Published online Jun 27, 2021. doi: 10.4254/wjh.v13.i6.662
Peer-review started: February 24, 2021
First decision: May 3, 2021
Revised: May 13, 2021
Accepted: June 4, 2021
Article in press: June 4, 2021
Published online: June 27, 2021
Processing time: 118 Days and 8.3 Hours
Chromosome 1q often has been observed to be amplified in hepatocellular carcinoma. This review summarizes literature reports of multiple genes that have been proposed as possible 1q amplification drivers. These largely fall within 1q21-1q23. In addition, publicly available copy number alteration data from The Cancer Genome Atlas project were used to identify additional candidate genes involved in carcinogenesis. The most frequent location for gene amplification was 1q22, consistent with the results of the literature search. The genes TPM3 and NUF2 were found to be candidates whose amplification and/or mRNA up-regulation was most highly associated with poorer hepatocellular carcinoma outcomes.
Core Tip: A list of candidate chromosome 1q amplification driver genes was compiled from the existing literature by PubMed search. Bioinformatics tools were used to identify additional candidates using publicly available genomics and transcriptomics data. Genes identified this way were largely distinct from those identified from the literature. Thus, these two strategies can be used in a complementary manner.
- Citation: Jacobs NR, Norton PA. Role of chromosome 1q copy number variation in hepatocellular carcinoma. World J Hepatol 2021; 13(6): 662-672
- URL: https://www.wjgnet.com/1948-5182/full/v13/i6/662.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i6.662
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer related deaths worldwide. Liver cancers are the fourth most common cause of cancer related deaths (the sixth most commonly diagnosed type of cancer), and HCC accounts for between 75% and 85% of primary liver cancer cases[1]. About 54% of HCC cases worldwide are attributed to the hepatitis B virus (HBV) while 31% of cases are attributed to hepatitis C virus (HCV) infections[2]. Given the fact that chronic HBV infection presents as a significant risk factor for HCC, vaccination against HBV is recommended as a way to prevent HCC[3].
More recently, technological advances have permitted the sequencing of the genomes and transcriptomes of numerous cancers. Mutations in several genes have been detected repeatedly in HCC[4]. Common somatic changes include mutations to beta-catenin and p53, resulting in activation of the Wnt signaling pathway and dysregulation of the cell cycle, respectively. Mutations activating TERT gene expression are also common. Patterns of genetic alterations in individual tumors have been examined with the goal of classifying them, to predict outcome and potentially guide therapeutic decisions[5].
Over the past few decades, a significant amount of research has shown an association between HCC and specific chromosomal abnormalities. In particular, chromosomal gains have been noted for 1q, 6p, 8q, 17q, and 20q. Similarly, chromosomal losses have been detected for 1p, 4q, 6q, 8p, 13q 16p, and 17q[6-8]. Amplification of chromosome 1q21-23 has been identified as the most frequent chromosomal alteration associated with HCC[9]. Thus, we were interested in considering the evidence for which gene or genes is critical for driving this chromosomal abnormality.
During the past two decades, several genes within or near the 1q21-23 range have been highlighted as potentially significant to HCC[10]. Many of these are highlighted in Table 1. In 2003, Wong et al[11] studied the 1q21-1q22 region using positional mapping by interphase cytogenetics. They identified significantly increased levels of gene expression of the JTB, SHC1, CCT3, and COPA genes in five cases of HCC compared to paired adjacent non-malignant liver tissues, and they concluded that these genes may represent targets in HCC progression[11]. More recently, JTB (Jumping Translocation Breakpoint) has been identified as a protein that negatively regulates the apoptotic process by affecting the activation of caspase 9[12]. SHC1 is involved in signal transduction from receptor tyrosine kinases to various downstream proteins and has been identified in mitogenic signaling[13-15]. CCT3 is involved in cell cycle regulation[16]. COPA is the α-subunit of the coatomer protein complex I which plays a role in retrograde protein trafficking from the Golgi to the endoplasmic reticulum[17].
| Gene1 | Location2 | Description of protein product | Ref. |
| JTB | 1q21.3 | Promotes cell resistance to apoptosis | Wong et al[11] and Kanome et al[12] |
| SHC1 | 1q21.3 | Downstream signaling from receptor | Wong et al[11], Midorikawa et al[8], Pelicci et al[13], |
| CCT3 | 1q22 | Associated with cell cycle regulation | Wong et al[11], Midorikawa et al[8], Won et al[16] |
| COPA | 1q23.2 | Assists in retrograde vesicular transport | Wong et al[11] and Vece et al[17] |
| CKS1B | 1q21.2 | Associated with cell cycle regulation | Midorikawa et al[8] and Ganoth et al[19] |
| HAX-1 (HAX1) | 1q21.3 | Plays a role in the activation of receptor | Midorikawa et al[8] and Suzuki et al[18] |
| CREB3L4 | 1q21.3 | Associated with androgen receptor signaling | Inagaki et al[21] and Qi et al[22] |
| INTS3 | 1q21.3 | Associated with RNA polymerase II | Inagaki et al[21] and Baillat et al[24] |
| SNAPAP | 1q21.3 | Part of SNARE complex (docking and | Inagaki et al[21] and Ilardi et al[25] |
| ALC1 (CHD1L) | 1q21.1 | Facilitates DNA synthesis and cell cycle | Ma et al[26] |
| ASH1L | 1q22 | Histone methyltransferase involved in | Elsemman et al[27] and An et al[29] |
| METTL13 (EEF1AKNMT) | 1q24.3 | Regulates protein synthesis in cancer cells; | Elsemman et al[27]; Liu et al[30], and Li et al[31] |
| TARBP1 | 1q42.2 | Double-stranded RNA binding protein; | Elsemman et al[27], Zhang et al[50], and Christensen et al[32] |
| SMYD2 | 1q32.2 | Part of the protein lysine methyltransferase | Elsemman et al[27] and Leinhart and Brown[34] |
| SMYD3 | 1q44 | Part of the protein lysine methyltransferase | Elsemman et al[27] and Leinhart and Brown[34] |
In 2004, Midorikawa et al[8] used an expression imbalance map analysis [which they confirmed using genomic quantitative real-time polymerase chain reaction (qPCR)] to demonstrate amplification of the 1q21-12 region in HCC tumor samples. Moreover, they identified two new genes (HAX-1 and CKS1B) as being as being highly expressed in HCC tissue compared with noncancerous tissues. They also described the amplification of SHC1 and CCT3 (previously identified by Wong et al[11]). HAX-1 (HCLS1 associated protein X-1, gene name HAX1) has been associated with activation of tyrosine kinases[18]. Like CCT3, CKS1B (CDC28 protein kinase regulatory subunit) plays an essential role in mediating a cell’s progression through the cell cycle[19]. To further support the conclusions of Midorikawa et al[8], Shen et al[20] demonstrated that HCC cells had increased levels of CKS1B mRNA and protein compared to adjacent non-tumor liver tissue. Elevated CKS1B expression was also positively associated with poor differentiation features[20].
In 2008, Inagaki et al[21] analyzed a 700-kb DNA region located at 1q21 in 19 HCC-derived cell lines. Using high-density SNP microarray analysis, fluorescence in situ hybridization (FISH), and real-time quantitative PCR, they identified a significant increase in copy number at the 1q21 region. Using reverse transcriptase PCR, they identified three genes (CREB3L4, INTS3, and SNAPAP) that were significantly overexpressed in samples taken from HCC tumors[21]. Based on these findings, they concluded that these three genes are likely targets for the amplification mechanism, and they may be involved in HCC progression. CREB3L4 (cyclic amplification responsive element binding protein 3-like 4) is part of the CREB/ATF family of transcriptional factors, and it is primarily expressed in the prostate gland in humans as well as prostate and breast cancer cell lines[22]. CREB3L4 has been shown (by immunostaining) to have a higher expression level in cancerous prostate cells than in adjacent noncancerous cells[22] and it has also been shown to contribute to the progression of breast cancer[23]. INTS3 (integrator complex subunit 3) is part of the Integrator complex which is associated with the C-terminal domain of RNA polymerase II[24]. SNAPAP (snare-associated protein, gene name SNAPIN) is part of the SNARE complex of proteins that is involved in the docking and fusion of synaptic vessel[25]. At this point, little is known about the relationship of either INTS3 or SNAPAP with tumorigenesis.
Later in 2008, Ma et al[26] used microdissected DNA from 1q21 and hybrid selection to isolate ALC1 (also known as CHD1L) as a candidate oncogene. After confirming the amplification of ALC1 using FISH, they transfected it into human liver cell lines resulting in the cells being able to form more colonies than vector-transfected cells when grown in soft agar[26]. They also demonstrated that ALC1 overexpression plays a role in facilitating DNA synthesis, down-regulating p53 expression, promoting G1/S phase transition, and inhibiting apoptosis.
More recently, in 2016 Elsemman et al[27] were interested in S-adenosylmethionine (SAMe) which has been described by Lu et al[28] as playing a significant role in hepatic diseases including HCC. SAMe is synthesized from ATP and methionine by methionine adenosyl transferase genes including MAT1A which is significantly downregulated in HCC. Elsemman et al[27] analyzed reactions containing SAMe, and using copy number variation analysis they identified five methyltransferase genes (ASH1L, METTL13, TARBP1, SMYD2, and SMYD3) located on chromosome 1q, all of which were amplified in samples of HCC relative the healthy tissue samples. ASH1L is a histone methyltransferase protein which is involved in the regulation of gene expression[29]. METTL13 (gene name EEF1AKNMT) has been shown repeatedly to promote tumor growth and metastasis and is negatively associated with survival among lung and pancreatic cancer patients[30,31]. TARBP1 is a double-stranded RNA binding protein that promotes the replication of human immunodeficiency virus-1 and -2 as well as HCV[32]. It has also been directly correlated with decreased survival rates in patients with HCC[33]. SMYD2 and SMYD3 are both members of the protein lysine methyltransferase family of proteins[34], and each has been associated with a variety of cancer types. SMYD2 has been shown to be overexpressed in esophageal squamous carcinoma, gastric cancer, and pediatric acute lymphoblastic leukemia[35-37]. SMYD3 is overexpressed in cancers including breast, liver, and colorectal cancer[38,39].
We were interested in what more recent genomic and transcriptomic studies have revealed about chromosome 1q amplification and HCC. The Cancer Genome Atlas (TCGA) Project has accumulated an important, publicly available genomic and mRNA expression data set which includes multiple cancers types including HCC (data set Liver Hepatocellular Carcinoma, LIHC)[40]. There is also a more recent version of this data, which is part of TCGA Pan-Cancer Clinical Data Resource[41], a subset of the LIHC data set that has been curated to include four major clinical outcome endpoints. We chose to use this data set to try to identify additional candidate amplification driver genes. This version of the LIHC patient cohort (PanCan-LIHC) has the following patient characteristics: 251 males/121 female with 241 living, and 131 deceased. Most individuals had a total of 10-140 mutations genome wide; 23 had 140-190, 18 had greater than 190, and 2 had fewer than 10 (14 did not have data available). Most PanCan-LIHC individuals exhibited genome alterations, with gains in 1q being the most common alteration: 225 individuals (60.5%) exhibited 1q gains, with 23.7% called as diploid and 15.9% with data not available).
The original publication reporting the LIHC cohort analyses identified copy number alterations (CNAs) in several likely driver genes spread across several chromosomes[40]. However, the only driver gene listed for 1q is MCL1 at 1q21.3. They also reported a short stretch of four genes that were significantly amplified at 1q22, but no candidate genes were indicated. In a report on the analysis of aneuploidy across TCGA cancer types, strong 1q amplification was noted in the PanCan-LIHC cohort (as well as in other epithelial breast and lung tumors)[42]. Using the Oncoprint tool at the cBioPortal for Cancer Genomics (https://www.cbioportal.org/), we could see that all of the genes listed in Table 1 were amplified in 7%-13% of tumors, with mRNAs overexpressed in 9%-41% of tumors (data not shown), consistent with the earlier reports described above.
To further explore possible 1q amplification driver candidates, the frequency of CNAs in the Pan-Cancer version of LIHC sample set was explored using the cBioPortal suite of tools[43,44]. First, the CNA data set for all genes in the PanCan-LIHC was downloaded and imported into in an Excel spreadsheet. Second, all genes that had been scored as having an amplification or homozygous deletion with a frequency of at least 5% of tumor samples were sorted from those with lower frequency. This resulted in a list of 1871 genes meeting these criteria. Finally, this set of 1871 altered genes was sorted by chromosome and further restricted to those that were annotated as Cancer genes according OncoKB[45].
These steps produced a list of 49 candidate genes localized to chromosome 1q (not shown). These fell into two groups, a centromere proximal group spanning intervals 1q21.2-1q25.2 (28 genes), and a second group covering the distal interval of 1q31.1-1q44 (21 genes). Across the 1q region, the gene amplified in the highest percentage of tumors was MUC1 located at 1q22 (11.7% amplification). This might correspond to the short stretch identified at 1q22 by the TCGA-LIHC paper referred to above. The overall frequency of amplification was greater in the proximal group of genes (mean of 10.29%, range of 8.2%-11.7%) vs the distal set (mean of 6.41%, range of 5.4%-7.4%). Of the 15 genes listed in Table 1, only two were present in the list of 49, CKS1B and SMYD3.
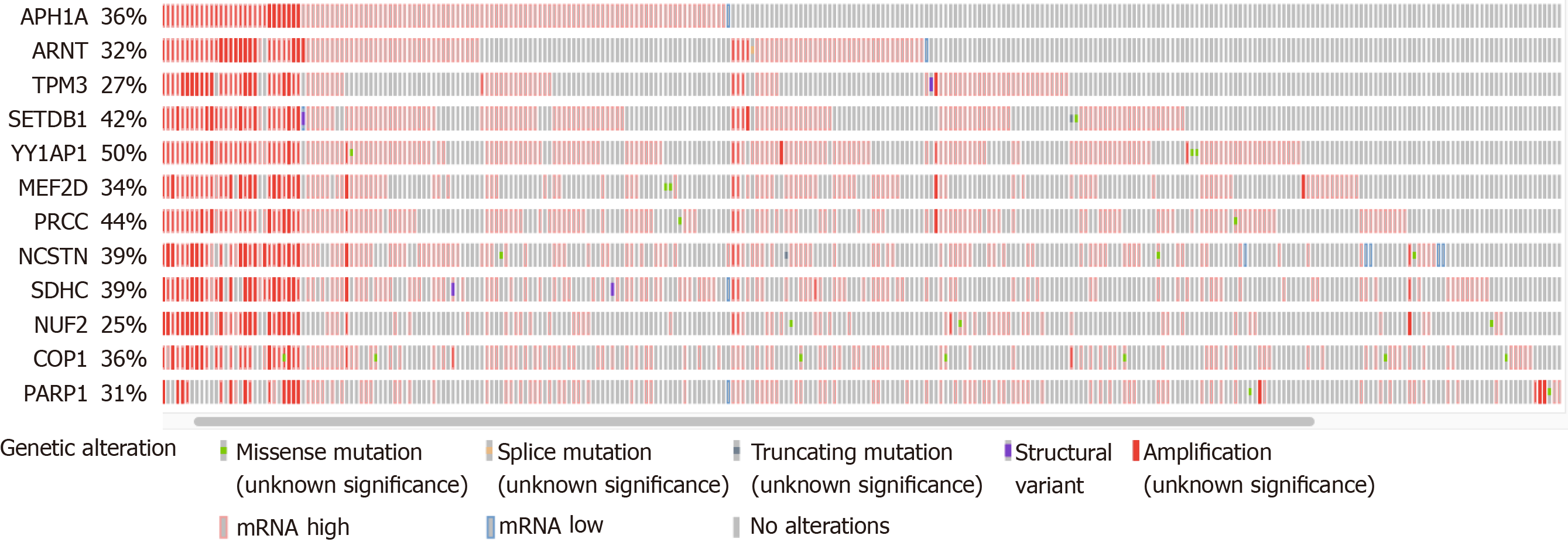
Using the Oncoprint visualization tool at cBioPortal, all 49 genes were examined to determine the putative CNAs from GISTIC2.0 calls[46], as well as the presence of non-synonymous mutations and altered mRNA expression (z-score threshold of +/- 2.0 relative to diploid samples). The total alteration percentages ranged from < 10% to 50% for the individual genes, with few non-synonymous mutations (not shown). The total number of genes under consideration was narrowed down to 12 by focusing on those with at least 25% of samples with one or more of the various alterations (Figure 1). All but one of these genes was derived from the centromere proximal half of the 1q arm (the exception was PARP1 at 1q42.12). All 12 genes exhibited numerous instances of mRNA upregulation, both with and without DNA amplification. Note that COP1 in Figure 1 at 1q25.1 is not the same as COPA at 1q23.3 (Table 1).
Each of the 12 genes was examined individually using the cBioPortal Comparison and Survival tools to determine whether the presence of alterations was associated with survival outcomes. There were only two genes where amplification, or mRNA increase, or both were associated with reduced survival compared with the samples without either type of alteration. These two were TPM3 at 1q21.3 and NUF2 at 1q23.3 (Table 2, scores designated “all”). However, when the CNAs were examined separately from increased mRNA levels, amplification alone was not associated with any survival or outcome measure (not shown). Instead, the mRNA elevations clearly had a more significant correlation with patient outcome, as can be seen from the Logrank test q-values (Table 2, “mRNA”). Patients with TPM3 mRNA elevation had an overall median survival of 25.15 mo vs 80.74 mo for those without the elevation. Patients with NUF2 mRNA elevation had an overall median survival of 23.38 mo vs 70.06 mo for the unaltered group. Thus, altered expression of these two genes may contribute to clinical outcome.
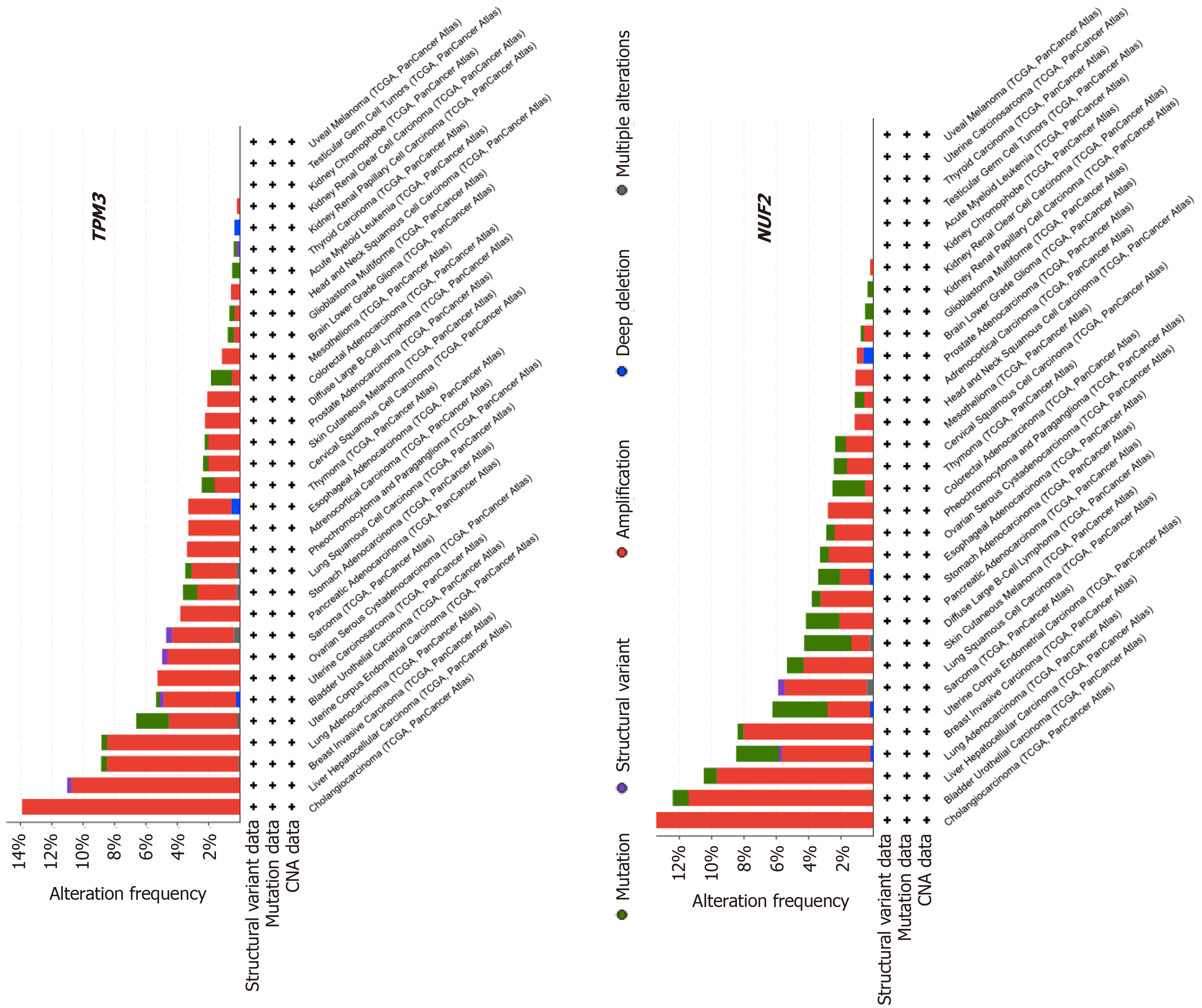
We were interested whether TPM3 and NUF2 alterations were common in other types of cancer besides HCC. To explore the alteration frequencies in other cancer types, the entire Pan-Cancer patient cohort was analyzed using the cBioPortal suite of tools[41]. All 32 cancer types included in the Pan-Cancer sample set were selected, and the TPM3 and NUF2 genes were searched individually. The Cancer Types Summary produced a display showing the frequency of gene alterations (amplifications, deep deletions, non-synonymous mutations, structural variants) in all 32 types of cancer as well as the types of alterations identified (Figure 2). The PanCan-LIHC HCC dataset had the second highest percentage of TPM3 alterations and the third highest percentage of NUF2 alterations. In the case of both genes, amplification of TPM3 and NUF2 was the most common type of alteration seen in the HCC patient sample. Interestingly, NUF2 had a relatively higher frequency of mutations than amplifications in some cancer types.
Despite the low q-values, it remains possible that the association between TPM3 and NUF2 gene expression and patient survival is random. Therefore, we searched the literature to find whether either TPM3 or NUF2 genes had been associated previously with HCC. Kim et al[47] examined chromosomal alterations in 76 HCC, finding frequent gain of 1q. They found TPM3 mRNA was elevated in tumors compared to normal tissue, and proposed that it might represent an oncogene in HCC, consistent with our analysis. A follow up study found that knock down of TPM3 in HCC cells reduced migration and invasion capabilities[48].
NUF2 elevation was reported in micro-dissected malignant hepatocytes derived from HBV-associated tumors[49]. Analysis of the Gene Expression Omnibus HCC data also revealed upregulation of NUF2 in HCC compared with healthy colon epithelial cells[50]. An analysis of the original TCGA-LIHC data set, which has substantial overlap with the PanCan-LIHC samples that we explored, also found that NUF2 was overexpressed compared with normal liver samples[51], and that overexpression was significantly associated with overall median survival. Other independent analyses of the same data set also reported NUF2 upregulation and association with poorer prognosis[52-54]. It has been suggested that NUF2 may represent a biomarker for early recurrence after HCC resection[55], and that it might represent a potential therapeutic target[56].
The product of the TPM3 gene is tropomyosin3, an actin binding protein. The four TPM genes TPM genes produce 40 distinct protein isoforms by use of alternative promoters and extensive alternative mRNA splicing[57]. Changes in isoform production have been associated with cellular transformation[48,58]. The specific role of increased TPM3 in cancer cells is unclear, as the protein is involved in numerous activities related to the actin cytoskeleton. Despite this, it is worth noting that small molecules that block the binding of isoform TPM3.1 to actin showed promise in perturbing the growth of cancer cells[59,60].
The protein encoded by the NUF2 gene, along with those encoded NDC80, SPC24 and SPC25 form the Nuclear Division Cycle 80 complex. This complex plays an important role in mitotic spindle formation and chromosome segregation[61]. Over expression of other complex members, especially NDC80, has also been observed frequently in multiple cancers, and it has been proposed that overexpression of NDC80 complex proteins leads to defective mitosis and may promote aneuploidy[62]. Screening in epithelial ovarian carcinoma cells of an siRNA library has identified NUF2 as one of four genes that reduced cell viability and increased apoptosis when knocked down[63]. This study also found a correlation between NUF2 mRNA elevation and poorer prognosis in ovarian carcinoma patients. NDC80 (also known as Hec1) interacts directly with NUF2 and may represent a therapeutic target. A screen of a small molecule library for inhibitors of the interaction between NDC80 and mitotic kinase Nek2 identified a compound named INH1 as being able to disrupt the protein-protein interaction[64]. This study also showed that INH1 decreased proliferation of breast cancer cells in culture and in a mouse xenograft assay.
In conclusion, our review of the literature and independent analysis of the TCGA-LIHC PanCancer data set identified two non-overlapping sets of genes that reside on chromosome 1q and frequently undergo amplification in HCC (compare Figure 1 and Table 1). We found what appears to be a significant correlation between amplification and/or increased expression of TPM3 and NUF2 and poorer prognosis, which is consistent with previous reports in the literature. Amplification of 1q also is observed frequently in other cancers. One limitation to our strategy to identify additional driver genes is that only genes previously identified as involved in cancer by OncoKB were considered. The absence of many genes in Table 1 suggests more candidate genes may still be identified. In the case of large chromosomal CNAs such as seen with 1q, it is truly challenging to identify the critical driver mutations involved. Further studies will be needed to understand the contributions of numerous genes amplified on chromosome 1q so as to effectively target therapeutics.
We thank Danielle Gerken for some of the initial research that led to this project. We are also grateful for the tools provided by the cBioPortal for Cancer Genomics (https://www.cbioportal.org/) for the analysis of the TCGA data.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gomez-Quiroz L S-Editor: Gao CC L-Editor: A P-Editor: Wang LL
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55842] [Article Influence: 7977.4] [Reference Citation Analysis (132)] |
| 2. | European Association For The Study Of The Liver; European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4059] [Cited by in RCA: 4521] [Article Influence: 347.8] [Reference Citation Analysis (2)] |
| 3. | Mak LY, Cruz-Ramón V, Chinchilla-López P, Torres HA, LoConte NK, Rice JP, Foxhall LE, Sturgis EM, Merrill JK, Bailey HH, Méndez-Sánchez N, Yuen MF, Hwang JP. Global Epidemiology, Prevention, and Management of Hepatocellular Carcinoma. Am Soc Clin Oncol Educ Book. 2018;38:262-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 157] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 4. | Schulze K, Nault JC, Villanueva A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J Hepatol. 2016;65:1031-1042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 225] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 5. | Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, Watanabe G, Gabriel S, Friedman SL, Kumada H, Llovet JM, Golub TR. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385-7392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 956] [Cited by in RCA: 944] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 6. | Marchio A, Meddeb M, Pineau P, Danglot G, Tiollais P, Bernheim A, Dejean A. Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer. 1997;18:59-65. [PubMed] |
| 7. | Sakakura C, Hagiwara A, Taniguchi H, Yamaguchi T, Yamagishi H, Takahashi T, Koyama K, Nakamura Y, Abe T, Inazawa J. Chromosomal aberrations in human hepatocellular carcinomas associated with hepatitis C virus infection detected by comparative genomic hybridization. Br J Cancer. 1999;80:2034-2039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 58] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 8. | Midorikawa Y, Tsutsumi S, Nishimura K, Kamimura N, Kano M, Sakamoto H, Makuuchi M, Aburatani H. Distinct chromosomal bias of gene expression signatures in the progression of hepatocellular carcinoma. Cancer Res. 2004;64:7263-7270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 9. | Steinemann D, Skawran B, Becker T, Tauscher M, Weigmann A, Wingen L, Tauscher S, Hinrichsen T, Hertz S, Flemming P, Flik J, Wiese B, Kreipe H, Lichter P, Schlegelberger B, Wilkens L. Assessment of differentiation and progression of hepatic tumors using array-based comparative genomic hybridization. Clin Gastroenterol Hepatol. 2006;4:1283-1291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Chen L, Chan TH, Guan XY. Chromosome 1q21 amplification and oncogenes in hepatocellular carcinoma. Acta Pharmacol Sin. 2010;31:1165-1171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 11. | Wong N, Chan A, Lee SW, Lam E, To KF, Lai PB, Li XN, Liew CT, Johnson PJ. Positional mapping for amplified DNA sequences on 1q21-q22 in hepatocellular carcinoma indicates candidate genes over-expression. J Hepatol. 2003;38:298-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 73] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 12. | Kanome T, Itoh N, Ishikawa F, Mori K, Kim-Kaneyama JR, Nose K, Shibanuma M. Characterization of Jumping translocation breakpoint (JTB) gene product isolated as a TGF-beta1-inducible clone involved in regulation of mitochondrial function, cell growth and cell death. Oncogene. 2007;26:5991-6001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Pelicci G, Lanfrancone L, Grignani F, McGlade J, Cavallo F, Forni G, Nicoletti I, Pawson T, Pelicci PG. A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell. 1992;70:93-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1012] [Cited by in RCA: 1066] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 14. | Kavanaugh WM, Williams LT. An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science. 1994;266:1862-1865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 395] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 15. | van der Geer P, Wiley S, Gish GD, Pawson T. The Shc adaptor protein is highly phosphorylated at conserved, twin tyrosine residues (Y239/240) that mediate protein-protein interactions. Curr Biol. 1996;6:1435-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 192] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 16. | Won KA, Schumacher RJ, Farr GW, Horwich AL, Reed SI. Maturation of human cyclin E requires the function of eukaryotic chaperonin CCT. Mol Cell Biol. 1998;18:7584-7589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 125] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 17. | Vece TJ, Watkin LB, Nicholas S, Canter D, Braun MC, Guillerman RP, Eldin KW, Bertolet G, McKinley S, de Guzman M, Forbes L, Chinn I, Orange JS. Copa Syndrome: a Novel Autosomal Dominant Immune Dysregulatory Disease. J Clin Immunol. 2016;36:377-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 141] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 18. | Suzuki Y, Demoliere C, Kitamura D, Takeshita H, Deuschle U, Watanabe T. HAX-1, a novel intracellular protein, localized on mitochondria, directly associates with HS1, a substrate of Src family tyrosine kinases. J Immunol. 1997;158:2736-2744. [PubMed] |
| 19. | Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, Hershko A. The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat Cell Biol. 2001;3:321-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 379] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 20. | Shen DY, Fang ZX, You P, Liu PG, Wang F, Huang CL, Yao XB, Chen ZX, Zhang ZY. Clinical significance and expression of cyclin kinase subunits 1 and 2 in hepatocellular carcinoma. Liver Int. 2010;30:119-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 21. | Inagaki Y, Yasui K, Endo M, Nakajima T, Zen K, Tsuji K, Minami M, Tanaka S, Taniwaki M, Itoh Y, Arii S, Okanoue T. CREB3L4, INTS3, and SNAPAP are targets for the 1q21 amplicon frequently detected in hepatocellular carcinoma. Cancer Genet Cytogenet. 2008;180:30-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 22. | Qi H, Fillion C, Labrie Y, Grenier J, Fournier A, Berger L, El-Alfy M, Labrie C. AIbZIP, a novel bZIP gene located on chromosome 1q21.3 that is highly expressed in prostate tumors and of which the expression is up-regulated by androgens in LNCaP human prostate cancer cells. Cancer Res. 2002;62:721-733. [PubMed] |
| 23. | Pu Q, Lu L, Dong K, Geng WW, Lv YR, Gao HD. The Novel Transcription Factor CREB3L4 Contributes to the Progression of Human Breast Carcinoma. J Mammary Gland Biol Neoplasia. 2020;25:37-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 24. | Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 419] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 25. | Ilardi JM, Mochida S, Sheng ZH. Snapin: a SNARE-associated protein implicated in synaptic transmission. Nat Neurosci. 1999;2:119-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 192] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 26. | Ma NF, Hu L, Fung JM, Xie D, Zheng BJ, Chen L, Tang DJ, Fu L, Wu Z, Chen M, Fang Y, Guan XY. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology. 2008;47:503-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 119] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 27. | Elsemman IE, Mardinoglu A, Shoaie S, Soliman TH, Nielsen J. Systems biology analysis of hepatitis C virus infection reveals the role of copy number increases in regions of chromosome 1q in hepatocellular carcinoma metabolism. Mol Biosyst. 2016;12:1496-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 28. | Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92:1515-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 415] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 29. | An S, Yeo KJ, Jeon YH, Song JJ. Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. J Biol Chem. 2011;286:8369-8374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 108] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 30. | Liu S, Hausmann S, Carlson SM, Fuentes ME, Francis JW, Pillai R, Lofgren SM, Hulea L, Tandoc K, Lu J, Li A, Nguyen ND, Caporicci M, Kim MP, Maitra A, Wang H, Wistuba II, Porco JA Jr, Bassik MC, Elias JE, Song J, Topisirovic I, Van Rechem C, Mazur PK, Gozani O. METTL13 Methylation of eEF1A Increases Translational Output to Promote Tumorigenesis. Cell 2019; 176: 491-504. e21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 137] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 31. | Li L, Zheng YL, Jiang C, Fang S, Zeng TT, Zhu YH, Li Y, Xie D, Guan XY. HN1L-mediated transcriptional axis AP-2γ/METTL13/TCF3-ZEB1 drives tumor growth and metastasis in hepatocellular carcinoma. Cell Death Differ. 2019;26:2268-2283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 32. | Christensen HS, Daher A, Soye KJ, Frankel LB, Alexander MR, Lainé S, Bannwarth S, Ong CL, Chung SW, Campbell SM, Purcell DF, Gatignol A. Small interfering RNAs against the TAR RNA binding protein, TRBP, a Dicer cofactor, inhibit human immunodeficiency virus type 1 Long terminal repeat expression and viral production. J Virol. 2007;81:5121-5131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 33. | Ye J, Wang J, Tan L, Yang S, Xu L, Wu X, Deng H, Tan H. Expression of protein TARBP1 in human hepatocellular carcinoma and its prognostic significance. Int J Clin Exp Pathol. 2015;8:9089-9096. [PubMed] |
| 34. | Leinhart K, Brown M. SET/MYND Lysine Methyltransferases Regulate Gene Transcription and Protein Activity. Genes (Basel). 2011;2:210-218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Sakamoto LH, Andrade RV, Felipe MS, Motoyama AB, Pittella Silva F. SMYD2 is highly expressed in pediatric acute lymphoblastic leukemia and constitutes a bad prognostic factor. Leuk Res. 2014;38:496-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 36. | Komatsu S, Ichikawa D, Hirajima S, Nagata H, Nishimura Y, Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Imoto I, Inazawa J, Otsuji E. Overexpression of SMYD2 contributes to malignant outcome in gastric cancer. Br J Cancer. 2015;112:357-364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 37. | Komatsu S, Imoto I, Tsuda H, Kozaki KI, Muramatsu T, Shimada Y, Aiko S, Yoshizumi Y, Ichikawa D, Otsuji E, Inazawa J. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis. 2009;30:1139-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 137] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 38. | Hamamoto R, Silva FP, Tsuge M, Nishidate T, Katagiri T, Nakamura Y, Furukawa Y. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 219] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 39. | Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 549] [Cited by in RCA: 569] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 40. | Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017; 169: 1327-1341. e23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1578] [Cited by in RCA: 1732] [Article Influence: 216.5] [Reference Citation Analysis (1)] |
| 41. | Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V; Cancer Genome Atlas Research Network; Hu H. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018; 173: 400-416. e11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2534] [Cited by in RCA: 2364] [Article Influence: 337.7] [Reference Citation Analysis (0)] |
| 42. | Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, Lazar AJ; Cancer Genome Atlas Research Network; Cherniack AD, Beroukhim R, Meyerson M. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018; 33: 676-689. e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 846] [Cited by in RCA: 716] [Article Influence: 102.3] [Reference Citation Analysis (0)] |
| 43. | Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8187] [Cited by in RCA: 11261] [Article Influence: 938.4] [Reference Citation Analysis (0)] |
| 44. | Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9144] [Cited by in RCA: 12814] [Article Influence: 985.7] [Reference Citation Analysis (0)] |
| 45. | Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, Chang MT, Chandarlapaty S, Traina TA, Paik PK, Ho AL, Hantash FM, Grupe A, Baxi SS, Callahan MK, Snyder A, Chi P, Danila D, Gounder M, Harding JJ, Hellmann MD, Iyer G, Janjigian Y, Kaley T, Levine DA, Lowery M, Omuro A, Postow MA, Rathkopf D, Shoushtari AN, Shukla N, Voss M, Paraiso E, Zehir A, Berger MF, Taylor BS, Saltz LB, Riely GJ, Ladanyi M, Hyman DM, Baselga J, Sabbatini P, Solit DB, Schultz N. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;2017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1079] [Cited by in RCA: 1229] [Article Influence: 153.6] [Reference Citation Analysis (0)] |
| 46. | Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1825] [Cited by in RCA: 2511] [Article Influence: 179.4] [Reference Citation Analysis (0)] |
| 47. | Kim TM, Yim SH, Shin SH, Xu HD, Jung YC, Park CK, Choi JY, Park WS, Kwon MS, Fiegler H, Carter NP, Rhyu MG, Chung YJ. Clinical implication of recurrent copy number alterations in hepatocellular carcinoma and putative oncogenes in recurrent gains on 1q. Int J Cancer. 2008;123:2808-2815. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 80] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 48. | Choi HS, Yim SH, Xu HD, Jung SH, Shin SH, Hu HJ, Jung CK, Choi JY, Chung YJ. Tropomyosin3 overexpression and a potential link to epithelial-mesenchymal transition in human hepatocellular carcinoma. BMC Cancer. 2010;10:122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 49. | Melis M, Diaz G, Kleiner DE, Zamboni F, Kabat J, Lai J, Mogavero G, Tice A, Engle RE, Becker S, Brown CR, Hanson JC, Rodriguez-Canales J, Emmert-Buck M, Govindarajan S, Kew M, Farci P. Viral expression and molecular profiling in liver tissue vs microdissected hepatocytes in hepatitis B virus-associated hepatocellular carcinoma. J Transl Med. 2014;12:230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 50. | Zhang D, Liu E, Kang J, Yang X, Liu H. MiR-3613-3p affects cell proliferation and cell cycle in hepatocellular carcinoma. Oncotarget. 2017;8:93014-93028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 51. | Agarwal R, Narayan J, Bhattacharyya A, Saraswat M, Tomar AK. Gene expression profiling, pathway analysis and subtype classification reveal molecular heterogeneity in hepatocellular carcinoma and suggest subtype specific therapeutic targets. Cancer Genet. 2017;216-217:37-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 52. | Chen X, Li W, Xiao L, Liu L. Nuclear division cycle 80 complex is associated with malignancy and predicts poor survival of hepatocellular carcinoma. Int J Clin Exp Pathol. 2019;12:1233-1247. [PubMed] |
| 53. | Guo L, Wang Z, Du Y, Mao J, Zhang J, Yu Z, Guo J, Zhao J, Zhou H, Wang H, Gu Y, Li Y. Random-forest algorithm based biomarkers in predicting prognosis in the patients with hepatocellular carcinoma. Cancer Cell Int. 2020;20:251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 54. | Jiang X, Jiang Y, Luo S, Sekar K, Koh CKT, Deivasigamani A, Dong Q, Zhang N, Li S, Hao F, Goh BKP, Ooi LL, Wang Y, Hui KM. Correlation of NUF2 Over-expression with Poorer Patient Survival in Multiple Cancers. Cancer Res Treat. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 55. | Wang Y, Tan PY, Handoko YA, Sekar K, Shi M, Xie C, Jiang XD, Dong QZ, Goh BKP, Ooi LL, Gao Z, Hui KM. NUF2 is a valuable prognostic biomarker to predict early recurrence of hepatocellular carcinoma after surgical resection. Int J Cancer. 2019;145:662-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 56. | Liu Q, Dai SJ, Li H, Dong L, Peng YP. Silencing of NUF2 inhibits tumor growth and induces apoptosis in human hepatocellular carcinomas. Asian Pac J Cancer Prev. 2014;15:8623-8629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 57. | Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 374] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 58. | Choi C, Kim D, Kim S, Jeong S, Song E, Helfman DM. From skeletal muscle to cancer: insights learned elucidating the function of tropomyosin. J Struct Biol. 2012;177:63-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 59. | Stehn JR, Haass NK, Bonello T, Desouza M, Kottyan G, Treutlein H, Zeng J, Nascimento PR, Sequeira VB, Butler TL, Allanson M, Fath T, Hill TA, McCluskey A, Schevzov G, Palmer SJ, Hardeman EC, Winlaw D, Reeve VE, Dixon I, Weninger W, Cripe TP, Gunning PW. A novel class of anticancer compounds targets the actin cytoskeleton in tumor cells. Cancer Res. 2013;73:5169-5182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 131] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 60. | Currier MA, Stehn JR, Swain A, Chen D, Hook J, Eiffe E, Heaton A, Brown D, Nartker BA, Eaves DW, Kloss N, Treutlein H, Zeng J, Alieva IB, Dugina VB, Hardeman EC, Gunning PW, Cripe TP. Identification of Cancer-Targeted Tropomyosin Inhibitors and Their Synergy with Microtubule Drugs. Mol Cancer Ther. 2017;16:1555-1565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 61. | Tooley J, Stukenberg PT. The Ndc80 complex: integrating the kinetochore's many movements. Chromosome Res. 2011;19:377-391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 62. | Tang NH, Toda T. MAPping the Ndc80 Loop in cancer: A possible link between Ndc80/Hec1 overproduction and cancer formation. Bioessays. 2015;37:248-256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 63. | Sethi G, Pathak HB, Zhang H, Zhou Y, Einarson MB, Vathipadiekal V, Gunewardena S, Birrer MJ, Godwin AK. An RNA interference lethality screen of the human druggable genome to identify molecular vulnerabilities in epithelial ovarian cancer. PLoS One. 2012;7:e47086. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 64. | Wu G, Qiu XL, Zhou L, Zhu J, Chamberlin R, Lau J, Chen PL, Lee WH. Small molecule targeting the Hec1/Nek2 mitotic pathway suppresses tumor cell growth in culture and in animal. Cancer Res. 2008;68:8393-8399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 90] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 65. | Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4732] [Cited by in RCA: 7150] [Article Influence: 310.9] [Reference Citation Analysis (0)] |