Published online Mar 27, 2010. doi: 10.4254/wjh.v2.i3.127
Revised: January 14, 2010
Accepted: January 21, 2010
Published online: March 27, 2010
AIM: To identify new markers of hepatocellular carcinoma (HCC) using a proteomic analysis.
METHODS: Patients with liver cirrhosis of the three most frequent etiologies: hepatitis C virus, hepatitis B virus and alcoholic liver disease, were included in the study. The samples were analysed by 2D-electrophoresis in order to determine the differential protein expression. The proteins were separated according to the charge in immobilized pH 3-10 gradient strips and then by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Proteins of interest were excised, digested with trypsin and the resulting peptides were separated and identified.
RESULTS: Three differentially expressed apolipoproteins (Apo) were identified based on the protein profile using proteomic techniques: Apo-A1, Apo-A4 and Apo-E. Apo-A4 levels were significantly lower in HCC than in non-HCC patients regardless of etiology (P < 0.01). Multivariate logistic regression showed that Apo-A4 and Apo-A1 were the only independent factors related to HCC diagnosis (P < 0.05). The receiver operating characteristic (ROC) curve including both Apo-A4 and Apo-A1 showed an area under the ROC of 0.944 (P < 0.001), a sensitivity of 0.89 and a specificity of 0.81 for diagnosis of HCC.
CONCLUSION: Apo-A4 and Apo-A1 may be used clinically as biomarkers of HCC with a high sensibility and specificity. These findings may provide additional insights into the mechanism of HCC development and progression.
- Citation: Pleguezuelo M, Lopez-Sanchez LM, Rodriguez-Ariza A, Montero JL, Briceno J, Ciria R, Muntane J, Mata ML. Proteomic analysis for developing new biomarkers of hepatocellular carcinoma. World J Hepatol 2010; 2(3): 127-135
- URL: https://www.wjgnet.com/1948-5182/full/v2/i3/127.htm
- DOI: https://dx.doi.org/10.4254/wjh.v2.i3.127
Hepatocellular carcinoma (HCC) is the 6th cancer in incidence worldwide and the 3rd leading cause of cancer death. Overall, the survival of patients diagnosed with HCC remains very poor, with 1-year and 3-year survival rates of 36% and 17%, respectively[1]. The high mortality associated with HCC is primarily because by the time it is diagnosed, it is often unresponsive to treatment. Only 12% of cases receive potentially curative therapy (resection or transplantation).
Major common risk factors of HCC include hepatitis C virus (HCV), hepatitis B virus (HBV) and alcoholic liver disease (ALD). Rare risk factors include hemochromatosis, α1 anti-trypsin deficiency, and Wilson's disease. HBV or HCV infection is responsible for at least 80% of all HCC. With increase of HCV infection and immigration from HBV endemic populations, the number of deaths due to HCC in the United States is expected to rise over the next 20 years[2]. Most HCC cases develop in patients with advanced chronic liver disease; the increase in the number of patients living with cirrhosis may be another cause of the increased incidence of HCC. Therefore, methods to improve early detection and diagnosis of HCC as well as stratification of prognosis would be of great clinical benefit.
Although HCC meets the criteria of a tumor that would benefit from surveillance programs, the poor sensitivity and specificity of currently available tools have hindered their implementation. Ultrasound is particularly subject to low sensitivity and specificity when applied to cirrhotic patients and depends on the skills of ultrasonographist. Due to its high incidence and poor prognosis when diagnosed at a symptomatic stage, early HCC diagnosis has become a priority nowadays.
The outcome of HCC patients still remains dismal due to the difficulty in detecting the disease at its early stage, partly because of our limited knowledge of the molecular pathogenesis. There is a need to search for more serologic markers that are specifically associated with HCC, especially in the presence of cirrhosis. Therefore, studies aimed to improve the knowledge of the mechanisms associated with HCC development and to identify new biomarkers are urgently needed for its early diagnosis and the application of more effective therapeutic interventions[3].
Hepatocarcinogenesis is a slow multistep and multifactorial process, usually the consequence of long-term inflammation and fibrosis, which involves the accumulation of changes in the genome. At early stages, these alterations lead to the disruption of several genes that act in different regulatory pathways. The accumulation of irreversible structural alterations in genes and chromosomes result in the development of dysplastic hepatocytes, nodules and eventually HCC[4].
Therefore, the application of new technologies to improve our knowledge about the molecular pathogenesis of HCC, to identify biomarkers leading to an early diagnosis, and to define new therapeutic targets, is of great interest. Biomarkers are defined as indicators of genetic, cellular, biochemical or molecular alterations which can distinguish normal from abnormal biological processes. The ideal biomarker for HCC must be specific, traceable at a very early stage and not detectable in pre-malignant hepatic disease. With recent advances in genomics and proteomics, a great number of potential markers have been identified and developed as new candidate markers for HCC. Tumor markers may be useful for the detection of HCC in early stages, and also may provide information about its prognosis. Their use might be extended to therapeutic assessment and detection of recurrence[5].
There are different strategies for searching tumor markers. One is based on direct analysis of serum or other biological fluid, the other one is based on analysis of the tissue[6]. However, it is a challenging task due to the complexity of the proteome. Hundreds of thousands of different protein species present in the biological fluid or tissues must be separated, identified, and characterized, and it cannot be fully accomplished by a single experimental approach[3].
Although alpha fetoprotein (AFP) has been the most widely used marker for HCC, its sensitivity and specificity are poor[7] and the false-negative or positive rate with AFP level alone can reach 40%, especially for early HCC (< 3 cm in diameter). Its positive predictive value depends on the cut-off value, ethnicity, treatment and tumor stage. Its sensitivity and specificity for HCC diagnosis are 41%-65% and 80%-94%, respectively with a cut-off of 20 ng/mL[8]. A fucosylated variant of the AFP glycoprotein (AFP-L3) has shown better specificity than AFP for HCC diagnosis (63%-91.6%), but similar sensitivity (36%-71%)[9,10]. However, its specificity is limited, since its concentration can also increase in non-tumor, extrahepatic diseases such as diabetes, pancreatitis and hypothyroidism.
Although other numerous biomarkers with potential diagnostic or prognostic significance for HCC have been identified, most of them are considered non-specific as they can also be abnormally expressed in patients with non-malignant liver diseases. Furthermore, some of them have never reached general use due to lack of reagents, reproducibility and a good and clear system of development[11]. There have been only two FDA-approved tumor markers until now (AFP and AFP-L3). Therefore, a standardized approach is required to assess the tumor markers, and validation in large patient cohorts and preferably from multiple centres is necessary.
Proteins perform and regulate most biological functions. The systematic analysis of the whole proteome (proteinic complement of cells) may provide a functional meaning to the information provided by genome expression studies. Expression of proteins or their isoforms can be detected by proteomic analysis, and it also allows the detection of post-translational modifications. These data provide us with precious information to understand the molecular basis of HCC and to follow the course of the disease. Eventually, it could lead to earlier diagnosis of HCC that is essential in determining the best course of treatment options and possible outcomes. The plasma is an excellent target for proteomic approaches since it is readily available from patients on a regular basis and it is in contact with all tissues in the body, thus may reveal differences in the proteins expressed in these tissues. The new techniques for proteomic analysis allow several strategies for the identification of marker proteins for HCC.
One of the most common applications of proteomics is the development of novel biomarkers of disease, particularly cancer. A challenge for successful biomarker identification is to obtain appropriate samples. In spite of many recent technological advances in methods for the separation and analysis of proteins, two-dimensional gel electrophoresis (2DGE) is still the ‘‘gold standard’’ technique in this area[12]. This technique allows the separation of thousand proteins on the basis of both size and charge from a tissue or biological fluid. The high-resolution study of proteins by 2DGE is performed using immobilized pH gradients (IPG) in gels; this approach provides better resolution, reproducibility and loading capacity. The high potential of 2DGE on biomarker discovery is related to its ability to get information from the separated protein spots. Thus, in-gel digestion of proteins with specific endoproteases such as trypsin, enables us to obtain protein fingerprints, which can be analyzed by Matrix-assisted laser desorption / ionization time-of-flight mass spectrometry (MALDI-TOF). A protein spot can be identified by comparison of the mass spectrometric peptide map with that theoretically calculated in a database.
This study was aimed to identify potential biomarkers of HCC in patients with cirrhosis. Plasma samples from patients with cirrhosis and HCC were compared with those from patients with cirrhosis but without HCC. Proteomic analysis was performed to compare the profile of protein expression. We hypothesize that those markers may be useful for the diagnosis of HCC.
Patients with liver cirrhosis of the three most frequent aetiologies currently (HBV, HCV and ALD) were included in the study. Those patients were classified into two groups on the basis of the diagnosis of HCC. Histopathological classification for cirrhosis was performed according to the Ishak grading system[13]. HCC diagnosis was established according to the Barcelona-2000 criteria[7]. Written consent was obtained from each patient in the study and approval from Institutional Review Board.
The samples were analyzed by 2D-electrophoresis in order to determine the differential protein expression. The proteins were separated according to the charge in immobilized pH 3-10 gradient strips and then by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE). Proteins of interest were excised, digested with trypsin and the resulting peptides were separated and identified. The protein expression profile of patients with HCC was compared with that of patients without HCC for the identification of potential circulating biomarkers of HCC.
Peripheral blood samples (4.0 mL) were collected in sterile tubes containing 5.4 mg EDTA. Blood was immediately cooled on ice and within 15 min centrifuged at 3000 xg for 10 min to separate the plasma which was aliquoted and stored at -80°C.
The plasma was depleted of high abundant proteins using the ProteoPrepTM kit (Sigma-Aldrich). This affinity chromatography removes the 20 most abundant proteins in plasma, effectively enriching the other plasma proteins and maximizing their resolution by electrophoresis. Protein concentration of the depleted plasma was estimated with the Bradford assay using bovine serum albumin as standard.
2D gel electrophoresis was performed on selected samples using IPG strips (3-10 pH range, Biorad). Briefly, 1 mg plasma protein was mixed with 300 μL sample buffer (7 mol/L urea, 2mol/L thiourea, 4% CHAPS, 20mmol/L DTT, 0.5% TRITONx-100, 0.5% pharmalyte 3-10 and 0.001% blue bromophenol) and rehydrated into IPG strips overnight.
Isoelectric focusing was carried out for 34 000 VHs using a PROTEAN IEF system (BioRad). The IPG strips were then soaked into equilibration buffer (50mmol/L Tris-HCL, pH 8.8, 6 mol/L urea, 30% glycerol, 2% SDS and 0.001% bromophenol blue), containing 7.5 mg/mL DTT for 15 min. Thereafter, they were soaked into equilibration buffer containing 45 mg/mL iodoacetamide for 15 min. Second dimension was carried out in 12% polyacrylamide gel at 35 mA/gel (PROTEAN Xi Cell, BioRad).
The gels were stained with fluorescent dye (SYPRO Ruby, BioRad) according to the instruction of the manufacturer. Subsequently, the gels were imaged using LAS3000 (Fuji photo film) and analyzed with 2D PDQuest software (BioRad). To accurately compare the spots between gels, image spot intensity was normalized dividing the raw intensity of each spot in a gel by the total intensity of all the valid spots in that gel.
Protein spots of interest were excised from the polyacrylamide gels using a robotic workstation (Investigatore Propice, Genomics Solutions, Ann Arbor, MI, USA) and were trypsin-digested using a robotic digestion system (ProGeste, Genomic Solutions). Finally, peptides were analyzed on a MALDI-ToF/ToF 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA, USA). Mass spectrometry data were searched against the human protein database from MSDB (mass spectrometry protein sequence DataBase), using Mascot search engine (Matrix Science Inc., Boston, MA, USA).
Protein was confirmed using either Western Blot or nephelometry. For the Western Blot, the proteins separated by 2D-electrophoresis were immobilized on a nitrocellulose membrane using the semidry transfer system Transblot (BioRad). Subsequently, the proteins were detected using a primary polyclonal antibody (Santa Cruz Biotechnology diluted 1:200 and Chemicon International diluted 1:1 000) and the ECL Advance detection system (Amersham Biosciences, Uppsala, Sweden).
Demographical and clinical data were evaluated for all the patients. Differences between patients with and without HCC were assessed by univariate analysis using Chi-square tests for categorical variables and t tests for continuous variables. In the multivariate analysis, Odds ratios (OR) and 95% confidence intervals (CI) as well as P values were calculated for each risk factor. The P value ≤ 0.05 was considered statistically significant.
Statistical analysis was carried out using Statistical Package for Social Sciences (SPSS) 12.0 for Windows (release 12.0 SPSS Chicago, IL).
From January 2005 to January 2006, 22 consecutive patients with cirrhosis and 18 consecutive patients with HCC, were recruited from the outpatient clinic for liver diseases at the Reina Sofia University Hospital (Cordoba, Spain). The characteristics of the patients are shown in Table 1. Most of them were male, with a mean age of 56 years. HCV was the most frequent etiological factor of cirrhosis. Univariate analysis comparing patients with and without HCC (Table 2) showed that they were similar except for Child-Pugh score (5.6 ± 0.9 in HCC group vs 10 ± 3.2 in non-HCC group, P = 0.01) and INR (1.2 ± 0.1 in HCC patients vs 1.4 ± 0.3 in non-HCC patients, P = 0.04). Interestingly, AFP levels in patients with HCC were not significantly different from those without HCC (14.7 ± 15.8 vs 698.4 ± 1391.1; P = 0.13). In patients with HCC, the mean number of nodules was 1.7 (range 1-4), including 33% multinodules. The size of principal nodules ranged from 10 to 50 mm (mean 34.7 mm). There was no macrovascular invasion and extrahepatic spread in all cases.
| n = 40 | n (%) | Mean/median | SD/range |
| Gender | |||
| Male | 29 (72.5) | ||
| Female | 11 (27.5) | ||
| Age | 55.6 | 9.5 | |
| Etiology of cirrhosis | |||
| HBV | 10 (25) | ||
| HCV | 16 (40) | ||
| ALD | 14 (35) | ||
| Child-Pugh | 5.5 | (5-10) |
| NO HCC | HCC | P | |
| Male | 72.70% | 72.20% | 0.97 |
| Age (yrs) | 54.6 ± 10.1 | 56.8 ± 8.7 | 0.46 |
| Etiology of cirrhosis | |||
| HBV | 22.70% | 27.80% | 0.69 |
| HCV | 36.40% | 44.40% | 0.69 |
| ALD | 40.90% | 27.80% | 0.69 |
| Child-Pugh | 10 ± 3.2 | 5.6 ± 0.9 | 0.01 |
| HBV-DNA | 81.2 ± 94.8 | 146 ± 206.4 | 0.56 |
| HCV viral load | 1.8 × 106± 2.9 × 106 | 6.1 × 106± 4.2 × 106 | 0.28 |
| ALT | 53.9 ± 66.1 | 83.5 ± 95.7 | 0.39 |
| AST | 86.7 ± 111.1 | 92.2 ± 80.6 | 0.89 |
| Bilirubin | 3.4 ± 2.9 | 3.1 ± 3.6 | 0.81 |
| GGT | 69.5 ± 38.8 | 156.1 ± 159.6 | 0.09 |
| AP | 156 ± 74.5 | 101.2 ± 40.5 | 0.05 |
| Albumin | 3.3 ± 0.7 | 3.3 ± 0.4 | 0.99 |
| INR | 1.4 ± 0.3 | 1.2 ± 0.1 | 0.04 |
| Creatinine | 1.7 ± 2.1 | 1.1 ± 0.3 | 0.28 |
| LDH | 414 ± 52.7 | 399 ± 52.3 | 0.97 |
| AFP | 14.7 ± 15.8 | 698.4 ± 1391.1 | 0.13 |
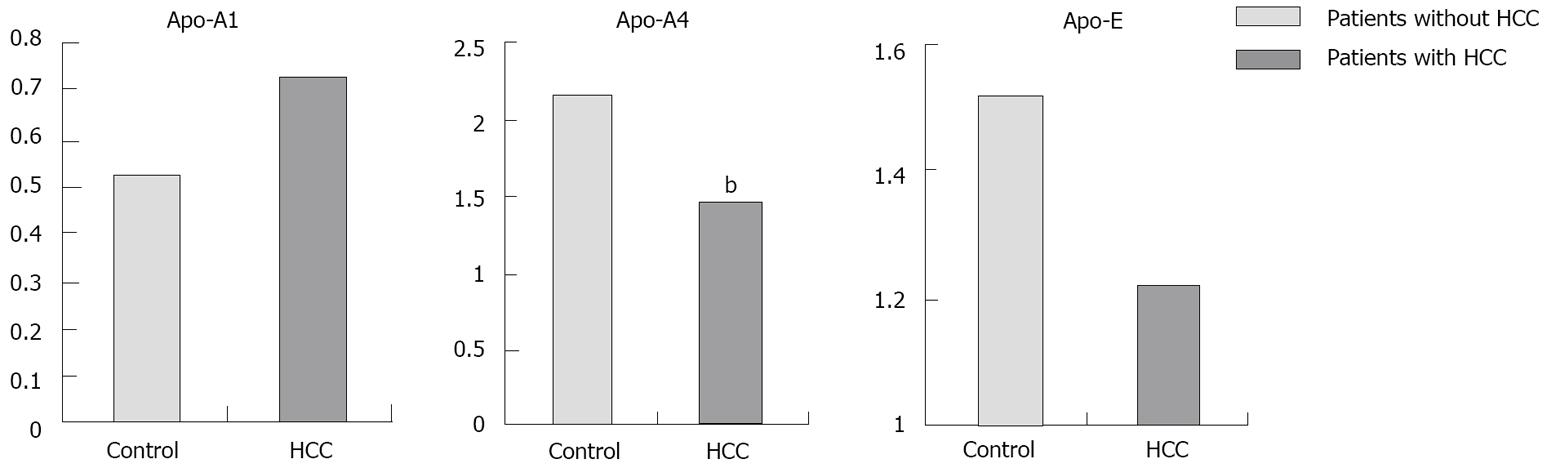
Proteomic analysis of the plasma samples and the comparison between patients with and without HCC revealed differential expression of 8 spots (Figure 1). These spots were identified as 3 different apolipoproteins:apolipoproteins (Apo-A1, Apo-A4 and Apo-E). Levels of Apo-A4 were significantly higher in patients without HCC than in patients with HCC (2.1 ± 0.4 vs 1.4 ± 0.5; P < 0.01) (Figure 2).
We performed a multivariate analysis in which HCC diagnosis was the dependent variable (all variables are shown in Table 2) and Apos levels were independent variables. Logistic multivariate regression revealed that levels of Apo-A1 (OR 1.45; 95% CI: 1.11-1.89, P = 0.006) and Apo-A4 (OR 0.21; 95% CI: 0.09-0.50, P < 0.001) were the only factors independently associated with HCC. Interestingly, Apo-A1 was associated with an elevated risk of HCC where as Apo-A4 was associated with lower risk of HCC; and AFP was not associated with risk of HCC. From these data the resulting logistic equation is:
P (HCC) = 1/(1 + e-z); in which z = logit (P) = 7.877 + (0.37 × Apo-A1.2) – (1.54 × Apo-A4.1)
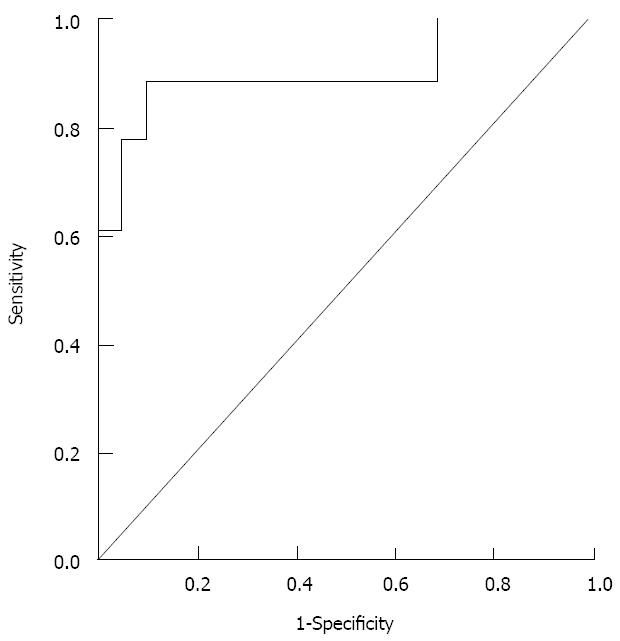
The receiver operating characteristic (ROC) curve for this equation using a cut-off level of 0.35 showed an area under the ROC of 0.91; with an 89% sensitivity and a 91% specificity for diagnosis of HCC (Figure 3).
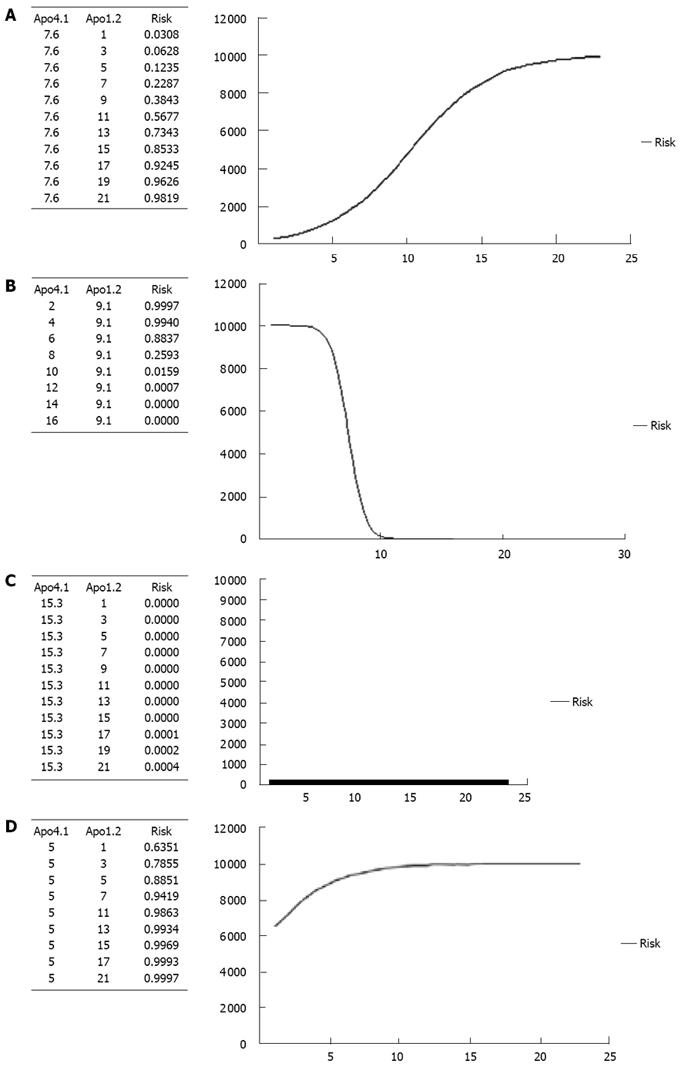
In order to define the real impact of Apos in HCC, we calculated the risk of HCC for different levels of Apo-A1 and a fixed level of Apo-A4 and vice versa. Risk curves are shown in Figure 4A-D. Figure 4A represents the risk of HCC for different levels of Apo-A1 (from its minimum level 0 to its maximum 20.3); adjusted for a fixed level of Apo-A4 (defined as its mean: 7.6). It reveals that the risk of HCC increases following the increase in Apo-A1 level and it is higher than 50% when Apo-A1 > 11. Figure 4B represents the risk of HCC for different levels of Apo-A4 (from its minimum level 5 to its maximum 15.3); and adjusted for a fixed level of Apo-A1 (defined as its mean: 9.1). It is shown that the risk of HCC decreases following the increase in Apo-A1 level. The risk of HCC is less than 25% when Apo-A4 < 8. Figure 4C shows the risk of HCC for different levels of Apo-A1, adjusted for a fixed level of Apo-A4 defined as its maximum in our study: 15.3. Interestingly, in this situation the risk of HCC is very low (close to null) regardless of Apo-A1 level. Finally, Figure 4D represents the risk of HCC for different levels of Apo-A1, adjusted for a fixed level of Apo-A4 defined as its minimum in our study: 5. As it is shown, the risk of HCC is high (> 60%) regardless of Apo-A1 level.
The increasing incidence and the poor prognosis of patients with HCC urge the identification of tumor-specific markers for the early detection of the disease and the discovery of potential therapeutic targets. Considerable efforts are being extended toward development of non-invasive methods for HCC detection. The ideal biomarker for this type of application should be detected with a high sensitivity in biological samples in a non-invasive manner; and blood represents the best source for detection of HCC related biomarkers.
Proteomics is a rapidly expanding discipline with a tremendous potential to extend our understanding of the molecular pathogenesis of human diseases and to identify biomarkers improving patient diagnosis, treatment, and prognosis. Hopefully, this knowledge will allow individualized approaches to patient care with the development of selective treatment modalities to benefit the patients, however there are still some limitations that must be overcome before they are put into clinical applications. New analytical strategies are expected to increase our capability to detect target proteins with clinical impact. In this field, several approaches have recently been taken in order to simplify the analysis of serum proteins, including removal of albumin and other high abundance proteins by affinity columns prior to analysis. This strategy provides gains in the number of lower abundance proteins, but it also results in the loss of small proteins bound to albumin.
Complexity of liver function hinders the development of a laboratory test to evaluate each clinical situation. Hepatocarcinogenesis is a slow and multifactorial process that involves the progressive accumulation of changes at the level of gene and protein expression. Up to now, dissimilar profiles of up- and down-regulated proteins have been reported; this discrepancy might result from the distinct etiology and differentiation of the analyzed HCC. However, it may also suggest that HCC may progress through different pathways resulting in the molecular heterogeneity denoted by proteomic studies. All these factors make the identification of universal HCC biomarkers difficult.
This study was aimed to identify plasma protein markers for HCC in cirrhotic patients. We compared the protein profiles in plasma between cirrhotic patients with and without HCC. This analysis revealed that 3 proteins were differentially expressed: Apo-A1, Apo-A4 and Apo-E.
Univariate analysis comparing patients with and without HCC showed no differences in AFP plasma concentration, whereas Apo-A4 was significantly higher in patients without HCC (2.1 ± 0.4 vs 1.4 ± 0.5; P < 0.01). The multivariate analysis confirmed Apo-A4 and Apo-A1 as the only independent factors related to HCC risk. Apo-A1 is associated with a higher risk of HCC, as against to Apo-A4 which is related to lower risk of HCC, while AFP was not associated with risk of HCC. The logistic equation from these data allows us to estimate the risk of HCC for a single patient with an 89% sensitivity and a 91% specificity. Apo-A1 is associated with higher risk of HCC (OR 1.45; 95% CI: 1.11-1.89, P = 0.006). On the contrary, Apo-A4 is inversely correlated with HCC risk (OR 0.21; 95% CI: 0.09-0.50, P < 0.001), i.e. when its level is high, the risk of HCC is very low independent of Apo-A1 levels.
Metabolism and homeostasis of carbohydrates, amino acids and lipids depend on liver function. Most Apo, lipids and lipoproteins, are synthesized in the liver. Thus hepatocellular injury or chronic liver diseases including HCC may result in abnormal pattern of these molecules in plasma[14]. The mechanisms leading to this alteration may be related to cytokines, metabolic cellular substances, or tumor factors, however they are not fully known. Patients with HCC frequently have other liver diseases such as chronic hepatitis and cirrhosis, which are often associated with plasma lipid and lipoprotein alterations[15]. Most Apos are synthesized in the liver[16] and some of them have been identified as serum markers in different types of cancer[17]. Furthermore, an Apo-A1 isoform has been identified as a pathological hallmark that may help understand the molecular pathogenesis of HCC[18].
Plasma triglycerides concentration in HCC patients was compared with controls in several studies. It was found to be decreased in one study[19], while increased[20] or not significantly different[21] in other studies. These data emphasize the lack of specificity of these findings, so that the results must be interpreted with caution. Lipoprotein-a together with ferritin and AFP may be a sensitive marker of liver function, since it has been found to increase in patients with acute hepatitis[22] and in those with HCC[23]. Liver is also the main organ for the synthesis, storage, transportation and degradation of some Apo[24]. Each Apo may be influenced by liver disease in a different way. To date, few data have been reported concerning changes in Apo concentration related to liver diseases or HCC. Hyperexpression of HBx in liver cells could inhibit Apo-B secretion[25]. Patients with metastatic liver cancer, showed an increase of Apo-E levels during slight bile stagnation[21]. Apo-M mRNA levels were significantly lower in HCC tissues than in the surrounding normal hepatic tissues. However, these data have to be confirmed in further studies[26].
The identification of biological targets leading to an early diagnosis of HCC is considered a priority of clinical hepatology. State of the art technologies such as genomics and proteomics have opened new frontiers in modern biomedical research. The methodological breakthrough that has taken place within proteomics over the last decade creates a major impact on clinical practice by promoting new ways in disease diagnosis, treatment, and surveillance. It is expected that the discovery of new biomarkers from differential protein/peptide profiling will benefit the clinical management of HCC in the near future. However, the complexity of HCC is still challenging for this still young science.
We compared protein profiles of cancerous and non-cancerous plasma samples in order to identify new biomarkers of HCC. Two apolipoproteins were identified: Apo-A4 and Apo-A1, which may be considered as tumor markers. This may extend our knowledge of the molecular pathogenesis of HCC. These findings may have important implications for the screening for HCC, since Apo-A4 and Apo-A1 may be used in combination with other traditional markers such as AFP, for an earlier and more efficient diagnosis of this cancer. However, further studies of large cohorts of patients are needed to determine their clinical use. Assessment of the relationship between these biomarkers and specific features of HCC such as size, presence of vascular invasion or extrahepatic spread, may help determine their prognostic usefulness. Analysis of plasma and tissue samples from patients with HCC by proteomic and genomic approaches, may allow discovery of potential targets for therapeutic intervention.
Our results provide additional confirmation that proteomic approaches can accurately identify HCC in patients with cirrhosis. These findings may have important implications for the screening and diagnosis of the HCC. They also provide some valuable information to recognize changes in molecular pathways that might participate in HCC development. However, further studies of large cohorts of patients are needed in order to define their clinical use.
The identification of biological targets leading to an early diagnosis of hepatocellular carcinoma (HCC) is considered a priority of clinical hepatology. State of the art technologies such as genomics and proteomics have opened new frontiers in modern biomedical research. Protein profiles of cancerous and non-cancerous plasma samples can be compared in order to identify new biomarkers of HCC. Those markers may led to an earlier diagnosis and application of more effective therapeutic interventions, thus improving the HCC patients prognosis.
Although numerous biomarkers with potential diagnostic or prognostic significance for HCC have been identified, most of them have never reached general use, in part due to lack of availability of reagents, lack of reproducibility or lack of a good and clear system of development. There have been only two FDA-approved tumor markers until now [alpha fetoprotein (AFP) and AFP-L3]. Therefore, a standardized approach is required to assess tumor markers, and further studies in large patient cohorts and preferably from multiple centers are necessary. Proteins perform and regulate most biological functions. The systematic analysis of the whole proteome may provide a functional meaning to the information provided by genome expression studies. One of the most common applications of proteomics is the development of novel biomarkers of disease, particularly cancer. In spite of many recent technological advances in methods for the separation and analysis of proteins, two-dimensional gel electrophoresis is still the ‘‘gold standard’’ technique in this area.
The present study identified two apolipoproteins (Apo-A4 and Apo-A1) that are differentially expressed in HCC and non-tumor serum samples. Our results provide additional confirmation that proteomic approaches can accurately identify HCC in patients with cirrhosis. These findings may extend our knowledge of the molecular pathogenesis of HCC. Since two-dimensional gel electrophoresis is an expensive technology, most studies are based on a modest sample size, thus it is critical that the statistical power should be sufficient to detect protein expression differences of interest. Our findings have clear statistical significance and may have important clinical implications, since Apo-A4 and Apo-A1 may be used in combination with other traditional markers such as AFP for an earlier and more efficient diagnosis of liver cancer. However, further studies of large cohorts of patients are needed to determine their clinical use.
These findings may have important implications for the screening for HCC, since Apo-A4 and Apo-A1 may be used in combination with other traditional markers such as AFP, for an earlier and more efficient diagnosis of this cancer. They also provide valuable information for the investigation of the molecular pathways that might participate in HCC development. Assessment of the relationship between these biomarkers and specific features of HCC such as size, presence of vascular invasion or extrahepatic spread, may help determine their prognostic usefulness. Analysis of plasma and tissue samples from patients with HCC by proteomic and genomic approaches, may allow discovery of potential targets for therapeutic intervention.
The authors evaluated biomarkers of HCC using proteomic approach. Differential expression of plasma protein between patients with and without HCC was analyzed. The results of this study concluded that Apo-A4 and Apo-A1 may be used clinically as biomarkers of HCC with a high sensibility and specificity. This is an interesting study, and the results may be important to the clinical field.
Peer reviewer: Ajith T A, PhD, Assistant Professor of Biochemistry, Department of Biochemistry, Amala Institute of Medical Sciences, Amala Nagar, Thrissur 680555, India
| 1. | El-Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127:S27-S34. |
| 2. | Block TM, Marrero J, Gish RG, Sherman M, London WT, Srivastava S, Wagner PD. The degree of readiness of selected biomarkers for the early detection of hepatocellular carcinoma: notes from a recent workshop. Cancer Biomark. 2008;4:19-33. |
| 3. | Santamaría E, Muñoz J, Fernández-Irigoyen J, Prìeto J, Corrales FJ. Toward the discovery of new biomarkers of hepatocellular carcinoma by proteomics. Liver Int. 2007;27:163-173. |
| 4. | Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339-346. |
| 5. | Yoon SK, Lim NK, Ha SA, Park YG, Choi JY, Chung KW, Sun HS, Choi MJ, Chung J, Wands JR. The human cervical cancer oncogene protein is a biomarker for human hepatocellular carcinoma. Cancer Res. 2004;64:5434-5441. |
| 6. | Chignard N, Beretta L. Proteomics for hepatocellular carcinoma marker discovery. Gastroenterology. 2004;127:S120 S125. |
| 7. | Bruix J, Sherman M, Llovet JM, Beaugrand M, Lencioni R, Burroughs AK, Christensen E, Pagliaro L, Colombo M, Rodés J. Clinical management of hepatocellular carcinoma. Conclusions of the Barcelona-2000 EASL conference. European Association for the Study of the Liver. J Hepatol. 2001;35:421-430. |
| 8. | Gupta S, Bent S, Kohlwes J. Test characteristics of alpha-fetoprotein for detecting hepatocellular carcinoma in patients with hepatitis C. A systematic review and critical analysis. Ann Intern Med. 2003;139:46-50. |
| 9. | Leerapun A, Suravarapu SV, Bida JP, Clark RJ, Sanders EL, Mettler TA, Stadheim LM, Aderca I, Moser CD, Nagorney DM. The utility of Lens culinaris agglutinin-reactive alpha-fetoprotein in the diagnosis of hepatocellular carcinoma: evaluation in a United States referral population. Clin Gastroenterol Hepatol. 2007;5:394-402; quiz 267. |
| 10. | Sterling RK, Jeffers L, Gordon F, Sherman M, Venook AP, Reddy KR, Satomura S, Schwartz ME. Clinical utility of AFP-L3% measurement in North American patients with HCV-related cirrhosis. Am J Gastroenterol. 2007;102:2196-2205. |
| 11. | Pleguezuelo M, Germani G, Marelli L, Xiruochakis E, Misseri M, Pinelopi M, Arvaniti V, Burroughs AK. Evidence-based diagnosis and locoregional therapy for hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol. 2008;2:761-784. |
| 12. | Elrick MM, Walgren JL, Mitchell MD, Thompson DC. Proteomics: recent applications and new technologies. Basic Clin Pharmacol Toxicol. 2006;98:432-441. |
| 13. | Ishak KG. Chronic hepatitis: morphology and nomenclature. Mod Pathol. 1994;7:690-713. |
| 14. | Jiang JT, Wu CP, Xu N, Zhang XG. Mechanisms and significance of lipoprotein (a) in hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2009;8:25-28. |
| 15. | Cicognani C, Malavolti M, Morselli-Labate AM, Zamboni L, Sama C, Barbara L. Serum lipid and lipoprotein patterns in patients with liver cirrhosis and chronic active hepatitis. Arch Intern Med. 1997;157:792-796. |
| 16. | Tietge UJ, Boker KH, Bahr MJ, Weinberg S, Pichlmayr R, Schmidt HH, Manns MP. Lipid parameters predicting liver function in patients with cirrhosis and after liver transplantation. Hepatogastroenterology. 1998;45:2255-2260. |
| 17. | Zhang Z, Bast RC Jr, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng XY. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64:5882-5890. |
| 18. | Fernández-Irigoyen J, Santamaría E, Sesma L, Muñoz J, Riezu JI, Caballería J, Lu SC, Prieto J, Mato JM, Avila MA. Oxidation of specific methionine and tryptophan residues of apolipoprotein A-I in hepatocarcinogenesis. Proteomics. 2005;5:4964-4972. |
| 19. | Motta M, Giugno I, Ruello P, Pistone G, Di Fazio I, Malaguarnera M. Lipoprotein (a) behaviour in patients with hepatocellular carcinoma. Minerva Med. 2001;92:301-305. |
| 21. | Ooi K, Shiraki K, Sakurai Y, Morishita Y, Nobori T. Clinical significance of abnormal lipoprotein patterns in liver diseases. Int J Mol Med. 2005;15:655-660. |
| 22. | Geiss HC, Ritter MM, Richter WO, Schwandt P, Zachoval R. Low lipoprotein (a) levels during acute viral hepatitis. Hepatology. 1996;24:1334-1337. |
| 23. | Basili S, Andreozzi P, Vieri M, Maurelli M, Cara D, Cordova C, Alessandri C. Lipoprotein (a) serum levels in patients with hepatocarcinoma. Clin Chim Acta. 1997;262:53-60. |
| 24. | Lewis GF, Rader DJ. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ Res. 2005;96:1221-1232. |
| 25. | Kang SK, Chung TW, Lee JY, Lee YC, Morton RE, Kim CH. The hepatitis B virus X protein inhibits secretion of apolipoprotein B by enhancing the expression of N-acetylglucosaminyltransferase III. J Biol Chem. 2004;279:28106-28112. |
| 26. | Jiang J, Nilsson-Ehle P, Xu N. Influence of liver cancer on lipid and lipoprotein metabolism. Lipids Health Dis. 2006;5:4. |