Published online Feb 27, 2015. doi: 10.4254/wjh.v7.i2.150
Peer-review started: August 29, 2014
First decision: October 14, 2014
Revised: October 23, 2014
Accepted: November 17, 2014
Article in press: November 19, 2014
Published online: February 27, 2015
Processing time: 169 Days and 16.3 Hours
Acute and chronic hepatitis B virus (HBV) infections remain to present a major global health problem. The infection can be associated with acute symptomatic or asymptomatic hepatitis which can cause chronic inflammation of the liver and over years this can lead to cirrhosis and the development of hepatocellular carcinomas. Currently available therapeutics for chronically infected individuals aim at reducing viral replication and to slow down or stop the progression of the disease. Therefore, novel treatment options are needed to efficiently combat and eradicate this disease. Here we provide a state of the art overview of gene therapeutic approaches to inhibit HBV replication. We discuss non-viral and viral approaches which were explored to deliver therapeutic nucleic acids aiming at reducing HBV replication. Types of delivered therapeutic nucleic acids which were studied since many years include antisense oligodeoxynucleotides and antisense RNA, ribozymes and DNAzymes, RNA interference, and external guide sequences. More recently designer nucleases gained increased attention and were exploited to destroy the HBV genome. In addition we mention other strategies to reduce HBV replication based on delivery of DNA encoding dominant negative mutants and DNA vaccination. In combination with available cell culture and animal models for HBV infection, in vitro and in vivo studies can be performed to test efficacy of gene therapeutic approaches. Recent progress but also challenges will be specified and future perspectives will be discussed. This is an exciting time to explore such approaches because recent successes of gene therapeutic strategies in the clinic to treat genetic diseases raise hope to find alternative treatment options for patients chronically infected with HBV.
Core tip: With various successful clinical trials ongoing, gene therapeutic approaches gained increasing attention in the community over the recent years. Here we introduce gene therapy as a versatile platform for treatment of hepatitis B (HBV) virus infection. Newest delivery methods based on non-viral and viral techniques combined with most advanced technologies for inhibition of HBV replication based on DNA, RNA and designer nucleases are discussed. An overview of various gene therapeutic systems which were explored in vitro and in vivo is provided. Advantages but also limitations of the different strategies to inhibit HBV replication are mentioned.
- Citation: Gebbing M, Bergmann T, Schulz E, Ehrhardt A. Gene therapeutic approaches to inhibit hepatitis B virus replication. World J Hepatol 2015; 7(2): 150-164
- URL: https://www.wjgnet.com/1948-5182/full/v7/i2/150.htm
- DOI: https://dx.doi.org/10.4254/wjh.v7.i2.150
The hepatitis B virus (HBV) is an enveloped, partially double-stranded DNA virus which replicates through an RNA intermediate. Upon cell entry the DNA containing core particle is transported to the nucleus where the DNA is released. Next the partial double-stranded DNA is repaired by host enzymes to form the covalently closed circular DNA (cccDNA). The cccDNA serves as a template for transcription of viral proteins and reverse transcription of new viral genomes by the viral polymerase. The large 3.5 kb HBV transcript represents the pregenomic RNA serving as a template for virus replication. Furthermore, there are three additional mRNAs with a length of 2.4, 2.1, and 0.9 kb. The HBV genome is 3.2 kb in size and contains four overlapping major open reading frames tightly arranged that encode polymerase, surface (HBsAg), core (HBcAg) and X proteins (HBx)[1,2]. In addition, especially early during infection the HBV early antigen (HBeAg) can be detected which is a proteolytic product of the pre-core protein.
HBV infection counts as a major global health problem since more than two billion people show evidence of a past or present infection with the virus. This hepatotropic virus can cause acute and chronic infection of the liver. Fortunately, for most people the infection proceeds nearly without symptoms when taking an acute course of disease and complete recovery is likely. However, 240 million people suffer from chronic HBV infection and more than 780000 people die every year because of hepatitis B related secondary diseases. Mostly newborns and infants are prone to develop the chronic type of the infection[3] and so far no treatment is available that reliable cures those patients.
Current therapeutics for chronic HBV infection are intended to reduce viral replication and slow down or stop the progression of the disease. To date there are seven Food and Drug Administration (FDA) approved compounds for the treatment of chronic hepatitis B. These include interferon alpha and pegylated interferon alpha, nucleoside analogues (lamivudine, entecavir and telbivudine) and nucleotide analogues (adefovir, dipivoxil and tenofovir)[4].
Interferon alpha has an antiviral effect by inhibiting the synthesis of viral DNA and activating antiviral enzymes and additionally, acts in an immunomodulatory way by enhancing the cellular immune response against infected cells[5]. It has to be administered daily or three times a week as unmodified version and once in a week in the pegylated form. The main disadvantages of interferon alpha are the parenteral administration causing discomfort to the patients and potential adverse effects such as flu-like symptoms in the beginning of treatment and later on for instance fatigue and low blood counts. It is only given to selected patients because under certain conditions administration of interferon alpha is contraindicated[4]. In a long-term follow-up study of HBeAg-positive patients, 11% lost HBsAg after treatment with interferon alpha[6] which is considered as a cure of the disease.
Nucleos(t)ide analogues interfere with the HBV replication primarily by targeting the HBV polymerase functions such as reverse transcriptase and DNA polymerase activity[5]. These drugs are administered orally as a daily dose. The major limitation associated with nucleos(t)ide analogues is the emergence of antiviral drug resistance and that life-long treatment can be indicated in the presence of chronic infection. In this context failure of medication adherence is another problem, because viral relapse is common when ending the treatment. Another prognostic marker of HBV infection is the presence of HBeAg which correlates with high viral replication rates. However, HBeAg seroconversion can be achieved with nucleos(t)ide treatment. In addition, HBV DNA levels can be decreased to an undetectable level but at the same time HBsAg is not lost[4]. These features demonstrate another peculiarity of the hepatitis B virus. After entry into a cell the cccDNA is maintained as an episomally maintained template in the nucleus. It is not attacked by nucleos(t)ide analogues nor by interfon in general, so that it is able to serve as a reservoir from which previously cleared or treated infections can recur[4]. The clinical management of chronic hepatitis B infection is reviewed in detail by Santantonio and Fasano[7].
The viral reservoir and potential reactivation of the virus represents a major problem when developing novel HBV treatments options and this is a challenge that could be faced by gene therapy. Researchers seek to inhibit viral replication in a long-lasting manner without the need for continuous drug administration through gene therapeutic approaches. The more ambitious goal is to completely eradicate the viral cccDNA depot and hence find a true cure for chronic hepatitis B virus infection. Here we discuss gene therapeutic approaches as a versatile platform to combat HBV infection.
Gene therapy is a strategy to transfer therapeutic nucleic acids into the desired target cell for treatment of a variety of different diseases. To efficiently deliver the genetic payload, multiple gene transfection techniques were explored which can be subdivided into two major groups: virus-based and non-viral vector systems for delivery of respective therapeutic nucleic acid. Both delivery techniques were also utilized in gene therapeutic approaches to treat chronic infectious diseases such as HBV infection. Since HBV infection resides in liver, the majority of gene transfer approaches were focused on targeting hepatocytes.
Non-viral vectors are based on delivery of naked RNA or DNA which in combination with chemical and physical means can result in efficient delivery of the nucleic acid into the respective target cell[8,9]. Chemical methods in the context of non-viral vector delivery rely on various chemical formulations such as cationic lipids[10] and polymers including polyamidoamine dendrimers and polyethylenimine (PEI)[11]. All chemical reagents were explored in different approaches and there are several commercially available transfection reagents which are commonly used for transfection of DNA and RNA resulting in sufficient transfer efficiencies in many cell lines in vitro. Major constraints of these methods are transfection reagent-associated toxicity and the difficulty to cross the nuclear membrane. In addition to chemical transfer methods, physical transfer techniques were explored involving needle injection[12], gene gun[13], electroporation[14], sonoporation[15], magnetofection[16], and hydrodynamic gene transfer[17]. These methods directly deliver therapeutic nucleic acid into the cytosol of the target cell and compared to chemical methods these techniques harbor a reduced risk of transfection-mediated side effects due to dispersion of the transfection reagent. However, limitations of these methods are exposed by the difficulty to cross the nuclear membrane, potential cellular damage caused by the transfection method, and the requirement of costly instruments.
Virus-based transfection techniques were utilized in numerous pre-clinical and clinical gene therapeutic applications. Predominantly used viral vectors can be attributed to three viruses which were converted into viral vectors by deletion of essential viral genes: adenovirus, adeno-associated virus (AAV) and retrovirus. All viral vector systems display advantages and disadvantages which were discussed in more detail in previous reviews[18]. Adenoviruses combine a large transgene capacity of up to 36 kilo bases (kb), an episomal nature of the adenoviral genome reducing the risk of genotoxicity, the possibility to produce high viral titers and the ability to transduce dividing and non-dividing cells at high efficiencies in vitro and in vivo[19]. However, one major obstacle for in vivo applications are the innate and the adaptive immune responses induced by the incoming adenoviral particle. AAV vectors were explored in clinical trials, are non-pathogenic, lead to a reduced immune response and predominantly exist as extrachromosomal vector genomes in the transduced cell[20]. One major disadvantage, however, is the small transgene capacity which is below 5 kb. Lentiviral vectors[21] were broadly explored in clinical trials to treat rare genetic diseases in ex vivo gene therapeutic approaches. Various generations of lentiviral vectors are available which carry a transgene capacity of up to 8 kb. Although commonly used lentiviral vectors integrate their genetic cargo into the host genome, newest versions of these vectors can circumvent side effects associated with somatic integration by changing their integration profile.
Various non-viral and viral transfer techniques were exploited to combat HBV infection in vitro and in vivo which will be discussed in the following paragraphs.
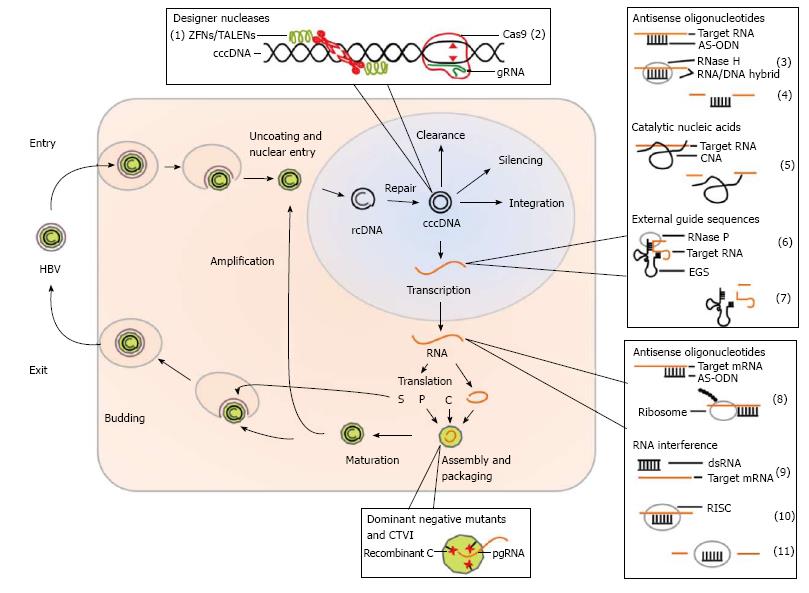
Various gene therapeutic approaches to treat HBV infection were studied in cell culture models and in animal models for HBV infection. Within the viral life cycle in an infected cell there are various points of attack which can serve as targets in gene therapeutic approaches to inhibit HBV replication. Figure 1 schematically shows the life cycle of HBV infection and indicates points of attack when considering a gene therapeutic treatment.
As shown in Figure 1 the mechanisms of viral inhibition in gene therapeutic approaches can be on the level of RNA (HBV derived transcripts), DNA (cccDNA) and proteins. On the RNA level antisense oligodeoxynucleotides and antisense RNA, catalytic nucleic acids such as ribozymes and DNAzymes, RNA interference and external guide sequences (EGS) can be considered. On the level of DNA (cccDNA) as a potential target designer nuclease such as zinc finger nuclease (ZFN), transcription activator-like effector nucleases (TALEN) and the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology can be used. On the level of proteins, dominant negative HBV mutants and a strategy based on capsid-targeted viral inactivation (CTVI) were studied. Another technology for prevention of infection but also for a potential treatment option of chronically infected patients, DNA vaccination can be considered as an attractive alternative. Although the majority of the described strategies for inhibition of HBV replication were not translated into the clinic so far, we believe that gene therapy may represent a valuable alternative in the future. Studies describing milestones of the various gene therapeutic approaches are listed in Table 1 and are described in more detail in the following sections.
| Year | Strategy | Milestone | Ref. |
| 1990 | AS-ODN | First in vitro application | [22] |
| 1992 | AS-ODN | First non-viral transfection (targeted polycation peptide complex) | [28] |
| Ribozyme | First in vitro application | [44] | |
| 1993 | AS-ODN | First in vivo application | [26] |
| DNA vaccination | [75] | ||
| 1994 | Dominant negative mutants | First in vitro application | [65] |
| 1997 | AS-ODN | First viral transduction (retroviral) | [39] |
| Ribozyme | [46] | ||
| 1998 | DNAzyme | First in vitro application | [60] |
| EGS | [87] | ||
| 2001 | CTVI | [69] | |
| 2003 | RNAi | First in vitro application | [93] |
| First in vivo application | [94] | ||
| 2004 | Ribozyme | First in vivo application | [56] |
| DNA vaccination | First clinical trial in chronic HBV carriers | [81] | |
| 2008 | CTVI | First in vivo application | [72] |
| 2010 | ZFN | First in vitro application | [138] |
| 2011 | RNAi | First clinical trial in chronic HBV carriers | [117] |
| 2013 | EGS | First in vivo application | [89] |
| TALEN | First in vitro/vivo application | [146] | |
| 2014 | CRISPR/Cas9 | First in vitro/vivo application | [148] |
The beginning of gene therapy against HBV can be linked to the first tests of antisense oligodeoxynucleotides (AS-ODNs) directed against the HBV genome[22-24]. With respect to antisense nucleic acids post-transcriptional inhibition is achieved by blockade of ribosomal access, inhibition of ribosomal assembly and induction of RNase H cleavage[25].
First in vivo studies showed applicability of this approach in duck-HBV-infected Peking ducklings. Infected animals were treated daily by intravenous injection of AS-ODNs for ten days. The treatment resulted in nearly complete inhibition of viral replication which was assessed by liver DNA analysis for DNA replicative intermediates and blockade of viral gene expression as demonstrated by disappearance of surface antigen in serum and core antigen in liver[26]. AS-ODNs proved to be most effective when directed against the initiation site of the HBsAg-gene[22] or at the encapsidation signal[27]. Except for a study published by Wu et al[28] in which already targeted DNA complexes were used, delivery of AS-ODNs was initially restricted to simple cellular uptake and binding of the unmodified antisense DNA to its target sites. Soon the system was improved by enhancing stability of the respective nucleic acids and by increasing uptake of AS-ODNs by the chosen target cell[29-32]. In other studies AS-ODNs were conjugated to ribonuclease H or manganese porphyrin which after binding to the target site can lead to cleavage of the desired target sequences[33,34]. Furthermore, DNA carrier systems were used and also enhanced by making them targetable to hepatocytes[28,35,36].
Other oligonucleotide based approaches include antisense RNA delivered by episomally replicating expression vectors[37,38] or retroviral vectors[39,40]. The advantage of these approaches is the fact that antisense RNAs can be expressed in the cells, whereas AS-ODNs have to be exogenously delivered. This allows for experimental settings with long-term effects. For instance efficacies lasting longer than ten months were observed after stable transfection of antisense RNA expression vectors[37]. Antisense RNA-mediated inhibition functions preferentially through destabilization of the sense RNA by targeting the antisense/sense-RNA duplex to dsRNase[41].
A completely different idea unrelated to complementary antisense RNA was introduced by Hafkemeyer et al[42] and is based on so-called “antisense-toxin-RNA”. In this approach the authors took advantage of the HBV reverse transcriptase in infected cells to selectively kill those cells through Pseudomonas exotoxin expression from reverse transcribed antisense-toxin-RNA.
There are two types of catalytically active nucleic acids, ribozymes and DNAzymes. Ribozymes are naturally occurring RNA molecules that can execute enzymatic activity on itself in the absence of proteins (in cis) or on extrinsic targets (in trans). DNAzymes were generated by in vitro evolution. They resemble ribozymes and do not exist in nature. The substrate for both species is RNA. There are various types of ribozymes known, hammerhead and hairpin ribozymes being the most popular ones. All catalytic nucleic acids have in common that they consist of an antisense sequence recognizing the target site and a catalytic domain mediating cleavage[43]. Scientists were able to manipulate the recognition of target sites, rendering such nucleic acids very attractive for enhanced transient knockdown of gene expression.
The first experiments with ribozymes targeting the HBV genome were performed in 1992 using a triple ribozyme construct. In this study three hammerhead ribozymes were encoded on a single DNA template and it was shown that they were simultaneously active in vitro. However, cleavage kinetics were similar to single ribozyme constructs. Nonetheless, this approach may still be favorable because it enables facing high target variability and emergence of viral resistance[44]. However, first studies performed in a cellular context disclosed a first drawback of the hammerhead ribozymes because they were active after in vitro transcription and in Mg2+-supplemented cell extracts, but not in intact cells. The authors suggested some non-viral block, or inappropriate target site selection being responsible for the lack of intracellular activity[45].
However, to overcome these problems novel strategies were pursued using other types of ribozymes. Welch and colleagues[46] designed several hairpin ribozymes against different conserved regions of the HBV genome and tested them in a human hepatoma cell line (Huh7) transfected with full-length HBV genomes. The ribozymes were transduced into target cells using a retroviral vector system. The HBV production could be inhibited to up to 83% assayed through an endogenous polymerase assay. In the following years further in vitro studies were performed using different types of modified ribozymes[47-51]. These were predominantly delivered via transfection of an expression plasmid into various cell lines which were additionally transfected with a HBV genome-containing plasmid resulting in varying effects on inhibition of HBV replication[52-55].
In 2004 the first in vivo experiment in a transgenic mouse model was conducted by Pan et al[56]. For this study a self-processing triple-ribozyme cassette was used, with ribozymes acting in cis and in trans. The constructs were packaged in liposomes that where targeted to hepatocytes in the presence of asialofetuin. Quantitative PCR analysis for quantification of HBV genome copy numbers in murine liver showed a more than 80% decrease of HBV genome copy numbers and immunohistochemistry revealed a robust reduction in the number of hepatocytes staining positive for HBV core antigen. This was the first proof of concept demonstrating in vivo feasibility of viral RNA degradation mediated by ribozymes.
Next, recombinant hepatitis D virus (HDV)[57] as well as lentiviral vectors were utilized as delivery vehicles for respective ribozymes which achieved effective reduction of HBV mRNA levels over four months[58]. Furthermore, HDV-derived ribozymes were also utilized which were delivered by a pseudotyped retroviral vector (Moloney murine leukemia virus)[59]. According to the authors the main advantages of these ribozymes are the natural activity of HDV ribozymes in human cells even at physiological Mg2+-ion concentrations and the comparably highest cleavage rates among all known ribozymes. Their results revealed significant reduction in the intracellular HBV DNA concentration in HepG2.2.15 cells, which secrete infectious HBV virions. Furthermore, decreased extracellular HBsAg and HBeAg levels were observed after treatment with HDV ribozymes in comparison to the negative control. The conclusion was that using this strategy, HBV can be effectively inhibited at post-transcription and replication levels.
DNAzymes cleave RNA substrates based on a similar mechanism also used by ribozymes. However, they may be superior to ribozymes because their production is comparably straight forward and DNAzymes are less sensitive to chemical and enzymatic degradation. The first DNAzyme directed against HBV mRNA was created by Asahina et al[60]. They targeted the direct repeat 1 (DR1) and polyadenylation signal regions of HBV. In this study the authors used stabilized forms of DNAzymes that on the one hand lost some degree of activity compared to unmodified versions but on the other hand the degradation level was less pronounced. The DNAzyme was tested in Huh7 cells on an HBV-luciferase fusion reporter system where it exhibited 48% suppression compared to untreated control groups. Further in vitro studies[61-64], however, revealed a major disadvantage of this system because intracellular expression of these DNA species is not feasible. DNAzymes have to be transfected directly because they act on post-transcriptional level which is clearly disadvantageous if long-term administration for instance in clinical applications is required.
Dominant negative mutants of viral proteins are able to inhibit viral replication by interfering with the function of the wild type protein. The first study on the molecular effects of dominant negative mutants on the HBV replication was conducted in 1994 by Scaglioni et al[65] They mutated the core protein and observed an inhibition of viral replication by 90%-95%[65] and it was concluded that this was the result of the disruption of the viral nucleocapsid assembly process. In a follow-up study the authors provided a delivery system using retroviral and adenoviral expression vectors[66]. von Weizsäcker et al[67] showed that carboxy-terminal, but not amino-terminal core mutants inhibit viral replication. Furthermore, it was discussed that rather the packaging of the viral pre-genome and the reverse transcription reaction within the particles than the nucleocapsid formation itself is restrained by dominant negative core mutants[68]. However, it was also shown that some mixed particles retain replication competency suggesting a possible mechanism of viral escape.
Using the principle of CTVI may represent an interesting variant of capsid modification, as the goal is to introduce a destructive element into the virus. The first time this technology was pursued was by fusing a Ca2+-dependent nuclease from Staphylococcus aureus to the HBV core protein. This led to inhibition of proper synthesis of viral DNA inside the capsid and rapid viral DNA degradation[69]. Later human eosinophil-derived neurotoxin was fused to the HBV core protein by another group and thoroughly studied also in vivo[70-72].
Initial considerations to use vaccination in a therapeutic manner against HBV infection were made in 1993 by a group around Christian Bréchot[73,74]. This approach was based on classical vaccination compounds. Also in 1993 the first genetic immunization with expression vectors for HBsAg was introduced in mice[75]. In a further step Mancini et al[76] proved that a combination of both strategies is feasible and they showed efficacy in a HBsAg transgenic mouse as a model for chronic HBV infection. A single intramuscular injection of HBV envelope encoding plasmid DNA resulted in the elimination of serum HBV antigen levels as early as 4 wk after administration of the therapeutic DNA in some mice. Notably no liver cell injury was detected. Soon genetic immunization employing retroviral delivery was tested first in rhesus monkeys[77] and then in chimpanzees chronically infected with HBV[78]. The first time a DNA vaccine encoding HBsAg delivered with a gene gun was tested in healthy humans was in 1999 and it turned out to be ineffective at the applied dosage (0.25 μg)[79]. However, the vaccine was well tolerated. A subsequent study used higher dosages (1, 2, or 4 μg) and this time antigen-specific CD8+ T cells, T helper cells, and protective levels of antibodies could be induced[80]. Therapeutic DNA vaccination for chronic HBV infection was first tested in 2004 and in other subsequent studies[81-83]. It was shown that HBV DNA vaccination was safe and immunologically effective because T-cell responses were activated in some chronic HBV carriers who did not respond to current antiviral therapies. These results indicated that in combination with conventional therapy, DNA vaccination may have a synergic effect leading to complete recovery from HBV infection.
The recruitment of ribonuclease P (RNase P) by EGS represents another interesting gene interference strategy. RNase P is an enzyme that removes the leader sequence of tRNA precursors by recognizing a common structure shared amongst all tRNAs. Using a custom-designed EGS that hybridizes with an mRNA to form a tRNA-like structure, RNase P can be recruited to cleave the target mRNA[84,85]. With respect to HBV infection, this strategy was first proposed by Werner et al[86] in 1997. They took advantage of EGS designed to target HBV sequences as proof-of-concept for the use of EGS as anti-viral or anti-cancer therapeutics[87,88]. The EGS technology gained more attraction in 2013, when EGS was considered as a sufficient method to antagonize HBV replication. One study was conducted in cell culture and in mice using Salmonella-mediated delivery. Oral inoculation of attenuated Salmonella carrying the EGS construct led to up to approximately 95% inhibition of HBV gene expression levels and a approximately 200000-fold reduction of viral DNA levels in the livers and sera of the treated mice transfected with a HBV plasmid[89]. In a follow-up study efficacy could be improved about 50-fold by modifying the EGS derived from natural tRNA through in vitro selection[90].
RNA interference (RNAi) is a natural intracellular antiviral immune response mechanism triggered by double-stranded RNA (dsRNA). After conversion of dsRNA into guide molecules (siRNA), dsRNA induces the degradation of RNA via the RNA-induced silencing complex (RISC). Synthetic siRNA can be delivered exogenously or produced endogenously in the form of precursor small hairpin RNAs (shRNA) from plasmid DNA or viral vectors[91]. When RNAi was first discovered[92] it released a wave of multiple studies and applications. Also in HBV therapy it gained attention. In 2003 several studies were published[93-97] and marked the beginning of the most intense exploitation of a gene therapy method as a valuable treatment option for HBV infection.
Shlomai et al[93] were the first to publish the use of RNAi in an in vitro cell culture model of HBV replication. They found that HBV gene suppression was achieved with different efficacies for different gene targets. Analysis of HBV transcripts revealed a reduction of about 68% on the level of all viral transcripts for RNAi targeting HBx protein. However, RNAi against two different targets on the same HBcAg open reading frame, which is exclusively encoded by the large 3.5 kb transcript, resulted in about 13% and 50% reduction on the level of the 3.5 kb transcript, respectively. This led to the conclusion, that RNAi target sequences have to be evaluated carefully.
In the next study McCaffrey et al[94] tested the RNAi-based approach in vivo in immunocompetent and immunodeficient mice. DNAs containing a HBV expression plasmid and a shRNA expression plasmid were co-transfected into mouse liver by hydrodynamic plasmid delivery. The authors could show that RNAi can inhibit all the steps of HBV replication that occur in cell culture and in mice. This was indicated by reduced secreted HBsAg levels in the supernatant of transfected cells and in mouse serum, reduced HBV RNAs levels in mouse liver, reduction of HBV genomic DNA to undetectable levels in mouse liver, and decreased numbers of cells stained positive for HBcAg.
The first viral vectors used to transduce RNAi sequence expression cassettes were either based on a prototype foamy virus (PFV) or an adeno-associated virus (AAV)[98]. The vectors expressing the respective RNAi molecule were assessed in 293T.HBs cells, a cell line stably expressing HBsAg and HepG2.2.15 cells. In 293T.HBs cells HBsAg was knocked down by approximately 90% if directly compared to controls cells. HBsAg expression was also inhibited in HepG2.2.15 cells even in the presence of HBV replication. Uprichard et al[99] introduced recombinant adenovirus vectors for the delivery of shRNAs. Additional, they used for the first time HBV-transgenic mice to show that ongoing HBV replication in vivo can be cleared by RNAi-targeted suppression of viral RNA for at least 26 d. Other viral vectors used included retroviral vectors[100,101], AAV-7, -8 and -9 pseudotyped vectors[102-104], recombinant human foamy virus[105], gene-deleted adenoviral vectors[106,107], lentiviral vectors[108,109], and recombinant baculovirus[110].
Further studies of the RNAi system included lipid-encapsulation of chemically modified siRNAs for non-viral delivery[111] and the assessment of efficacy and pharmacodynamic properties of different RNAi target sequences and constructs, including methyl-modified siRNAs and plasmid based DNA vectors[112]. Moreover, high-throughput generation and screening of siRNAs was established[113] and expression systems introducing multiple siRNAs were developed[105,114-117]. Ely et al[116] found that a Pol II promoter may be advantageous compared to a Pol III promoter, which was traditionally used for shRNA expression. The Pol III promoter can result in shRNA overexpression and saturation of the endogenous microRNA pathway leading to serious toxic effect in vivo[118]. This could be restricted with the Pol II promoter, which provides the possibility to control the production of RNAi activators[119].
Another non-viral vector system which was used in shRNA approaches is the episomal replicating plasmid vector pEPI-1. Herein, the transcription unit is linked to a scaffold/matrix attachment region (S/MAR) which ensures that the vector is mitotically stable in transfected cells. It was shown that it provides long-term expression of shRNAs which resulted in suppression of HBV gene expression, intracellular HBV DNA replication and release of progeny HBV over 8 mo[120].
Besides therapeutic DNA vaccination, siRNA is the only gene therapeutic approach that was translated into clinical trials. In 2006 a Phase Ib, first-in-human safety and tolerability study of an RNAi-based therapy (NUC B1000) in patients with mild to moderate chronic HBV infection was conducted[117]. NUC B1000 is composed out of four expressed shRNAs on one plasmid carried on a nanoparticle (cholesteryl spermine complex) and administered through intravenous infusion. The results revealed elevated cytokines and no HBV DNA or HBsAg decrease in the patients. However, the safety profile of RNAi therapy conducted among patients with HBV was considered as reasonable. A second compound, ARC-520, a liver-tropic cholesterol-conjugated siRNA (chol-siRNA), transported by the proprietary Dynamic Polyconjugate delivery system is just reaching a Phase II clinical study[121,122].
In summary the RNAi system was thoroughly exploited for treatment of chronic HBV infection in the past and research will be ongoing on this topic to overcome major hurdles like evocation of immune responses and maintenance of long-term suppression. Long-term suppression is required because it was shown that the HBV cccDNA depot in the host cells is not affected by this approach[110]. Interestingly, another limitation might be the manipulation of the host RNAi defense by the HBx protein that potentially functions as a RNA-silencing suppressor (RSS)[123].
For sequence-specific DNA targeting designer nucleases such as zinc finger nucleases (ZFNs)[124-126], transcription activator-like effector nucleases (TALENs)[127,128] and the clustered, regularly interspaced, short palindromic repeats (CRISPR)-CRISPR-associated protein (Cas) system[129-131] can be applied. They combine customizable DNA binding molecules for sequence-specific DNA-binding and a nuclease for introduction of doubled-strand DNA (dsDNA) breaks. The induced dsDNA breaks activate different cellular DNA repair pathways. The two most exploited pathways in gene therapy are homologous recombination (HR) and nonhomologous end-joining (NHEJ). In the presence of a respective homologous donor DNA, cells are able to repair the dsDNA break via HR by exchanging the respective sequence[132]. Without any homologous donor DNA cells repair dsDNA breaks via NHEJ. This error-prone repair mechanism can lead to insertions or deletions of one or several base pairs and may cause specific knockout of a gene[132]. Therefore, designer nucleases are valuable tools to specifically introduce knock out mutations at a desired DNA locus.
The DNA binding domains (DBD) of ZFNs commonly contains 3-4 zinc fingers. Each zinc finger consists of 30 amino acids and forms two β-sheets and one α-helix. Upon DNA-binding the α-helix is placed in the major groove of the dsDNA and directs contact with a certain base pair triplet. Depending on the amino acids within the α-helix and the number of zinc fingers, ZFNs can be designed specifically to target defined stretches of DNA triplets with high affinity[133-135]. The DBD is connected to a sequence independent cleavage domain of the type IIS restriction enzyme FokI which causes double strand breaks after dimerization[136]. Therefore two ZFN monomers are necessary to create the desired dsDNA break in the spacer between the binding sites of the ZFNs. The double strand breaks will be repaired via NHEJ, causing insertion and deletion mutations (indels) of several base pairs within the sequence. These indels can lead to frame shifts within the open reading frame or translation abortion by newly formed stop codons. Both options result in dysfunctional proteins.
There are some approaches using zinc fingers to target HBV cccDNA. Zimmerman et al[137] were the first to use zinc fingers in conjunction with HBV although they did not yet use ZFNs. Instead, they created several zinc finger proteins (ZFPs) targeting the enhancer region of duck HBV (DHBV), which probably form a steric hindrance for the RNA polymerase. After screening the candidates for binding efficiency, the two most efficient ZFPs were expressed in a special DHBV tissue culture system via transfection and both the transcription of viral genomic RNA and viral protein production was assessed via quantitative PCR and western blot analysis. They showed that both ZFPs significantly reduce transcription from cccDNA compared to controls. The authors concluded that ZFPs designed to target HBV DNA are able to substantially reduce viral transcription and interfere with viral replication of cccDNA.
In another study, Cradick et al[138] showed that ZFNs can mediate inhibition viral replication in vitro. They generated ZFN pairs targeting several conserved regions of HBV genomic DNA and chose the most robust pairs for further studies. Huh7 cells were transfected with plasmids encoding ZFNs under the control of a cytomegalovirus (CMV) immediate early promoter and the target plasmid pTHBV2. This target plasmid contains a 1.3-fold HBV genome which is capable of full transcription of the viral RNAs, translation of the proteins and production of infectious virus. Three days post transfection, Southern blotting analyses revealed about 26% linearized DNA and about 10% cleaved target plasmids being rejoined in a tail to tail orientation. Both DNA species indicate that ZFN-specific cleavage at the intended ZFN target site occurred. Linear genomes are the result of direct cleavage without any subsequent repair while concatamer formation is caused by NHEJ after ZFN cleavage. To investigate if NHEJ led to indels in recircularized genomes or head-to-tail concatamers, a XbaI site was inserted within the spacer region of the ZFN dimer. After NHEJ occurred this site should be destroyed. Target sites of treated samples were amplified by PCR, digested with XbaI and resistant amplicons were sequenced. 13 of 16 samples showed a frameshift which would lead to dysfunctional proteins. Furthermore this study indicated a 29% reduction of pregenomic RNA in northern blot analysis compared to controls. Besides these results it could also be demonstrated that the ZFN pairs caused only moderate toxicity. In conclusion, Cradick et al[138] revealed that specifically designed ZFNs targeting regions which are conserved among many different HBV serotypes can significantly reduce viral replication which is associated with negligible toxicity.
A third study by Weber et al[139] deals with the delivery of functional ZFNs into target cells via a viral vector system. Here, in contrast to the latter study, obligate heterodimeric ZFNs were designed, which are not able to form homodimers. This measure minimizes off-target effects because ZFNs that are able to form homodimers might cleave at unintended genomic loci by tolerating some mismatches. The three designed ZFN pairs targeted the open reading frames of HBx, HBcAg and polymerase. For delivery a self-complementary AAV-vector (scAAV) was used, which shows higher transduction efficiencies compared to single stranded AAV vectors[140]. Since scAAV vectors have a reduced transgene capacity each ZFN of a pair was delivered individually in co-transduction experiments. Transduction of each ZFN pair into HepAD38 cells, a model cell line for controllable HBV replication, revealed mutation rates ranging from 9.8% to 34%. Transduction of all three ZFN pairs simultaneously resulted in mutation rates of 8% to 20%. Off-target mutagenesis for seven potential off-target sites was investigated using single molecule real time (SMRT) sequencing which detected indels (> 1 nt) in four sequencing reads out of 9290 filtered reads. It is of note that indels of only one nucleotide were not taken into account because they could not be distinguished from sequencing artefacts. In order to test if ZFN treatment has an effect on HBV replication, ZFNs were transduced into HepAD38 cells in which HBV replication was shut down. After turning HBV replication on, the controls showed a 30-fold and 323-fold increase in HBV marker levels in cells and in the supernatant, respectively. In ZFNs treated cells no significant increase of cellular or supernatant HBV marker levels could be detected, leading to the conclusion that ZFN-mediated cleavage resulted in replication-deficient HBV. In summary, it was demonstrated that HBV specific ZFN can be delivered successfully via AAV vectors into target cells in vitro, resulting in an efficient inhibition of HBV replication.
TALENs are a promising new class of designer nucleases that can be specifically designed to bind DNA sequences of interest and to introduce dsDNA breaks. Comparable to ZFNs, TALENs are chimeric proteins consisting of a N-terminal nuclear localization signal, a central DNA binding domain and a C-terminal FokI nuclease domain. The DBD originates from transcription activator like effectors (TALEs) of bacterial plant pathogens of the genus Xanthomonas, which secrete these proteins to regulate gene expression within their host cells[141]. The DBD is comprised of a repeat region consisting of a number of incomplete tandem repeats containing repeat-variable diresidues (RVD)[127]. The RVD of each repeat is specific for binding a corresponding nucleotide in their contiguous target DNA sequence[127,141]. Similar to ZFNs, the TALEN-DNA binding domain is combined with a non-specific endonuclease activity which is dependent on dimerization of two FokI-cleavage domains. Binding of the DBDs upstream and downstream of a DNA target brings the FokI nuclease domains of a TALEN pair in close proximity and dimerization of the FokI monomers introduces dsDNA breaks. Note that the spacer between the binding sites of a pair needs to be considered in the TALEN design to ensure optimal cleavage[127,128]. Up to now several techniques are available to specifically design[127,142] and to assemble TALENs for the desired application[127,143-145]. Assembly can be performed without highly complex screening procedures or the need for special equipment, making it cheap, simple and fast and thereby with that respect superior to the ZFN technology.
Bloom et al[146] designed TALENs with target sequences in the HBsAg or HBcAg expressing region of the HBV genome. TALEN efficacy was determined after co-transfection of Huh7 cells with TALEN expression plasmids together with the pCH-9/3091 HBV replication-competent plasmid. They observed that the HBsAg production was diminished in cells expressing TALENs. In a more stringent experimental model of HBV replication, the HepG2.2.15 cell line was transfected with TALEN expression plasmids. After three subsequent transfections and culturing cells under hypothermic conditions, HBsAg-specific TALEN expression resulted in disruption of cccDNA molecules with efficiencies of approximately 31%. This was confirmed in a T7E1 mutation detection assay and correlated with a decrease of HBsAg secretion of these cells. Expression of the HBcAg-specific TALEN pair only mutated 12% of cccDNA molecules which was not sufficient to inhibit HBsAg secretion. After hydrodynamic tail vein injection of replication-competent HBV DNA together with HBsAg-specific TALEN expression plasmids, serum HBsAg levels were decreased by more than 90% and circulating viral particle equivalents (VPEs) were decreased by approximately 70%. The T7E1 assay demonstrated mutation rates of 58%-87% in HBV-DNA extracted from livers of TALEN treated mice.
Chen et al[147] used three TALEN pairs targeting regions of HBV genomic DNA conserved among HBV genotypes A-D. Huh7 cells were transfected with the monomeric linear full-length HBV DNA of subgroups A, B, C, or D, respectively and plasmids expressing one of the respective TALEN pairs. Cells containing HBV DNA simulated the complete HBV replication cycle, including the nuclear generation of cccDNA. Suppression of HBeAg and HBsAg production by the TALEN expression was observed for all four HBV genotypes. Additionally HBcAg–RNA as well as pregenomic HBV RNA levels were decreased in TALEN expressing cells. Furthermore T7E1 mutation detection assay confirmed that mutations were successfully induced at the respective TALEN target site within the HBV genome leading to a 10%-50% decrease of cccDNA levels. In combination with interferon alpha treatment TALENs expression led to synergistic effects further increasing the inhibition of HBV transcription in Huh7 cells. After delivery of monomeric linear full-length HBV DNA and TALEN expression plasmids into C3H/HeN mice by hydrodynamic tail vein injection, serum levels of HBeAg, HBsAg and cccDNA as well as liver pregenomic HBV RNA significantly decreased compared to control animals. In summary, HBV-targeting TALENs were shown to be active in cell culture models as well as in in vivo models and are capable of introducing mutations at their target sites reducing HBV gene expression levels and cccDNA genome numbers.
The most recent approach exploited the CRISPR/Cas9 system for genome engineering[131]. Because sequence specificity is achieved by a guide RNA (gRNA) which can be produced from an adaptable expression cassette, this system is easier to manipulate in comparison to ZFNs and TALENs where the DBD is based on a protein sequence. Lin et al[148] were the first to apply this system for achieving HBV genome degradation[148]. They co-expressed the Cas9 nuclease together with eight HBV specific guide RNAs individually and in addition combined two of them on one expression vector. They tested all constructs in Huh7 cells which were transfected with a HBV-expression vector. Inhibition varied among the different guide sequences. A maximum inhibition of 70% of intracellular HBsAg expression was reached after using one guide RNA and Cas9 nuclease expressed from individual vectors and up to 96% inhibition was reached when they were combined on one expression vector. Next, they multiplexed the two most effective gRNAs, an approach which proved to be even more effective. Finally, the expression vectors that contained expression cassettes for one gRNA and the Cas9 nuclease were tested in the HBV-hydrodynamic mouse model. Serum HBsAg levels were significantly reduced two days post-injection but increased again on day seven. However, Southern blot analyses revealed a reduction of intrahepatic HBV-expression levels of 20% to 60%. In conclusion the RNA-guided Cas9 nuclease system may be a useful technique for achieving inhibition of HBV replication.
Numerous and highly diverse gene therapeutic approaches were pursued to combat chronic HBV infection. Although strategies that solely rely on nucleic acids like antisense oligonucleotides and catalytic nucleic acids have a great advantage in their simplicity, these technologies are also limited due to their instability and imprecision. RNAi was most adopted and thoroughly investigated not only in the field of HBV therapy and this generated a deep knowledge regarding this technique. However, especially in the case of HBV, RNAi may be disadvantageous, because it does not affect the HBV cccDNA. It can rather be considered as an alternative to nucleos(t)ide analogues based therapy potentially associated with an improved ability to also respond to escape mutants. In addition, if long-term expression is required, RNAi-based approaches are superior compared to conventional therapy based on daily administration required for nucleos(t)ide analogues.
We believe that the most recent technology to combat HBV infection based on designer nucleases may be one of the most promising approaches to be explored in the future. This strategy bears the potential to actually eradicate cccDNA species in infected cells. Especially in combination with compounds that inhibit the HBV replication cycle this could be an attractive therapeutic option. However, major obstacles are the production and delivery of the designer nucleases and for translation of this approach into the clinic, this methodology needs to be further improved. The most promising results to date were obtained with genetic vaccines which were also pursued in the clinic. Also this strategy may hold great potential for eradicating chronic hepatitis B infection.
P- Reviewer: Kim ST, Song LT S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 805] [Cited by in RCA: 827] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 2. | Glebe D, Bremer CM. The molecular virology of hepatitis B virus. Semin Liver Dis. 2013;33:103-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 122] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 3. | WHO. Hepatitis B Fact Sheet No. 204. Available from: http://www.who.int/mediacentre/factsheets/fs204/en/. |
| 4. | Kwon H, Lok AS. Hepatitis B therapy. Nat Rev Gastroenterol Hepatol. 2011;8:275-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 211] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 5. | Thomas H, Foster G, Platis D. Mechanisms of action of interferon and nucleoside analogues. J Hepatol. 2003;39 Suppl 1:S93-S98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 6. | Buster EH, Flink HJ, Cakaloglu Y, Simon K, Trojan J, Tabak F, So TM, Feinman SV, Mach T, Akarca US, Schutten M, Tielemans W, van Vuuren AJ, Hansen BE, Janssen HL. Sustained HBeAg and HBsAg loss after long-term follow-up of HBeAg-positive patients treated with peginterferon alpha-2b. Gastroenterology. 2008;135:459-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 323] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 7. | Santantonio TA, Fasano M. Chronic hepatitis B: Advances in treatment. World J Hepatol. 2014;6:284-292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 8. | Jin L, Zeng X, Liu M, Deng Y, He N. Current progress in gene delivery technology based on chemical methods and nano-carriers. Theranostics. 2014;4:240-255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 272] [Cited by in RCA: 264] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 9. | Wang W, Li W, Ma N, Steinhoff G. Non-viral gene delivery methods. Curr Pharm Biotechnol. 2013;14:46-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 10. | Pezzoli D, Kajaste-Rudnitski A, Chiesa R, Candiani G. Lipid-based nanoparticles as nonviral gene delivery vectors. Methods Mol Biol. 2013;1025:269-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 11. | Demeneix B, Behr JP. Polyethylenimine (PEI). Adv Genet. 2005;53:217-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 12. | Davis BR, Brown DB, Prokopishyn NL, Yannariello-Brown J. Micro-injection-mediated hematopoietic stem cell gene therapy. Curr Opin Mol Ther. 2000;2:412-419. [PubMed] |
| 13. | Johnston SA, Tang DC. Gene gun transfection of animal cells and genetic immunization. Methods Cell Biol. 1994;43 Pt A:353-365. [PubMed] |
| 14. | Murakami T, Sunada Y. Plasmid DNA gene therapy by electroporation: principles and recent advances. Curr Gene Ther. 2011;11:447-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Escoffre JM, Zeghimi A, Novell A, Bouakaz A. In-vivo gene delivery by sonoporation: recent progress and prospects. Curr Gene Ther. 2013;13:2-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 16. | Schwerdt JI, Goya GF, Calatayud MP, Hereñú CB, Reggiani PC, Goya RG. Magnetic field-assisted gene delivery: achievements and therapeutic potential. Curr Gene Ther. 2012;12:116-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | Hodges BL, Scheule RK. Hydrodynamic delivery of DNA. Expert Opin Biol Ther. 2003;3:911-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 18. | Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1851] [Cited by in RCA: 1839] [Article Influence: 83.6] [Reference Citation Analysis (0)] |
| 19. | Cots D, Bosch A, Chillón M. Helper dependent adenovirus vectors: progress and future prospects. Curr Gene Ther. 2013;13:370-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 20. | Mingozzi F, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet. 2011;12:341-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 656] [Cited by in RCA: 697] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 21. | Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3710] [Cited by in RCA: 3697] [Article Influence: 127.5] [Reference Citation Analysis (0)] |
| 22. | Goodarzi G, Gross SC, Tewari A, Watabe K. Antisense oligodeoxyribonucleotides inhibit the expression of the gene for hepatitis B virus surface antigen. J Gen Virol. 1990;71:3021-3025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 54] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 23. | Blum HE, Galun E, von Weizsäcker F, Wands JR. Inhibition of hepatitis B virus by antisense oligodeoxynucleotides. Lancet. 1991;337:1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 24. | Reinis M, Reinisová M, Korec E, Hlozánek I. Inhibition of hepatitis B virus surface gene expression by antisense oligodeoxynucleotides in a human hepatoma cell line. Folia Biol (Praha). 1993;39:262-269. [PubMed] |
| 25. | Zhang YC, Taylor MM, Samson WK, Phillips MI. Antisense inhibition: oligonucleotides, ribozymes, and siRNAs. Methods Mol Med. 2005;106:11-34. [PubMed] |
| 26. | Offensperger WB, Offensperger S, Walter E, Teubner K, Igloi G, Blum HE, Gerok W. In vivo inhibition of duck hepatitis B virus replication and gene expression by phosphorothioate modified antisense oligodeoxynucleotides. EMBO J. 1993;12:1257-1262. [PubMed] |
| 27. | Korba BE, Gerin JL. Antisense oligonucleotides are effective inhibitors of hepatitis B virus replication in vitro. Antiviral Res. 1995;28:225-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 28. | Wu GY, Wu CH. Specific inhibition of hepatitis B viral gene expression in vitro by targeted antisense oligonucleotides. J Biol Chem. 1992;267:12436-12439. [PubMed] |
| 29. | Matsukura M, Koike K, Zon G. Antisense phosphorothioates as antivirals against human immunodeficiency virus (HIV) and hepatitis B virus (HBV). Toxicol Lett. 1995;82-83:435-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 30. | Yao ZQ, Zhou YX, Wang AL, Bai XF, Yang WS. Inhibition of hepatitis B viral gene expression by antisense phosphorothioate oligodeoxynucleotides. J Viral Hepat. 1995;2:85-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 31. | Robaczewska M, Narayan R, Seigneres B, Schorr O, Thermet A, Podhajska AJ, Trepo C, Zoulim F, Nielsen PE, Cova L. Sequence-specific inhibition of duck hepatitis B virus reverse transcription by peptide nucleic acids (PNA). J Hepatol. 2005;42:180-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 32. | Sun Z, Xiang W, Guo Y, Chen Z, Liu W, Lu D. Inhibition of hepatitis B virus (HBV) by LNA-mediated nuclear interference with HBV DNA transcription. Biochem Biophys Res Commun. 2011;409:430-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 33. | Walton CM, Wu CH, Wu GY. A ribonuclease H-oligo DNA conjugate that specifically cleaves hepatitis B viral messenger RNA. Bioconjug Chem. 2001;12:770-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 34. | Guang L, Yuan F, Xi M, Zhao C, Liu L, Wen E, Ai Y. Combination and cleavage of HBV DNA fragments by triple helix-forming oligonucleotides modified with manganese porphyrin in vitro. Chin Med J (Engl). 2003;116:1248-1252. [PubMed] |
| 35. | Madon J, Blum HE. Receptor-mediated delivery of hepatitis B virus DNA and antisense oligodeoxynucleotides to avian liver cells. Hepatology. 1996;24:474-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 36. | Nakazono K, Ito Y, Wu CH, Wu GY. Inhibition of hepatitis B virus replication by targeted pretreatment of complexed antisense DNA in vitro. Hepatology. 1996;23:1297-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 37. | Wu J, Gerber MA. The inhibitory effects of antisense RNA on hepatitis B virus surface antigen synthesis. J Gen Virol. 1997;78:641-647. [PubMed] |
| 38. | Zhang J, Chen F, Zhong S, Tang K, Shi X, Wang M, Peng J. Anti-HBV effect of targeted antisense RNA against HBV C gene. Zhonghua Ganzangbing Zazhi. 2000;8:169-170. [PubMed] |
| 39. | Ji W, St CW. Inhibition of hepatitis B virus by retroviral vectors expressing antisense RNA. J Viral Hepat. 1997;4:167-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 40. | Tung FY, Bowen SW. Targeted inhibition of hepatitis B virus gene expression: a gene therapy approach. Front Biosci. 1998;3:a11-a15. [PubMed] |
| 41. | Brantl S. Antisense-RNA regulation and RNA interference. Biochim Biophys Acta. 2002;1575:15-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 150] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 42. | Hafkemeyer P, Brinkmann U, Brinkmann E, Pastan I, Blum HE, Baumert TF. Pseudomonas exotoxin antisense RNA selectively kills hepatitis B virus infected cells. World J Gastroenterol. 2008;14:2810-2817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 43. | Schubert S, Kurreck J. Ribozyme- and deoxyribozyme-strategies for medical applications. Curr Drug Targets. 2004;5:667-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 73] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 44. | von Weizsäcker F, Blum HE, Wands JR. Cleavage of hepatitis B virus RNA by three ribozymes transcribed from a single DNA template. Biochem Biophys Res Commun. 1992;189:743-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 46] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 45. | Beck J, Nassal M. Efficient hammerhead ribozyme-mediated cleavage of the structured hepatitis B virus encapsidation signal in vitro and in cell extracts, but not in intact cells. Nucleic Acids Res. 1995;23:4954-4962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 46. | Welch PJ, Tritz R, Yei S, Barber J, Yu M. Intracellular application of hairpin ribozyme genes against hepatitis B virus. Gene Ther. 1997;4:736-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 50] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 47. | Zheng WC, Qi GR. tRNA-embedded Hammerhead Ribozymes Mediate Destruction of HBV (Subtype adr) in vitro. Shengwuhuaxue Yu Shengwuwuli Xuebao (Shanghai). 1998;30:432-438. [PubMed] |
| 48. | Bergeron LJ, Perreault JP. Development and comparison of procedures for the selection of delta ribozyme cleavage sites within the hepatitis B virus. Nucleic Acids Res. 2002;30:4682-4691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 49. | Tan TM, Zhou L, Houssais S, Seet BL, Jaenicke S, Peter F, Lim SG. Intracellular inhibition of hepatitis B virus S gene expression by chimeric DNA-RNA phosphorothioate minimized ribozyme. Antisense Nucleic Acid Drug Dev. 2002;12:257-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 50. | Hoeprich S, Zhou Q, Guo S, Shu D, Qi G, Wang Y, Guo P. Bacterial virus phi29 pRNA as a hammerhead ribozyme escort to destroy hepatitis B virus. Gene Ther. 2003;10:1258-1267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 62] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 51. | Goila R, Banerjea AC. Sequence-specific cleavage of hepatitis X RNA in cis and trans by novel monotarget and multitarget hammerhead motif-containing ribozymes. Oligonucleotides. 2004;14:249-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 52. | Ruiz J, Wu CH, Ito Y, Wu GY. Design and preparation of a multimeric self-cleaving hammerhead ribozyme. Biotechniques. 1997;22:338-345. [PubMed] |
| 53. | zu Putlitz J, Yu Q, Burke JM, Wands JR. Combinatorial screening and intracellular antiviral activity of hairpin ribozymes directed against hepatitis B virus. J Virol. 1999;73:5381-5387. [PubMed] |
| 54. | Passman M, Weinberg M, Kew M, Arbuthnot P. In situ demonstration of inhibitory effects of hammerhead ribozymes that are targeted to the hepatitis Bx sequence in cultured cells. Biochem Biophys Res Commun. 2000;268:728-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 55. | Zheng WC, Lu CD, Kong YY, Wang Y, Qi GR. Hammerhead Ribozymes Suppress HBV(adr) in HepG2 Cells. Shengwuhuaxue Yu Shengwuwuli Xuebao (Shanghai). 2001;33:25-29. [PubMed] |
| 56. | Pan WH, Xin P, Morrey JD, Clawson GA. A self-processing ribozyme cassette: utility against human papillomavirus 11 E6/E7 mRNA and hepatitis B virus. Mol Ther. 2004;9:596-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 57. | Li X, Kuang E, Dai W, Zhou B, Yang F. Efficient inhibition of hepatitis B virus replication by hammerhead ribozymes delivered by hepatitis delta virus. Virus Res. 2005;114:126-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 58. | Nash KL, Alexander GJ, Lever AM. Inhibition of hepatitis B virus by lentiviral vector delivered antisense RNA and hammerhead ribozymes. J Viral Hepat. 2005;12:346-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 59. | Wang CX, Lu YQ, Qi P, Chen LH, Han JX. Efficient inhibition of hepatitis B virus replication by hepatitis delta virus ribozymes delivered by targeting retrovirus. Virol J. 2010;7:61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 60. | Asahina Y, Ito Y, Wu CH, Wu GY. DNA ribonucleases that are active against intracellular hepatitis B viral RNA targets. Hepatology. 1998;28:547-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 61. | Goila R, Banerjea AC. Inhibition of hepatitis B virus X gene expression by novel DNA enzymes. Biochem J. 2001;353:701-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 62. | Hou W, Wo JE, Li MW, Liu KZ. In vitro cleavage of hepatitis B virus C mRNA by 10-23 DNA enzyme. Hepatobiliary Pancreat Dis Int. 2005;4:573-576. [PubMed] |
| 63. | Wo JE, Wu XL, Zhou LF, Yao HP, Chen LW, Dennin RH. Effective inhibition of expression of hepatitis B virus genes by DNAzymes. World J Gastroenterol. 2005;11:3504-3507. [PubMed] |
| 64. | Hou W, Ni Q, Wo J, Li M, Liu K, Chen L, Hu Z, Liu R, Hu M. Inhibition of hepatitis B virus X gene expression by 10-23 DNAzymes. Antiviral Res. 2006;72:190-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 65. | Scaglioni PP, Melegari M, Wands JR. Characterization of hepatitis B virus core mutants that inhibit viral replication. Virology. 1994;205:112-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 66. | Scaglioni P, Melegari M, Takahashi M, Chowdhury JR, Wands J. Use of dominant negative mutants of the hepadnaviral core protein as antiviral agents. Hepatology. 1996;24:1010-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 67. | von Weizsäcker F, Wieland S, Köck J, Offensperger WB, Offensperger S, Moradpour D, Blum HE. Gene therapy for chronic viral hepatitis: ribozymes, antisense oligonucleotides, and dominant negative mutants. Hepatology. 1997;26:251-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 46] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 68. | von Weizsäcker F, Köck J, Wieland S, Offensperger WB, Blum HE. Dominant negative mutants of the duck hepatitis B virus core protein interfere with RNA pregenome packaging and viral DNA synthesis. Hepatology. 1999;30:308-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 69. | Beterams G, Nassal M. Significant interference with hepatitis B virus replication by a core-nuclease fusion protein. J Biol Chem. 2001;276:8875-8883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 70. | Liu J, Li YH, Xue CF, Ding J, Gong WD, Zhao Y, Huang YX. Targeted ribonuclease can inhibit replication of hepatitis B virus. World J Gastroenterol. 2003;9:295-299. [PubMed] |
| 71. | Liu J, Li YH, Ding J, Gong WD, Xue CF, Zhao Y, Huang YX. Quantifying anti-HBV effect of targeted ribonuclease by real-time fluorescent PCR. World J Gastroenterol. 2004;10:2883-2885. [PubMed] |
| 72. | Zhao Y, Li Y, Liu J, Liu Z, Huang Y, Lei J, Li S, Xue C. Adenoviral-vector mediated transfer of HBV-targeted ribonuclease can inhibit HBV replication in vivo. Biochem Biophys Res Commun. 2008;371:541-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 73. | Pol S, Driss F, Carnot F, Michel ML, Berthelot P, Brechot C. [Efficacy of immunotherapy with vaccination against hepatitis B virus on virus B multiplication]. C R Acad Sci III. 1993;316:688-691. [PubMed] |
| 74. | Pol S. Immunotherapy of chronic hepatitis B by anti HBV vaccine. Biomed Pharmacother. 1995;49:105-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 75. | Davis HL, Michel ML, Whalen RG. DNA-based immunization induces continuous secretion of hepatitis B surface antigen and high levels of circulating antibody. Hum Mol Genet. 1993;2:1847-1851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 279] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 76. | Mancini M, Hadchouel M, Davis HL, Whalen RG, Tiollais P, Michel ML. DNA-mediated immunization in a transgenic mouse model of the hepatitis B surface antigen chronic carrier state. Proc Natl Acad Sci USA. 1996;93:12496-12501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 110] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 77. | Townsend K, Sällberg M, O’Dea J, Banks T, Driver D, Sauter S, Chang SM, Jolly DJ, Mento SJ, Milich DR. Characterization of CD8+ cytotoxic T-lymphocyte responses after genetic immunization with retrovirus vectors expressing different forms of the hepatitis B virus core and e antigens. J Virol. 1997;71:3365-3374. [PubMed] |
| 78. | Sällberg M, Hughes J, Javadian A, Ronlov G, Hultgren C, Townsend K, Anderson CG, O’Dea J, Alfonso J, Eason R. Genetic immunization of chimpanzees chronically infected with the hepatitis B virus, using a recombinant retroviral vector encoding the hepatitis B virus core antigen. Hum Gene Ther. 1998;9:1719-1729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 79. | Tacket CO, Roy MJ, Widera G, Swain WF, Broome S, Edelman R. Phase 1 safety and immune response studies of a DNA vaccine encoding hepatitis B surface antigen delivered by a gene delivery device. Vaccine. 1999;17:2826-2829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 141] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 80. | Roy MJ, Wu MS, Barr LJ, Fuller JT, Tussey LG, Speller S, Culp J, Burkholder JK, Swain WF, Dixon RM. Induction of antigen-specific CD8+ T cells, T helper cells, and protective levels of antibody in humans by particle-mediated administration of a hepatitis B virus DNA vaccine. Vaccine. 2000;19:764-778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 258] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 81. | Mancini-Bourgine M, Fontaine H, Scott-Algara D, Pol S, Bréchot C, Michel ML. Induction or expansion of T-cell responses by a hepatitis B DNA vaccine administered to chronic HBV carriers. Hepatology. 2004;40:874-882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 146] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 82. | Mancini-Bourgine M, Fontaine H, Bréchot C, Pol S, Michel ML. Immunogenicity of a hepatitis B DNA vaccine administered to chronic HBV carriers. Vaccine. 2006;24:4482-4489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 80] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 83. | Yang FQ, Yu YY, Wang GQ, Chen J, Li JH, Li YQ, Rao GR, Mo GY, Luo XR, Chen GM. A pilot randomized controlled trial of dual-plasmid HBV DNA vaccine mediated by in vivo electroporation in chronic hepatitis B patients under lamivudine chemotherapy. J Viral Hepat. 2012;19:581-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 84. | Forster AC, Altman S. External guide sequences for an RNA enzyme. Science. 1990;249:783-786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 240] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 85. | Yuan Y, Hwang ES, Altman S. Targeted cleavage of mRNA by human RNase P. Proc Natl Acad Sci USA. 1992;89:8006-8010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 114] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 86. | Werner M, Rosa E, George ST. Design of short external guide sequences (EGSs) for cleavage of target molecules with RNase P. Nucleic Acids Symp Ser. 1997;19-21. [PubMed] |
| 87. | Werner M, Rosa E, Nordstrom JL, Goldberg AR, George ST. Short oligonucleotides as external guide sequences for site-specific cleavage of RNA molecules with human RNase P. RNA. 1998;4:847-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 88. | Werner M, Rosa E, Al Emran O, Goldberg AR, George ST. Targeted cleavage of RNA molecules by human RNase P using minimized external guide sequences. Antisense Nucleic Acid Drug Dev. 1999;9:81-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 89. | Xia C, Chen YC, Gong H, Zeng W, Vu GP, Trang P, Lu S, Wu J, Liu F. Inhibition of hepatitis B virus gene expression and replication by ribonuclease P. Mol Ther. 2013;21:995-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 90. | Zhang Z, Vu GP, Gong H, Xia C, Chen YC, Liu F, Wu J, Lu S. Engineered external guide sequences are highly effective in inhibiting gene expression and replication of hepatitis B virus in cultured cells. PLoS One. 2013;8:e65268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 91. | Hannon GJ. RNA interference. Nature. 2002;418:244-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3058] [Cited by in RCA: 2900] [Article Influence: 126.1] [Reference Citation Analysis (0)] |
| 92. | Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10522] [Cited by in RCA: 10146] [Article Influence: 375.8] [Reference Citation Analysis (1)] |
| 93. | Shlomai A, Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 201] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 94. | McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, Wieland SF, Marion PL, Kay MA. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 458] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 95. | Klein C, Bock CT, Wedemeyer H, Wüstefeld T, Locarnini S, Dienes HP, Kubicka S, Manns MP, Trautwein C. Inhibition of hepatitis B virus replication in vivo by nucleoside analogues and siRNA. Gastroenterology. 2003;125:9-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 136] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 96. | Giladi H, Ketzinel-Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther. 2003;8:769-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 236] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 97. | Hamasaki K, Nakao K, Matsumoto K, Ichikawa T, Ishikawa H, Eguchi K. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 2003;543:51-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 109] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 98. | Moore MD, McGarvey MJ, Russell RA, Cullen BR, McClure MO. Stable inhibition of hepatitis B virus proteins by small interfering RNA expressed from viral vectors. J Gene Med. 2005;7:918-925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 99. | Uprichard SL, Boyd B, Althage A, Chisari FV. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc Natl Acad Sci USA. 2005;102:773-778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 150] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 100. | Jia F, Zhang YZ, Liu CM. A retrovirus-based system to stably silence hepatitis B virus genes by RNA interference. Biotechnol Lett. 2006;28:1679-1685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 101. | Jia F, Zhang YZ, Liu CM. Stable inhibition of hepatitis B virus expression and replication in HepG2.2.15 cells by RNA interference based on retrovirus delivery. J Biotechnol. 2007;128:32-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 102. | Chen CC, Ko TM, Ma HI, Wu HL, Xiao X, Li J, Chang CM, Wu PY, Chen CH, Han JM. Long-term inhibition of hepatitis B virus in transgenic mice by double-stranded adeno-associated virus 8-delivered short hairpin RNA. Gene Ther. 2007;14:11-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 103. | Giering JC, Grimm D, Storm TA, Kay MA. Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol Ther. 2008;16:1630-1636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 155] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 104. | Chen CC, Sun CP, Ma HI, Fang CC, Wu PY, Xiao X, Tao MH. Comparative study of anti-hepatitis B virus RNA interference by double-stranded adeno-associated virus serotypes 7, 8, and 9. Mol Ther. 2009;17:352-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 105. | Sun Y, Li Z, Li L, Li J, Liu X, Li W. Effective inhibition of hepatitis B virus replication by small interfering RNAs expressed from human foamy virus vectors. Int J Mol Med. 2007;19:705-711. [PubMed] |
| 106. | Rauschhuber C, Xu H, Salazar FH, Marion PL, Ehrhardt A. Exploring gene-deleted adenoviral vectors for delivery of short hairpin RNAs and reduction of hepatitis B virus infection in mice. J Gene Med. 2008;10:878-889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 107. | Crowther C, Mowa MB, Ely A, Arbuthnot PB. Inhibition of HBV replication in vivo using helper-dependent adenovirus vectors to deliver antiviral RNA interference expression cassettes. Antivir Ther. 2014;19:363-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 108. | Deng L, Li G, Xi L, Yin A, Gao Y, You W, Wang X, Sun B. Hepatitis B virus inhibition in mice by lentiviral vector mediated short hairpin RNA. BMC Gastroenterol. 2009;9:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 109. | Xiangji L, Feng X, Qingbao C, Weifeng T, Xiaoqing J, Baihe Z, Feng S, Hongyang W, Mengchao W. Knockdown of HBV surface antigen gene expression by a lentiviral microRNA-based system inhibits HBV replication and HCC growth. J Viral Hepat. 2011;18:653-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 110. | Starkey JL, Chiari EF, Isom HC. Hepatitis B virus (HBV)-specific short hairpin RNA is capable of reducing the formation of HBV covalently closed circular (CCC) DNA but has no effect on established CCC DNA in vitro. J Gen Virol. 2009;90:115-126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 111. | Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 880] [Cited by in RCA: 866] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 112. | Peng J, Zhao Y, Mai J, Pang WK, Wei X, Zhang P, Xu Y. Inhibition of hepatitis B virus replication by various RNAi constructs and their pharmacodynamic properties. J Gen Virol. 2005;86:3227-3234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 113. | Wu K, Gong Y, Zhang X, Zhang Y, Mu Y, Liu F, Song D, Zhu Y, Wu J. Inhibition of hepatitis B virus replication by recombinant small interfering RNAs. Acta Virol. 2005;49:235-241. [PubMed] |
| 114. | Wu KL, Zhang X, Zhang J, Yang Y, Mu YX, Liu M, Lu L, Li Y, Zhu Y, Wu J. Inhibition of Hepatitis B virus gene expression by single and dual small interfering RNA treatment. Virus Res. 2005;112:100-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 115. | Weinberg MS, Ely A, Barichievy S, Crowther C, Mufamadi S, Carmona S, Arbuthnot P. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Mol Ther. 2007;15:534-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 68] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 116. | Ely A, Naidoo T, Arbuthnot P. Efficient silencing of gene expression with modular trimeric Pol II expression cassettes comprising microRNA shuttles. Nucleic Acids Res. 2009;37:e91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 117. | Gish RG, Satishchandran C, Young M, Pachuk C. RNA interference and its potential applications to chronic HBV treatment: results of a Phase I safety and tolerability study. Antivir Ther. 2011;16:547-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 118. | Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1240] [Cited by in RCA: 1252] [Article Influence: 65.9] [Reference Citation Analysis (0)] |
| 119. | Ely A, Naidoo T, Mufamadi S, Crowther C, Arbuthnot P. Expressed anti-HBV primary microRNA shuttles inhibit viral replication efficiently in vitro and in vivo. Mol Ther. 2008;16:1105-1112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 78] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 120. | Jenke AC, Wilhelm AD, Orth V, Lipps HJ, Protzer U, Wirth S. Long-term suppression of hepatitis B virus replication by short hairpin RNA expression using the scaffold/matrix attachment region-based replicating vector system pEPI-1. Antimicrob Agents Chemother. 2008;52:2355-2359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 121. | Wooddell CI, Rozema DB, Hossbach M, John M, Hamilton HL, Chu Q, Hegge JO, Klein JJ, Wakefield DH, Oropeza CE. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol Ther. 2013;21:973-985. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 242] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 122. | Li Z. Arrowhead Completes Enrollment of First Cohort in Phase 2a Trial of Chronic Hepatitis B Candidate ARC-520. Available from: http://www.arrowheadresearch.com/press-releases/arrowhead-completes-enrollment-first-cohort-phase-2a-trial-chronic-hepatitis-b-0. |
| 123. | Chinnappan M, Singh AK, Kakumani PK, Kumar G, Rooge SB, Kumari A, Varshney A, Rastogi A, Singh AK, Sarin SK. Key elements of the RNAi pathway are regulated by hepatitis B virus replication and HBx acts as a viral suppressor of RNA silencing. Biochem J. 2014;462:347-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 124. | Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 430] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 125. | Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298-1306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 715] [Cited by in RCA: 655] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 126. | Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 882] [Cited by in RCA: 826] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 127. | Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1484] [Cited by in RCA: 1494] [Article Influence: 106.7] [Reference Citation Analysis (0)] |
| 128. | Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, Weeks DP, Yang B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315-6325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 328] [Cited by in RCA: 279] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 129. | Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11979] [Cited by in RCA: 11018] [Article Influence: 847.5] [Reference Citation Analysis (0)] |
| 130. | Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10336] [Cited by in RCA: 11173] [Article Influence: 931.1] [Reference Citation Analysis (0)] |
| 131. | Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6555] [Cited by in RCA: 7009] [Article Influence: 584.1] [Reference Citation Analysis (0)] |
| 132. | Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1246] [Cited by in RCA: 1258] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 133. | Blancafort P, Segal DJ, Barbas CF. Designing transcription factor architectures for drug discovery. Mol Pharmacol. 2004;66:1361-1371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 140] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 134. | Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 231] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 135. | Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463-1468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 169] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 136. | Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156-1160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1419] [Cited by in RCA: 1334] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 137. | Zimmerman KA, Fischer KP, Joyce MA, Tyrrell DL. Zinc finger proteins designed to specifically target duck hepatitis B virus covalently closed circular DNA inhibit viral transcription in tissue culture. J Virol. 2008;82:8013-8021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 138. | Cradick TJ, Keck K, Bradshaw S, Jamieson AC, McCaffrey AP. Zinc-finger nucleases as a novel therapeutic strategy for targeting hepatitis B virus DNAs. Mol Ther. 2010;18:947-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 146] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 139. | Weber ND, Stone D, Sedlak RH, De Silva Feelixge HS, Roychoudhury P, Schiffer JT, Aubert M, Jerome KR. AAV-mediated delivery of zinc finger nucleases targeting hepatitis B virus inhibits active replication. PLoS One. 2014;9:e97579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 88] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 140. | McCarty DM, Monahan PE, Samulski RJ. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8:1248-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 510] [Cited by in RCA: 538] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 141. | Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509-1512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2048] [Cited by in RCA: 1814] [Article Influence: 113.4] [Reference Citation Analysis (0)] |
| 142. | Neff KL, Argue DP, Ma AC, Lee HB, Clark KJ, Ekker SC. Mojo Hand, a TALEN design tool for genome editing applications. BMC Bioinformatics. 2013;14:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 262] [Cited by in RCA: 216] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 143. | Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460-465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 835] [Cited by in RCA: 917] [Article Influence: 70.5] [Reference Citation Analysis (0)] |
| 144. | Schmid-Burgk JL, Schmidt T, Kaiser V, Höning K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator–like effector genes. Nat Biotechnol. 2013;31:76-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 192] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 145. | Uhde-Stone C, Gor N, Chin T, Huang J, Lu B. A do-it-yourself protocol for simple transcription activator-like effector assembly. Biol Proced Online. 2013;15:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 146. | Bloom K, Ely A, Mussolino C, Cathomen T, Arbuthnot P. Inactivation of hepatitis B virus replication in cultured cells and in vivo with engineered transcription activator-like effector nucleases. Mol Ther. 2013;21:1889-1897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 170] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 147. | Chen J, Zhang W, Lin J, Wang F, Wu M, Chen C, Zheng Y, Peng X, Li J, Yuan Z. An efficient antiviral strategy for targeting hepatitis B virus genome using transcription activator-like effector nucleases. Mol Ther. 2014;22:303-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 129] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 148. | Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, Lin YY, Wang HY, Wang CC, Shen YC. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol Ther Nucleic Acids. 2014;3:e186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 299] [Article Influence: 27.2] [Reference Citation Analysis (0)] |