Published online Apr 8, 2015. doi: 10.4254/wjh.v7.i4.703
Peer-review started: August 30, 2014
First decision: November 1, 2014
Revised: November 5, 2014
Accepted: January 18, 2015
Article in press: January 20, 2015
Published online: April 8, 2015
Processing time: 227 Days and 19.9 Hours
AIM: To evaluate pre-treatment factors associated with sustained virological response (SVR) in patients with hepatitis C virus (HCV) genotype 3 treated with peginterferon and ribavirin (RBV).
METHODS: We retrospectively analyzed treatment naive, mono-infected HCV genotype 3 patients treated with peginterferon and RBV. Exclusion criteria included presence of other liver disease, alcohol consumption and African American or Asian ethnicity. The variables collected and compared between patients who achieved an SVR and patients who did not were as follows: gender, age, fibrosis stage, diabetes, body mass index, steatosis, INFL3 polymorphism, pre-treatment HCV-RNA, type of peginterferon, RBV dose and adherence.
RESULTS: A total of 107 patients treated between June, 2004 and March, 2013 were included. Mean treatment duration was 25.1 (± 1.8) wk. Overall, 58% (62/107) of the patients achieved an SVR and 42% (45/107) did not. In the multivariate logistic regression analysis, pre-treatment HCV-RNA ≥ 600000 UI/mL (OR = 0.375, 95%CI: 0.153-0.919, P = 0.032) and advanced fibrosis (OR = 0.278, 95%CI: 0.113-0.684, P = 0.005) were significantly associated with low SVR rates. In patients with pre-treatment HCV-RNA ≥ 600000 UI/mL and advanced fibrosis, the probability of achieving an SVR was 29% (95%CI: 13.1-45.2). In patients with pre-treatment HCV-RNA < 600000 UI/mL and mild to moderate fibrosis, the probability of achieving an SVR was 81% (95%CI: 68.8-93.4).
CONCLUSION: In patients with HCV genotype 3 infections the presence of advance fibrosis and high pre-treatment viral load might be associated with poor response to peginterferon plus RBV. These patients could benefit the most from new direct antiviral agents-based regimes.
Core tip: Our study evaluates pre-treatment factors associated with sustained virological response in patients with hepatitis C virus genotype 3 treated with peginterferon plus ribavirin. We identified a sub-group of patients with high pre-treatment viral load and advanced fibrosis whose chance of achieving a sustained virological response is as low as 29%. We believe these patients should be prioritized to access new treatment strategies.
- Citation: Marciano S, Borzi SM, Dirchwolf M, Ridruejo E, Mendizabal M, Bessone F, Sirotinsky ME, Giunta DH, Trinks J, Olivera PA, Galdame OA, Silva MO, Fainboim HA, Gadano AC. Pre-treatment prediction of response to peginterferon plus ribavirin in chronic hepatitis C genotype 3. World J Hepatol 2015; 7(4): 703-709
- URL: https://www.wjgnet.com/1948-5182/full/v7/i4/703.htm
- DOI: https://dx.doi.org/10.4254/wjh.v7.i4.703
Hepatitis C virus (HCV) is a major health problem affecting more than 180 million people worldwide[1]. It is estimated that at least 350000 HCV infected people die annually due to liver-related causes[2].
Six genotypes of HCV have been identified. In Latin America, genotype 1 is the most prevalent, followed by genotypes 2 and 3[3,4]. In some areas of this region the prevalence of HCV genotype 3 (HCV-3) is as high as 30%[3].
Traditionally, HCV-2 and HCV-3 have been grouped together as “easy to treat” genotypes. However we now know that sustained virological response (SVR) rates of HCV-3 patients treated with peginterferon plus ribavirin (RBV) are sub-optimal. The global SVR rates for HCV-3 patients treated with peginterferon plus RBV are around 65%-70%[5,6]. Since these data mainly arise from randomized trials, SVR rates are expected to be lower in real-life patients with adverse factors[7].
Several host and viral factors have an impact on the SVR rate of patients infected with HCV-3 treated with peginterferon plus RBV. Pre-treatment factors that have been proposed to have a negative impact on SVR are advanced fibrosis or cirrhosis, male gender, non-Caucasian race, high body weight, diabetes mellitus, and high pre-treatment HCV-RNA[7-12]. More recently, INFL3 (formerly IL28B) polymorphisms were evaluated, but a clear association between the favorable INFL3 genotypes and SVR could not be established[13-15].
A major limitation of the studies that assessed predictors of SVR in HCV-3 patients lies in the fact that they evaluated HCV-2 and HCV-3 together, and difficulties arise when trying to draw conclusions for HCV-3 individually.
By the end of 2013, sofosbuvir was approved for the treatment of chronic HCV-3, being the standard of care in a minority of countries at the moment this manuscript was submitted. However, in regions like Latin America and Middle East/Africa, payer-related barriers were reported[16], which might hamper adequate treatment delivery.
When new treatments become globally available, a careful selection of candidates will be mandatory, particularly for cost-related reasons. Therefore, it will be necessary to identify patients at higher risk of fibrosis progression and treatment failure to current therapies.
Thus, we decided to conduct this study that evaluates pre-treatment variables associated with SVR in patients with chronic HCV-3 treated with peginterferon plus RBV.
This is a retrospective multicenter study performed in 7 Liver Units from Argentina.
The centers involved in the study were, Hospital Italiano from Buenos Aires, Hospital R. Rossi from La Plata, Hospital F. J. Muñiz, Centro de Educación Médica e Investigaciones Clínicas Norberto Quirno (CEMIC), Hospital Universitario Austral, Sanatorio Parque from Rosario, and the HEPATOSUR Group representing the Patagonia.
The survey was discussed by all the participating centers. The local institutional review board of each center approved the study. The study was conducted according to the principles of the Declaration of Helsinki.
Patients included in this study were aged ≥ 18 years and received their first treatment with peginterferon plus RBV for chronic hepatitis C genotype 3. Patients received weekly peginterferon alfa-2a 180 μg or peginterferon alfa-2b 1.5 μg/kg of body weight. Ribavirin dose could be either fixed (800 mg/d) or adjusted to weight (1000 mg/d in patients ≤ 75 kg; 1200 mg/d in patients > 75 kg).
Exclusion criteria included alcohol intake greater than 20 g/d, history of organ transplantation, creatinine clearance < 50 mL/min, co-infection with hepatitis B virus or human immunodeficiency virus and African American or Asian ethnicity. Patients were also excluded if they presented evidence of other liver disease, such as autoimmune hepatitis, primary biliary cirrhosis, sclerosing cholangitis, Wilson’s disease or alfa-1-antitrypsin deficiency.
Each center provided detailed information of patients included in the study. Patient management and treatment candidacy were determined in each center. All data records were checked for missing values and inconsistencies, queries were sent to all participating institutions, and corrections were made at the data coordinating center, namely Hospital Italiano.
Pre-treatment and follow-up blood samples were collected for virologic testing in each participating center. Serum HCV-RNA was either qualitatively or quantitatively evaluated.
The primary end-point was SVR, defined as an undetectable serum HCV-RNA at 24 wk after the end of treatment. Rapid virological response was defined as an undetectable HCV-RNA by treatment week four. Virologic relapse was defined as a detectable HCV-RNA during follow-up in patients who had had undetectable HCV-RNA at the end of treatment. Those patients who never achieved a negative on-treatment HCV-RNA were classified as non-responders.
The variables that were collected and compared between patients who achieved an SVR and patients who did not were as follows: gender, age, fibrosis stage, diabetes, body mass index (BMI), steatosis, INFL3 polymorphism, pre-treatment HCV-RNA, type of peginterferon, RBV dose and adherence.
For the purpose of this study, high HCV-RNA was defined as ≥ 600000 UI/mL.
Fibrosis grade was staged either by biopsy or Transient Elastography (Fibroscan®). In patients without fibrosis evaluation, the aspartate aminotransferase to Platelet Ration Index (APRI) was calculated[17]. Patients were divided into two groups: patients with mild to moderate fibrosis, including patients without fibrosis and patients with up to METAVIR F2 fibrosis; and patients with advanced fibrosis, including patients with METAFIR F3 and cirrhosis[18]. Patients with clinical or endoscopic findings of cirrhosis were included in the advanced fibrosis group.
The presence of steatosis was evaluated either by ultrasound or histology.
Since most patients did not have data regarding INFL3 polymorphism, they were contacted and invited to participate in this study in order to determine the genotype. Patients signed an informed consent before INFL3 genotyping. Genomic DNA was extracted from oral swabs using QIAamp DNA Blood Mini Kit (QIAGEN, GmbH, Hilden, Germany) following the manufacturer’s protocol.
SNP rs12979860 (INFL3) was PCR-amplified from isolated genomic DNA with standard Taq polymerase (Inbio-Highway, Tandil, Argentina)[19]. The PCR-amplified fragments were bi-directionally sequenced using Big-Dye Termination chemistry system (Applied Biosystems, Life Technologies Corp., Foster City, CA, United States). The sequencing chromatogram was analyzed by using the BioEdit Sequence Alignment Editor version 7.1.3.0 in order to discriminate between homozygotes and heterozygotes. Patients were grouped as INFL3 CC and INFL3 non-CC (including patients with INFL3 TT and CT).
In order to generate a predictive model including four variables, at least 40 patients without SVR had to be included. Assuming an SVR rate of 65%-70% in patients with HCV-3 treated with peginterferon plus RBV, between 100 and 110 patients had to be included[5,6].
We presented data as percentages or medians and interquartile ranges. We evaluated the association of pre-treatment characteristics with SVR using the Mann-Whitney test for continuous variables and the χ2 test for categorical variables.
In order to identify independent predictors of SVR we used a logistic regression model for the variables that showed a level of significance lower than 0.1 in the univariate analysis. We compared different models by estimating the area under the receiver operating characteristic (ROC) curve as a measure of predictive accuracy. We chose the model with the greatest area under the ROC and we presented the estimated probabilities predicted by the regression model with their 95%CI. Tests were two-sided and significance was accepted at P < 0.05. Statistical analysis was performed using software SPSS, version 17.0 (Chicago, IL).
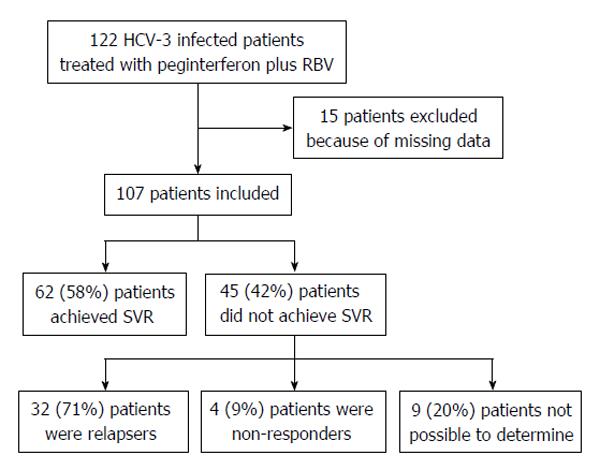
Table 1 provides an overview of the patients’ characteristics. A total of 122 HCV-3 patients treated with peginterferon plus RBV patients were identified. Fifteen were excluded because of missing data, whereas 107 were included (Figure 1). Patients were treated between June/2004 and March/2013. The mean age at treatment was 47.5 (± 8.8) years, and 68% (73/107) of the patients were male. In 93% (103/107) of the patients, a pre-treatment serum sample was collected. Of those patients, 55% (57/103) had high HCV-RNA. Pre-treatment fibrosis could be determined in 97% (104/107) of the patients, either by biopsy (62%), Fibroscan® (26%) or by a combination of clinical or endoscopic features and the APRI score (12%). Of these patients, 40% (41/104) had advanced fibrosis. Steatosis and a BMI > 27 were reported in 48% (51/107) and 54% (58/107) of the patients, respectively. Data regarding INFL3 polymorphism was available in 60% (64/107) of the patients, of whom 28% (18/64) were CC.
| Characteristic | Patients |
| (n = 107) | |
| Age, yr (mean ± SD)1 | 47.5 (± 8.8) |
| Gender | |
| Male | 68.2 (73) |
| Female | 31.8 (34) |
| INFL3 polymorphism2 | |
| CT | 56.2 (36) |
| CC | 28.1 (18) |
| TT | 15.7 (10) |
| Pre treatment HCV-RNA3 | |
| < 600000 UI/mL | 44.7 (46) |
| ≥ 6000000 UI/mL | 55.3 (57) |
| Pre treatment elevated ALT4 | 88.5 (92) |
| Fibrosis5 | |
| Mild to moderate6 | 59.8 (61) |
| Advanced7 | 40.2 (41) |
| Steatosis | 47.6 (51) |
| BMI (kg/m2) > 27 | 54.2 (58) |
| Diabetes | 5.6 (6) |
| Type of peginterferon | |
| alfa 2a | 75.7 (81) |
| alfa 2b | 24.3 (26) |
| RBV dose | |
| Fix8 | 63.6 (68) |
| Weight-Based9 | 36.4 (39) |
| 80/80/80 adherence10 | 94.4 (101) |
Mean treatment durations was 25.1 (± 1.8) wk. Fix dose RBV was prescribed in 64% (68/107) patients, while the remaining ones received a weight-adjusted approach. More than 80% of the peginterferon and RBV planned doses were received by 94% (101/107) of the patients.
Overall, 58% (62/107) of the patients achieved an SVR. Of the 45 patients who did not achieve an SVR, 71% (32/45) were relapsers and 9% (4/45) were non-responders. In 20% (9/45) of the cases, the type of treatment failure was not possible to characterize due to lack of precise viral kinetic information (Figure 1).
In the univariate analysis, SVR rate was significantly lower in patients with high baseline HCV-RNA (OR = 0.330, 95%CI: 0.145-0.755, P = 0.009), advanced fibrosis (OR = 0.274, 95%CI: 0.145-0.755, P = 0.001) and BMI > 27 (OR = 0.422, 95%CI: 0.149-0.892, P = 0.033) (Table 2).
| SVR (n = 62) | No-SVR (n = 45) | OR | 95%CI | P | |
| Age, yr (mean ± SD)1 | 47.8 (± 9.1) | 47.1 (± 8.3) | 1 | 0.965-1.055 | 0.696 |
| Male | 61.3 (38) | 77.8 (35) | 0.425 | 0.190-1.079 | 0.093 |
| INFL3 polymorphism CC2 | 31.4 (11) | 24.1 (7) | 1.44 | 0.475-4.372 | 0.585 |
| 3HCV-RNA ≥ 600000 UI/mL | 44.1 (26) | 70.5 (31) | 0.33 | 0.145-0.755 | 0.009 |
| Pre treatment elevated ALT4 | 86.7 (52) | 90.9 (40) | 0.65 | 0.183-2.312 | 0.553 |
| Advanced fibrosis56 | 26.7 (16) | 59.5 (25) | 0.247 | 0.107-0.573 | 0.001 |
| Steatosis | 46.8 (29) | 48.9 (22) | 0.919 | 0.426-1.981 | 0.847 |
| BMI (kg/m2) > 27 | 46.8 (29) | 64.4 (29) | 0.422 | 0.149-0.892 | 0.033 |
| Diabetes | 2 (1) | 11 (5) | 0.131 | 0.015-1.165 | 0.08 |
| Peginterferon alfa 2a | 75.8 (47) | 75.6 (34) | 0.986 | 0.403-2.413 | 1 |
| RBV fix-dose7 | 64.5 (40) | 62.2 (28) | 1.104 | 0.498-2.447 | 0.841 |
| 80/80/80 adherence8 | 95.2 (59) | 93.3 (42) | 0.75 | 0.113-2.552 | 0.919 |
These variables were included in the multivariate logistic regression analysis and only high HCV-RNA (OR = 0.375, 95%CI: 0.153-0.919, P = 0.032) and advanced fibrosis (OR = 0.278, 95%CI: 0.113-0.684, P = 0.005) had a statistically significant negative association with SVR.
The SVR rate was estimated according to the fibrosis grade and to the pre-treatment HCV-RNA. In patients with low pre-treatment HCV-RNA and mild to moderate fibrosis, the probability of achieving an SVR was 81% (95%CI: 68.8-93.4). In patients with high baseline HCV-RNA and advanced fibrosis, the probability of achieving an SVR was 29% (95%CI: 13.1-45.2) (Table 3).
In the present study, we assessed pre-treatment predictors of SVR in patients infected with HCV-3 who were treated with peginterferon and RBV. We identified high HCV-RNA and advanced fibrosis to be independently associated with treatment failure.
Advanced fibrosis or cirrhosis have consistently been associated with lower rates of SVR in patients with HCV-3 treated with peginterferon plus RBV[9-11,20]. The chances of achieving an SVR after a 24-wk treatment for these patients are 49%-57%[21-23]. Besides, it is estimated that the likelihood of achieving an SVR is reduced by 59% in comparison with patients with lower fibrosis grades[20]. The impact of the pre-treatment HCV-RNA on SVR is more difficult to determine, since different cut-off points were selected in prior studies (400.000-800.000 IU/mL). Even though it has not been consistently associated with lower SVR rates, HCV-3 patients with high pre-treatment viral load do have lower rapid virological response rates[24].
In our study, we found that by combining baseline HCV-RNA and fibrosis stage, SVR rates could be accurately predicted. In patients with low HCV-RNA, and without advanced fibrosis, the SVR rate was 81%. On the contrary, in patients with high baseline HCV-RNA and advanced fibrosis, the SVR rate was 29%.
Other variables that were assessed in our study included age, gender, INFL3 polymorphism, steatosis, adherence, RBV dose, type of peginterferon, diabetes and BMI. None of these were associated with SVR.
Steatosis is known to be more frequent in patients infected with HCV-3 than in other genotypes[25,26]. In our series, 48% of the patients presented steatosis, which was diagnosed either by biopsy or by ultrasonography. Even though HCV-3 is known to cause steatosis through specific viral mechanisms, in our study, 58% of the patients had a BMI > 27, reflecting an overweight population. In line with our results, most studies reported no impact of steatosis on SVR for patients with HCV-3[27,28]. Although the BMI was previously reported to have an impact on SVR[8], prior studies proposed different cut-off points and results were discordant. We selected a cut-off point of 27, and no association with SVR was found. Even though diabetes is associated with higher relapse rates in HCV G3 patients[20], we did not find this association, probably due to the low number of patients with diabetes that were included in the study.
INFL3 polymorphisms were more recently evaluated in patients with HCV-3 treated with peginterferon plus RBV. Putting together all the available information, it seems that INFL3-CC is not globally associated with SVR in HCV-3 patients. Nevertheless, it does predict SVR in the sub-group of patients who do not achieve a rapid virological response[13,15]. We did not find an association between INFL3-CC and SVR.
No differences in the SVR rates were found in patients who received fixed-dose or weight-adjusted RBV. This was an expected finding, since mean treatment duration was 25.7 (± 1.8) wk, and RBV dose does have an impact on SVR when treatment duration is reduced, but not for the 24-wk regimen[21].
In order to evaluate treatment adherence we used the 80/80/80 rule. Overall, 94% of the patients were adherent. No differences were found in SVR rates between patients who were adherent and patients who were not. This was most likely due to the low proportion of patients who were non-compliant.
Until the end of 2013, the only treatment for HCV-3 was peginterferon plus RBV. At that time, sofosbuvir-based regimens were approved and released. With this new approach, more than 90% of the HCV-3 treatment-naïve patients achieve an SVR, regardless of the fibrosis stage[29,30]. When this manuscript was submitted for its publication, sofosbuvir was only approved and available in a minority of countries. Numerous barriers related to patient, provider, government and payers are known to affect HCV treatment accessibility[31]. In developing countries, treatment-related costs constitute a major concern. In fact, in some Latin American countries, after more than three years of the release of Boceprevir and Telaprevir for the treatments of HCV genotype 1, accessibility is still limited.
In our study, we identified a sub-group of HCV-3 infected patients with very low chances of achieving an SVR after treatment peginterferon plus RBV. These patients with high baseline HCV-RNA and advanced fibrosis are in urgent need for new therapeutic approaches and should be prioritized when sofosbuvir or other antivirals become available. A similar approach was recently proposed to identify a sub-group of patients infected with HCV genotype 1 who might benefit from peginterferon plus RBV therapy[32].
Owing to its retrospective nature, our study has several limitations. First, since patients were not consecutive, selection bias is possible, and therefore the SVR rate of our population cannot be extrapolated to the general population. Second, on-treatment virologic kinetics was not evaluated. Rapid virological response is probably the most important predictor of SVR across all genotypes[24]. A major limitation of rapid virological response to predict SVR, lies in the fact that being an on-treatment variable, it is not useful to determine treatment candidacy. Third, even though the number of patients included is small, it adequately represents the sample size estimated to provide the specific power. Finally, although differences regarding treatment protocols between the participating centers might have existed, treatment duration and adherence were globally homogeneous.
In conclusion, our study identified a sub-group of patients with chronic HCV-3 with high baseline viral load and advanced fibrosis whose chances of achieving an SVR with peginterferon plus RBV were poor. These patients are in urgent need for direct antiviral agent-based regimes.
Hepatitis C virus (HCV) is a major health problem affecting more than 180 million people worldwide. It is estimated that at least 350000 HCV infected people die annually due to liver-related causes.
Several host and viral factors have an impact on the sustained virological response (SVR) rate of patients infected with HCV genotype 3 (HCV-3) treated with peginterferon plus ribavirin (RBV). Pre-treatment factors that have been proposed to have a negative impact on SVR are advanced fibrosis or cirrhosis, male gender, non-Caucasian race, high body weight, diabetes mellitus, and high pre-treatment HCV-RNA. More recently, INFL3 (formerly IL28B) polymorphisms were evaluated, but a clear association between the favorable INFL3 genotypes and SVR could not be established. A major limitation of the studies that assessed predictors of SVR in HCV-3 patients lies in the fact that they evaluated HCV-2 and HCV-3 together, and difficulties arise when trying to draw conclusions for HCV-3 individually.
This study was designed to evaluate pre-treatment variables associated with SVR particularly for patients with chronic HCV-3 treated with peginterferon plus RBV.
In the authors’ study, they identified a sub-group of HCV-3 infected patients with very low chances of achieving an SVR after treatment peginterferon plus RBV. In patients with both high baseline HCV-RNA and advanced fibrosis, the probability of achieving an SVR was 29%. They believe that these patients are in urgent need for new therapeutic approaches and should be prioritized when sofosbuvir or other antivirals become available.
Genotype refers to the genetic relatedness of the different HCV species. Six genotypes of HCV have been well characterized. Fibrosis is a process in which scarring occurs in the liver, ultimately leading to cirrhosis, which is the greatest degree of fibrosis. Sustained virological response means viral eradication or cure.
This study was intended to find some factors associated with SVR in patients with HCV-3 treated with peginterferon and ribavirin. Both high viral load and advanced fibrosis were concluded to be associated with low SVR rates. The authors think this manuscript is well written.
P- Reviewer: Lakatos PL, Lisotti A, Luo GH S- Editor: Tian YL L- Editor: A E- Editor: Liu SQ
| 1. | Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333-1342. [PubMed] [DOI] [Full Text] |
| 2. | Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529-538. [PubMed] [DOI] [Full Text] |
| 3. | Kershenobich D, Razavi HA, Sánchez-Avila JF, Bessone F, Coelho HS, Dagher L, Gonçales FL, Quiroz JF, Rodriguez-Perez F, Rosado B. Trends and projections of hepatitis C virus epidemiology in Latin America. Liver Int. 2011;31 Suppl 2:18-29. [PubMed] [DOI] [Full Text] |
| 4. | Szabo SM, Bibby M, Yuan Y, Donato BM, Jiménez-Mendez R, Castañeda-Hernández G, Rodríguez-Torres M, Levy AR. The epidemiologic burden of hepatitis C virus infection in Latin America. Ann Hepatol. 2012;11:623-635. [PubMed] |
| 5. | Tapper EB, Afdhal NH. Is 3 the new 1: perspectives on virology, natural history and treatment for hepatitis C genotype 3. J Viral Hepat. 2013;20:669-677. [PubMed] |
| 6. | Andriulli A, Mangia A, Iacobellis A, Ippolito A, Leandro G, Zeuzem S. Meta-analysis: the outcome of anti-viral therapy in HCV genotype 2 and genotype 3 infected patients with chronic hepatitis. Aliment Pharmacol Ther. 2008;28:397-404. [PubMed] [DOI] [Full Text] |
| 7. | Marcellin P, Cheinquer H, Curescu M, Dusheiko GM, Ferenci P, Horban A, Jensen D, Lengyel G, Mangia A, Ouzan D. High sustained virologic response rates in rapid virologic response patients in the large real-world PROPHESYS cohort confirm results from randomized clinical trials. Hepatology. 2012;56:2039-2050. [PubMed] [DOI] [Full Text] |
| 8. | Sarin SK, Kumar CK. Treatment of patients with genotype 3 chronic hepatitis C--current and future therapies. Liver Int. 2012;32 Suppl 1:141-145. [PubMed] [DOI] [Full Text] |
| 9. | Shiffman ML, Suter F, Bacon BR, Nelson D, Harley H, Solá R, Shafran SD, Barange K, Lin A, Soman A. Peginterferon alfa-2a and ribavirin for 16 or 24 weeks in HCV genotype 2 or 3. N Engl J Med. 2007;357:124-134. [PubMed] [DOI] [Full Text] |
| 10. | Mangia A, Santoro R, Minerva N, Ricci GL, Carretta V, Persico M, Vinelli F, Scotto G, Bacca D, Annese M. Peginterferon alfa-2b and ribavirin for 12 vs 24 weeks in HCV genotype 2 or 3. N Engl J Med. 2005;352:2609-2617. [PubMed] [DOI] [Full Text] |
| 11. | Yu ML, Dai CY, Huang JF, Hou NJ, Lee LP, Hsieh MY, Chiu CF, Lin ZY, Chen SC, Hsieh MY. A randomised study of peginterferon and ribavirin for 16 versus 24 weeks in patients with genotype 2 chronic hepatitis C. Gut. 2007;56:553-559. [PubMed] [DOI] [Full Text] |
| 12. | Shah SR, Patel K, Marcellin P, Foster GR, Manns M, Kottilil S, Healey L, Pulkstenis E, Subramanian GM, McHutchison JG. Steatosis is an independent predictor of relapse following rapid virologic response in patients with HCV genotype 3. Clin Gastroenterol Hepatol. 2011;9:688-693. [PubMed] [DOI] [Full Text] |
| 13. | Mangia A, Thompson AJ, Santoro R, Piazzolla V, Tillmann HL, Patel K, Shianna KV, Mottola L, Petruzzellis D, Bacca D. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821-827, 827.e1. [PubMed] [DOI] [Full Text] |
| 14. | Sarrazin C, Susser S, Doehring A, Lange CM, Müller T, Schlecker C, Herrmann E, Lötsch J, Berg T. Importance of IL28B gene polymorphisms in hepatitis C virus genotype 2 and 3 infected patients. J Hepatol. 2011;54:415-421. [PubMed] [DOI] [Full Text] |
| 15. | Moghaddam A, Melum E, Reinton N, Ring-Larsen H, Verbaan H, Bjøro K, Dalgard O. IL28B genetic variation and treatment response in patients with hepatitis C virus genotype 3 infection. Hepatology. 2011;53:746-754. [PubMed] [DOI] [Full Text] |
| 16. | McGowan CE, Monis A, Bacon BR, Mallolas J, Goncales FL, Goulis I, Poordad F, Afdhal N, Zeuzem S, Piratvisuth T. A global view of hepatitis C: physician knowledge, opinions, and perceived barriers to care. Hepatology. 2013;57:1325-1332. [PubMed] [DOI] [Full Text] |
| 17. | Lin ZH, Xin YN, Dong QJ, Wang Q, Jiang XJ, Zhan SH, Sun Y, Xuan SY. Performance of the aspartate aminotransferase-to-platelet ratio index for the staging of hepatitis C-related fibrosis: an updated meta-analysis. Hepatology. 2011;53:726-736. [PubMed] [DOI] [Full Text] |
| 18. | Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289-293. [PubMed] [DOI] [Full Text] |
| 19. | Ito K, Higami K, Masaki N, Sugiyama M, Mukaide M, Saito H, Aoki Y, Sato Y, Imamura M, Murata K. The rs8099917 polymorphism, when determined by a suitable genotyping method, is a better predictor for response to pegylated alpha interferon/ribavirin therapy in Japanese patients than other single nucleotide polymorphisms associated with interleukin-28B. J Clin Microbiol. 2011;49:1853-1860. [PubMed] [DOI] [Full Text] |
| 20. | Shoeb D, Rowe IA, Freshwater D, Mutimer D, Brown A, Moreea S, Sood R, Marley R, Sabin CA, Foster GR. Response to antiviral therapy in patients with genotype 3 chronic hepatitis C: fibrosis but not race encourages relapse. Eur J Gastroenterol Hepatol. 2011;23:747-753. [PubMed] [DOI] [Full Text] |
| 21. | Zeuzem S, Rizzetto M, Ferenci P, Shiffman ML. Management of hepatitis C virus genotype 2 or 3 infection: treatment optimization on the basis of virological response. Antivir Ther. 2009;14:143-154. [PubMed] |
| 22. | Cornberg M, Razavi HA, Alberti A, Bernasconi E, Buti M, Cooper C, Dalgard O, Dillion JF, Flisiak R, Forns X. A systematic review of hepatitis C virus epidemiology in Europe, Canada and Israel. Liver Int. 2011;31 Suppl 2:30-60. [PubMed] [DOI] [Full Text] |
| 23. | Lagging M, Langeland N, Pedersen C, Färkkilä M, Buhl MR, Mørch K, Dhillon AP, Alsiö A, Hellstrand K, Westin J. Randomized comparison of 12 or 24 weeks of peginterferon alpha-2a and ribavirin in chronic hepatitis C virus genotype 2/3 infection. Hepatology. 2008;47:1837-1845. [PubMed] [DOI] [Full Text] |
| 24. | Fried MW, Hadziyannis SJ, Shiffman ML, Messinger D, Zeuzem S. Rapid virological response is the most important predictor of sustained virological response across genotypes in patients with chronic hepatitis C virus infection. J Hepatol. 2011;55:69-75. [PubMed] [DOI] [Full Text] |
| 25. | Leandro G, Mangia A, Hui J, Fabris P, Rubbia-Brandt L, Colloredo G, Adinolfi LE, Asselah T, Jonsson JR, Smedile A. Relationship between steatosis, inflammation, and fibrosis in chronic hepatitis C: a meta-analysis of individual patient data. Gastroenterology. 2006;130:1636-1642. [PubMed] [DOI] [Full Text] |
| 26. | Rubbia-Brandt L, Quadri R, Abid K, Giostra E, Malé PJ, Mentha G, Spahr L, Zarski JP, Borisch B, Hadengue A. Hepatocyte steatosis is a cytopathic effect of hepatitis C virus genotype 3. J Hepatol. 2000;33:106-115. [PubMed] |
| 27. | Patton HM, Patel K, Behling C, Bylund D, Blatt LM, Vallée M, Heaton S, Conrad A, Pockros PJ, McHutchison JG. The impact of steatosis on disease progression and early and sustained treatment response in chronic hepatitis C patients. J Hepatol. 2004;40:484-490. [PubMed] [DOI] [Full Text] |
| 28. | Rodriguez-Torres M, Govindarajan S, Diago M, Morgan T, Anand B, Barange K, Suter F, Lin A, Hooper G, Shiffman M. Hepatic steatosis in patients with chronic hepatitis C virus genotype 2 or 3 does not affect viral response in patients treated with peginterferon alpha-2a (40KD) (PEGASYS) plus ribavirin (COPEGUS) for 16 or 24 weeks. Liver Int. 2009;29:237-241. [PubMed] [DOI] [Full Text] |
| 29. | Zeuzem S, Dusheiko GM, Salupere R, Mangia A, Flisiak R, Hyland RH, Illeperuma A, Svarovskaia E, Brainard DM, Symonds WT. Sofosbuvir and ribavirin in HCV genotypes 2 and 3. N Engl J Med. 2014;370:1993-2001. [PubMed] [DOI] [Full Text] |
| 30. | European Association for Study of Liver. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol. 2014;60:392-420. [PubMed] [DOI] [Full Text] |
| 31. | Mcgowan CE, Fried MW. Barriers to hepatitis C treatment. Liver International. 2012;57:151-156. [DOI] [Full Text] |
| 32. | Andriulli A, Nardi A, Di Marco V, Ippolito AM, Gavrila C, Aghemo A, Di Paolo D, Squadrito G, Grassi E, Calvaruso V. An a priori prediction model of response to peginterferon plus ribavirin dual therapy in naïve patients with genotype 1 chronic hepatitis C. Dig Liver Dis. 2014;46:818-825. [PubMed] [DOI] [Full Text] |