Copyright
©The Author(s) 2025.
World J Gastrointest Oncol. May 15, 2025; 17(5): 104076
Published online May 15, 2025. doi: 10.4251/wjgo.v17.i5.104076
Published online May 15, 2025. doi: 10.4251/wjgo.v17.i5.104076
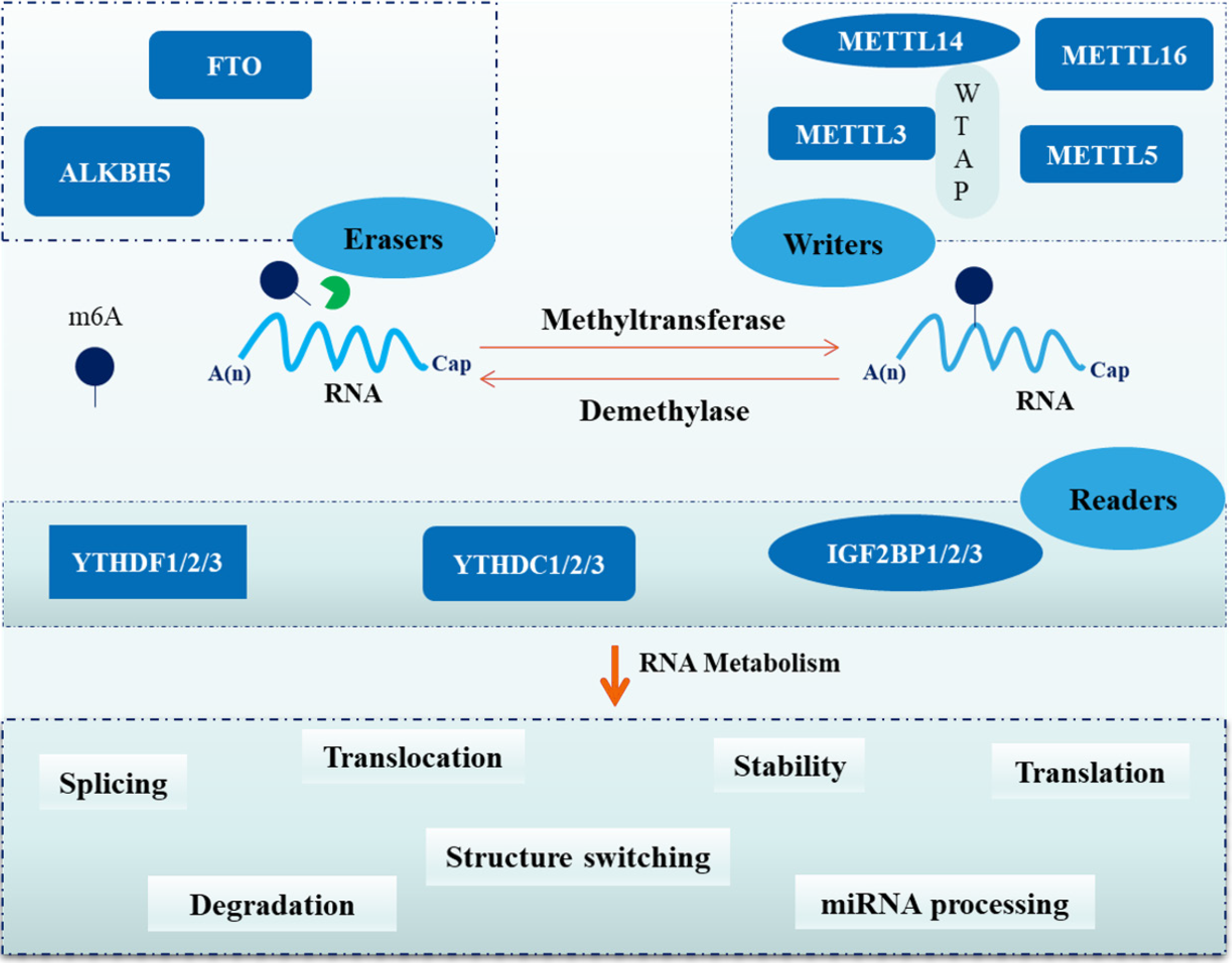
Figure 1 Dynamic regulation of N6-methyladenosine modification.
N6-methyladenosine (m6A) writers, erasers, and readers control the process of m6A modification. Writers (METTL3, METTL14, WTAP, METTL5, and METTL16) catalyze m6A deposition, erasers (FTO and ALKBH5) remove methyl groups, and readers (YTHDF1/YTHDF2/YTHDF3, YTHDC1/YTHDC2/YTHDC3, IGF2BP1/IGF2BP2/IGF2BP3) decode these marks to influence RNA stability, messenger RNA translation, RNA degradation, translocation, RNA splicing, structure switching, and primary microRNA processing. M6A: N6-methyladenosine.
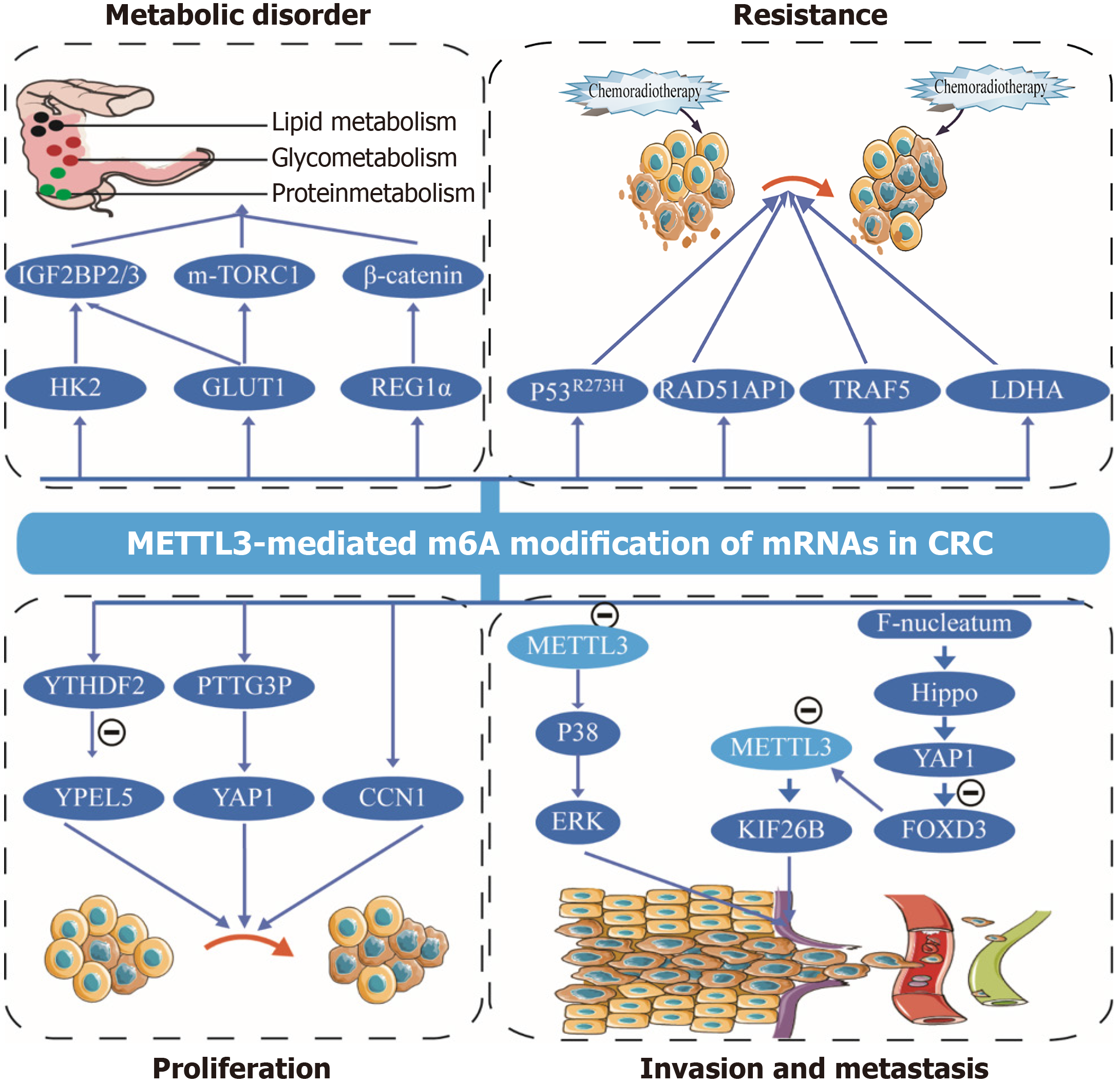
Figure 2 METTL3-mediated N6-methyladenosine modification of messenger RNAs in colorectal cancer.
METTL3 participates in colorectal cancer (CRC) by regulating N6-methyladenosine modification of messenger RNAs (mRNAs) involved in proliferation, invasion and metastasis, metabolic disorder, and resistance of CRC cells. Every mRNA was detailed with a name. HK2: Hexokinase 2; GLUT1: Glucose transporter type 1; mRNAs: Messenger RNAs; m6A: N6-methyladenosine; CRC: Colorectal cancer.
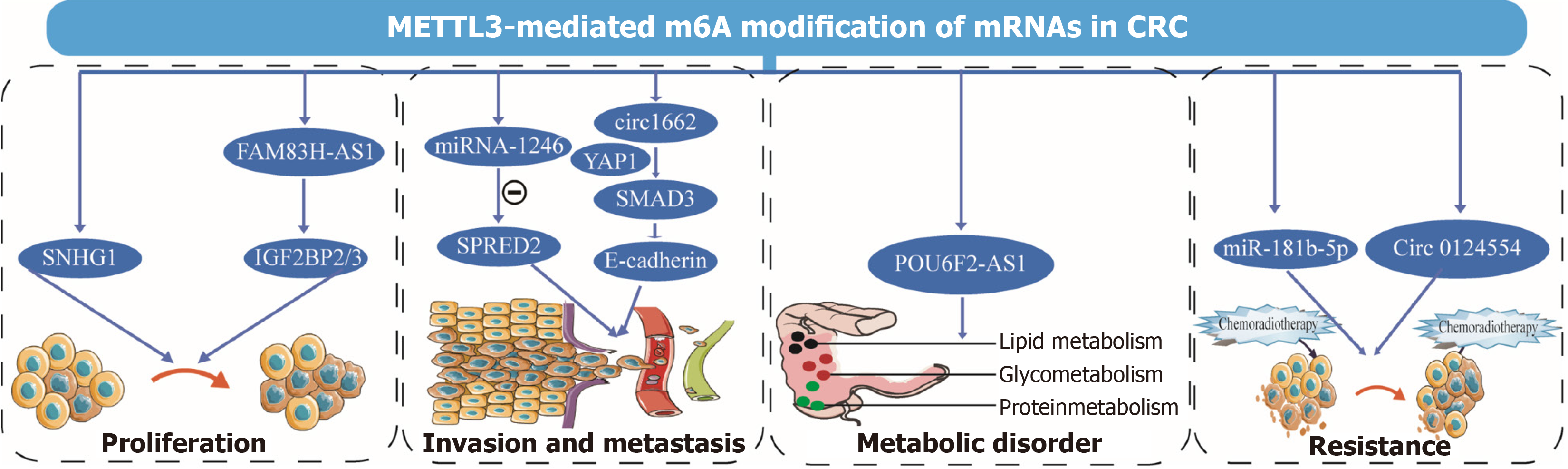
Figure 3 METTL3-mediated N6-methyladenosine modification of noncoding RNAs in colorectal cancer.
METTL3 participates in colorectal cancer (CRC) by regulating N6-methyladenosine modification of noncoding RNAs (ncRNAs) involved in proliferation, invasion and metastasis, metabolic disorder, and resistance of CRC cells. Each ncRNA was detailed with a name. ncRNAs: Noncoding RNAs; m6A: N6-methyladenosine; CRC: Colorectal cancer.
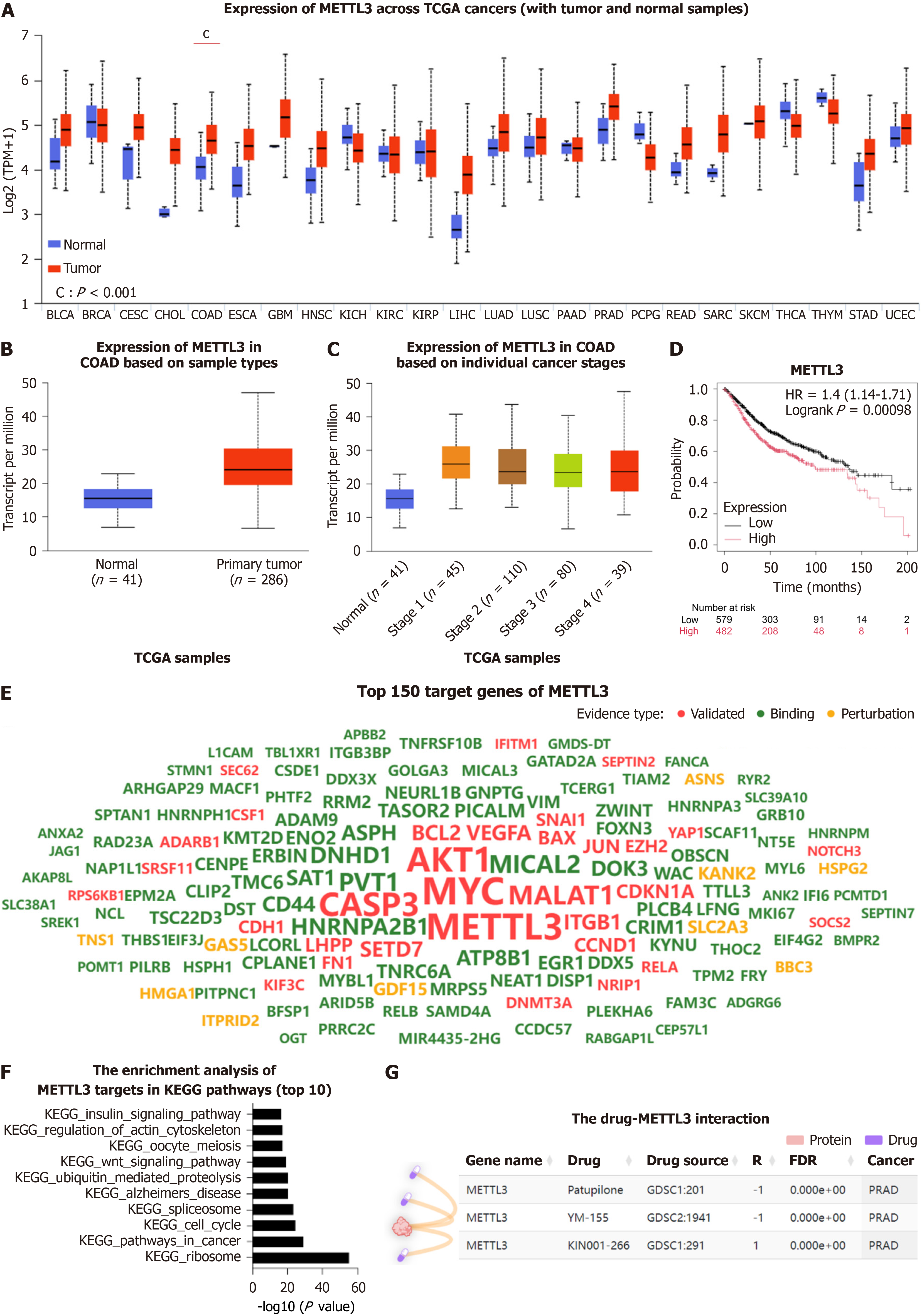
Figure 4 Expression and function analysis of METTL3.
A: Pan-cancer analysis of METTL3; B: Expression of METTL3 in colon adenocarcinoma (COAD); C: METTL3 expression across COAD stages; D: Prognosis of METTL3 in COAD; E: Integration of the top 150 METTL3 target genes; F: Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of METTL3 targets; G: Drug-protein interaction analysis of Patupilone, YM-155, KIN001-266, and METTL3 in prostate adenocarcinoma. TCGA: The Cancer Genome Atlas; TPM: Transcripts per million; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; ESCA: Esophageal carcinoma; GBM: Glioblastoma multiforme; HNSC: Head and neck squamous cell carcinoma; KICH: Kidney chromophobe; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; PAAD: Pancreatic adenocarcinoma; PRAD: Prostate adenocarcinoma; PCPG: Pheochromocytoma and paraganglioma; READ: Rectum adenocarcinoma; SARC: Sarcoma; SKCM: Skin cutaneous melanoma; STAD: Stomach adenocarcinoma; UCEC: Uterine corpus endometrial carcinoma; KEGG: Kyoto Encyclopedia of Genes and Genomes.
- Citation: Liao JN, Ni WJ, Wu PH, Yang YD, Yang Y, Long W, Xie MZ, Zhu XZ, Xie FH, Leng XM. Switching from messenger RNAs to noncoding RNAs, METTL3 is a novel colorectal cancer diagnosis and treatment target. World J Gastrointest Oncol 2025; 17(5): 104076
- URL: https://www.wjgnet.com/1948-5204/full/v17/i5/104076.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i5.104076