reference Citation Analysis (0)] |
| 2. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 4810] [Article Influence: 437.3] [Reference Citation Analysis (2)] |
| 3. | Bourreille J, Metayer P, Sauger F, Matray F, Fondimare A. [Existence of alpha feto protein during gastric-origin secondary cancer of the liver]. Presse Med. 1970;78:1277-1278. [PubMed] |
| 4. | Chang YC, Nagasue N, Abe S, Kohno H, Yamanoi A, Uchida M, Nakamura T. [The characters of AFP-producing early gastric cancer]. Nihon Geka Gakkai Zasshi. 1990;91:1574-1580. [PubMed] |
| 5. | Motoyama T, Aizawa K, Watanabe H, Fukase M, Saito K. alpha-Fetoprotein producing gastric carcinomas: a comparative study of three different subtypes. Acta Pathol Jpn. 1993;43:654-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 33] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | Chang YC, Nagasue N, Abe S, Taniura H, Kumar DD, Nakamura T. Comparison between the clinicopathologic features of AFP-positive and AFP-negative gastric cancers. Am J Gastroenterol. 1992;87:321-325. [PubMed] |
| 7. | Koide N, Nishio A, Igarashi J, Kajikawa S, Adachi W, Amano J. Alpha-fetoprotein-producing gastric cancer: histochemical analysis of cell proliferation, apoptosis, and angiogenesis. Am J Gastroenterol. 1999;94:1658-1663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 8. | Kono K, Amemiya H, Sekikawa T, Iizuka H, Takahashi A, Fujii H, Matsumoto Y. Clinicopathologic features of gastric cancers producing alpha-fetoprotein. Dig Surg. 2002;19:359-365; discussion 365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 95] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 9. | Chang YC, Nagasue N, Kohno H, Taniura H, Uchida M, Yamanoi A, Kimoto T, Nakamura T. Clinicopathologic features and long-term results of alpha-fetoprotein-producing gastric cancer. Am J Gastroenterol. 1990;85:1480-1485. [PubMed] |
| 10. | Chun H, Kwon SJ. Clinicopathological characteristics of alpha-fetoprotein-producing gastric cancer. J Gastric Cancer. 2011;11:23-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 11. | Liu X, Cheng Y, Sheng W, Lu H, Xu Y, Long Z, Zhu H, Wang Y. Clinicopathologic features and prognostic factors in alpha-fetoprotein-producing gastric cancers: analysis of 104 cases. J Surg Oncol. 2010;102:249-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 107] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 12. | McIntire KR, Waldmann TA, Moertel CG, Go VL. Serum alpha-fetoprotein in patients with neoplasms of the gastrointestinal tract. Cancer Res. 1975;35:991-996. [PubMed] |
| 13. | Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3368] [Cited by in RCA: 4102] [Article Influence: 372.9] [Reference Citation Analysis (1)] |
| 14. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5384] [Cited by in RCA: 5599] [Article Influence: 294.7] [Reference Citation Analysis (0)] |
| 15. | Chen Q, Ge X, Zhang Y, Xia H, Yuan D, Tang Q, Chen L, Pang X, Leng W, Bi F. Plasma miR-122 and miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncol Rep. 2014;31:1863-1870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 16. | Hiramoto H, Muramatsu T, Ichikawa D, Tanimoto K, Yasukawa S, Otsuji E, Inazawa J. miR-509-5p and miR-1243 increase the sensitivity to gemcitabine by inhibiting epithelial-mesenchymal transition in pancreatic cancer. Sci Rep. 2017;7:4002. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 17. | Hiyoshi Y, Akiyoshi T, Inoue R, Murofushi K, Yamamoto N, Fukunaga Y, Ueno M, Baba H, Mori S, Yamaguchi T. Serum miR-143 levels predict the pathological response to neoadjuvant chemoradiotherapy in patients with locally advanced rectal cancer. Oncotarget. 2017;8:79201-79211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 18. | Imamura T, Komatsu S, Ichikawa D, Miyamae M, Okajima W, Ohashi T, Kiuchi J, Nishibeppu K, Konishi H, Shiozaki A. Depleted tumor suppressor miR-107 in plasma relates to tumor progression and is a novel therapeutic target in pancreatic cancer. Sci Rep. 2017;7:5708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 19. | Matsushita R, Seki N, Chiyomaru T, Inoguchi S, Ishihara T, Goto Y, Nishikawa R, Mataki H, Tatarano S, Itesako T. Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br J Cancer. 2015;113:282-289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 20. | Yang R, Fu Y, Zeng Y, Xiang M, Yin Y, Li L, Xu H, Zhong J, Zeng X. Serum miR-20a is a promising biomarker for gastric cancer. Biomed Rep. 2017;6:429-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 21. | Roderburg C, Benz F, Vargas Cardenas D, Koch A, Janssen J, Vucur M, Gautheron J, Schneider AT, Koppe C, Kreggenwinkel K. Elevated miR-122 serum levels are an independent marker of liver injury in inflammatory diseases. Liver Int. 2015;35:1172-1184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 100] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 22. | Koyama S, Kuragaichi T, Sato Y, Kuwabara Y, Usami S, Horie T, Baba O, Hakuno D, Nakashima Y, Nishino T. Dynamic changes of serum microRNA-122-5p through therapeutic courses indicates amelioration of acute liver injury accompanied by acute cardiac decompensation. ESC Heart Fail. 2017;4:112-121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 23. | Satishchandran A, Ambade A, Rao S, Hsueh YC, Iracheta-Vellve A, Tornai D, Lowe P, Gyongyosi B, Li J, Catalano D. MicroRNA 122, Regulated by GRLH2, Protects Livers of Mice and Patients From Ethanol-Induced Liver Disease. Gastroenterology. 2018;154:238-252.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 24. | Wang N, Wang Q, Shen D, Sun X, Cao X, Wu D. Downregulation of microRNA-122 promotes proliferation, migration, and invasion of human hepatocellular carcinoma cells by activating epithelial-mesenchymal transition. Onco Targets Ther. 2016;9:2035-2047. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 25. | Qin H, Sha J, Jiang C, Gao X, Qu L, Yan H, Xu T, Jiang Q, Gao H. miR-122 inhibits metastasis and epithelial-mesenchymal transition of non-small-cell lung cancer cells. Onco Targets Ther. 2015;8:3175-3184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 26. | Lu W, Zhang Y, Zhou L, Wang X, Mu J, Jiang L, Hu Y, Dong P, Liu Y. miR-122 inhibits cancer cell malignancy by targeting PKM2 in gallbladder carcinoma. Tumour Biol. 2015;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Wang Y, Xing QF, Liu XQ, Guo ZJ, Li CY, Sun G. MiR-122 targets VEGFC in bladder cancer to inhibit tumor growth and angiogenesis. Am J Transl Res. 2016;8:3056-3066. [PubMed] |
| 28. | Ergün S, Ulasli M, Igci YZ, Igci M, Kırkbes S, Borazan E, Balik A, Yumrutaş Ö, Camci C, Cakmak EA, Arslan A, Oztuzcu S. The association of the expression of miR-122-5p and its target ADAM10 with human breast cancer. Mol Biol Rep. 2015;42:497-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Rao M, Zhu Y, Zhou Y, Cong X, Feng L. MicroRNA-122 inhibits proliferation and invasion in gastric cancer by targeting CREB1. Am J Cancer Res. 2017;7:323-333. [PubMed] |
| 30. | Xu X, Gao F, Wang J, Tao L, Ye J, Ding L, Ji W, Chen X. MiR-122-5p inhibits cell migration and invasion in gastric cancer by down-regulating DUSP4. Cancer Biol Ther. 2018;19:427-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 31. | Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 303] [Cited by in RCA: 340] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 32. | Hirajima S, Komatsu S, Ichikawa D, Kubota T, Okamoto K, Shiozaki A, Fujiwara H, Konishi H, Ikoma H, Otsuji E. Liver metastasis is the only independent prognostic factor in AFP-producing gastric cancer. World J Gastroenterol. 2013;19:6055-6061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 33] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 33. | Lu S, Ma Y, Sun T, Ren R, Zhang X, Ma W. Expression of α-fetoprotein in gastric cancer AGS cells contributes to invasion and metastasis by influencing anoikis sensitivity. Oncol Rep. 2016;35:2984-2990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |