Published online Dec 27, 2022. doi: 10.4254/wjh.v14.i12.1985
Peer-review started: September 5, 2022
First decision: October 20, 2022
Revised: October 24, 2022
Accepted: November 22, 2022
Article in press: November 22, 2022
Published online: December 27, 2022
Processing time: 109 Days and 4.5 Hours
Among the most common cancers, hepatocellular carcinoma (HCC) has a high rate of tumor recurrence, tumor dormancy, and drug resistance after initial successful chemotherapy or radiotherapy. A small subset of cancer cells, cancer stem cells (CSCs), exhibit stem cell characteristics and are present in various cancers, including HCC. The dysregulation of microRNAs (miRNAs) often acc
Core Tip: The liver cancer stem cells (LCSCs) play a crucial role in the development of hepatocellular carcinomas (HCCs) and play a significant role in the development of drug resistance and cancer recurrence. LCSCs are regulated by many factors, of which microRNAs (miRNAs) are an important part. miRNAs can influence the development of HCC by regulating the stem cell properties of LCSCs.
- Citation: Li L, Xun C, Yu CH. Role of microRNA-regulated cancer stem cells in recurrent hepatocellular carcinoma. World J Hepatol 2022; 14(12): 1985-1996
- URL: https://www.wjgnet.com/1948-5182/full/v14/i12/1985.htm
- DOI: https://dx.doi.org/10.4254/wjh.v14.i12.1985
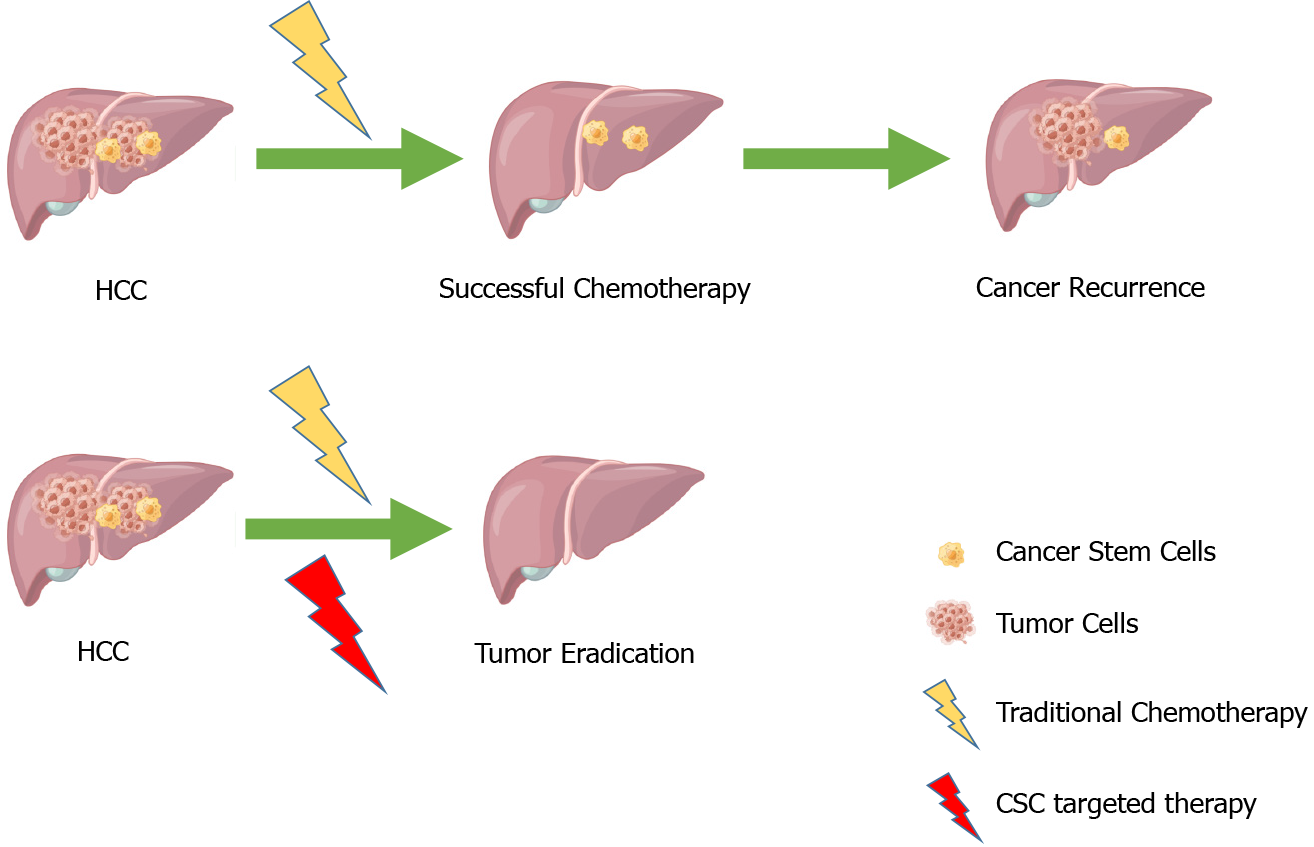
Hepatocellular carcinoma (HCC) is one of the most prevalent cancers in the liver, accounting for about 75% of all liver cancers, with a poor clinical prognosis, resulting in 500000-600000 deaths each year[1-4]. In recent years, there has been substantial progress in the diagnosis and treatment of HCC, but the high recurrence and metastasis rates of HCC still pose a headache for doctors and patients. The proposal of cancer stem cell (CSC) theory provides us with a direction. CSCs are considered one of the very small cell types in tumor cells with unlimited proliferative potential, which can drive tumorigenesis, and development. They can confer unique drug resistance, recurrence, and metastasis capabilities to tumors[5-8]. Conventional cancer treatments only kill common cancer cells, but CSCs remain. When in the right microenvironment, CSCs begin to proliferate and differentiate, leading to cancer recurrence. In recent years, many studies have focused on liver cancer stem cells (LCSCs) and achieved satisfactory results. Therefore, targeting CSCs is considered a more promising approach to improving the outcomes of conventional treatments (Figure 1).
An example of a microRNA (miRNA) is a small, non-coding RNA that is produced by endogenous cells and can be used to regulate gene expression by binding to the 3' untranslated region (UTR) of genes to inhibit their translation[9,10]. It has been shown that miRNAs can regulate tumorigenesis, progression, invasion, and even tumor recurrence in HCC by acting as tumor promoters or suppressors[11,12]. Another important finding is that miRNAs can modulate the stemness profile of LCSCs to combat conventional therapy further. Pollutri et al[13] reported that miR-494 induces sorafenib resistance in HCC and is associated with stem cell phenotypes. Further research has demonstrated that miR-181 family members play a critical role in maintaining the stem cell characteristics of HCC cells in a study by Ai et al[14]. Therefore, we believe miRNAs play a key role in LCSCs, and understanding this information will help our further research and development of HCC therapies. This review summarizes recent years' research findings and reports, outlines the role of miRNAs in LCSCs, and discusses potential therapeutic strategies for HCC recurrence, intending to provide clinical practitioners with information about how to treat HCC patients effectively.
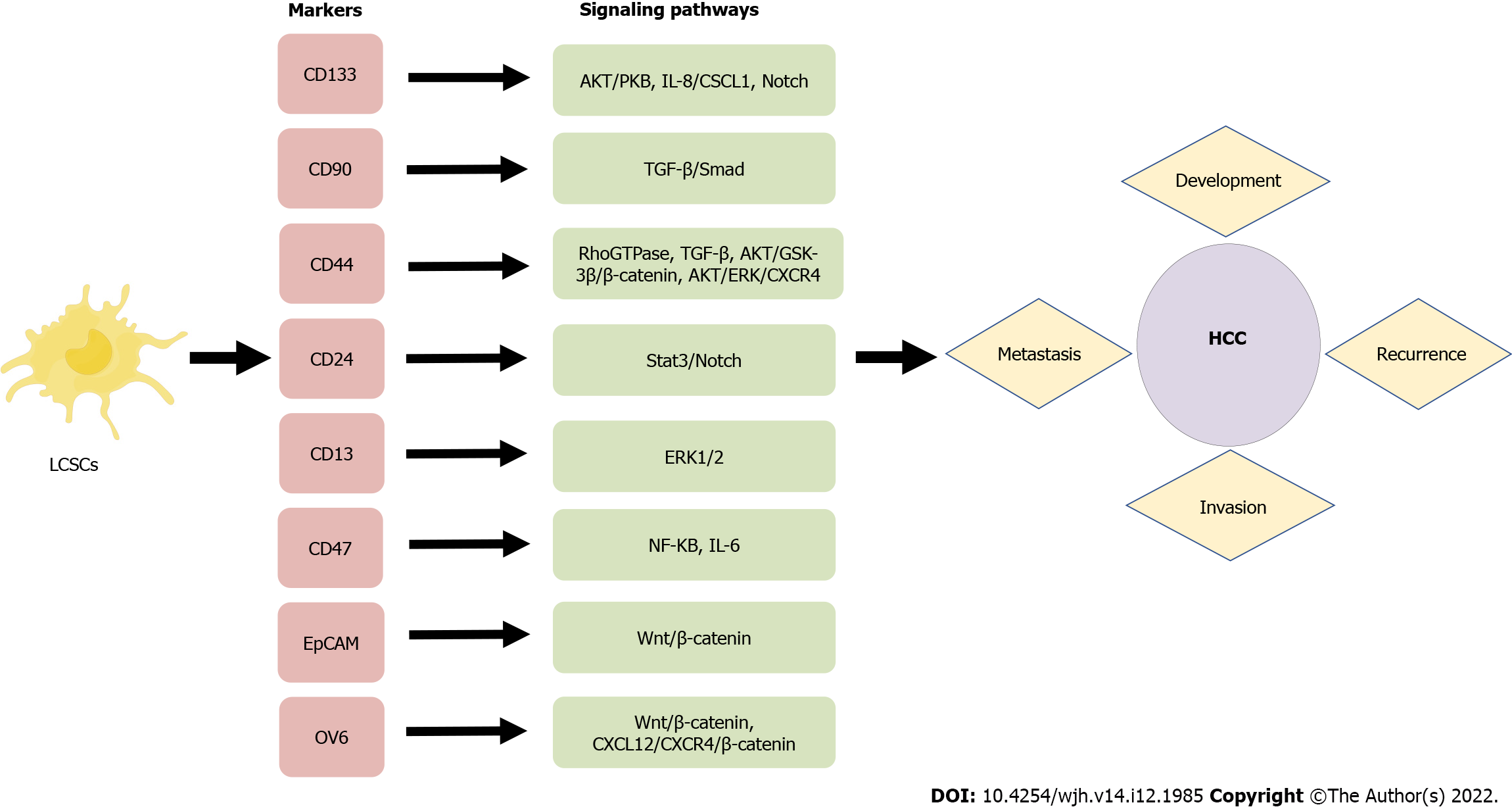
A number of characteristics of LCSCs are similar to those of normal stem cells, including their ability to self-renew and differentiate. LCSCs are more prevalent in vivo than other tumor cell types. They can promote the growth of primary cancer cells and facilitate the metastasis of transplanted secondary tumors, and they are crucial in the recurrence of HCC. In order to identify and isolate CSCs effectively, it is mostly necessary to take advantage of surface markers. Common LCSCs are CD133, CD90, CD44, CD13, CD47, etc. During the past few decades, a growing body of evidence has been generated concerning the properties of specific surface markers on LCSCs, which has provided opportunities for investigating potential biological functions, signaling pathways, and therapeutic approaches for HCC (Figure 2). Table 1 summarizes the major surface molecular markers of LCSCs and their potential roles in HCC.
| Markers | Biological functions in LCSCs | Signaling pathways | Recurrence | Ref. |
| CD133 | Tumor angiogenesis, growth, self-renewal, invasion, and chemoresistance | AKT/PKB, IL-8/CXCL1, Notch | High recurrence | [16-23] |
| CD90 | Preferably in poorly differentiated HCC, inflammation, circulation, drug resistance, and lipid metabolism | TGF-β/Smad | A shorter time to recurrence | [25-28] |
| CD44 | Extensive proliferation, self-renewal, invasion, and tunorigenicity | TGF-β, AKT/GSK-3β/β-catenin, AKT/ERK/CXCR4 | The significant risk factors of recurrence | [30-36] |
| CD24 | Cell surface glycoprotein, drives CSC genesis | Stat3/Notch | A prognostic predictor for recurrence-free survival | [37-41] |
| CD13 | Tumorigenicity, cell proliferation, cell cycle, self-renewal, and chemoresistance | ERK1/2 | Early recurrence | [42-44] |
| CD47 | Tumor initiation, self-renewal, and metastasis | CTSS/PAR2, NF-κB, IL-6 | Shorter recurrence-free survival | [45-47] |
| OV6 | Invasive and metastatic potential, form tumors, invasiveness, metastasis, substantial chemoresistance | Wnt/β-catenin, CXCL12/CXCR4/β-catenin | [48,49] | |
| EpCAM | An early biomarker for HCC, self-renewal, differentiation, chemoresistance, highly invasion and tumorigenisis | Wnt/β-catenin | High recurrence | [50-54] |
In 1997, CD133 was discovered as the first protein on the surface of neuroepithelial stem cells[15]. A transmembrane glycoprotein consisting of five transmembrane domains, two extracellular glycosylation chains, and three transmembrane domains is an important surface glycoprotein that serves as a cell surface marker. CD133 is expressed in embryonic epithelial stem cells, colon cancer, prostate cancer, pancreatic cancer, brain tumor, HCC, hematopoietic stem cells, and the like. CD133 was identified as a liver CSC marker in 2007[16-18]. According to studies conducted by our laboratory, the expression of CD133 in HCC cells is negatively related to the overall survival rate of patients with HCC and the rate of recurrence[19]. HCC patients with higher CD133 expression levels in the primary lesion tend to live shorter and have a higher recurrence rate postoperatively than those with lower CD133 expression levels[20]. HCC patients with higher CD133 expression levels also responded poorly to the conventional chemotherapy drug sorafenib. Several molecular mechanisms have been involved in the action of CD133 on tumors, including angiogenesis, self-renewal, growth, invasion, and chemoresistance. CD133+ cells in HCC contribute to chemoresistance by preferentially activating the Akt/PKB and Bcl-2 cell survival receptors during the chemoresistance response[21]. As a result of the interaction between neurotensin and interleukin-8 and CXCL1 signals in the liver, CD133 controls tumorigenesis, growth, and self-renewal of liver tumor-initiating cells[22]. The expression of iNOS in CD24+CD133+ LCSCs, but not CD24-CD133- LCSCs, enhanced Notch1 signaling, and accelerated HCC initiation in the mouse xenograft tumor model[23].
CD90+ cells from HCC Cell Lines were reported to have higher tumorigenic and metastatic potential than CD90− cells in 2008, suggesting that CD90+ cells can be used as a marker of metastatic HCC[24,25]. Consistent with these findings, CD90 expression is positively correlated with HCC progression and poor prognosis[26-28]. CD90 is involved in varies molecular mechanisms, including inflammation, circulation, drug resistance, and lipid metabolism. In HCC 97H cells, the cyclin D1-mediated activation of Smad2/3 and Smad4 is an important regulatory mechanism in enhancing single sphere formation, enhancing the CD90+ population, increasing stemness gene expression, and increasing chemoresistance[29]. Therefore, CD90 may also be a surface marker for poor prognosis of HCC and a potential therapeutic target.
A transmembrane glycoprotein named CD44 has been found to be expressed on numerous cells, including hepatocytes, endothelial cells, lymphocytes, and mesenchymal stem cells. It plays a role in extensive proliferation, self-renewal, invasion, and tumorigenicity[30]. It is possible to isolate cancer cells with stem cell markers by using CD44 alone or in combination with other markers. CD44v6, a variant of CD44, participates in the proliferation of HCC cells by stimulating the Ras/MAPK signaling cascade through interaction with c-Met[31]. Several studies have indicated that CD44s are associated with poor prognoses in hepatocellular carcinoma patients and regulate the TGF-β-mediated mesenchymal phenotype[32]. TGF-β1 and CD44 are synergistic in that they contribute to epithelial mesenchymal transition (EMT) induction and the development of CSC properties in tumor cells by interacting via the AKT/GSK-3β/β-catenin pathway in HCC cells[33]. In addition, CD44 is known to enhance HCC migration and local metastases by triggering the AKT/ERK pathway via the CXCR4 receptor[34]. Therefore, CD44 may be a potential treatment target for HCC and a marker of poor prognosis for HCC[35,36].
It is known that CD24 is a glycoprotein that is expressed on the surface of stem cells, mature granulocytes, and B cells, as well as in malignant tumors, such as HCC, breast cancer, colon cancer, and small cell lung carcinoma[37,38]. As well as driving CSC development, CD24 is involved in the differentiation of progenitor and stem cells in the liver and in metastatic development, self-renewal, and chemotherapy resistance of HCC cells[39]. CD24+ liver tumor-initiating cells are driven to self-renew and initiate tumors via STAT3-mediated NANOG signaling[40]. An IL-6/STAT3 axis regulates CD24 and epithelial cell adhesion molecule (EpCAM) expression in liver cancer stem cells through long non-coding RNA DILC[41].
A membranous glycoprotein called CD13 is associated with the progression of cancer and drug resistance. Cell cycle, self-renewal, and tumorigenicity are all regulated by CD13, which is involved in tumorigenesis, cell proliferation, and chemoresistance[42]. The combination of CD13 with other surface markers could lead to prostate cancer tumorigenesis. The CD13 gene is expressed in LCSCs that are slow-growing or semi-quiescent, which contributes to the formation of HCC tumors[43]. Quiescent CD13+ CSCs accumulate after chemotherapy in HCCs, serving as a source of recurrence[44].
CD47 is a transmembrane member of immunoglobulin associated with immune evasion, tumor apoptosis, metastasis, tumor-initiating ability, chemoresistance, and proliferation in various cancers. In addition to tumor initiation and self-renewal, CD47 also plays an important role in metastasis in HCC. HCC growth can be inhibited by suppression of CD47, which inhibits CTSS/PAR2 signaling in vivo and causes chemosensitization[45]. There is a positive correlation between CD47 and NF-κB expression in HCC samples from clinical trials[46]. Patients with HCC with upregulated CD47 expression had poor overall survival and recurrence-free survival, and IL-6 derived from macrophages infiltrating the tumor was shown to activate STAT3 and upregulate CD47 expression on hepatoma cells[47].
OV6, a monoclonal antibody raised against hepatic progenitor cells isolated from rat livers treated with carcinogens, was shown to be a marker for such cells. An HCC cell line expressing OV6+ tumor-initiating cells has a greater potential for invasiveness and metastatic spread, both in vitro and in vivo, which promotes the metastasis and progression of HCC[48]. There was an association between higher levels of OV6+ tumor cells, aggressive clinicopathologic features, and a poor prognosis. Inhibition of β-catenin signaling leads to a decrease in the proportion of OV6+ cells in HCC cell lines and primary HCC tissues, which indicates the role of Wnt/β-catenin signaling in OV6+ HCC cells[49].
As another transmembrane glycoprotein found in most epithelial tissues, the EpCAM plays a role in signal transduction, cell adhesion, migration, proliferation, and differentiation[50-54]. EpCAM was discovered as a biomarker early in the diagnosis of HCC. A strong correlation was found between EpCAM expression in LCSCs and differentiation, chemoresistance, high invasion, and tumorigenesis in HCC. EpCAM is a target gene for Wnt-beta-catenin signaling that may help improve HCC prognosis.
Dysregulated miRNAs contribute to many critical processes in HCC, ranging from growth, proliferation, apoptosis, and differentiation to migration, invasion, and progress. Moreover, miRNAs are important in tumor recurrence and metastasis. Understanding miRNAs' biological roles and specific targets will help further research and development of HCC therapies. Table 2 summarizes the major miRNAs in HCC and their potential roles in HCC.
| miRNA | Target genes/pathways | Effects | Expression | Clinical relevance | Ref. |
| miR-9-3p | HBGF5, lncRNA SAMMSON, ERK1/2 pathway | Cell proliferation, migration, and invasion | Down | Lower levels in HCC than in healthy donors | [66] |
| miR-21 | KLF5, CAMSAP1, DDX1, MARCKSL1, PTEN, AKT, D24 RECK, PDCD4, TETs/PTENp1/PTEN pathway, TGF-β1/smad3 pathway | Cell proliferation, migration, invasion, and metastasis | Up | Higher in HCC than in CHB and in healthy volunteers, early diagnosis | [55-58] |
| miR-26 | ULK1, EphA2, TAK1, TAB3, NF-κB pathway | Apoptosis | Down | Poor survival | [67-69] |
| miR-30a | Beclin1, Atg5, Snail1, FOXA1, ADAMTS14, Ras/Raf/MEK/ERK pathway | Proliferation, apoptosis, metastasis, migration, invasion, and EMT | Down | Prevention of HCC recurrence | [70-73] |
| miR-122 | ADAM10, ADAM17, IGF1R, SRF, SNAI1, SNAI2, WNT1, CREB1, BCL9, Cyclin G1, NMPDK4, LDHA, and CD133, Wnt/β-catenin pathway, IGF-1R pathway | Cell growth, proliferation, differentiation, metabolism, invasion, and EMT | Down | More sensitive to chemotherapeutic agents and improves the anti-tumor effect of sorafenib on HCC in vivo | [74-78] |
| miR-125b | MCL1, BCLw, IL-6R, SIRT7, SMAD2, SMAD4 | Proliferation, metastasis, migration, and apoptosis | Down | A significantly longer time to recurrence and longer overall survival time | [79-82] |
| miR-130b-3p | HOXA5 | Up | Poor prognosis, higher in patients with recurrence | [59] | |
| miR-142 | TGFβ, THBS4, LDHA, CD-133, HMGB1 | Cell growth, metastasis, migration, and invasion | Down | [83-85] | |
| miR-155 | ZHX2, TP53INP1, TGF-β1 pathway | Cell proliferation, migration, invasion, and EMT | Up | Diagnostic biomarkers for HCC | [62, 62] |
| miR-182-5p | FOXO3, AKT, Wnt/β-catenin | Proliferation, motility, invasion, and metastasis | Up | Poor prognosis and early recurrence | [63] |
| miR-199b-5p | TGFβ, MAP4K3, DDR1 | Metastasis, migration, invasion, and EMT | Down | [86, 87] | |
| miR-200a | GAB1, FOXA2 | Proliferation, invasion, migration, and EMT | Down | Biomarkers for early-stage HCC | [87, 89] |
| miR-203 | Ki67, CAPNS1 | Proliferation, invasion, migration, and metastasis | Down | Tumor recurrence and poor survival of patients with early-stage HCC | [90, 91] |
| miR-221 | p53, PUMA, NF-kB, STAT3, AAV8, PTEN, TIMP3, TRAIL, RAS/RAF/ERK, AKT | Apoptosis, and proliferation | Up | [64, 65] | |
| miR-449a | Notch1, FOS, Met, Calpain6, POU2F1, Notch pathway | Metastasis, apoptosis, proliferation, migration, invasion, and EMT | Down | Short-term recurrence | [92-94] |
| miR-541 | ATG2A, RAB1B | Inhibited the growth, metastasis, and autophagy | Down | Associated with malignant clinicopathologic phenotypes, recurrence and survival | [95] |
Cells from HCC cell lines and patients express high levels of miR-21. There is a positive correlation between miR-21 expression and HCC migration and invasion. As a result of silencing miR-21, the protein levels of PTEN, RECK, PDCD4, and KLF5, as well as the protein and mRNA levels of KLF5, increase, leading to a reduction in HCC cell migration and invasion[55,56]. Hepatocellular carcinoma growth is promoted by exosomal miR-21 regulation of the TETs/PTENp1/PTEN pathway, and three novels predicted miR-21 targets (CAMSAP1, DDX1, and MARCKSL1) correlate with HCC patient survival[57,58]. There is an association between miR-130b-3p up-regulation in HCC and a poor prognosis[59]. Patients who undergo HCC resection are at an increased risk of recurrence if their miR-135a expression is high[60]. A direct target of TP53INP1 is MiR-155, which regulates the migration and invasion of liver cancer cells, EMT, and CSC acquisition (which is positively correlated with CD90 and CD133)[61,62]. Patients with HCC who express MiR-182-5p in tumor tissues are more likely to experience poor prognosis and recurrence of the disease at an earlier stage. miR-182-5p activates AKT/FOXO3a pathway and Wnt/β-catenin signaling by targeting FOXO3a, enhancing HCC proliferation, motility, and invasion both in vitro and in vivo[63]. As miR-221 targets PTEN and TIMP3 tumor suppressors through activation of the AKT pathway, liver cancer cells express high levels of miR-221[64]. Upon Fas-induced fulminant liver failure, miR-221 is upregulated, which regulates liver expression of the p53 upregulated modulator of apoptosis[65].
Several miRNAs like miR-9-3p, miR-26, miR-30a, miR-122, miR-125b, miR-142, miR-142-3p, miR-199b-5p, miR-200a, miR-203, miR-449a, and miR-541 showed lower levels in HCC than in healthy donors. HBGF-5 expression is significantly downregulated by miR-9-3p overexpression, HCC viability and proliferation are reduced, and ERK1/2 is strongly downregulated[66]. Apoptosis is promoted by MiR-26 by targeting ULK1, EphA2, TAK1, and TAB3, which enhance chemosensitivity and radiosensitivity in HCC cells[67-69]. MiR-30a inhibits HCC cell proliferation by targeting FOXA1 via the Ras/ Raf/MEK/ERK signaling pathway, suppressing autophagy-mediated resistance and metastasis[70-72]. It facilitates tumor cell invasion, migration, and EMT when miR-30a is downregulated[73]. By downregulating miR-122, HCC cells proliferate, colonize, migrate, invade, metastasize, and activate IGF-1R and RAS/RAF/ERK pathways[74-77]. When miR-122 expression levels are elevated in HCC cells, it inhibits the EMT process by upregulating the expression of E-cadherin and downregulating ZEB1/2, Snail1/2, N-cadherin, and Vimentin[78]. miR-125b is correlated with cell proliferation, differentiation, metastasis, apoptosis migration, and EMT[79-81]. miR-125b overexpression attenuates EMT-associated chemoresistance, migration, and stemness and negatively correlated with CSC marker, EpCAM and CD13 expressions in HCC specimens by targeting SMAD2 and SMAD4[82]. Increasing the amount of miR-142 in the cells results in a decrease in vitality, proliferation, and EMT outcomes, as well as an increase in THBS4 which is overexpressed by cancer cells, resulting in more rapid migration and vascular invasion[83,84]. As a result of miR-142-3p inhibiting self-renewal, initiating tumor growth, invasion, migration, inducing angiogenesis and resisting chemotherapy in HCC cells, miR-142-3p is directly targeting CD133 to control the ability to confer cancer and stem cell-like characteristics[85]. It was found that overexpression of miR-19b-5p increases cell aggregation, suppresses migration and invasion in HCC cells, and inhibits the metastasis of xenograft tumors in nude mice. Akt phosphorylation is inhibited by miR-199b-5p overexpression, and N-cadherin and DDR1 are directly targeted and inhibited by miR-199b-5p overexpression[86,87]. In HCC, microRNA-200a directly targets GAB1 and FOXA2 to suppress cell invasion, migration, and metastasis[88,89]. MiR-203 expression is significantly associated with tumor recurrence and poor survival in HCC patients with early-stage tumors. In contrast, miR-203 overexpression suppresses Ki67 and CAPNS1 expression to inhibit proliferation, invasion, and metastasis of hepatic residual HCC[90,91]. Activating EMT via the Notch pathway promotes invasiveness in vitro by downregulating Calpain 6 and POU2F1; mir-449a induces apoptosis in liver cancer cells by downregulating Calpain 6 and POU2F1, it inhibits Met signaling and Snail accumulation in cells by targeting its 3'-UTR, and miR-449a contributes to short-term HCC recurrence[92-94]. HCC cells in vitro and in vivo are inhibited by miR-541 by inhibiting growth, metastasis, and autophagy, and the target genes are ATG2A and RAB1B[95].
It has been demonstrated that miRNAs could be therapeutic targets for HCC, but miRNA-based therapies have not been well developed for clinical applications. CSC therapies targeting miRNA are considered to be one of the most promising cancer treatments. In this way, miRNAs can regulate multiple genes at once, contributing to the regulation of CSC-related pathways. For example, miR-365 can regulate LCSCs through the RAC1 pathway[96]; miR-520f-3p is involved in altering the sensitivity of HCC cells to sorafenib treatment under hypoxic conditions by increasing stem cell phenotype[97]; miR-4320 inhibited epithelial-mesenchymal transition and reduced stemness characteristics in HCC cells by targeting FOXQ1 expression[98]; miR-206 inhibited LCSCs expansion by regulating EGFR expression[99]; Li et al[100] found that miR-613 inhibits LCSC proliferation and differentiation through regulation of SOX9; therapeutic delivery of miR-125b in a mouse model reduces the expression of CSC markers and inhibits HCC metastasis[82]. The findings of these studies suggest that miRNA therapy combined with targeting CSCs can treat HCC. However, the development of miRNA therapy remains challenging. The development of miRNA delivery systems in vivo has always been an area of interest for clinical treatment research. A specific, stable, low toxicity and durable delivery system is our hope, but currently, in the clinical treatment of HCC, there is still no very suitable in vivo delivery system. Furthermore, CSCs have great heterogeneity between patients, and how to accurately target CSCs is also a problem that needs to be addressed further.
In recent years, although research focusing on CSC has entered a trend of rapid growth, there are still many problems to be solved in clinical translation and practical application, especially in HCC patients. Targeting CSCs is considered as a potential therapeutic approach that can overcome the shortcomings of traditional treatments and significantly inhibit tumor recurrence. miRNAs play key roles in the post-transcriptional regulation of genes, and miRNAs are involved in various biological processes, including tumorigenesis. miRNA therapy has been used in some tumors and has entered the clinical stage, such as miR-34a has been used in a phase 1 study in patients with advanced solid tumors[101]. In clinical treatment, miRNAs can enhance the sensitivity of LCSCs to treatment, and targeting the deregulated key miRNAs in LCSCs can effectively reduce the role of LCSCs in metastasis and recurrence[102-104]. El-Mahdy et al[105] summarized the key signaling pathways associated with miRNAs (such as TP53, PI3k/AKT/mTOR, JAK/STAT, Wnt/β-catenin, and MAPK pathways), through which miRNAs can further affect the cellular processes and responses of HCC to clinical treatment. Therefore, investigating the role of miRNAs in LCSCs can help improve the prognosis of HCC patients and inform the development of new therapies.
Thanks to the China Scholarship Council for the scholarship to Dr. Li L. Thanks to FIGDRAW for providing the picture material.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: New Zealand
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Roohvand F, Iran; Xue F, China S-Editor: Liu GL L-Editor: A P-Editor: Liu GL
| 1. | Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology. 2019;156:477-491.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 754] [Cited by in RCA: 1216] [Article Influence: 202.7] [Reference Citation Analysis (1)] |
| 2. | Wang W, Wei C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 2020;7:308-319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 292] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 3. | Jiang YF, Yang ZH, Hu JQ. Recurrence or metastasis of HCC: predictors, early detection and experimental antiangiogenic therapy. World J Gastroenterol. 2000;6:61-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 57] [Cited by in RCA: 63] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 4. | Monto A, Wright TL. The epidemiology and prevention of hepatocellular carcinoma. Semin Oncol. 2001;28:441-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 93] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 5. | Jones RJ. Cancer stem cells-clinical relevance. J Mol Med (Berl). 2009;87:1105-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | Sukowati CHC. Heterogeneity of Hepatic Cancer Stem Cells. Adv Exp Med Biol. 2019;1139:59-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 7. | Eun K, Ham SW, Kim H. Cancer stem cell heterogeneity: origin and new perspectives on CSC targeting. BMB Rep. 2017;50:117-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 253] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 8. | Dzobo K, Senthebane DA, Ganz C, Thomford NE, Wonkam A, Dandara C. Advances in Therapeutic Targeting of Cancer Stem Cells within the Tumor Microenvironment: An Updated Review. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 9. | Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs' Action through miRNA Editing. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 438] [Cited by in RCA: 601] [Article Influence: 100.2] [Reference Citation Analysis (0)] |
| 10. | Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 853] [Cited by in RCA: 1821] [Article Influence: 227.6] [Reference Citation Analysis (0)] |
| 11. | Sartorius K, Sartorius B, Winkler C, Chuturgoon A, Makarova J. The biological and diagnostic role of miRNA's in hepatocellular carcinoma. Front Biosci (Landmark Ed). 2018;23:1701-1720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 12. | Qi W, Liang W, Jiang H, Miuyee Waye M. The function of miRNA in hepatic cancer stem cell. Biomed Res Int. 2013;2013:358902. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 13. | Pollutri D, Patrizi C, Marinelli S, Giovannini C, Trombetta E, Giannone FA, Baldassarre M, Quarta S, Vandewynckel YP, Vandierendonck A, Van Vlierberghe H, Porretti L, Negrini M, Bolondi L, Gramantieri L, Fornari F. The epigenetically regulated miR-494 associates with stem-cell phenotype and induces sorafenib resistance in hepatocellular carcinoma. Cell Death Dis. 2018;9:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 14. | Ai J, Gong C, Wu J, Gao J, Liu W, Liao W, Wu L. MicroRNA181c suppresses growth and metastasis of hepatocellular carcinoma by modulating NCAPG. Cancer Manag Res. 2019;11:3455-3467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 15. | Yang MH, Chen CL, Chau GY, Chiou SH, Su CW, Chou TY, Peng WL, Wu JC. Comprehensive analysis of the independent effect of twist and snail in promoting metastasis of hepatocellular carcinoma. Hepatology. 2009;50:1464-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 303] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 16. | Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, Zheng BJ, Guan XY. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542-2556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 899] [Cited by in RCA: 922] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 17. | Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, Castilho A, Ng I, Man K, Wong N, To KF, Zheng BJ, Lai PB, Lo CM, Chan KW, Guan XY. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7:694-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 322] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 18. | Piao LS, Hur W, Kim TK, Hong SW, Kim SW, Choi JE, Sung PS, Song MJ, Lee BC, Hwang D, Yoon SK. CD133+ liver cancer stem cells modulate radioresistance in human hepatocellular carcinoma. Cancer Lett. 2012;315:129-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 154] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 19. | Wang Y, Wu G, Fu X, Xu S, Wang T, Zhang Q, Yang Y. Aquaporin 3 maintains the stemness of CD133+ hepatocellular carcinoma cells by activating STAT3. Cell Death Dis. 2019;10:465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 20. | Song Y, Kim S, Lee H, No JH, Ryu HC, Kim J, Lim JW, Kim M, Choi I, Seo HR. Chromenopyrimidinone Controls Stemness and Malignancy by suppressing CD133 Expression in Hepatocellular Carcinoma. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 21. | Ma S, Lee TK, Zheng BJ, Chan KW, Guan XY. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749-1758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 605] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 22. | Tang KH, Ma S, Lee TK, Chan YP, Kwan PS, Tong CM, Ng IO, Man K, To KF, Lai PB, Lo CM, Guan XY, Chan KW. CD133(+) liver tumor-initiating cells promote tumor angiogenesis, growth, and self-renewal through neurotensin/interleukin-8/CXCL1 signaling. Hepatology. 2012;55:807-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 196] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 23. | Wang R, Li Y, Tsung A, Huang H, Du Q, Yang M, Deng M, Xiong S, Wang X, Zhang L, Geller DA, Cheng B, Billiar TR. iNOS promotes CD24+CD133+ liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc Natl Acad Sci U S A. 2018;115:E10127-E10136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 125] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 24. | Yang ZF, Ngai P, Ho DW, Yu WC, Ng MN, Lau CK, Li ML, Tam KH, Lam CT, Poon RT, Fan ST. Identification of local and circulating cancer stem cells in human liver cancer. Hepatology. 2008;47:919-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 270] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 25. | Lu JW, Chang JG, Yeh KT, Chen RM, Tsai JJ, Hu RM. Overexpression of Thy1/CD90 in human hepatocellular carcinoma is associated with HBV infection and poor prognosis. Acta Histochem. 2011;113:833-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 26. | Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu PW, Lam CT, Poon RT, Fan ST. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 883] [Cited by in RCA: 927] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 27. | Lingala S, Cui YY, Chen X, Ruebner BH, Qian XF, Zern MA, Wu J. Immunohistochemical staining of cancer stem cell markers in hepatocellular carcinoma. Exp Mol Pathol. 2010;89:27-35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 88] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 28. | Luong AB, Do HQ, Tarchi P, Bonazza D, Bottin C, Cabral LKD, Tran LDC, Doan TPT, Crocè LS, Pham HLT, Tiribelli C, Sukowati CHC. The mRNA Distribution of Cancer Stem Cell Marker CD90/Thy-1 Is Comparable in Hepatocellular Carcinoma of Eastern and Western Populations. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 29. | Xia W, Lo CM, Poon RYC, Cheung TT, Chan ACY, Chen L, Yang S, Tsao GSW, Wang XQ. Smad inhibitor induces CSC differentiation for effective chemosensitization in cyclin D1- and TGF-β/Smad-regulated liver cancer stem cell-like cells. Oncotarget. 2017;8:38811-38824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 30. | Vermeulen L, De Sousa E Melo F, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H, Sprick MR, Kemper K, Richel DJ, Stassi G, Medema JP. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1301] [Cited by in RCA: 1446] [Article Influence: 96.4] [Reference Citation Analysis (1)] |
| 31. | Dang H, Steinway SN, Ding W, Rountree CB. Induction of tumor initiation is dependent on CD44s in c-Met⁺ hepatocellular carcinoma. BMC Cancer. 2015;15:161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 32. | Mima K, Okabe H, Ishimoto T, Hayashi H, Nakagawa S, Kuroki H, Watanabe M, Beppu T, Tamada M, Nagano O, Saya H, Baba H. CD44s regulates the TGF-β-mediated mesenchymal phenotype and is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Res. 2012;72:3414-3423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 168] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 33. | Park NR, Cha JH, Jang JW, Bae SH, Jang B, Kim JH, Hur W, Choi JY, Yoon SK. Synergistic effects of CD44 and TGF-β1 through AKT/GSK-3β/β-catenin signaling during epithelial-mesenchymal transition in liver cancer cells. Biochem Biophys Res Commun. 2016;477:568-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 34. | Xie P, Yan J, Wu M, Li H, Chen Z, Yu M, Zhang B, Chen L, Jin L, Zhou B, Li X, Xiao Y, Xu Y, Long J, Zhang J, Guo L. CD44 potentiates hepatocellular carcinoma migration and extrahepatic metastases via the AKT/ERK signaling CXCR4 axis. Ann Transl Med. 2022;10:689. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 35. | Zhu Z, Hao X, Yan M, Yao M, Ge C, Gu J, Li J. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int J Cancer. 2010;126:2067-2078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 226] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 36. | Tovuu LO, Imura S, Utsunomiya T, Morine Y, Ikemoto T, Arakawa Y, Mori H, Hanaoka J, Kanamoto M, Sugimoto K, Iwahashi S, Saito Y, Yamada S, Asanoma M, Miyake H, Shimada M. Role of CD44 expression in non-tumor tissue on intrahepatic recurrence of hepatocellular carcinoma. Int J Clin Oncol. 2013;18:651-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 37. | Tsai SC, Lin CC, Shih TC, Tseng RJ, Yu MC, Lin YJ, Hsieh SY. The miR-200b-ZEB1 circuit regulates diverse stemness of human hepatocellular carcinoma. Mol Carcinog. 2017;56:2035-2047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 38. | Maimaitiming A, Zhou X, Ma X, Huang Y, Wang Q, Deng R, Ren Y, Chai X, Zhang P. Clinicopathological and Prognostic Value of Plasma CD24 Level in Hepatocellular Carcinoma. J Invest Surg. 2020;33:536-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 39. | Qiu Q, Hernandez JC, Dean AM, Rao PH, Darlington GJ. CD24-positive cells from normal adult mouse liver are hepatocyte progenitor cells. Stem Cells Dev. 2011;20:2177-2188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 40. | Lee TK, Castilho A, Cheung VC, Tang KH, Ma S, Ng IO. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell. 2011;9:50-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 487] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 41. | Wang X, Sun W, Shen W, Xia M, Chen C, Xiang D, Ning B, Cui X, Li H, Li X, Ding J, Wang H. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J Hepatol. 2016;64:1283-1294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 247] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 42. | Yamanaka C, Wada H, Eguchi H, Hatano H, Gotoh K, Noda T, Yamada D, Asaoka T, Kawamoto K, Nagano H, Doki Y, Mori M. Clinical significance of CD13 and epithelial mesenchymal transition (EMT) markers in hepatocellular carcinoma. Jpn J Clin Oncol. 2018;48:52-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 43. | Haraguchi N, Ishii H, Mimori K, Tanaka F, Ohkuma M, Kim HM, Akita H, Takiuchi D, Hatano H, Nagano H, Barnard GF, Doki Y, Mori M. CD13 is a therapeutic target in human liver cancer stem cells. J Clin Invest. 2010;120:3326-3339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 491] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 44. | Sun L, Zhang L, Chen J, Li C, Sun H, Wang J, Xiao H. Activation of Tyrosine Metabolism in CD13+ Cancer Stem Cells Drives Relapse in Hepatocellular Carcinoma. Cancer Res Treat. 2020;52:604-621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 45. | Lee TK, Cheung VC, Lu P, Lau EY, Ma S, Tang KH, Tong M, Lo J, Ng IO. Blockade of CD47-mediated cathepsin S/protease-activated receptor 2 signaling provides a therapeutic target for hepatocellular carcinoma. Hepatology. 2014;60:179-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 172] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 46. | Lo J, Lau EY, Ching RH, Cheng BY, Ma MK, Ng IO, Lee TK. Nuclear factor kappa B-mediated CD47 up-regulation promotes sorafenib resistance and its blockade synergizes the effect of sorafenib in hepatocellular carcinoma in mice. Hepatology. 2015;62:534-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 154] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 47. | Chen J, Zheng DX, Yu XJ, Sun HW, Xu YT, Zhang YJ, Xu J. Macrophages induce CD47 upregulation via IL-6 and correlate with poor survival in hepatocellular carcinoma patients. Oncoimmunology. 2019;8:e1652540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 48. | Yang W, Wang C, Lin Y, Liu Q, Yu LX, Tang L, Yan HX, Fu J, Chen Y, Zhang HL, Zheng LY, He YQ, Li YQ, Wu FQ, Zou SS, Li Z, Wu MC, Feng GS, Wang HY. OV6⁺ tumor-initiating cells contribute to tumor progression and invasion in human hepatocellular carcinoma. J Hepatol. 2012;57:613-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 49. | Yang W, Yan HX, Chen L, Liu Q, He YQ, Yu LX, Zhang SH, Huang DD, Tang L, Kong XN, Chen C, Liu SQ, Wu MC, Wang HY. Wnt/beta-catenin signaling contributes to activation of normal and tumorigenic liver progenitor cells. Cancer Res. 2008;68:4287-4295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 296] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 50. | Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, Tang ZY, Wang XW. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451-1461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 545] [Cited by in RCA: 593] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 51. | Matsumoto T, Takai A, Eso Y, Kinoshita K, Manabe T, Seno H, Chiba T, Marusawa H. Proliferating EpCAM-Positive Ductal Cells in the Inflamed Liver Give Rise to Hepatocellular Carcinoma. Cancer Res. 2017;77:6131-6143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 52. | Zeng SS, Yamashita T, Kondo M, Nio K, Hayashi T, Hara Y, Nomura Y, Yoshida M, Oishi N, Ikeda H, Honda M, Kaneko S. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. J Hepatol. 2014;60:127-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 53. | Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, Reid LM, Minato H, Honda M, Kaneko S, Tang ZY, Wang XW. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012-1024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 936] [Cited by in RCA: 958] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 54. | Yamashita T, Budhu A, Forgues M, Wang XW. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007;67:10831-10839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 360] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 55. | Zhou L, Yang ZX, Song WJ, Li QJ, Yang F, Wang DS, Zhang N, Dou KF. MicroRNA-21 regulates the migration and invasion of a stem-like population in hepatocellular carcinoma. Int J Oncol. 2013;43:661-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 56. | Wang J, Chu Y, Xu M, Zhang X, Zhou Y. miR-21 promotes cell migration and invasion of hepatocellular carcinoma by targeting KLF5. Oncol Lett. 2019;17:2221-2227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 57. | Koenig AB, Barajas JM, Guerrero MJ, Ghoshal K. A Comprehensive Analysis of Argonaute-CLIP Data Identifies Novel, Conserved and Species-Specific Targets of miR-21 in Human Liver and Hepatocellular Carcinoma. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 58. | Cao LQ, Yang XW, Chen YB, Zhang DW, Jiang XF, Xue P. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol Cancer. 2019;18:148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 139] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 59. | Liao Y, Wang C, Yang Z, Liu W, Yuan Y, Li K, Zhang Y, Wang Y, Shi Y, Qiu Y, Zuo D, He W, Qiu J, Guan X, Li B. Dysregulated Sp1/miR-130b-3p/HOXA5 axis contributes to tumor angiogenesis and progression of hepatocellular carcinoma. Theranostics. 2020;10:5209-5224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 60. | von Felden J, Heim D, Schulze K, Krech T, Ewald F, Nashan B, Lohse AW, Wege H. High expression of micro RNA-135A in hepatocellular carcinoma is associated with recurrence within 12 months after resection. BMC Cancer. 2017;17:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 61. | Liu F, Kong X, Lv L, Gao J. TGF-β1 acts through miR-155 to down-regulate TP53INP1 in promoting epithelial-mesenchymal transition and cancer stem cell phenotypes. Cancer Lett. 2015;359:288-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 80] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 62. | Ji J, Zheng X, Forgues M, Yamashita T, Wauthier EL, Reid LM, Wen X, Song Y, Wei JS, Khan J, Thorgeirsson SS, Wang XW. Identification of microRNAs specific for epithelial cell adhesion molecule-positive tumor cells in hepatocellular carcinoma. Hepatology. 2015;62:829-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 63. | Cao MQ, You AB, Zhu XD, Zhang W, Zhang YY, Zhang SZ, Zhang KW, Cai H, Shi WK, Li XL, Li KS, Gao DM, Ma DN, Ye BG, Wang CH, Qin CD, Sun HC, Zhang T, Tang ZY. miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J Hematol Oncol. 2018;11:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 157] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 64. | Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 650] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 65. | Sharma AD, Narain N, Händel EM, Iken M, Singhal N, Cathomen T, Manns MP, Schöler HR, Ott M, Cantz T. MicroRNA-221 regulates FAS-induced fulminant liver failure. Hepatology. 2011;53:1651-1661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 60] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 66. | Tang J, Li Y, Liu K, Zhu Q, Yang WH, Xiong LK, Guo DL. Exosomal miR-9-3p suppresses HBGF-5 expression and is a functional biomarker in hepatocellular carcinoma. Minerva Med. 2018;109:15-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 67. | Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang C, Wang F, Zhang CY, Zen K, Li L. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017;8:e2540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 176] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 68. | Jin Q, Li XJ, Cao PG. miR-26b enhances radiosensitivity of hepatocellular carcinoma cells by targeting EphA2. Iran J Basic Med Sci. 2016;19:851-857. [PubMed] |
| 69. | Zhao N, Wang R, Zhou L, Zhu Y, Gong J, Zhuang SM. MicroRNA-26b suppresses the NF-κB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3. Mol Cancer. 2014;13:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 139] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 70. | Fu XT, Shi YH, Zhou J, Peng YF, Liu WR, Shi GM, Gao Q, Wang XY, Song K, Fan J, Ding ZB. MicroRNA-30a suppresses autophagy-mediated anoikis resistance and metastasis in hepatocellular carcinoma. Cancer Lett. 2018;412:108-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 71. | Zhou K, Luo X, Wang Y, Cao D, Sun G. MicroRNA-30a suppresses tumor progression by blocking Ras/Raf/MEK/ERK signaling pathway in hepatocellular carcinoma. Biomed Pharmacother. 2017;93:1025-1032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 72. | Li Y, Fu L, Fu XY, Li RH, Peng SF. [MicroRNA-30a inhibits proliferation of hepatocellular carcinoma cells via targeted regulation of forkhead-box protein A1]. Zhonghua Ganzangbing Zazhi. 2017;25:706-711. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 73. | Liu Z, Tu K, Liu Q. Effects of microRNA-30a on migration, invasion and prognosis of hepatocellular carcinoma. FEBS Lett. 2014;588:3089-3097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 74. | Xu Y, Huang J, Ma L, Shan J, Shen J, Yang Z, Liu L, Luo Y, Yao C, Qian C. MicroRNA-122 confers sorafenib resistance to hepatocellular carcinoma cells by targeting IGF-1R to regulate RAS/RAF/ERK signaling pathways. Cancer Lett. 2016;371:171-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 137] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 75. | Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526-3536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 585] [Cited by in RCA: 590] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 76. | Song K, Kwon H, Han C, Zhang J, Dash S, Lim K, Wu T. Active glycolytic metabolism in CD133(+) hepatocellular cancer stem cells: regulation by MIR-122. Oncotarget. 2015;6:40822-40835. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 94] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 77. | Jopling C. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012;9:137-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 322] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 78. | Jin Y, Wang J, Han J, Luo D, Sun Z. MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing WNT/β-cadherin signaling pathway. Exp Cell Res. 2017;360:210-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 79. | Ren WW, Li DD, Chen X, Li XL, He YP, Guo LH, Liu LN, Sun LP, Zhang XP. MicroRNA-125b reverses oxaliplatin resistance in hepatocellular carcinoma by negatively regulating EVA1A mediated autophagy. Cell Death Dis. 2018;9:547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 80. | Gong J, Zhang JP, Li B, Zeng C, You K, Chen MX, Yuan Y, Zhuang SM. MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene. 2013;32:3071-3079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 163] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 81. | Wei X, Zhao L, Ren R, Ji F, Xue S, Zhang J, Liu Z, Ma Z, Wang XW, Wong L, Liu N, Shi J, Guo X, Roessler S, Zheng X, Ji J. MiR-125b Loss Activated HIF1α/pAKT Loop, Leading to Transarterial Chemoembolization Resistance in Hepatocellular Carcinoma. Hepatology. 2021;73:1381-1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 82. | Zhou JN, Zeng Q, Wang HY, Zhang B, Li ST, Nan X, Cao N, Fu CJ, Yan XL, Jia YL, Wang JX, Zhao AH, Li ZW, Li YH, Xie XY, Zhang XM, Dong Y, Xu YC, He LJ, Yue W, Pei XT. MicroRNA-125b attenuates epithelial-mesenchymal transitions and targets stem-like liver cancer cells through small mothers against decapentaplegic 2 and 4. Hepatology. 2015;62:801-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 129] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 83. | Yu Q, Xiang L, Yin L, Liu X, Yang D, Zhou J. Loss-of-function of miR-142 by hypermethylation promotes TGF-β-mediated tumour growth and metastasis in hepatocellular carcinoma. Cell Prolif. 2017;50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 84. | Su F, Zhao J, Qin S, Wang R, Li Y, Wang Q, Tan Y, Jin H, Zhu F, Ou Y, Cheng Z, Su W, Zhao F, Yang Y, Zhou Z, Zheng J, Li Z, Wu Q. Over-expression of Thrombospondin 4 correlates with loss of miR-142 and contributes to migration and vascular invasion of advanced hepatocellular carcinoma. Oncotarget. 2017;8:23277-23288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 85. | Chai S, Tong M, Ng KY, Kwan PS, Chan YP, Fung TM, Lee TK, Wong N, Xie D, Yuan YF, Guan XY, Ma S. Regulatory role of miR-142-3p on the functional hepatic cancer stem cell marker CD133. Oncotarget. 2014;5:5725-5735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 61] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 86. | Zhou SJ, Liu FY, Zhang AH, Liang HF, Wang Y, Ma R, Jiang YH, Sun NF. MicroRNA-199b-5p attenuates TGF-β1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Br J Cancer. 2017;117:233-244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 87. | Shen Q, Cicinnati VR, Zhang X, Iacob S, Weber F, Sotiropoulos GC, Radtke A, Lu M, Paul A, Gerken G, Beckebaum S. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Mol Cancer. 2010;9:227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 158] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 88. | Wang J, Song W, Shen W, Yang X, Sun W, Qu S, Shang R, Ma B, Pu M, Tao K, Dou K, Li H. MicroRNA-200a Suppresses Cell Invasion and Migration by Directly Targeting GAB1 in Hepatocellular Carcinoma. Oncol Res. 2017;25:1-10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 89. | Chen SY, Ma DN, Chen QD, Zhang JJ, Tian YR, Wang ZC, Cai H, Lin Y, Sun HC. MicroRNA-200a inhibits cell growth and metastasis by targeting Foxa2 in hepatocellular carcinoma. J Cancer. 2017;8:617-625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 90. | Chen H, Kong M, Chen Y, Jiang Y, Wen M, Zhang X. Prognostic significance of miR-203 and ZEB1 expression in early-stage hepatocellular carcinoma. J Cancer. 2021;12:4810-4818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 91. | Zheng XB, Chen XB, Xu LL, Zhang M, Feng L, Yi PS, Tang JW, Xu MQ. miR-203 inhibits augmented proliferation and metastasis of hepatocellular carcinoma residual in the promoted regenerating liver. Cancer Sci. 2017;108:338-346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 92. | Chen SP, Liu BX, Xu J, Pei XF, Liao YJ, Yuan F, Zheng F. MiR-449a suppresses the epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by multiple targets. BMC Cancer. 2015;15:706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 93. | Liu Y, Wang Y, Sun X, Mei C, Wang L, Li Z, Zha X. miR-449a promotes liver cancer cell apoptosis by downregulation of Calpain 6 and POU2F1. Oncotarget. 2016;7:13491-13501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 94. | Han B, Huang J, Yang Z, Zhang J, Wang X, Xu N, Meng H, Wu J, Huang Q, Yang X, Shen R, Sun C. miR-449a Is Related to Short-Term Recurrence of Hepatocellular Carcinoma and Inhibits Migration and Invasion by Targeting Notch1. Onco Targets Ther. 2019;12:10975-10987. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 95. | Xu WP, Liu JP, Feng JF, Zhu CP, Yang Y, Zhou WP, Ding J, Huang CK, Cui YL, Ding CH, Zhang X, Lu B, Xie WF. miR-541 potentiates the response of human hepatocellular carcinoma to sorafenib treatment by inhibiting autophagy. Gut. 2020;69:1309-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 87] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 96. | Jiang ZB, Ma BQ, Liu SG, Li J, Yang GM, Hou YB, Si RH, Gao P, Yan HT. miR-365 regulates liver cancer stem cells via RAC1 pathway. Mol Carcinog. 2019;58:55-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 97. | Xiao Y, Sun Y, Liu G, Zhao J, Gao Y, Yeh S, Gong L, Chang C. Androgen receptor (AR)/miR-520f-3p/SOX9 signaling is involved in altering hepatocellular carcinoma (HCC) cell sensitivity to the Sorafenib therapy under hypoxia via increasing cancer stem cells phenotype. Cancer Lett. 2019;444:175-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 98. | Han S, Shi Y, Sun L, Liu Z, Song T, Liu Q. MiR-4319 induced an inhibition of epithelial-mesenchymal transition and prevented cancer stemness of HCC through targeting FOXQ1. Int J Biol Sci. 2019;15:2936-2947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 99. | Liu C, Li J, Wang W, Zhong X, Xu F, Lu J. miR-206 inhibits liver cancer stem cell expansion by regulating EGFR expression. Cell Cycle. 2020;19:1077-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 100. | Li B, Liu D, Yang P, Li HY, Wang D. miR-613 inhibits liver cancer stem cell expansion by regulating SOX9 pathway. Gene. 2019;707:78-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 101. | Hong DS, Kang YK, Borad M, Sachdev J, Ejadi S, Lim HY, Brenner AJ, Park K, Lee JL, Kim TY, Shin S, Becerra CR, Falchook G, Stoudemire J, Martin D, Kelnar K, Peltier H, Bonato V, Bader AG, Smith S, Kim S, O'Neill V, Beg MS. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer. 2020;122:1630-1637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 409] [Cited by in RCA: 591] [Article Influence: 118.2] [Reference Citation Analysis (0)] |
| 102. | Nasr MA, Salah RA, Abd Elkodous M, Elshenawy SE, El-Badri N. Dysregulated MicroRNA Fingerprints and Methylation Patterns in Hepatocellular Carcinoma, Cancer Stem Cells, and Mesenchymal Stem Cells. Front Cell Dev Biol. 2019;7:229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 103. | Jayachandran A, Dhungel B, Steel JC. Epithelial-to-mesenchymal plasticity of cancer stem cells: therapeutic targets in hepatocellular carcinoma. J Hematol Oncol. 2016;9:74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 158] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 104. | Loosen SH, Schueller F, Trautwein C, Roy S, Roderburg C. Role of circulating microRNAs in liver diseases. World J Hepatol. 2017;9:586-594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 105. | El-Mahdy HA, Sallam AM, Ismail A, Elkhawaga SY, Elrebehy MA, Doghish AS. miRNAs inspirations in hepatocellular carcinoma: Detrimental and favorable aspects of key performers. Pathol Res Pract. 2022;233:153886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 57] [Article Influence: 19.0] [Reference Citation Analysis (0)] |