Published online Oct 28, 2015. doi: 10.4254/wjh.v7.i24.2535
Peer-review started: June 29, 2015
First decision: July 25, 2015
Revised: August 24, 2015
Accepted: September 25, 2015
Article in press: September 28, 2015
Published online: October 28, 2015
Processing time: 124 Days and 22.2 Hours
Infection by hepatitis C virus (HCV), a plus-stranded RNA virus that can cause cirrhosis and hepatocellular carcinoma, is one of the major health problems in the world. HCV infection is considered as a multi-step complex process and correlated with abnormal metabolism of lipoprotein. In addition, virus attacks hepatocytes by the initial attaching viral envelop glycoprotein E1/E2 to receptors of lipoproteins on host cells. With the development of HCV model system, mechanisms of HCV cell entry through lipoprotein uptake and its receptor have been extensively studied in detail. Here we summarize recent knowledge about the role of lipoprotein receptors, scavenger receptor class B type I and low-density lipoprotein receptor in the entry of HCV, providing a foundation of novel targeting therapeutic tools against HCV infection.
Core tip: As cirrhosis and hepatocellular carcinoma caused by hepatitis C virus (HCV) is one of the major health problems in the world, the investigation of HCV infection becomes more and more important. HCV entry is the initial step to start infection and is a multiple process involved in abnormal metabolism of lipid. Hence, here we summarize recent knowledge about the role of lipoprotein receptors for better understanding of HCV.
- Citation: Lyu J, Imachi H, Fukunaga K, Yoshimoto T, Zhang H, Murao K. Roles of lipoprotein receptors in the entry of hepatitis C virus. World J Hepatol 2015; 7(24): 2535-2542
- URL: https://www.wjgnet.com/1948-5182/full/v7/i24/2535.htm
- DOI: https://dx.doi.org/10.4254/wjh.v7.i24.2535
Hepatitis C virus (HCV) mainly affects liver and causes infectious disease hepatitis C in the world[1]. As a RNA virus, HCV infects about 2%-4% of people all over the world and induces kinds of liver diseases, including about 343000 deaths due to liver cancer from HCV occurred in 2013 up from 198000 in 1990 and an additional 358000 in 2013 occurred due to cirrhosis[2]. Different from hepatitis A virus and hepatitis B virus, there is no available vaccine against HCV until now, and current therapy for HCV infection is based on direct-acting antivirals with or without peginterferon plus ribavirin[3,4]. Hence, knowing the mechanism of HCV infection is becoming more and more important.
The HCV belongs to the family Flaviviridae and is a kind of the genus hepacivirus. Based on the differences of nucleotide sequence, which is 30%-35% varying over the complete genome, it is classified into seven genotypes[5]. Sixty percent of all cases are caused by subtypes 1a and 1b and both types of HCV are able to be found all around the world. For further study on HCV entry and the elucidation of the mechanisms of HCV infection, HCV-like particles (HCV-LP), HCV pseudotyped particles (HCVpp) and HCV cell culture (HCVcc) system is widely developed recently[6-8]. HCV-LPs, production of baculovirus expression systems, can infect both hepatoma cells and human primary hepatocytes by the mediation of its receptors[9]. However, they are lack of reporter to reflect the earliest stages of infection[10]. The disadvantage of HCV-LPs is made up by HCVpp, which is produced by lentiviral particles incorporating unmodified HCV glycoproteins into the lipid envelope[11,12]. HCVpp mimics the very early stage of cell entry by carrying a marker gene. Only one deficiency of HCVpp system, they cannot be associated with lipoproteins, because it is lack of lipoproteins in the producing cells, 293T kidney cells[10]. Following, HCVcc system is developed to represent the complete replication cycle of virus and to release the production of authentic virus particles that are able to infect in vitro and in vivo[13,14]. Until now, HCVcc system is the only model system which can completely mimic a natural HCV infection, although its recipient cells are limited to two specific cell lines, LH86 and Huh-7[10]. Unfortunately, there are few suitable small animal models for the research of HCV and only used for certain aspects of HCV infection in vivo[15,16].
In a typical HCV particle, a core of genetic material (a positive single-standard RNA), which consists of a single open reading frame of 9600 nucleotide bases long, is surrounded by a protective shell of nonstructural protein (NS2, NS3, NS4A, NS4B, NS5A and NS5B), and further encased in a lipid envelope[10]. There are two kinds of viral envelope glycoproteins, E1 and E2, which are embedded in the lipid envelope. Since nonstructural proteins play important roles in viral self-replication, both envelope proteins are necessary and serve as the fusogenic subunit during the process of HCV entry; particularly, E2 acts as the receptor binding protein[17,18]. Based on this, soluble form of recombinant E2 glycoprotein (sE2) was synthesized to study the receptors of HCV in cell entry[19,20].
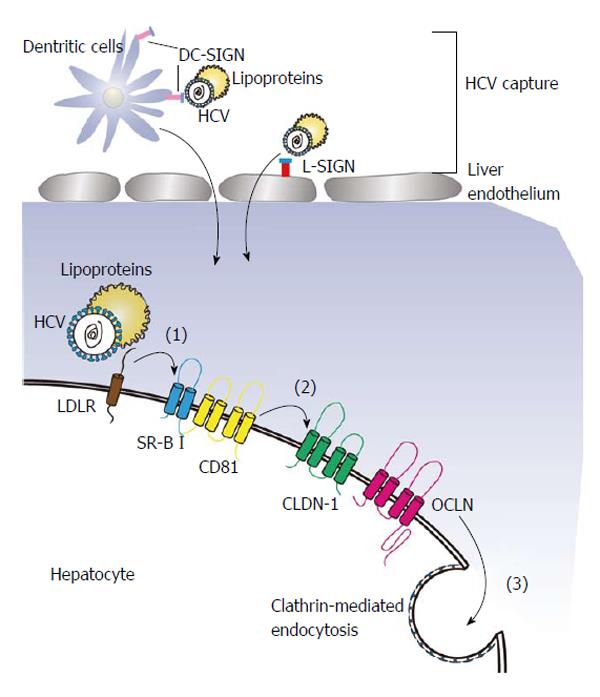
Infecting a target cell by HCV entails an orchestrated process which can be described into several steps starting from the binding of the viral particles to receptors with co-receptors[21]. Usually, the interaction of glycoproteins (E1 and E2) on the viral surface and specific receptors on the surface of target cell determines the association of a HCV with a target cell. Here, we define the process of viral entry into cells into a three-step process (Figure 1). Initially, HCV recognizes a target cell by binding to the mannose-binding lectins L-SIGN, which is mainly expressed on the endothelium of liver and DC-SIGN, which is expressed on dendritic cells. Both of the cell surface proteins are believed to function as HCV capture receptors[22]. Laterly, the viral glycoproteins interacts with the CD81 tetraspanin[23] and lipoprotein receptors[24-26], transferring the virus from the surface to side gradually. Finally, tight junction proteins may be utilized to help HCV entry by inducing clathrin-mediated endocytosis.
During the process of HCV cell entry, CD81 teraspanin, which contains a small extracellular loop, a large extracellular loop (LEL), four transmembrane domains and intracellular N- and C-terminal domains, plays an important role. It was firstly reported that CD81 interacted with a soluble HCV glycoprotein E2 and blockade of CD81 by a specific antibody or silencing of CD81 inhibited the HCV entry and decreased HCV infectivity, demonstrating that CD81 is necessary for the entry of HCV[19,27,28]. In more detailed, the initial step of HCV binding to CD81 is actually the linking between HCV glycoprotein E2 and the LEL of CD81[29], showing LEL served directly in HCV entry. Subsequent research pointed out the relation between CD81 expression on cell surface and membrane lipid composition, that ceramide enrichment of the plasma membrane strongly inhibited the expression of CD81. As lipids organization on the membrane of host cells is essential for HCV entry, internalization of CD81 induced by ceramide inhibited HCV entry[30]. In HepG2 cells and Huh-7, which are derived from hepatoma, CD81 was also demonstrated to affect the susceptibility to HCV infection and the efficiency of HCV entry[31-33]. In addition, the dynamic of CD81, which is dependent on the hepatocytes polarization, could regulate HCV infection[34] and the trafficking of CD81 on the host cell membrane promoted claudin-1-dependent HCV particle internalization[35]. Recently, it was demonstrated that the expression of CD81 also modulated HCV RNA replication[36], suggesting that the HCV life-cycle also requires CD81.
Recent study pointed out that multiple RTKs could mediate HCV entry by regulating CD81-claudin-1 and viral glycoprotein-dependent membrane fusion[37]. Liu et al[38] also found that HCV transiently activates the phosphatidylinositol-3-kinase/AKT pathway to facilitate its entry. These findings may contribute to a new approach to prevention and treatment of HCV infection.
The metabolism of apolipoproteins, lipids and lipoproteins is mainly regulated by the liver and HCV attacks liver, leading to abnormal serum lipoproteins and accumulation of lipids in hepatic cells in a chronic mode[39-41]. In recent years, the relationship between cholesterol metabolism and fatty acid biosynthetic pathways in target cells and HCV infection has gained much attention. As a result, the role of lipoprotein receptor in the HCV entry is extensively investigated in detail. Hence, we will focus on roles of the two lipoprotein receptors, scavenger receptor class B type I (SR-BI) and low-density lipoprotein receptor (LDLR) in this review.
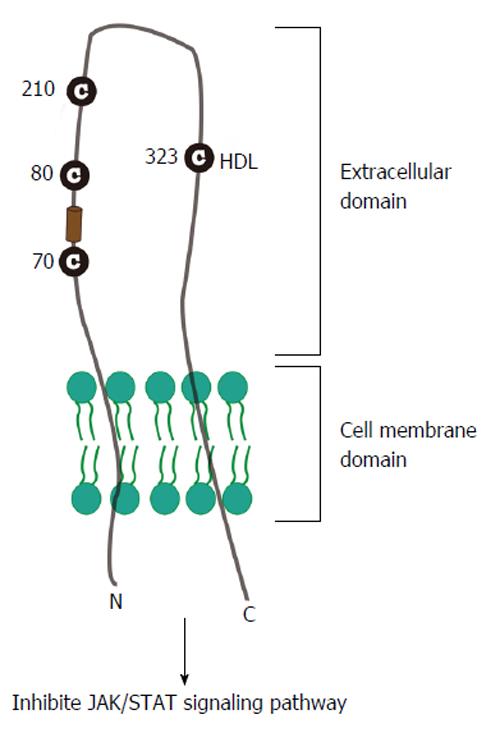
SR-BI, as a 509 amino acid glycoprotein, is an integral membrane receptor with cytoplasmic C-terminal and N-terminal domains separated by a large extracellular domain (Figure 2); and is found in numerous cell types and tissues, including the liver and adrenal. There are evidences that SR-BI selectively mediates uptake of high-density lipoprotein (HDL) cholesterol ester (CE) into transfected Chinese hamster ovary cells[42] and C323 of SR-BI is critical for SR-BI-mediated cholesterol ester uptake[43]. Previous study also proves that the human homologue of SR-BI, CD36 and LIMPII Analogous-1 (hSR-BI/CLA-1), serves as a receptor of HDL and regulates cholesterol efflux to HDL during the process of reverse cholesterol transport[44-47].
Recent reports indicate that HDL promoted HCV entry and this enhancement was mediated by the formation of SR-BI, HDL and HCV envelope glycoproteins complex[20,48,49]. Many groups demonstrated that the glycoprotein E2 could bind SR-BI in hepatoma cells: Scarselli et al[20] demonstrated that extracellular domain of SR-BI interacts with E2 hypervariable region 1 (HVR1)[20,26]; Catanese et al[50] found out that amino acids 70-87 and the single residue E210 of SR-BI are required for E2 recognition, raising a possibility for new therapeutic strategies targeting virus/SR-BI recognition. Based on the detailed internship between virus and SR-BI, Murao et al[25] point out that interferon alpha decreases the efficiency of HCV infection by down regulating the binding of SR-BI with both synthesized E2 region-I (4931) and E2 region-II (4938) peptides in HepG2 cells. Vercauteren et al[51] recently found a new anti-SR-BI antibody, small molecule inhibitors monoclonal antibody1671 (mAb1671), significantly inhibited infection of hepatoma cells with wild-type HCV by inhibiting the function of SR-BI, suggesting that mAb1671 could be used as a therapeutic antibody.
SR-BI not only acts as a binding receptor of HCV, but also plays a critical role in the post-binding steps. Catanese et al[50] and Zeisel et al[52] showed that the susceptibility of human hepatoma cells to HCVcc infection is markedly reduced by silencing of SR-BI with specific siRNA, SR-BI specific antibody or mutation of SR-BI and the effect is independent of lipoprotein, pointing out the role of SR-BI in the post-binding process.
HCV particles associated with plasma lipoproteins like HDL can be found in the viral particles isolated from patients and the abnormal metabolism of lipid influences the HCV infection, suggesting that HCV entry might also potentially involve the interactions with SR-BI ligands, HDL. Although HCVpp is not able to interact with HDL apolipoprotein, the increase of HDL still markedly induces the enhancement of HCVpp entry, while inhibition of the transfer of HDL CE reduces the entry of HCVpp into cells[49,53]. However, the ability of HDL to facilitate HCV entry is largely in a SR-BI-dependent manner since silencing of this receptor cancelled the effect of HDL on enhancement of viral entry[49].
Another goal of recent researches was to examine the signaling pathways involved in the HCV infection in more detail. After binding of glycoprotein E2 to SR-BI, multiple signaling pathways will be inactivated to facility HCV entry in host cells. There is a report points out that HCV selectively decreased the abundance of signal transducer and activator of transcription 1 (STAT1) and reduced the phosphorylation of STAT1 in the nucleus by binding its core protein to STAT1 in a proteasome-dependent manner to defense the immunity induced by JAK/STAT pathway[54]. In turn, the treatment with interferon alpha was proved to phosphorylate STAT1 to protect the host cells from infection of HCV[25]. STAT3 activation in human hepatocytes was also confirmed to resist an attack from HCV infection in vitro[55]. Clinically, treatment with interferon alpha and ribavirin is one of the therapies for chronic HCV infection. Type 1 interferon is a production from host cells infected with virus and constitute the primary defense mechanism against viral infection and replication[56]. Secreted interferon acts through an autocrine and paracrine loop that requires intact interferon receptor and JAK/STAT pathways involving STAT family members[57].
In summary, all of these evidences point out the critical role of SR-BI in enhancing HCV entry into hepatic cells and the complicated process requires the complex between lipoproteins, SR-BI, and HCV envelope glycoproteins. The SR-BI gene is able to transcript into two mRNA splice variants, SR-BI and SR-BII and the two variants are different from their C-termini. Although there is evidence that HCV soluble envelope glycoprotein E2 is able to interact with not only human SR-BI but also SR-BII[58], the role of SR-BII in the HCV entry is rarely reported.
In the family of scavenger receptors, there is another member named scavenger receptor class A (SR-A), which is mainly expressed in macrophage. It is composed of a cytosol domain, a transmembrane domain, a spacer domain, an alpha-helical coiled-coil domain, a collagen-like domain and a cysteine-rich domain and has two different types, SR-AI and SR-AII[59]. Different from the SR-B, the main function of SR-A in innate immunity is defense of bacteria. Recently, it was reported that SR-AI could bind to the non-structural protein NS3 of HCV in dendritic cells, pointing out that SR-A may serve as endocytic innate receptors in NS3 recognition[60].
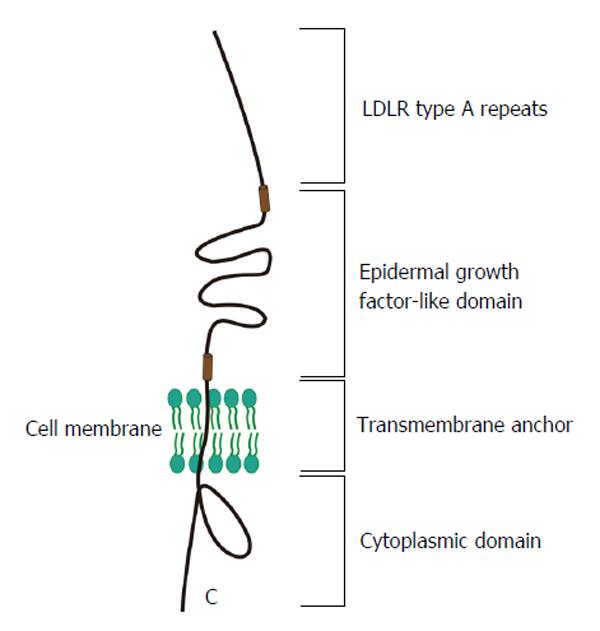
LDLR is another potential lipoprotein receptor involved in HCV infection of hepatocytes. LDLR (Figure 3), an 893 amino acids transmembrane protein, is a cell surface receptor that mediates uptake of cholesterol-rich low-density lipoprotein (LDL)[61,62]. When the main ligand cholesterol-LDL binds to the receptor, it is transferred into hepatic cells by clathrin-mediated endocytosis and then the receptor will release the bound LDL particle because of the conformational change induced by change in pH. Accumulation of serum LDL directly leads to the development of atherosclerosis.
Since Agnello et al[63] firstly suggested the role of LDL-R in HCV entry in 1999, most studies focus on its role as a receptor of HCV or facilitating initial attachment to cell surface[10]. André et al[64] reported that lipo-viro-particles isolated from patients with hepatoma infect hepatic cells in an LDLR-dependent manner, indicating the important role of LDLR in HCV infection. In coincidence with this report, LDLR is confirmed to take part in an early stage in infection of normal human hepatocytes by serum-derived HCV virions in vitro[65]. A human study that LDLR expression of 68 patients with HCV chronic infection was significantly associated with HCV-viral load, supplies the evidence that the LDLR may be one of the receptors implicated in HCV replication[66].
While HDL facilities the entry of HCV into hepatic cells, LDL could significantly inhibit the cell entry of serum HCV and HCVpp via LDLR. Some studies also reported that ApoE-containing very LDL, as a ligands of LDLR, mediates the HCV entry in vitro. Recently, Ficolin-2, as a lectin-complement pathway activator, inhibited the chronic HCV infection by inhibition the function of LDLR and SR-BI and this effect was blocked by ApoE3-mediated immune escape[67]. It was confirmed by a recent report that ApoE3 and ApoE4 rescue the production of infectious virus and it requires both the LDLR and SR-BI[68]. By contrast, Prentoe et al[26] found in the process of HCV entry, the function of LDLR is in an ApoE-independent but E2 HVR1-dependent manner. Although there are lots of evidences to prove that LDLR, the same as SR-BI and CD81, plays a critical role in the initial step of HCV entry, one of studies suggested that LDLR is not necessary for HCV entry and implied the physiological function of LDLR in HCV replication[69]. Recently, it was demonstrated that HCV upregulates the expression of LDLR via SREBPs and PCSK9 at both transcriptional and posttranslational level to increase the uptake of lipid and to promote viral proliferation[70]. Until now, there is no doubt that LDLR is able to mediate the HCV infection. However, the detailed mechanism how it really works during this complex process needs to be further investigated.
By using screening cDNA library, two kinds of tight junction proteins, claudin-1 (CLDN-1) and occludin (OCLN), were identified as factors that are able to affect the HCV entry in the later phase[71,72]. Either CLDN-1 or OCLN contains four transmembrane domains and two extracellular loops with the N-terminus and C-terminus in the cytoplasm. Interestingly, there is no evidence to confirm that there is direct interaction between CLDN-1 or OCLN and HCV particles. However, it was proved that CLDN-1 directly interacts to CD81 and the association increases the virus entry in the later phase[73]. Laterly, Krieger et al[74] produced CLDN-1 specific antibody and found it inhibited HCV infection by reducing the binding of E2 with host cell surface and disrupting the formation of CD-81-CLDN-1 complex. OCLN is also able to interact directly with E2, and silence of CLDN-1 and OCLN by specific siRNA reduced both HCVpp and HCVcc cell entry[75].
Besides the receptors we talked above, there are some other factors on host cell surface, which are believed to be functional in HCV entry. Lupberger et al[37] pointed out the important role of epidermal growth factor receptor (EGFR) and ephrin receptor A2 (EphA2) as cofactors in HCV entry. EGF accelerated HCV entry by activating signaling pathways and inhibition of EGFR or EphA2 activity reduced CD81-CLDN1 association. Following, Diao et al[76] confirmed that EGFR internalization and activation are critical for HCV entry and firstly identified a hitherto-unknown association between CD81 and EGFR by using HCVcc system. Based on these theories, Meyer et al[77] recently supposed a model that interferon-α inducible protein 6 inhibits HCV entry by impairing EGFR mediated CD81/CLDN1 interactions. Niemann-Pick C1-like 1 (NPC1L1), as a cholesterol uptake receptor was firstly identified as an HCV entry factor by Sainz et al[78] and they also proved clinically available FDA-approved NPC1L1 antagonist ezetimibe potently blocks HCV uptake in vitro via a virion cholesterol-dependent step, discovering a new antiviral target and potential therapeutic agent. Furthermore, transferrin receptor 1 has also been reported as a receptor for HCV entry[79]. However, the roles of these new factors in HCV entry remain to be determined in detailed.
The process of HCV entry is a multi-step process and the major steps have already been described as the combination of HCV glycoprotein and targeting cell-surface molecules, such as CD81 and lipoprotein receptor SR-BI and LDLR. With the development of HCV model system, the role of lipoprotein and its receptor in HCV infection is more and more detailed understood. However, since all the model system has their own limitations, the results obtained by using system in vitro do not completely reflect the in vivo situation. Further studies are required, especially by using engineering new animal models, for HCV infection, and a detailed understanding of the mechanism of HCV entry will give a sufficient groundwork for the development of new therapeutic drugs and tools.
P- Reviewer: Kanda T, Larrubia JR S- Editor: Ji FF L- Editor: A E- Editor: Liu SQ
| 1. | Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1770] [Cited by in RCA: 1847] [Article Influence: 153.9] [Reference Citation Analysis (3)] |
| 2. | GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5495] [Cited by in RCA: 5243] [Article Influence: 524.3] [Reference Citation Analysis (0)] |
| 3. | Kanda T, Imazeki F, Yokosuka O. New antiviral therapies for chronic hepatitis C. Hepatol Int. 2010;4:548-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 4. | Kanda T, Yokosuka O, Omata M. Treatment of hepatitis C virus infection in the future. Clin Transl Med. 2013;2:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 5. | Ohno O, Mizokami M, Wu RR, Saleh MG, Ohba K, Orito E, Mukaide M, Williams R, Lau JY. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol. 1997;35:201-207. [PubMed] |
| 6. | Zeisel MB, Fofana I, Fafi-Kremer S, Baumert TF. Hepatitis C virus entry into hepatocytes: molecular mechanisms and targets for antiviral therapies. J Hepatol. 2011;54:566-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 140] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 7. | Meyer K, Basu A, Przysiecki CT, Lagging LM, Di Bisceglie AM, Conley AJ, Ray R. Complement-mediated enhancement of antibody function for neutralization of pseudotype virus containing hepatitis C virus E2 chimeric glycoprotein. J Virol. 2002;76:2150-2158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | Lagging LM, Meyer K, Westin J, Wejstål R, Norkrans G, Lindh M, Ray R. Neutralization of pseudotyped vesicular stomatitis virus expressing hepatitis C virus envelope glycoprotein 1 or 2 by serum from patients. J Infect Dis. 2002;185:1165-1169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 9. | Baumert TF, Ito S, Wong DT, Liang TJ. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J Virol. 1998;72:3827-3836. [PubMed] |
| 10. | Burlone ME, Budkowska A. Hepatitis C virus cell entry: role of lipoproteins and cellular receptors. J Gen Virol. 2009;90:1055-1070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 153] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 11. | Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633-642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 885] [Cited by in RCA: 879] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 12. | Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci USA. 2003;100:7271-7276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 655] [Cited by in RCA: 644] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 13. | Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2323] [Cited by in RCA: 2275] [Article Influence: 113.8] [Reference Citation Analysis (0)] |
| 14. | Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1843] [Cited by in RCA: 1850] [Article Influence: 92.5] [Reference Citation Analysis (0)] |
| 15. | Barth H, Robinet E, Liang TJ, Baumert TF. Mouse models for the study of HCV infection and virus-host interactions. J Hepatol. 2008;49:134-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 16. | Boonstra A, van der Laan LJ, Vanwolleghem T, Janssen HL. Experimental models for hepatitis C viral infection. Hepatology. 2009;50:1646-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 17. | Kobayashi M, Bennett MC, Bercot T, Singh IR. Functional analysis of hepatitis C virus envelope proteins, using a cell-cell fusion assay. J Virol. 2006;80:1817-1825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Lavillette D, Pécheur EI, Donot P, Fresquet J, Molle J, Corbau R, Dreux M, Penin F, Cosset FL. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J Virol. 2007;81:8752-8765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 140] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 19. | Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G. Binding of hepatitis C virus to CD81. Science. 1998;282:938-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1572] [Cited by in RCA: 1554] [Article Influence: 57.6] [Reference Citation Analysis (0)] |
| 20. | Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017-5025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 888] [Cited by in RCA: 891] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 21. | Smith AE, Helenius A. How viruses enter animal cells. Science. 2004;304:237-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 558] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 22. | Lozach PY, Burleigh L, Staropoli I, Amara A. The C type lectins DC-SIGN and L-SIGN: receptors for viral glycoproteins. Methods Mol Biol. 2007;379:51-68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 23. | Zona L, Lupberger J, Sidahmed-Adrar N, Thumann C, Harris HJ, Barnes A, Florentin J, Tawar RG, Xiao F, Turek M. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe. 2013;13:302-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 24. | Schwarz AK, Grove J, Hu K, Mee CJ, Balfe P, McKeating JA. Hepatoma cell density promotes claudin-1 and scavenger receptor BI expression and hepatitis C virus internalization. J Virol. 2009;83:12407-12414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Murao K, Imachi H, Yu X, Cao WM, Nishiuchi T, Chen K, Li J, Ahmed RA, Wong NC, Ishida T. Interferon alpha decreases expression of human scavenger receptor class BI, a possible HCV receptor in hepatocytes. Gut. 2008;57:664-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 26. | Prentoe J, Serre SB, Ramirez S, Nicosia A, Gottwein JM, Bukh J. Hypervariable region 1 deletion and required adaptive envelope mutations confer decreased dependency on scavenger receptor class B type I and low-density lipoprotein receptor for hepatitis C virus. J Virol. 2014;88:1725-1739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 27. | Brimacombe CL, Grove J, Meredith LW, Hu K, Syder AJ, Flores MV, Timpe JM, Krieger SE, Baumert TF, Tellinghuisen TL. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J Virol. 2011;85:596-605. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 198] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 28. | Zhang J, Randall G, Higginbottom A, Monk P, Rice CM, McKeating JA. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J Virol. 2004;78:1448-1455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 283] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 29. | Masciopinto F, Campagnoli S, Abrignani S, Uematsu Y, Pileri P. The small extracellular loop of CD81 is necessary for optimal surface expression of the large loop, a putative HCV receptor. Virus Res. 2001;80:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 30. | Voisset C, Lavie M, Helle F, Op De Beeck A, Bilheu A, Bertrand-Michel J, Tercé F, Cocquerel L, Wychowski C, Vu-Dac N. Ceramide enrichment of the plasma membrane induces CD81 internalization and inhibits hepatitis C virus entry. Cell Microbiol. 2008;10:606-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 66] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 31. | Mee CJ, Harris HJ, Farquhar MJ, Wilson G, Reynolds G, Davis C, van IJzendoorn SC, Balfe P, McKeating JA. Polarization restricts hepatitis C virus entry into HepG2 hepatoma cells. J Virol. 2009;83:6211-6221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 101] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 32. | Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol. 2007;81:588-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 184] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 33. | Akazawa D, Date T, Morikawa K, Murayama A, Miyamoto M, Kaga M, Barth H, Baumert TF, Dubuisson J, Wakita T. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J Virol. 2007;81:5036-5045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 103] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 34. | Harris HJ, Clerte C, Farquhar MJ, Goodall M, Hu K, Rassam P, Dosset P, Wilson GK, Balfe P, Ijzendoorn SC. Hepatoma polarization limits CD81 and hepatitis C virus dynamics. Cell Microbiol. 2013;15:430-445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 35. | Farquhar MJ, Hu K, Harris HJ, Davis C, Brimacombe CL, Fletcher SJ, Baumert TF, Rappoport JZ, Balfe P, McKeating JA. Hepatitis C virus induces CD81 and claudin-1 endocytosis. J Virol. 2012;86:4305-4316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 100] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 36. | Zhang YY, Zhang BH, Ishii K, Liang TJ. Novel function of CD81 in controlling hepatitis C virus replication. J Virol. 2010;84:3396-3407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 37. | Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 603] [Cited by in RCA: 569] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 38. | Liu Z, Tian Y, Machida K, Lai MM, Luo G, Foung SK, Ou JH. Transient activation of the PI3K-AKT pathway by hepatitis C virus to enhance viral entry. J Biol Chem. 2012;287:41922-41930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 39. | Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci USA. 2007;104:5848-5853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 429] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 40. | Ye J. Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus. PLoS Pathog. 2007;3:e108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 109] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 41. | André P, Perlemuter G, Budkowska A, Bréchot C, Lotteau V. Hepatitis C virus particles and lipoprotein metabolism. Semin Liver Dis. 2005;25:93-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 134] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 42. | Cao WM, Murao K, Imachi H, Yu X, Dobashi H, Yoshida K, Muraoka T, Kotsuna N, Nagao S, Wong NC. Insulin-like growth factor-i regulation of hepatic scavenger receptor class BI. Endocrinology. 2004;145:5540-5547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 43. | Guo L, Chen M, Song Z, Daugherty A, Li XA. C323 of SR-BI is required for SR-BI-mediated HDL binding and cholesteryl ester uptake. J Lipid Res. 2011;52:2272-2278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 44. | Cao WM, Murao K, Imachi H, Yu X, Abe H, Yamauchi A, Niimi M, Miyauchi A, Wong NC, Ishida T. A mutant high-density lipoprotein receptor inhibits proliferation of human breast cancer cells. Cancer Res. 2004;64:1515-1521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 102] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 45. | Vergeer M, Korporaal SJ, Franssen R, Meurs I, Out R, Hovingh GK, Hoekstra M, Sierts JA, Dallinga-Thie GM, Motazacker MM. Genetic variant of the scavenger receptor BI in humans. N Engl J Med. 2011;364:136-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 267] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 46. | Imachi H, Murao K, Sato M, Hosokawa H, Ishida T, Takahara J. CD36 LIMPII analogous-1, a human homolog of the rodent scavenger receptor B1, provides the cholesterol ester for steroidogenesis in adrenocortical cells. Metabolism. 1999;48:627-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 47. | Murao K, Imachi H, Cao W, Yu X, Li J, Yoshida K, Ahmed RA, Matsumoto K, Nishiuchi T, Wong NC. High-density lipoprotein is a potential growth factor for adrenocortical cells. Biochem Biophys Res Commun. 2006;344:226-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 48. | Meuleman P, Catanese MT, Verhoye L, Desombere I, Farhoudi A, Jones CT, Sheahan T, Grzyb K, Cortese R, Rice CM. A human monoclonal antibody targeting scavenger receptor class B type I precludes hepatitis C virus infection and viral spread in vitro and in vivo. Hepatology. 2012;55:364-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 111] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 49. | Voisset C, Callens N, Blanchard E, Op De Beeck A, Dubuisson J, Vu-Dac N. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J Biol Chem. 2005;280:7793-7799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 183] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 50. | Catanese MT, Ansuini H, Graziani R, Huby T, Moreau M, Ball JK, Paonessa G, Rice CM, Cortese R, Vitelli A. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J Virol. 2010;84:34-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 121] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 51. | Vercauteren K, Van Den Eede N, Mesalam AA, Belouzard S, Catanese MT, Bankwitz D, Wong-Staal F, Cortese R, Dubuisson J, Rice CM. Successful anti-scavenger receptor class B type I (SR-BI) monoclonal antibody therapy in humanized mice after challenge with HCV variants with in vitro resistance to SR-BI-targeting agents. Hepatology. 2014;60:1508-1518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 52. | Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, Blum HE, Cosset FL, Wakita T, Jaeck D, Doffoel M, Royer C. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 2007;46:1722-1731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 190] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 53. | Dreux M, Pietschmann T, Granier C, Voisset C, Ricard-Blum S, Mangeot PE, Keck Z, Foung S, Vu-Dac N, Dubuisson J. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J Biol Chem. 2006;281:18285-18295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 172] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 54. | Lin W, Choe WH, Hiasa Y, Kamegaya Y, Blackard JT, Schmidt EV, Chung RT. Hepatitis C virus expression suppresses interferon signaling by degrading STAT1. Gastroenterology. 2005;128:1034-1041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 121] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 55. | Zhu H, Shang X, Terada N, Liu C. STAT3 induces anti-hepatitis C viral activity in liver cells. Biochem Biophys Res Commun. 2004;324:518-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 56. | Decker T, Stockinger S, Karaghiosoff M, Müller M, Kovarik P. IFNs and STATs in innate immunity to microorganisms. J Clin Invest. 2002;109:1271-1277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 147] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 57. | Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778-809, table of contents. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2029] [Cited by in RCA: 2003] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 58. | Grove J, Huby T, Stamataki Z, Vanwolleghem T, Meuleman P, Farquhar M, Schwarz A, Moreau M, Owen JS, Leroux-Roels G. Scavenger receptor BI and BII expression levels modulate hepatitis C virus infectivity. J Virol. 2007;81:3162-3169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 116] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 59. | Prabhudas M, Bowdish D, Drickamer K, Febbraio M, Herz J, Kobzik L, Krieger M, Loike J, Means TK, Moestrup SK. Standardizing scavenger receptor nomenclature. J Immunol. 2014;192:1997-2006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 155] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 60. | Beauvillain C, Meloni F, Sirard JC, Blanchard S, Jarry U, Scotet M, Magistrelli G, Delneste Y, Barnaba V, Jeannin P. The scavenger receptors SRA-1 and SREC-I cooperate with TLR2 in the recognition of the hepatitis C virus non-structural protein 3 by dendritic cells. J Hepatol. 2010;52:644-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 61. | Goldstein JL, Brown MS. The LDL receptor. Arterioscler Thromb Vasc Biol. 2009;29:431-438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1037] [Cited by in RCA: 913] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 62. | Go GW. Low-Density Lipoprotein Receptor-Related Protein 6 (LRP6) Is a Novel Nutritional Therapeutic Target for Hyperlipidemia, Non-Alcoholic Fatty Liver Disease, and Atherosclerosis. Nutrients. 2015;7:4453-4464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 63. | Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766-12771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 702] [Cited by in RCA: 705] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 64. | André P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Bréchot C, Paranhos-Baccalà G, Lotteau V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76:6919-6928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 517] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 65. | Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 219] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 66. | Petit JM, Minello A, Duvillard L, Jooste V, Monier S, Texier V, Bour JB, Poussier A, Gambert P, Verges B. Cell surface expression of LDL receptor in chronic hepatitis C: correlation with viral load. Am J Physiol Endocrinol Metab. 2007;293:E416-E420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 67. | Zhao Y, Ren Y, Zhang X, Zhao P, Tao W, Zhong J, Li Q, Zhang XL. Ficolin-2 inhibits hepatitis C virus infection, whereas apolipoprotein E3 mediates viral immune escape. J Immunol. 2014;193:783-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 68. | Hishiki T, Shimizu Y, Tobita R, Sugiyama K, Ogawa K, Funami K, Ohsaki Y, Fujimoto T, Takaku H, Wakita T. Infectivity of hepatitis C virus is influenced by association with apolipoprotein E isoforms. J Virol. 2010;84:12048-12057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 109] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 69. | Albecka A, Belouzard S, Op de Beeck A, Descamps V, Goueslain L, Bertrand-Michel J, Tercé F, Duverlie G, Rouillé Y, Dubuisson J. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology. 2012;55:998-1007. [PubMed] |
| 70. | Syed GH, Tang H, Khan M, Hassanein T, Liu J, Siddiqui A. Hepatitis C virus stimulates low-density lipoprotein receptor expression to facilitate viral propagation. J Virol. 2014;88:2519-2529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 71. | Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 991] [Cited by in RCA: 943] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 72. | Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 774] [Cited by in RCA: 732] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 73. | Harris HJ, Davis C, Mullins JG, Hu K, Goodall M, Farquhar MJ, Mee CJ, McCaffrey K, Young S, Drummer H. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem. 2010;285:21092-21102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 161] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 74. | Krieger SE, Zeisel MB, Davis C, Thumann C, Harris HJ, Schnober EK, Mee C, Soulier E, Royer C, Lambotin M. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology. 2010;51:1144-1157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 138] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 75. | Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83:2011-2014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 267] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 76. | Diao J, Pantua H, Ngu H, Komuves L, Diehl L, Schaefer G, Kapadia SB. Hepatitis C virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J Virol. 2012;86:10935-10949. [PubMed] |
| 77. | Meyer K, Kwon YC, Liu S, Hagedorn CH, Ray RB, Ray R. Interferon-α inducible protein 6 impairs EGFR activation by CD81 and inhibits hepatitis C virus infection. Sci Rep. 2015;5:9012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 78. | Sainz B, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18:281-285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 353] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 79. | Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci USA. 2013;110:10777-10782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 167] [Article Influence: 13.9] [Reference Citation Analysis (0)] |