INTRODUCTION
Due to its unique and distinct cellular composition with predominant abundance of Kupffer cells (KCs), natural killer (NK) cells and natural killer T (NKT) cells, the liver is considered to be an organ with special innate immune features[1,2]. While being constantly exposed to microbial products, (toxic) environmental substances and food antigens from the portal stream draining the intestine, the liver plays a pivotal role in the induction and maintenance of immune tolerance[3,4]. Nonetheless, the liver is involved in many systemic diseases[5] and can become the target of adverse immune reactions in chronic inflammatory liver diseases[6] like primary biliary cirrhosis (PBC), autoimmune hepatitis (AIH) and primary sclerosing cholangitis (PSC).
PBC, AIH and PSC are the three major immune-mediated hepatopathies. Variant forms of these diseases are generally called overlap syndromes although no standardized definition exists. Patients with overlap syndromes present with both hepatitic (AIH) and cholestatic (PBC, PSC) serum liver tests and exhibit histological features of AIH and PBC or PSC. A similar genetic predisposition may play a role in the development of these overlap syndromes as all three disorders share some common genetic susceptibility factors[7-9]. Poor responses to immune suppression suggest that these three diseases are immunopathogenic rather than autoimmune. Association of PSC with inflammatory bowel disease (IBD)[10] where commensal bacterial flora has been implicated in maintaining/triggering intestinal inflammation[11] and the specific association of PBC with an ubiquitous alphaproteobacterium[12-15] suggest that bacterial triggers may be involved in the pathogenesis of the two chronic cholestatic liver diseases.
ASSOCIATION OF PBC WITH NOVOSPHINGOBIUM AROMATICIVORANS
Chronic cholestasis is the main pathophysiological feature of PBC, one of the two most common chronic cholestatic liver diseases in adults. PBC usually progresses slowly to cirrhosis due to the immune-mediated destruction of small intrahepatic bile ducts, liver failure and death, unless liver transplantation is performed. Signature auto-antibodies to mitochondrial antigens (AMAs) with reactivity to the E2 subunit of the pyruvate dehydrogenase complex (PDC-E2) represent the serological hallmark for the diagnosis of PBC. A curative therapy is not available[16,17].
Several clinical reports have incriminated Novosphingobium aromaticivorans, an ubiquitous alpha proteobacterium[18,19] and intestinal commensal as a likely etiology for most cases of PBC in humans[12-14]. Based on these clinical studies, we have established a mouse model of autoimmune liver disease triggered by infection with this bacterium that strikingly resembles PBC in humans[15]. The onset and severity of liver lesions and anti-PDC-E2 antibody responses in this model were dependent on: (1) mouse genetic background; (2) hepatic persistence of Novosphingobium bacteria; and (3) hepatic presence of NKT cells activated by Novosphingobium glycosphingolipids (GSLs)[15,20]. These GSLs replace lipopolysaccharide (LPS) and constitute the unusual gram-negative cell wall of the Sphingomonodaceae[21,22]. NKT cells specifically recognize Sphingomonodaceae GSLs and, in the absence of toll-like receptor 4 (TLR4) engagement by LPS, dominate the innate immune response[20,23]. The preferential activation of NKT cells in the liver where NKT cells are abundant and Novosphingobium persists suggest, therefore, the basis for the biased autoreactivity towards autoantigens exposed in the liver environment and, ultimately, the severe, organ-specific manifestations of Novosphingobium infection resembling human PBC. A latent, unrecognized infection with Novosphingobium may also account for the striking redistribution of NKT cells from the blood to the livers of PBC patients and the expression of CD1d on biliary epithelial cells[24-26].
SPONTANEOUS AND INFECTION-INDUCED MOUSE MODELS OF PBC
Infection alone is not likely to be sufficient to confer disease; genetic predisposition plays an important role as well. Based on previous reports describing defined susceptibility loci in distinct autoimmune disorders[27,28], we evaluated the role of these loci in mediating and/or inhibiting Novosphingobium-infection in mice. Commonly used mouse strains, including C57BL/6 (B6), non-obese diabetic (NOD) and SJL, all exhibited chronic anti-mitochondrial autoantibodies including anti-PDC-E2 IgG as well as liver lesions after intravenous inoculation of Novosphingobium. However, while some developed severe liver lesions, others exhibited only milder pathology. In order to dissect the genetic susceptibility and resistance alleles within the different genetic backgrounds, we performed infection experiments in NOD congenic mice that were generated by the introgression of diabetes susceptibility loci from chromosome 3 and 4 of B6 and B10 mice onto the NOD background[29-31]. Utilizing these mouse strains with their concisely defined genetic regions, we aim to uncover the candidate alleles therein predisposing for severe liver disease and to understand the underlying mechanisms of genetic susceptibility to PBC by studying the regulation, expression and function of these candidate alleles. In all experiments, one NOD congenic strain, NOD 1101, consistently developed the most severe biliary pathology[15]. NOD 1101 is a congenic strain with a restricted B6 chromosomal segment obtained from the NOD.c3c4 mouse[29-31] that corresponds to the type 1 diabetes susceptibility loci Idd10 and Idd18 on chromosome 3. Infected, but not uninfected, NOD 1101 mice progressively develop massive enlargement of the liver in contrast to parental NOD.c3c4 mice that develop spontaneous liver autoimmunity and hepatomegaly[30]. Currently, we are dissecting these Idd10 and Idd18 regions in order to find the candidate gene(s) therein that promote(s) severe PBC.
Although NOD.c3c4 mice do not require an exogenous infection with Novosphingobium to exhibit liver lesions and develop autoantibody titers, we reasoned that re-circulation of bacteria or bacterial products from the intestine triggers this process in NOD.c3c4 mice as Novosphingobium has been detected in the feces of mice[32], similar as at mucosal surfaces of humans[12]. The combination of impaired tolerance and enhanced inflammation promoted by molecules relevant for the immune response encoded in these defined Idd regions may render these mice susceptible to lower bacterial innocula. In the context of genetic predisposition to autoimmunity in combination with bacterial infection, the immune reaction has to be evaluated from two additional perspectives: (1) as an insufficient immune reaction that allows a systemic infection with commensal bacteria; and/or (2) as an overzealous immune reaction that may allow bacterial eradication but only for the cost of collateral tissue damage; and, next to the traditional ones: (3) as an aberrant immune reaction towards self; and/or (4) as a failure to eliminate autoreactive lymphocytes before their migration into the periphery.
Here, we discuss these possibilities with respect to one candidate gene that has been associated with PBC and also with AIH and PSC in humans. Having unique tools for the dissection of these genetic loci with NOD congenics mice at hand, our studies in the mouse model allow not only the identification of susceptibility genes but also open the unique tool for directly testing them in translational studies for human disease.
GENETIC PREDISPOSITION FOR IMMUNE-MEDIATED LIVER PATHOLOGIES
Naturally occurring genetic polymorphisms determine the susceptibility of an individual to autoimmune diseases. Single nucleotide polymorphisms (SNPs) most likely evolved due to microbial pressure and reveal a consequence of natural selection for enhanced resistance/susceptibility to certain pathogens. Association studies mainly focused on immune genes that affect the immune system belonging to both the human leukocyte antigen (HLA) family and non-HLA immune modulator genes. Allelic variations in MHC class II (DQ, DR) and in components of the innate (C4*Q0, C4B*2, MBL, NRAMP1/SLC11A1, VDR) and of the adaptive (Cytotoxic T-Lymphocyte Antigen 4, [CTLA4, interleukin-1beta (IL-1beta), IL-12A, IL-12RB2, Tumor necrosis factor (TNF) alpha] immune system have been associated with susceptibility to PBC[33-37]. Next, with several associations with HLA genes in PSC[7,38,39] and AIH[40,41], SNPs within the CTLA-4 gene have been implicated with PSC[7] and AIH[8]. The potential role of allelic variations within several genes encoding components of the innate and adaptive immune system suggests some disturbances of host resistance to microbial infection and their implication in the initiation and/or perpetuation of inflammatory processes. This may apply in particular for genetic variations of the IL-12 pathway since several data link inherited deficiencies of IL-12, IL-12R and Interferon-gamma to increased susceptibility to microbial infection and severity of infectious diseases, in particular mycobacterial diseases[42]. As SNPs within CTLA-4 have been also associated with PSC and AIH[7-9], we will focus our discussion of genetic susceptibility to autoimmunity in the context of bacterial infection on this negative costimulatory molecule.
CTLA-4 is one of the leading examples of genetic variants that confer risk for developing diverse human autoimmune diseases as underscored by genome-wide association studies[43] on the one hand. On the other hand, it has been associated with an increased risk of infections with parasites[44-46], viruses[47-50] and invasive bacterial infections[51]. CTLA-4 and CD28 belong to the best-characterized co-receptors for T cells. Whereas CD28 activation propagates T cell activation, engagement of CTLA-4 can protect against the development of autoproliferative and/or autoimmune disease due to the inhibition of T cell responses. Indeed, CTLA-4 is an indispensable negative regulator of peripheral T cell function[52]. CTLA-4-/- mice display a lymphoproliferative disorder and a severe autoimmune phenotype with the development of myocarditis and pancreatitis and die within the first month of life[53,54]. No CTLA-4 deficient human has been reported so far. Two of the single nucleotide polymorphisms (SNPs) found within the CTLA4 gene, 49AG (rs231775) and CT60 (rs3087243), have been associated with susceptibility to PBC and/or correlated with autoantibody titers[9,55], although some controversy exists[56]. The 49AG SNP lies within the coding region of the CTLA4 signal peptide and is characterized by a threonine (A allele) to alanine (G allele) substitution, which has been shown to reduce the cell surface expression of CTLA4[57,58]. Thus, the G allele is proposed to contribute to autoimmune risk, resulting in increased T-cell proliferation in response to immune activation. The CT60 SNP of CTLA4 is located in the 3’ untranslated region of the gene and the slightly less common A allele is suggested to be protective for autoimmunity[43]. Conversely, the G allele is thought to impart autoimmune risk by interfering with splicing processes, resulting in reduced production of a soluble form of CTLA4[43] that has been shown to inhibit T-cell activation in vitro[59].
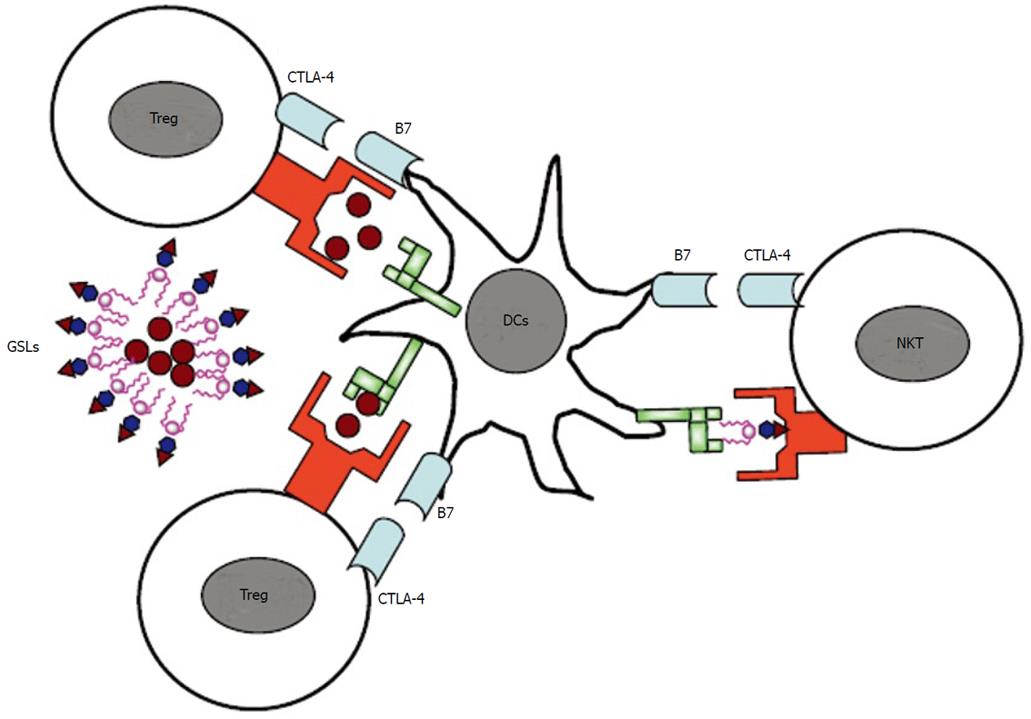
Whereas CD28 is constitutively expressed at moderate levels, CTLA-4 expression on most T cell populations is dependent on T cell activation although some subsets of regulatory T cells (Tregs) have been reported to constitutively express CTLA-4[60,61]. Whereas it is well-established that CTLA-4 is required for the optimal function of Tregs[62], its role on natural killer T (NKT) cells, conventional (T conv cells) or effector T (Teff) cells is less clear (Figure 1). A recent study showed that self-reactive T cells which escape negative selection must either express CTLA-4 by themselves or become subject to peripheral control by regulatory T cells that also depend on CTLA-4 for their function[63]. This may at least partially explain the differential expression of CTLA-4 among T cell subsets[64], not considering differences in the kinetics of its induction on different T cell subsets. Nonetheless, there is an important role for CTLA-4 expression by effector T cells in restraining tissue-specific CD4+ T cells from infiltrating, expanding their populations and/or surviving in target organs and provides evidence that CTLA-4 can control the pathogenicity of self-reactive T cell at multiple levels. It is not yet clear whether CTLA-4 acts directly on the T cell that expresses it or acts on the antigen-presenting cell, either by binding of the ligand B7 by CTLA-4 which leads to back-signaling into antigen-presenting cells[65,66] or by down-modulating B7 expression[62].
Figure 1 Regulation of T cell activation by cytotoxic T lymphocyte antigen 4.
While cytotoxic T lymphocyte antigen 4 (CTLA-4) is constitutively expressed on regulatory T cells (Tregs), its expression is induced on effector T cells (Teff) upon activation. While it is well-established that CTLA-4 is required for the optimal function of Tregs, its role on natural killer T cells or effector T cells is less clear. DCs: dentritic cells; NKT: natural killer T; GSLs: glycosphingolipids.
Considering the fact that (1) PBC can be induced in our model due to the infection with Novosphingobium; (2) Tregs promote infection; and (3) CTLA-4 is indispensable for Treg activity, expression of CTLA-4 variants may also influence the susceptibility of an individual towards infection. SNPs within CTLA-4 that may dampen the expression and/or function of the CTLA-4 protein may enhance autoreactive T cell responses due to the lack of intrinsic T cell or Treg control while decreasing bacterial infection at the same time. On the other hand, gain of function mutations may create a tolerogenic environment that allows the outgrowth of bacteria and, subsequently, tissue damage due to recruited macrophages that are independently of IFN-gamma activated. Therefore, it has to be determined if CTLA-4 activation affects T cell activation in general or affects the activation of defined T cell populations. Accordingly, (1) enhanced CTLA-4 activation decreases T cell activation in general, promoting bacterial infection and subsequently tissue damage due to impaired bacterial clearance and/or prolonged bacterial persistence; and/or (2) specific activation in T cell subpopulations, for example, regulatory T cells, allows tissue damage due to autoreactive T cells (that may be even less controlled by the respective CTLA-4 allele). This needs to be addressed in further studies. In addition, infection may modulate the surface expression of the CTLA-4 ligands, CD80 and CD86 on APCs.
CONCLUSION
Although genetic susceptibility factors have been often associated with aberrant immune responses to self-antigens and/or the inability of the immune system to eliminate autoreactive lymphocytes before emigration from the thymus, genetic susceptibility in light of bacterial infection may also refer to the susceptibility of an individual to develop an infection and/or to exhibit an inappropriate immune response causing collateral tissue damage and/or to prevent the development of an appropriate immune response to clear bacterial infection. In order to understand these mechanisms, animal models need to be developed which complement clinical and epidemiological studies. Uncovering the etiologies for these devastating liver diseases in the context of genetic susceptibility requires therefore further attention and research efforts. NOD congenic mice not only provide a unique tool for the identification of genetic susceptibility region, but also allow the translation into human disease as the regulation of candidate molecules in humans and mice can be directly compared.
Peer reviewers: Ali Sazci, MSc, PhD, Professor, Kocaeli University, Faculty of Medicine, Department of Medical Biology and Genetics, Umuttepe, Kocaeli 41380, Turkey; Julia Peinado Onsurbe, Assistant Professor, Department of Biochemistry and Molecular Biiology, Faculty of Biology, University of Barcelona, Avda Diagonal, Barcelona 64508028, Spain
S- Editor Zhang HN L- Editor Roemmele A E- Editor Liu N