Published online May 16, 2022. doi: 10.4253/wjge.v14.i5.311
Peer-review started: May 13, 2021
First decision: July 4, 2021
Revised: July 15, 2021
Accepted: April 27, 2022
Article in press: April 27, 2022
Published online: May 16, 2022
Processing time: 368 Days and 9.4 Hours
Esophagitis is an inflammatory and damaging process of the esophageal mucosa, which is confirmed by endoscopic visualization and may, in extreme cases, result in stenosis, fistulization and esophageal perforation. The use of deep learning (a field of artificial intelligence) techniques can be considered to determine the presence of esophageal lesions compatible with esophagitis.
To develop, using transfer learning, a deep neural network model to recognize the presence of esophagitis in endoscopic images.
Endoscopic images of 1932 patients with a diagnosis of esophagitis and 1663 patients without any pathological diagnosis provenient from the KSAVIR and HyperKSAVIR datasets were splitted in training (80%) and test (20%) and used to develop and evaluate a binary deep learning classifier built using the DenseNet-201 architecture, a densely connected convolutional network, with weights pretrained on the ImageNet image set and fine-tuned during training. The classifier model performance was evaluated in the test set according to accuracy, sensitivity, specificity and area under the receiver operating characteristic curve (AUC).
The model was trained using Adam optimizer with a learning rate of 0.0001 and applying binary cross entropy loss function. In the test set (n = 719), the classifier achieved 93.32% accuracy, 93.18% sensitivity, 93.46% specificity and a 0.96 AUC. Heatmaps for spatial predictive relevance in esophagitis endoscopic images from the test set were also plotted. In face of the obtained results, the use of dense convolutional neural networks with pretrained and fine-tuned weights proves to be a good strategy for predictive modeling for esophagitis recognition in endoscopic images. In addition, adopting the classification approach combined with the subsequent plotting of heat maps associated with the classificatory decision gives greater explainability to the model.
It is opportune to raise new studies involving transfer learning for the analysis of endoscopic images, aiming to improve, validate and disseminate its use for clinical practice.
Core Tip: Considering the clinical relevance of esophagitis, we proposed a deep learning model for its diagnosis from endoscopic images of the Z-line, via binary classification of the images according to the presence or absence of esophageal inflammation signs. The excellent accuracy and area under the receiver operating characteristic curve achieved demonstrate the potential of the adopted strategy, consisting of the conjunction of densely connected neural networks and transfer learning. With this, we contribute to the improvement and methodological advancement in the development of automated diagnostic tools for the disease, which reveal great potential in optimizing the management of these patients.
- Citation: Caires Silveira E, Santos Corrêa CF, Madureira Silva L, Almeida Santos B, Mattos Pretti S, Freire de Melo F. Recognition of esophagitis in endoscopic images using transfer learning. World J Gastrointest Endosc 2022; 14(5): 311-319
- URL: https://www.wjgnet.com/1948-5190/full/v14/i5/311.htm
- DOI: https://dx.doi.org/10.4253/wjge.v14.i5.311
Esophagitis is an inflammatory and damaging process of the esophageal mucosa, that can be the outcome of different pathological processes, which share, however, the same clinical presentation: retrosternal pain, dysphagia, odynophagia and heartburn[1,2]. Different pathological processes may lead to esophagitis, with possible etiologies embracing gastroesophageal reflux disease (GERD), infectious processes, in eosinophilic esophagitis, medications or even radiation. In extreme cases, it can result in stenosis, fistulization and esophageal perforation[3]. These complications, however, may be prevented with precoce diagnosis.
Esophagitis can be suspected based on the clinical history, with a confirmation performed through endoscopic visualization. The differentiation of its etiopathogenesis may be determined from endoscopic and histological study of the esophagus. The endoscopic presentation of eosinophilic esophagitis is characterized by exudates, strictures and concentric rings. In colonization by Candida sp. there are small and diffuse yellow-white plaques; in cytomegalovirus infection there are large ulcerations; Herpes Virus, in turn, may cause multiple small ulcerations[3-5]. GERD, on the other hand, has a better-defined endoscopic classification with the Los Angeles classification, which has four gradations based on the presence, size and distribution of esophageal[6].
Machine learning, main exponent of artificial intelligence, has gained space and attention in healthcare and medical research, especially after the development and validation by Beam and Kohane[7] and Gulshan et al[8] of a deep learning algorithm capable of detecting the presence of diabetic retinopathy in studies of the retina[7,8]. In the context of esophagitis, the use of machine learning, especially deep learning, may be considered to determine, among others, the presence of esophageal lesions compatible with esophagitis.
Deep learning - which comprehends deep artificial neural network-based algorithms capable of learning from large amounts of data - is considered the state of the art in the field of artificial intelligence for computer vision[9]. Among the possible uses of such applications, there is the binary classification of images according to the presence or absence of a given finding. In these cases, a dataset comprising examples of the image type to be classified is divided into two distinct subsets: one to train the model (from which the weights will be learned) and the other to evaluate its performance[10]. It is important that the two subsets obtained are representative, in terms of labels proportion, of the original dataset.
Traditionally, algorithms for deep learning use large volumes of data for training. However, obtaining databases large enough to accurately train them can prove to be a highly expensive process. As a way of mitigating this situation, one can choose to apply a technique called transfer learning, which is based on the use of external data to perform a training step mentioned above[10]. The use of this technique makes it possible to obtain a scale of pretrained weights in computational models for analyzing, among others, medical images. It should be noted, however, that the use of pretrained weights does not exempt the need to carry out a training stage with data that are representative of the base to be tested, with this second training step (called fine tuning) aiming to improve, principally, the deep layers of the algorithm in order to obtain results with greater accuracy[11].
This study aims to develop a supervised deep learning model using a fine-tuned transfer learning dense convolutional neural network (DCNN) to recognize, in a binary way, the presence of changes compatible with esophagitis in images from endoscopic studies. Thus, it seeks to contribute to the advancement and methodological improvement of a cost-effective and accurate automated technology for the diagnosis of esophagitis, optimizing the management of patients who present this condition.
Endoscopic images of 1932 patients with a diagnosis of esophagitis and 1663 patients without any pathological diagnosis (in both cases being z line the image topography) were obtained from the publicly available KSAVIR Dataset[12] and HyperKSAVIR Dataset[13]. Were included in this study the images in both datasets labeled as “normal z line” and the images labeled as “esophagitis”. From these data, we set out to develop a binary deep learning classifier using the DenseNet-201 architecture, a densely connected convolutional network which connects each layer to every other layer in a feed-forward fashion[14], pretrained on the ImageNet image set.
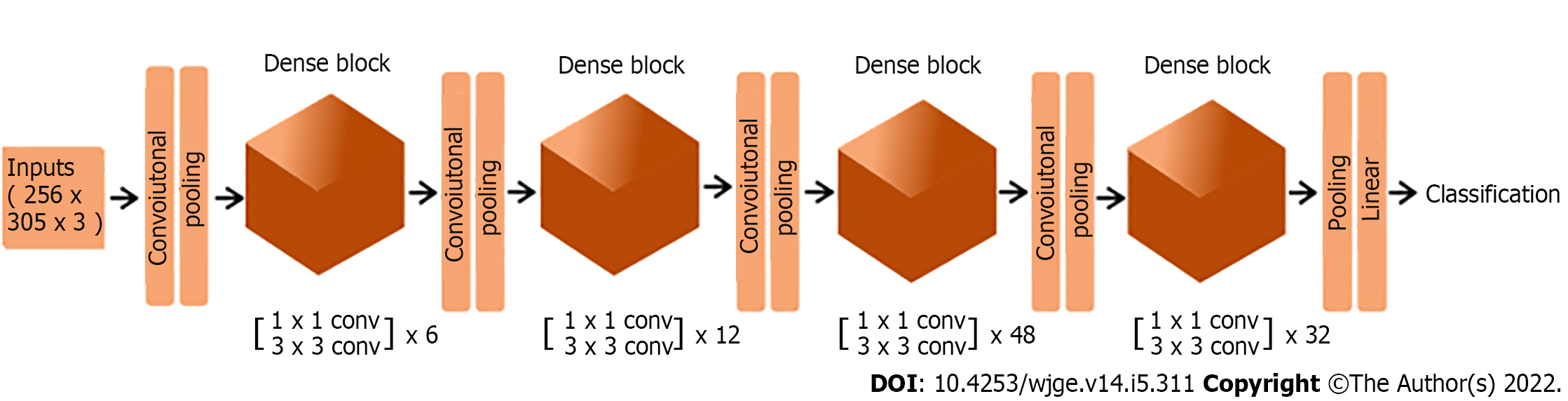
The top layer of the DenseNet-201 architecture was not included in our model, and its output (that is, the output of the final convolutional block) was converted from a 4 dimensional to a 2 dimensional tensor using global average pooling. As the final layer, we added a dense layer with one unit and sigmoid activation. The structure of the final deep neural network predictive model is summarized in Table 1, and its architecture is illustrated in Figure 1.
| Type of layer | Brief description | Number of parameters |
| Functional | Instantiates the DenseNet-201 architecture with average pooling of the output | 18321984 |
| Dense | One unit with sigmoid activation | 1921 |
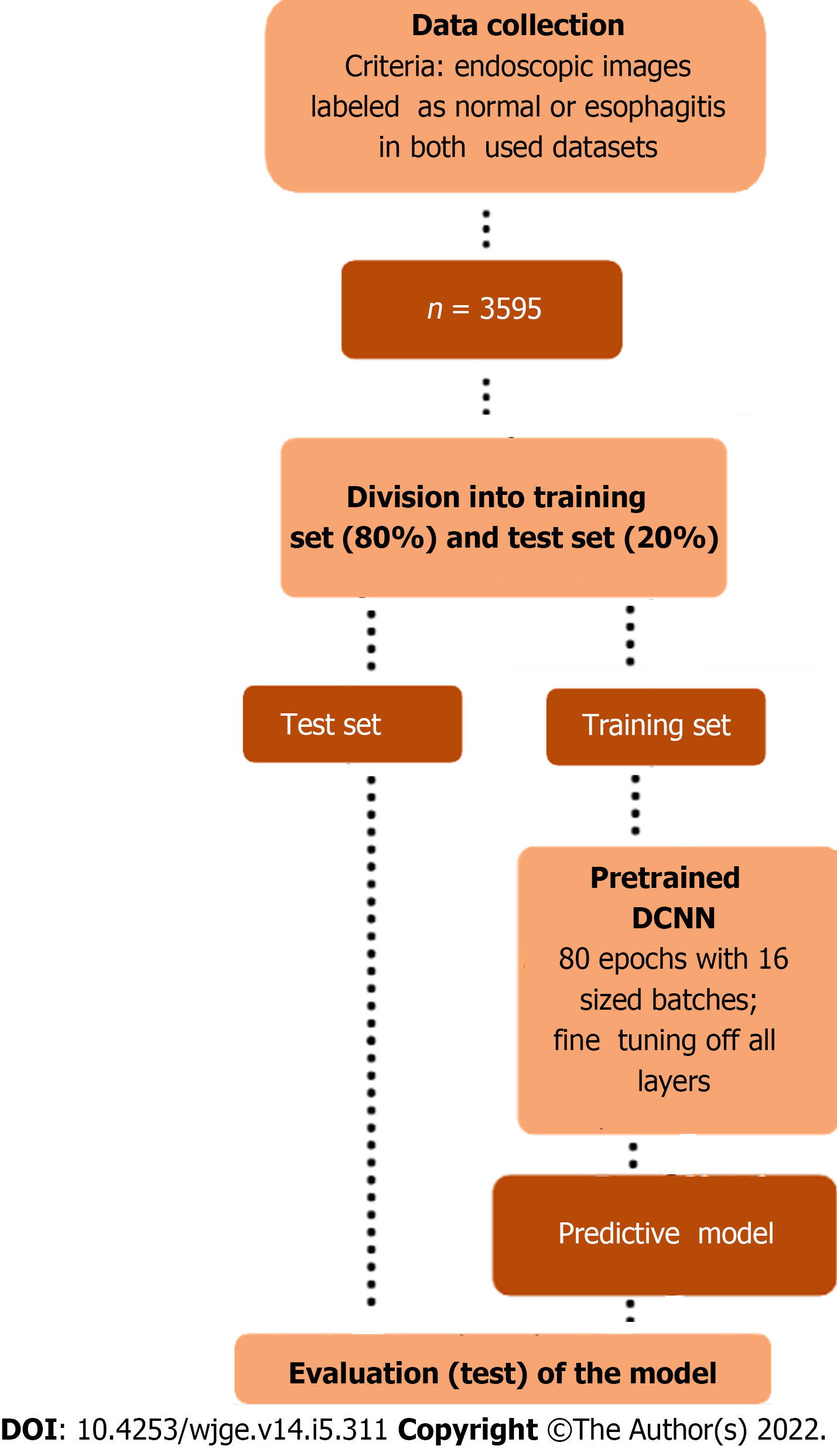
For this purpose, the images were converted to arrays of dimension 256 × 305 × 3, whose units were rescaled using the densenet preprocessor, and divided into training set (80%) and test set (20%). The training set (n = 2876) was divided in batches of size 16 and used to train, throughout 80 epochs, the transfer learning based neural network whose structure is shown in Table 1. The test set (n = 719) was used to evaluate the model according to the following metrics: accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve (AUC).
The adopted methodology is schematically summarized in Figure 2. All steps of the predictive model development were performed in Python (version 3.6.9), using Keras library.
As previously stated, all the imaging data was obtained from the public datasets KSAVIR Dataset[12] and HyperKSAVIR Dataset[13] that were released for both educational and research purposes. Therefore, it was not necessary to submit this study to the ethics committee, being in accordance with all the established precepts by the Committee on Publication Ethics.
The model was trained using Adam optimizer with a learning rate of 0.0001 and applying binary cross entropy loss function. All layers of the DenseNet architecture incorporated in the model were set as trainable (that is, we fine-tuned all weights).
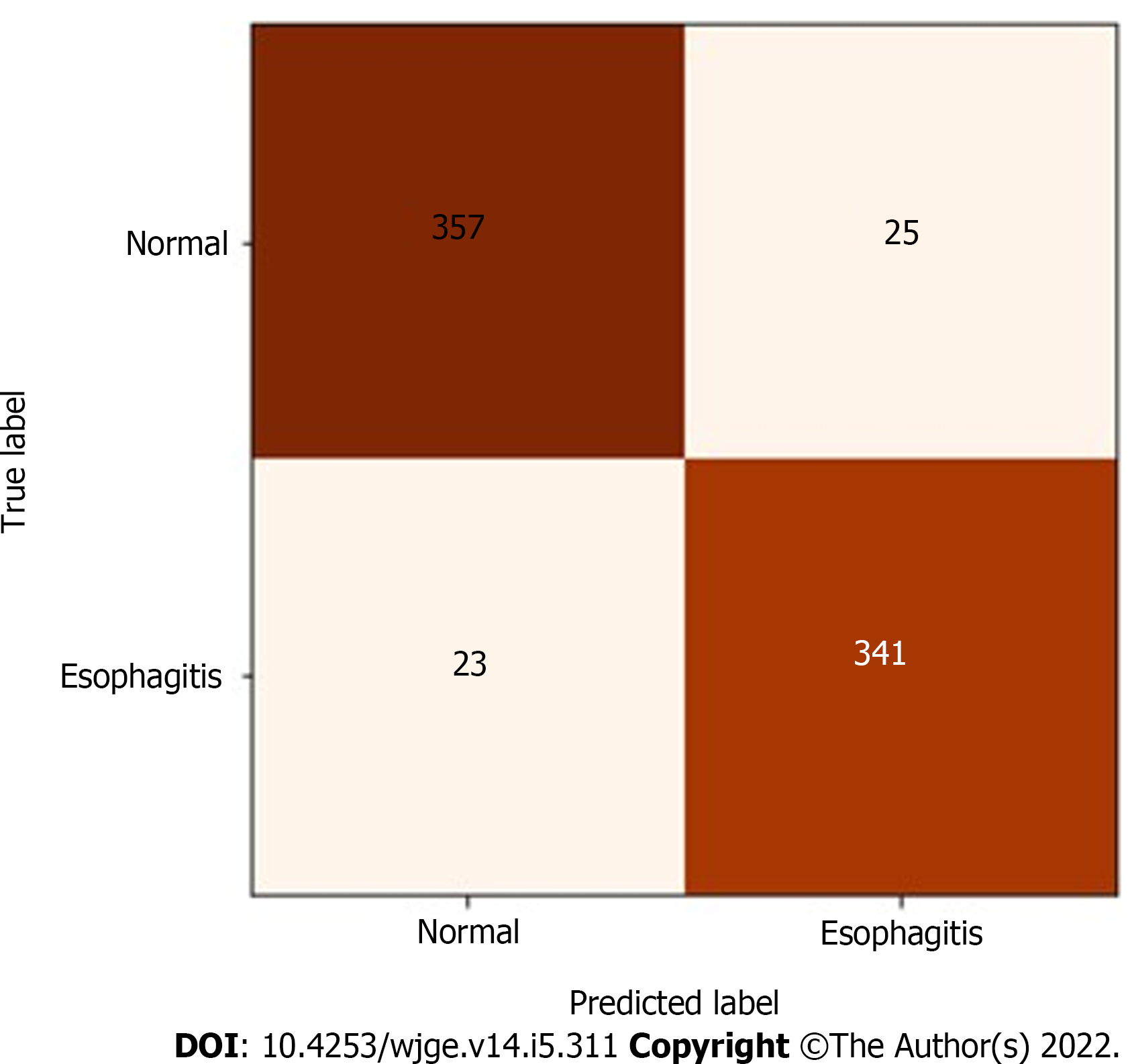
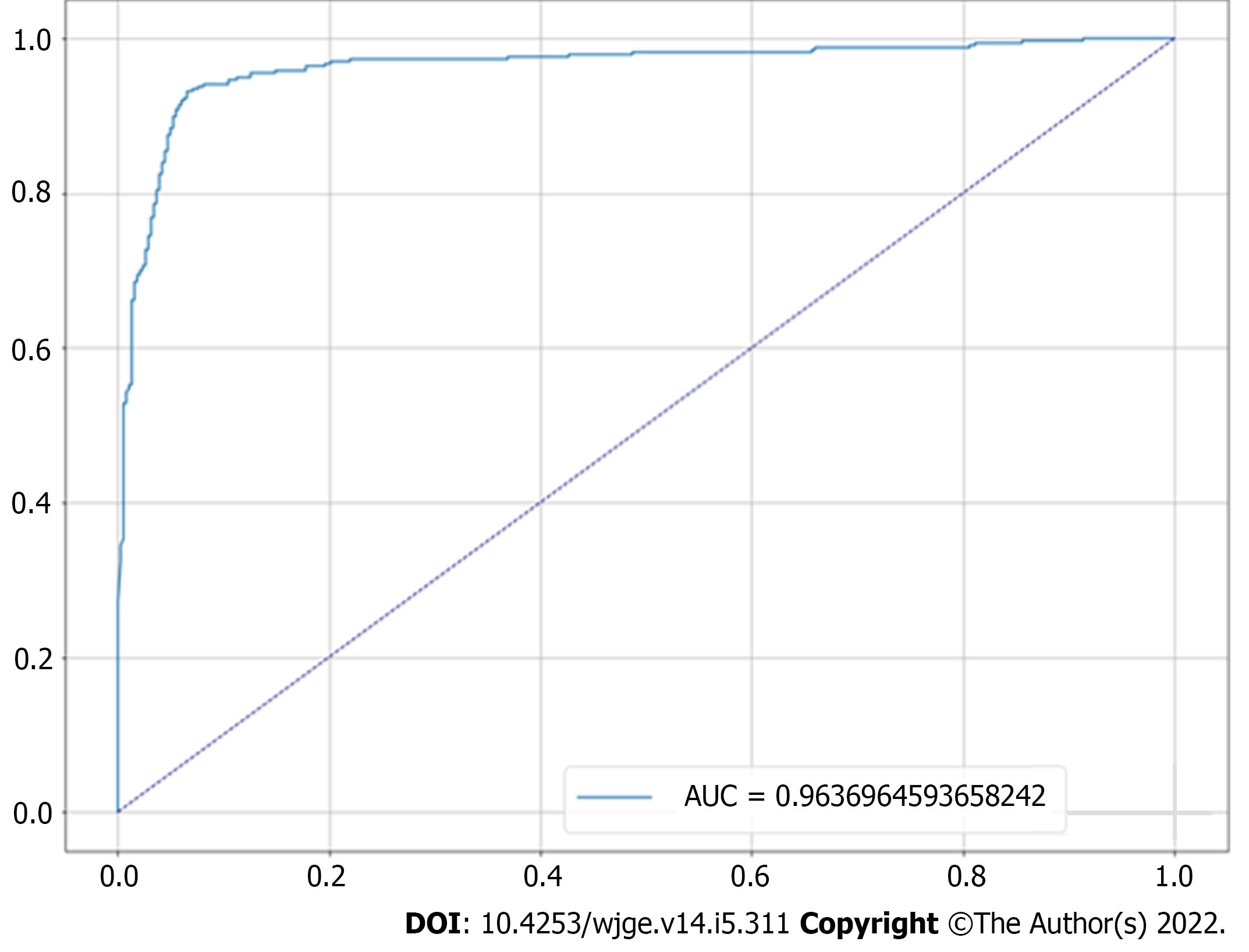
In the test set, which was designated to model evaluation, the classifier achieved 93.32% accuracy, 93.18% sensitivity, 93.46% specificity and a 0.96 AUC. The confusion matrix between true labels and labels predicted by the model is presented in Figure 3, while its receiver operating characteristic curve is presented in Figure 4.
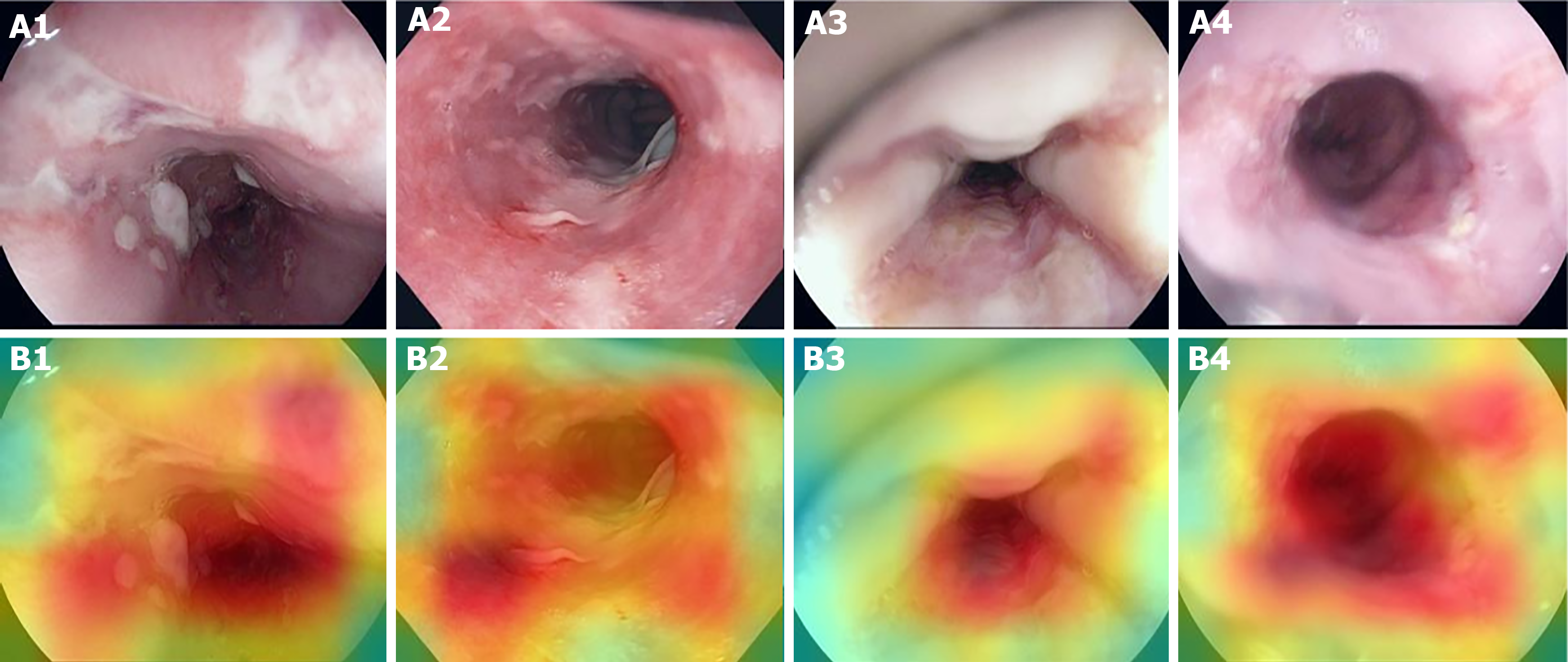
In order to identify the imagery aspects related to the predictive decision, it is possible to plot heatmaps that indicate, colorimetrically, the areas with the greatest influence on the prediction. Examples of such heatmaps for esophagitis images contained in the test set are shown in Figure 5.
This study understands that transfer learning associated with DCNN has great potential to aid and improve the quality and rate of esophagitis diagnosis through endoscopic imaging. Improving workflow, providing faster preliminary reports, relieving the burden of the increasing patient population associated with the intensive and repetitive mechanical work is some of the promises of the integration of CNN-based algorithms to medical practice[15].
Once the mark of at least 93% in the parameters of accuracy, sensitivity, and specificity has been reached, we were able to demonstrate the potential of these algorithms to assist in the premature recognition of pathological predecessor endoscopic abnormalities, and as a consequence, to intervene positively in the management of these. Thus, the use of DCNN with pretrained and fine-tuned weights proves to be a good strategy for predictive modeling of this type (and potentially other types) of medical images. In addition, adopting the classification approach combined with the subsequent plotting of heat maps associated with the classificatory decision gives greater explainability to the model.
In consistency with findings described by Wimmer et al[16], when they established the potential of the association of transfer learning with CNN in the classification of endoscopic images, previously used focused on celiac disease, or also described by Song et al[17] when they reported a deep learning-based model with the ability to histologically classify polyps with a higher accuracy than trained endoscopists, the performance of our algorithmic model reaffirms the potential of deep learning for computer vision in the field of gastrointestinal diagnostics. In line with the mentioned studies, our study demonstrates the already defended potential of CNN-based artificial intelligence systems to diagnose esophageal disease, and can contribute with methodological insights for the development and improvement of such systems[18].
By recognizing changes in the mucosa of the esophageal Z-line, the binary transfer learning classifier presented in this study aims to demonstrate the effectiveness of these algorithms to differentiate endoscopic images of the same topography with and without changes characteristic of esophagitis. Unlike other studies that aimed at automatic detection of anatomical landmarks and diverse diseases affecting different anatomical sites using the KVASIR database[19-22], we employed state-of-the-art deep learning to specifically target Z-line related changes, bringing great accuracy to its analysis.
However, as it is well settled in applications of deep learning in medical image analytics[23], a major limitation of the technical capability of the proposed classifier is the lack of large-scale labeled data. As already shown by Sun et al[24], the performance on artificial intelligence in visual tasks increases logarithmically based on volume of training data size. Coupled with this factor, we cannot ensure how the binary classifier would behave in patients with the presence of other diseases. In both cases, however, training on more plural datasets should optimize performance on the parameters evaluated.
Concerning the predictive behavior towards other possible esophageal Z-line abnormalities, assuming that the algorithm was able to differentiate with high accuracy normal images from images with different degrees of inflammation - and consequently different mucosal lesion configurations - it is reasonable to assume that other esophageal lesions would be differentiated from the healthy aspect, and thus categorized together with the esophagitis images. Among the possible clinical differential situations, esophageal and esophagogastric junction cancers are of particular relevance. Upper endoscopies are considered by the Society of Thoracic Surgeons and the National Comprehensive Cancer Network as the initial diagnostic evaluation to exclude esophageal cancer[25]; although techniques such as chromoendoscopy and narrow band imaging are often used to increase the sensitivity of detection of lesions suggestive of malignancy, traditional endoscopic imaging still plays an important role in the investigational flowchart, and can demonstrate suspicious findings incidentally[26].
In view of this, in order to extend the clinical utility of our proposed algorithm to the investigation of potentially malignant endoscopic findings, two main approaches are possible: (1) Propose an adaptation of the model to multiclass classification and, to this end, retrain the model including endoscopic images of esophageal cancer, fine-tuning, if necessary, only the final layers, making appropriate changes in the final dense layer and in the loss function to accommodate 3 classes (thus, the final layer would now have 3 neurons with softmax activation function, and the sparse categorical crossentropy loss function would be adopted); and (2) Preserve the binary classification structure, but proposing to change the labels for normal and abnormal findings (thus, the model would be used to triage any endoscopic abnormalities, ranging from inflammatory findings to lesions suggestive of malignancy) and, for this purpose, retrain the model including endoscopic images representative of other types of lesions (including neoplastic lesions). In either situation, the incorporation of images representative of lesions suspicious for malignancy would be necessary, and the weights derived from training with normal endoscopic images and with esophagitis findings already performed would be used (same domain fine-tuning).
Convolutional neural networks with transfer learning for automated analysis of endoscopic images, as proposed in this study, may be incorporated into daily practice as a clinical decision support tool - screening abnormalities and indicating the need for further specialized evaluation or double checking medical reports. This application would add value especially in contexts of scarce resources, in which the number of endoscopists is limited and they are often poorly trained - increasing, thus, the likelihood of diagnostic errors. Moreover, it is especially promising as an adjunct tool to telemedicine, favoring rural and remote areas.
The use of deep learning, especially the transfer learning technique, has great potential field for the analysis of clinical images, including endoscopic records. Observing this great potential, this paper applied such technique, associated with retraining of all layers, to classify, with a 93.3% accuracy, esophageal mucosa images obtained from endoscopic studies according to the presence or absence of esophagitis. It then becomes evident the potential of transfer learning with fine-tuning for the analysis of images obtained by endoscopic method and recognition of esophageal lesions.
In view of this, it is opportune to raise new studies involving transfer learning for the analysis of related data, with the aim of improving, disseminating and validating its use for the daily routine of clinical practice. Furthermore, the composition and dissemination high-quality endoscopic image sets representative of various clinical conditions (especially esophageal cancer, given its high clinical and epidemiological relevance) is essential for new studies to be developed and algorithms already proposed to be improved.
Computer vision allied with deep learning, especially through the use of deep convolutional neural networks, has been increasingly employed in the automation of medical image analysis. Among these are endoscopic images, which are of great importance in the evaluation of a number of gastroenterological diseases.
Endoscopic findings constitute the diagnostic definition for esophagitis, a multietiological condition with significant impacts on quality of life and the possibility of evolution to a series of complications. Automating the identification of findings suggestive of esophageal inflammation using artificial intelligence could add great value to the evaluation and management of this clinical condition.
To identify whether a densely connected convolutional neural network with pre-trained and fine-tuned weights is able to binary classify esophageal Z-line endoscopic images according to the presence or absence of esophagitis.
Endoscopic images of 1932 patients with a diagnosis of esophagitis and 1663 patients were splitted in training (80%) and test (20%) and used to develop and evaluate a binary deep learning classifier built using a pre-trained DenseNet-201 architecture. The classifier model performance was evaluated in the test set according to accuracy, sensitivity, specificity and area under the receiver operating characteristic curve.
The proposed model was able to diagnose esophagitis in the validation set with sensitivity of 93.18 and specificity of 93.46, demonstrating the feasibility of using deep transfer learning to discriminate normal from damaged mucosa in endoscopic images of the same anatomical segment. It remains to be investigated whether, by means of a more diverse set of images, this technique can be proposed to identify different types of esophageal abnormalities, and potentially in other organs.
Convolutional neural networks with transfer learning for automated analysis of endoscopic images, as proposed in this study, demonstrate potential for incorporation into clinical practice as a clinical decision support tool, mainly benefiting scarce resources settings.
Sets of endoscopic images representative of various clinical conditions should be published, in order to allow the findings of this study to be externally validated and for new models with different classificatory approaches to emerge.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Morya AK, India S-Editor: Gao CC L-Editor:A P-Editor: Cai YX
| 1. | Gomez Torrijos E, Gonzalez-Mendiola R, Alvarado M, Avila R, Prieto-Garcia A, Valbuena T, Borja J, Infante S, Lopez MP, Marchan E, Prieto P, Moro M, Rosado A, Saiz V, Somoza ML, Uriel O, Vazquez A, Mur P, Poza-Guedes P, Bartra J. Eosinophilic Esophagitis: Review and Update. Front Med (Lausanne). 2018;5:247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 2. | Habbal M, Scaffidi MA, Rumman A, Khan R, Ramaj M, Al-Mazroui A, Abunassar MJ, Jeyalingam T, Shetty A, Kandel GP, Streutker CJ, Grover SC. Clinical, endoscopic, and histologic characteristics of lymphocytic esophagitis: a systematic review. Esophagus. 2019;16:123-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 3. | Hoversten P, Kamboj AK, Katzka DA. Infections of the esophagus: an update on risk factors, diagnosis, and management. Dis Esophagus. 2018;31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 4. | Kim HP, Dellon ES. An Evolving Approach to the Diagnosis of Eosinophilic Esophagitis. Gastroenterol Hepatol (N Y). 2018;14:358-366. [PubMed] |
| 5. | O'Rourke A. Infective oesophagitis: epidemiology, cause, diagnosis and treatment options. Curr Opin Otolaryngol Head Neck Surg. 2015;23:459-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 6. | Lundell LR, Dent J, Bennett JR, Blum AL, Armstrong D, Galmiche JP, Johnson F, Hongo M, Richter JE, Spechler SJ, Tytgat GN, Wallin L. Endoscopic assessment of oesophagitis: clinical and functional correlates and further validation of the Los Angeles classification. Gut. 1999;45:172-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1518] [Cited by in RCA: 1649] [Article Influence: 63.4] [Reference Citation Analysis (1)] |
| 7. | Beam AL, Kohane IS. Big Data and Machine Learning in Health Care. JAMA. 2018;319:1317-1318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 759] [Cited by in RCA: 900] [Article Influence: 128.6] [Reference Citation Analysis (1)] |
| 8. | Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016;316:2402-2410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3669] [Cited by in RCA: 3323] [Article Influence: 369.2] [Reference Citation Analysis (0)] |
| 9. | Stead WW. Clinical Implications and Challenges of Artificial Intelligence and Deep Learning. JAMA. 2018;320:1107-1108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 135] [Article Influence: 19.3] [Reference Citation Analysis (4)] |
| 10. | Weiss K, Khoshgoftaar TM, Wang D. A survey of transfer learning. J Big Data. 2016;3:9. [DOI] [Full Text] |
| 11. | Raghu M, Zhang C, Kleinberg J, Bengio S. Transfusion: Understanding transfer learning for medical imaging. Adv Neural Inf Process Syst. 2019;3347-3357. |
| 12. | Pogorelov K, Randel K, Griwodz C, Eskeland S, Lange T, Johansen D, Spampinato C, Dang-Nguyen D, Lux M. , Schmidt P, Riegler M, Halvorsen P. Kvasir: A Multi-Class Image Dataset for Computer Aided Gastrointestinal Disease Detection. MMSys'17 Proceedings of the 8th ACM on Multimedia Systems Conference (MMSYS); 2017 June 20-23; Taipei, Taiwan. New York: Association for Computing Machinery, 2017: 164-169. |
| 13. | Borgli H, Thambawita V, Smedsrud PH, Hicks S, Jha D, Eskeland SL, Randel KR, Pogorelov K, Lux M, Nguyen DTD, Johansen D, Griwodz C, Stensland HK, Garcia-Ceja E, Schmidt PT, Hammer HL, Riegler MA, Halvorsen P, de Lange T. HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci Data. 2020;7:283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 297] [Cited by in RCA: 128] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 14. | Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely Connected Convolutional Networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21-26; Honolulu, HI, USA. New York: IEEE, 2017: 2261-2269. |
| 15. | Choi J, Shin K, Jung J, Bae HJ, Kim DH, Byeon JS, Kim N. Convolutional Neural Network Technology in Endoscopic Imaging: Artificial Intelligence for Endoscopy. Clin Endosc. 2020;53:117-126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (1)] |
| 16. | Wimmer G, Vécsei A, Uhl A. CNN transfer learning for the automated diagnosis of celiac disease. 2016 Sixth International Conference on Image Processing Theory, Tools and Applications (IPTA); 2016 Dec 12-15; Oulu, Finland. New York: IEEE, 2016: 1-6. |
| 17. | Song EM, Park B, Ha CA, Hwang SW, Park SH, Yang DH, Ye BD, Myung SJ, Yang SK, Kim N, Byeon JS. Endoscopic diagnosis and treatment planning for colorectal polyps using a deep-learning model. Sci Rep. 2020;10:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 18. | Namikawa K, Hirasawa T, Yoshio T, Fujisaki J, Ozawa T, Ishihara S, Aoki T, Yamada A, Koike K, Suzuki H, Tada T. Utilizing artificial intelligence in endoscopy: a clinician's guide. Expert Rev Gastroenterol Hepatol. 2020;14:689-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 19. | Cogan T, Cogan M, Tamil L. MAPGI: Accurate identification of anatomical landmarks and diseased tissue in gastrointestinal tract using deep learning. Comput Biol Med. 2019;111:103351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 20. | Majid A, Khan MA, Yasmin M, Rehman A, Yousafzai A, Tariq U. Classification of stomach infections: A paradigm of convolutional neural network along with classical features fusion and selection. Microsc Res Tech. 2020;83:562-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 21. | Safarov S, Whangbo TK. A-DenseUNet: Adaptive Densely Connected UNet for Polyp Segmentation in Colonoscopy Images with Atrous Convolution. Sensors (Basel). 2021;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 22. | Jha D, Smedsrud PH, Johansen D, de Lange T, Johansen HD, Halvorsen P, Riegler MA. A Comprehensive Study on Colorectal Polyp Segmentation With ResUNet++, Conditional Random Field and Test-Time Augmentation. IEEE J Biomed Health Inform. 2021;25:2029-2040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 84] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 23. | Ker J, Wang L, Rao J, Lim T. Deep Learning Applications in Medical Image Analysis. IEEE Access. 2018;6:9375-9389. [DOI] [Full Text] |
| 24. | Sun C, Shrivastava A, Singh S, Gupta A. Revisiting Unreasonable Effectiveness of Data in Deep Learning Era. 2017 IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22-29; Venice, Italy. Washington: IEEE Computer Society, 2017: 843-852. |
| 25. | Varghese TK Jr, Hofstetter WL, Rizk NP, Low DE, Darling GE, Watson TJ, Mitchell JD, Krasna MJ. The society of thoracic surgeons guidelines on the diagnosis and staging of patients with esophageal cancer. Ann Thorac Surg. 2013;96:346-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 26. | Short MW, Burgers KG, Fry VT. Esophageal Cancer. Am Fam Physician. 2017;95:22-28. [PubMed] |