Published online Mar 27, 2021. doi: 10.4254/wjh.v13.i3.270
Peer-review started: October 23, 2020
First decision: January 7, 2021
Revised: January 20, 2021
Accepted: March 12, 2021
Article in press: March 12, 2021
Published online: March 27, 2021
Processing time: 147 Days and 4.6 Hours
The liver is a unique parenchymal organ with a regenerative capacity allowing it to restore up to 70% of its volume. Although knowledge of this phenomenon dates back to Greek mythology (the story of Prometheus), many aspects of liver regeneration are still not understood. A variety of different factors, including inflammatory cytokines, growth factors, and bile acids, promote liver regeneration and control the final size of the organ during typical regeneration, which is performed by mature hepatocytes, and during alternative regeneration, which is performed by recently identified resident stem cells called “hepatic progenitor cells”. Hepatic progenitor cells drive liver regeneration when hepatocytes are unable to restore the liver mass, such as in cases of chronic injury or excessive acute injury. In liver maintenance, the body mass ratio is essential for homeostasis because the liver has numerous functions; therefore, a greater understanding of this process will lead to better control of liver injuries, improved transplantation of small grafts and the discovery of new methods for the treatment of liver diseases. The current review sheds light on the key molecular pathways and cells involved in typical and progenitor-dependent liver mass regeneration after various acute or chronic injuries. Subsequent studies and a better understanding of liver regeneration will lead to the development of new therapeutic methods for liver diseases.
Core Tip: The liver is a unique parenchymal organ with a regenerative capacity that can restore up to 70% of its volume. A variety of different factors and signaling pathways are involved in the process of liver mass regeneration during the priming, proliferative and termination phases. This review describes the types of liver regeneration, the phases of typical liver regeneration, the cell types involved in liver regeneration, the process of alternative liver regeneration, and the stem cells and micro ribonucleic acids that play roles in liver mass regeneration.
- Citation: Kiseleva YV, Antonyan SZ, Zharikova TS, Tupikin KA, Kalinin DV, Zharikov YO. Molecular pathways of liver regeneration: A comprehensive review. World J Hepatol 2021; 13(3): 270-290
- URL: https://www.wjgnet.com/1948-5182/full/v13/i3/270.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i3.270
The capacity of the liver to regenerate has been well known since the myths of Prometheus, who was banished from Olympus by Zeus. Legend has it that eagles pecked out half of his liver every day, but because the liver regrew during the night, the hero endured never-ending torture[1]. Today, many centuries after these events, liver regeneration is a universally known phenomenon that has been studied at the molecular and cellular levels. However, many aspects remain unclear[2].
Liver regeneration is a complex process regulated by the interaction between growth factors and cytokines secreted near the site of injury or transferred to the liver by the blood. This strictly orchestrated process is divided into 3 phases: Priming, proliferation, and termination[3]. The sum of all signals that sense the physiologically necessary liver mass is called the "hepatostat”, which can initiate and terminate liver regeneration[4]. This phenomenon reflects the correlation between the needs of organisms and the organ mass that is required for homeostasis[5].
A better understanding of liver regeneration mechanisms will help improve the methods used to treat various organ diseases, prevent hepatic failure in high-risk patients, control liver grafts for transplantation, and more[6]. Importantly, the term "liver regeneration" is used improperly because during actual regeneration, not only the function of the organ but also the morphology is restored whereas only compensatory hypertrophy occurs in the liver. Second, mature hepatocytes are the source of new liver cells, not stem cells; however, stem cells play an important role in some cases of liver regeneration[7]. However, the term "liver regeneration" is widely accepted and the most commonly used term[8].
Until recently, it was believed that the liver mass after partial hepatectomy (PH) or injury recovers via hepatocyte proliferation for 1–2 cell cycles; however, recent studies have shown that different stimuli define the type of liver regeneration that occurs[9]. There are two known types of liver regeneration: The first is conducted through the hypertrophy and/or hyperplasia of hepatocytes and biliary epithelial cells (BECs) and is called typical regeneration. Typical regeneration is specific to a healthy liver that was exposed to resection or an acute liver injury; conversely, progenitor-dependent regeneration requires the reprogramming of specific hepatic cells, whose activation depends on the volume of the residual liver mass. Progenitor-dependent regeneration is specific to chronic liver diseases and massive acute liver injuries[10,11]. Thus, a 2/3 hepatectomy leads to the immediate hypertrophy of hepatocytes and further hyperplasia, whereas a 1/3 hepatectomy only triggers cell hypertrophy. Various chronic diseases and massive injuries initiate the activation of hepatic progenitor cells (HPCs), which are responsible for liver regeneration[9]. Consequently, typical liver regeneration is driven by mature hepatocytes and BECs, whereas the alternative regeneration method is performed by HPCs[11].
PH causes a hemodynamic disturbance, expressed as a portal pressure escalation, which serves as a regeneration stimulus. Consequently, hepatocytes, BECs, Ito cells, Kupffer cells (KC) and sinusoid endothelial cells (SECs) are proliferated. Interestingly, hepatocytes proliferate first, whereas BECs start to proliferate only 2-3 d after PH. After a 2/3 PH, the hepatocytes go through one cycle of DNA synthesis, which is required for the restoration of 60% of the liver mass. In the following stages, several but not all hepatocytes continue to proliferate to achieve complete liver recovery. Afterward, apoptotic activity increases with the purpose of correcting an excessive regenerative response[12].
The beginning of each phase is initiated by a certain molecule set released in response to organ damage[13]. The earliest regeneration drivers are portal pressure changes and an increasing level of urokinase plasminogen activator (uPA)[8,14].
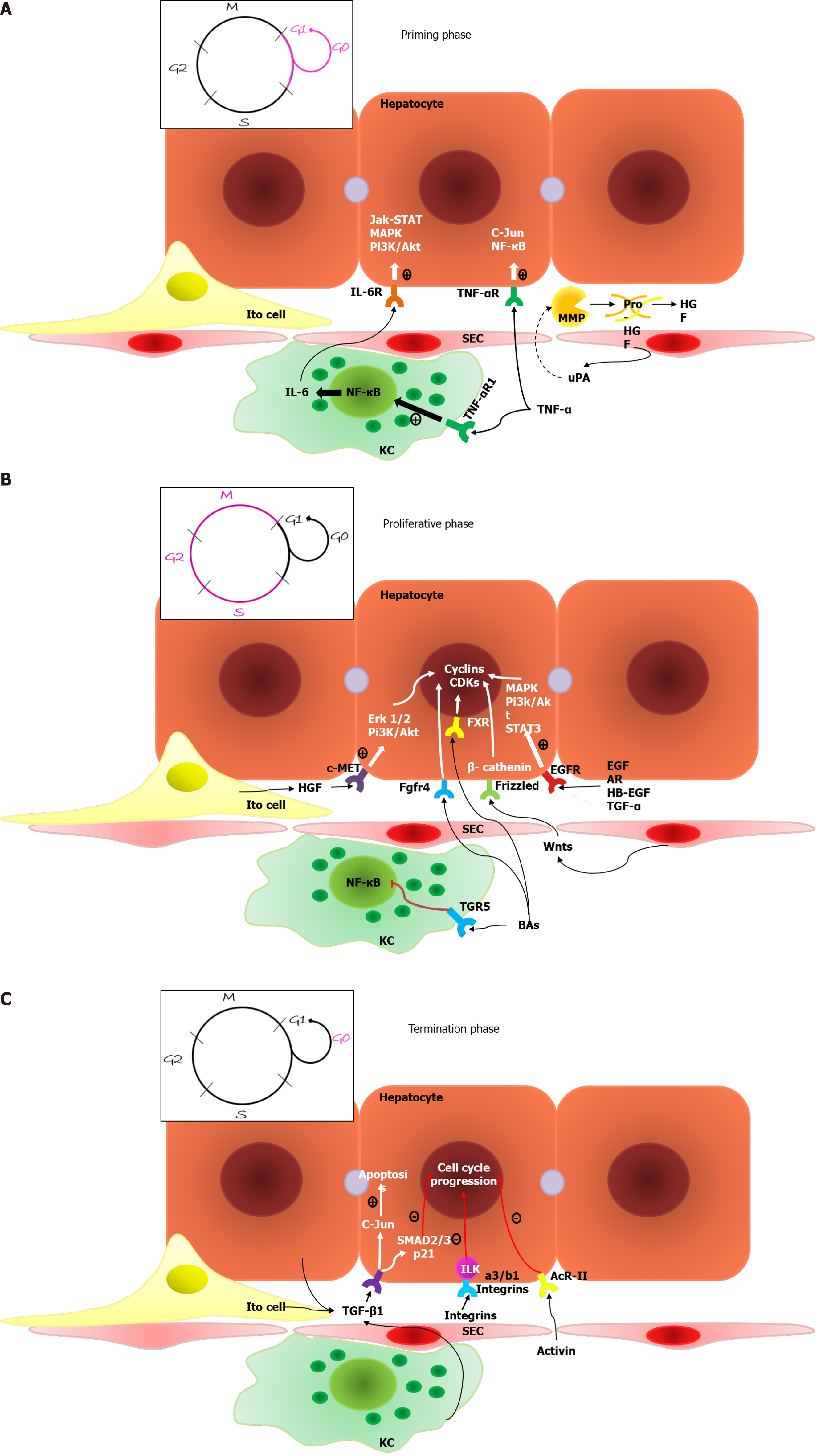
Priming phase: During the first phase of regeneration, hepatocytes, driven by various cytokines, simultaneously enter the G1 phase of the cell cycle[10].
The increasing blood pressure in the hepatic sinusoids is conditioned by the incompatibility between the volume of the liver and the volume of inflowing venous blood[15], which results in a turbulent flow and mechanically stimulates SECs to secrete large amounts of uPA. uPA promotes plasminogen-plasmin transformation, leading to matrix metalloproteinase (MMP) activation and fibrinogen degradation. Plasmin and MMPs are involved in extracellular matrix (ECM) remodeling, resulting in the release of growth factors, such as hepatocyte growth factor (HGF)[8].
Two proinflammatory cytokines are the main mediators of the first phase: Tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6); these cytokines are secreted primarily by liver macrophages under the influence of bacterial lipopolysaccharide and the C3a and C5a components of the complement system[16]. IL-6 drives the acute phase response and initiates cytoprotection and the proliferation of hepatocytes via the IL-6-IL-6R interaction and the activation of coreceptor glycoprotein 130 (gp130), which activates the Janus kinase (JAK)/signal transducer and activator of transcription (STAT), Mitogen-activated protein kinase (MAPK) and PI3K/AKT signaling pathways[5,17]. Although gp130 is present on the surface of most cells, IL-6R is primarily located on hepatocytes. However, there are also soluble IL-6Rs that initiate the trans-signaling pathway within cells lacking IL-6R and enhance the regenerative response of hepatocytes[3]. Fazel Modares et al[18] elicited the crucial role of the trans-signaling pathway in liver regeneration after PH because hepatocyte IL-6R activation alone was not sufficient to initiate cell proliferation. TNF-α has two main functions: It activates the NF-κβ signaling pathway through direct interaction with TNF-R1 on Kupffer cell surfaces and through the indirect induction of inhibitory KB kinase; it also stimulates hepatocyte c-Jun N-terminal kinase (JNK). JNK phosphorylates the c-Jun transcription factor in the nucleus to induce cyclin-dependent kinase 1 transcription, which activates hepatocyte proliferation[8].
The augmenter of liver regeneration (ALR) protein, which has three isoforms (15, 21 and 23 kDa) and is expressed primarily in the liver, testes, kidneys and brain, plays a crucial role in liver regeneration. Each isoform of ALR has a different location within the cell and thus plays a different role[19]. For example, mitochondrial long-form ALR translocates proteins and initiates MitoNEEt release, which leads to cell proliferation. Long-form ALR expression increases in cases of pathology and reduces liver damage, protects against oxidative stress and endoplasmic reticulum stress by decreasing Ca++ levels, and has an antimetastatic effect on hepatocellular carcinoma (HCC). Cytoplasmic short-form ALR enhances the hepatocyte response to IL-6 by inducing the phosphorylation of STAT3; it also has an antimetastatic effect on hepatoma by inhibiting the migrative and invasive capacity of cells[20]. After PH, the ALR concentration increases immediately and activates MAPK signaling; enhances IL-6, TNF-α and inducible nitricoxide synthase production by Kupffer cells; and inhibits NK cell activity. Short-form ALR protects hepatocytes by inhibiting apoptosis stimuli[21].
During the second phase of liver regeneration, the G1/M phase transition occurs, which is driven by two groups of mitogens: Complete mitogens, including HGF, TGF-α, epidermal growth factor (EGF), and HB-EGF; and the stimulation of DNA synthesis and cell proliferation via Ras-MAPK and PI3K/AKT signaling activation and auxiliary mitogens, including bile acids, vascular endothelial growth factor (VEGF), noradrenalin, insulin-like growth factors (IGFs), estrogen and serotonin[3].
HGF is produced by mesenchymal liver cells and interacts with the methionine (MET) receptor, leading to PI3K and MAPK signaling protein phosphorylation followed by PI3K/AKT and extracellular-signal-regulated kinase 1/2 signaling activation. This process results in the proliferation, migration, and differentiation of liver cells and antiapoptotic effects[22,23]. Epidermal growth factor receptor (EGFR)-transmembrane receptors with tyrosine kinase activity interact with EGF, TGFa, amphiregulin (AR), epigen, and HB-EGF, leading to MAPK, PI3K/AKT–mammalian target of rapamycin (mTOR) and STAT signaling activation, which drives hepatocyte proliferation[24]. Natarajan et al[25] identified impaired liver regenerative capacity and delayed cyclin D1 expression in mice lacking EGFR.
Nuclear factor erythroid 2-related factor 2 (NRF2) transcription factors, which regulate a wide range of genes including antioxidant proteins and detoxifying enzymes, are activated in response to increased reactive oxygen species levels. The expression of this molecule increases in the earliest stages of liver regeneration as a result of cellular damage[26]. Zou et al[27] discovered the important role of Nrf2 in the regulation of cell cycle progression in mice. Nfr2 is a transcriptional suppressor of Cyclin A2 and a regulator of the Wee1/Cdc2/Cyclin B1 pathway, which controls the beginning of the M phase[27]. Nfr2 also regulates hepatocyte proliferation by modulating the insulin/IGF-1 and Notch1 signaling activities and facilitates the capability of hepatocyte nuclear factor 4 alpha (HNF4α) to keep newly formed hepatocytes in a differentiated state[28].
Bile acids are the main end products of cholesterol metabolism and are synthesized exclusively in the liver, where they function as signaling molecules that activate membrane G-protein-coupled BA receptor 1 (or TGR5) and nuclear farnesoid X receptor (FXR)[29]. After the loss of liver mass due to PH, the bile acids concentration increases during the first minute, which leads to FXR activation, resulting in inhibited BA synthesis and induction of the FOXM1B gene[30]. FOXM1B is a transcription factor that regulates DNA synthesis and mitosis via cyclin-dependent kinase 2 (CDK2) activation, which is required for the G1/S transition and CDK1 activation and is responsible for the S/M transition[31]. FXR activation also appears in enterocytes and leads to the induction of fibroblast growth factor (FGF)15/FGF19 expression. The Fgfr4/β-Klotho receptor, which is located on the hepatocyte surface, inhibits BA synthesis and activates the cell cycle via FOXM1B induction when activated[32]. Fgfr4/β-Klotho activation also regulates the termination of liver regeneration and terminal organ size. Kong et al[33] showed that mice with enhanced Fgf15 expression have the most active Hippo signaling pathway, which induces cellular senescence and suppresses transcriptional activation. TGR5, which is located on KC, SEC and BEC surfaces, leads to cAMP induction and nuclear factor kappa B (NF-κB)-signaling inhibition[34]. As a result, decreased proinflammatory cytokine synthesis occurs in KCs and bone marrow macrophages via the protein kinase B-dependent activation of the mTOR[35]. TGR5 protects the liver from BA overload by increasing its excretion with urine; it also enhances the secretion of HCO3ˉ and Clˉ and controls BA polarity because inordinately hydrophobic molecules can damage the regenerating liver[36].
Wnt ligands are glycoproteins secreted by nonparenchymal liver cells, mostly KCs and SECs, and are crucial molecules of liver regeneration[37]. Wnt ligands lead to the integration of Axin into the cytoplasmic membrane through interaction with the Frizzled receptor and the coreceptors LRP5/6, resulting in impaired function of the β-catenin degradation complex. Therefore, Wnt ligands lead to β-catenin accumulation, followed by its translocation to the nucleus and interaction with members of the transcriptional T cell factor family, resulting in target gene transcription, for example, of cyclin D1, leading to hepatocyte proliferation[38]. Preziosi et al[39] identified the constitutional secretion of Wnt2 and Wnt9b by central vein endotheliocytes and the essential role of these molecules in the basal activation of β-catenin and metabolic zonation of hepatocytes. PH leads to increased Wnt2 and Wnt4 expressions within all zones of the hepatic acinus and the additional secretion of Wnt9b and Wnt5b within the pericentral zone during the first 12 h. This leads to a 7–8-fold increase in cyclin D1 expression within the periportal and intermediate zones and 20-and 100-fold increases in glutamine synthetase expression within the intermediate zone and the pericentral zone, respectively. The role of increased glutamine synthetase expression remains unknown but is thought to be an enhancer of pericentral detoxification since the other 2/3 of hepatocytes restore organ mass[39].
The Hedgehog (Hh) signaling pathway is a morphogenic pathway that regulates embryonic development and is implicated in homeostasis maintenance[40,41]. Among vertebrates, this pathway is activated within a special organelle, the primary cilium (PC), via the interaction of Hh ligands Sonic hedgehog, Indian hedgehog and Desert hedgehog and the Ptched receptor[42]. After that, phosphatidylinositol 4-phosphate[43], sumoylated molecules and cholesterol[44] form a complex with smoothened (Smo), which leads to its activation. Activated Smo dislocates to the apex of the PC and activates Glis (including Gli1, Gli2 and Gli3), which then translate to the nucleus and regulate gene transcription[45]. The said pathway is canonical, but there are also different types of noncanonical Hh signaling pathways; for example, the Smo-free activation of Glis or the Hh pathway arises beyond the PC[45-47]. Ochoa et al[48] identified a meaningful role of Hh signaling in liver regeneration. PH leads to Hip inhibition, thus activating the Hh pathway via an increase in the Indian hedgehog level in the replicative period and an increase in the Sonic hedgehog level in the postreplicative period[48]. Platelet-derived growth factor, TGF-β, and EGF are secreted in response to liver damage induced by JNK-dependent Hh ligand synthesis[49,50]. Hh signaling activation occurs within hepatocytes, Ito cells[51] and BECs[52], leading to ECM remodeling, progenitor cell expansion and liver epithelial cell proliferation[45]. Additionally, Hh signaling controls Yes-associated protein 1 (YAP) of activated Ito cells[53]. The Hh-YAP signaling pathway induces the glutaminolysis required for Ito cell activation to regulate liver regeneration[54]. Furthermore, Hh signaling facilitates cell survival via inhibiting hepatocytes, BECs, Ito cells and progenitor cell apoptosis[55].
Notch signaling is an important pathway in embryonic development, homeostasis maintenance, and liver regeneration[56]. Mammals have 4 types of receptors for this pathway (Notch1, Notch2, Notch3, and Notch4); Notch1 and Notch2 are located primarily on BECs and HPCs whereas Notch3 and Notch4 are expressed by the mesenchymal compartment of the liver and are poorly represented on epithelial liver cells. JAG-1 and DLL-4 are ligands of Notch signaling that are expressed in the liver[57]. The main role of this pathway in liver development is the JAG1-NOTCH2 interaction, which results in the differentiation of hepatoblasts to BECs and the development of the intrahepatic biliary tree[58]. Lu et al[59] showed that the role of the Notch–RBPJ interaction is to drive HPC differentiation to BECs via Yap inactivation after PH in mice. The direction of HPC differentiation is defined by the balance of NOTCH signaling and Wnt ligands[60]. Ortica et al[61] pointed out the important role of Notch3 in HPC differentiation to hepatocytes. Zhang et al[62] elicited the regulatory role of Notch signaling in hepatocyte proliferation via the NICD/Akt/Hif-1α pathway after PH, whereas its inhibition leads to delayed S phase entry, impaired S phase and M phase progression, and the loss of the hepatocyte mitotic rhythm due to cyclin E1, A2 and B1 dysregulation. Yang et al[63] demonstrated the involvement of Notch signaling in the regeneration of 8 types of liver cells, which is performed by the activity of 9 different pathways and regulates cellular proliferation, apoptosis, the cell cycle, etc.
When the needed liver mass: Body mass ratio is achieved, cellular proliferation stops due to inhibitory molecules that control the rapidity and direction of liver regeneration. Among the inhibitors of cell proliferation, IL-1, which is synthesized by nonparenchymal liver cells, inhibits the DNA synthesis induced by HGF, EGF and TGF-α. IL-6 is multifunctional and plays a role as both a liver regeneration inducer and inhibitor; its effect depends on the time and dose of the molecule. The IL-6-dependent inhibition of proliferation is likely to occur by increasing p21 expression[64]. The JAK/STAT signaling pathway is inhibited by 8 members of the SOCS family of proteins; hereafter, only SOCS1 and SOCS3 contain the extended SH2 and kinase inhibitory region. SOCS1 directly binds and inhibits JAK, whereas SOCS3 binds to cytokine receptors, forms a complex with JAK and inhibits the STAT3 signaling pathway. SOCS3 is the main suppressor of the signaling pathway activated by IL-6; it inhibits the phosphorylation of coreceptor gp130, JAK and STAT3. SOCS1 negatively regulates the hepatocyte proliferation induced by HGF via c-MET signaling inhibition and likely regulates the TNF-α levels because it interacts with toll-interleukin 1 receptor domain-containing adaptor protein, which drives the synthesis of a current mediator[65].
Some TGF-β family members function as inhibitors of proliferation. In particular, TGF-β1 plays a special role in binding to receptor types 1 and 2 and inducing cell apoptosis to correct an excessive liver mass. Outside of the liver, TGF-β1 is synthesized in platelets and the spleen. The spleen might be involved in the termination phase for it inhibits HGF and its c-MET receptor expression. In this regard, splenectomy leads to increased hepatocyte proliferation in the first 48 h after PH. Other members of the TGF-β family are involved in the termination phase of liver regeneration, including activin A-hepatocyte proliferation inhibitors and bone morphogenetic proteins (BMPs)[3]. BMP9 is expressed exclusively by liver tissues and in part by hepatocytes. BMP9 regulates a variety of biological functions such as glucose and lipid metabolism, angiogenesis, oncogenesis, and fibrogenesis, and it affects liver regeneration after acute injuries[66]. Addante et al[67] reported a regulatory function of BMP9 over HPCs that is affected by anaplastic lymphoma kinase 2 type I receptor activation, resulting in SMAD 1, 5, and 8 induction, HPC apoptosis stimulation, and a reduction in HPCs. Apart from its negative influence on liver regeneration, BMP9 also has profibrogenic activity and promotes HCC proliferation and invasion. Additionally, BMP9 enhances the expression of TLR4 on the SEC surface, leading to inflammatory cell recruitment. Therapy with anti-BMP-9/ anaplastic lymphoma kinase 1 can potentially enhance hepatocyte proliferation among patients with chronic liver diseases and decrease the probability of fibrosis and HCC development[68].
HNF4-α regulates hepatocyte differentiation and, according to Huck et al[69], promotes the termination of liver regeneration. The expression of the current molecule significantly decreased during the priming phase and increased during the following phases, which is necessary for termination and hepatocyte function recovery after PH[69]. HNF4-α is a YAP and TGF-β/SMAD3 antagonist; therefore, decreased expression of this molecule stimulates promitogenic functions and activates connective tissue growth factor. Increased HNF4-α expression during the subsequent phases of regeneration prevents the excessive synthesis of connective tissue and therefore fibrosis[70]. Hnf4-α also leads to the inhibition of HPC proliferation and migration in rats[71].
Integrin-linked kinase is a suppressor of hepatocyte proliferation that is located under the cytoplasmic membrane and is associated with a3/b1 integrins of the ECM. Interruption of this connection results in hepatostat imbalance and excessive liver mass. Focal adhesion kinase is also associated with a3/b1 integrin and promotes hepatocyte proliferation[72].
The Hippo signaling pathway is a crucial regulator of the terminal organ size within mammals. The key component of the mammalian Hippo pathway is a kinase cascade in which the Ste20-like kinases 1/2 phosphorylate and activate large tumor suppressor 1/2, its adapter protein Mps one binder 1, and the transcriptional coactivators Yap and Taz[73]. Phosphorylated Yap and Taz emerge from the nucleus, where they are bound to transcription factors that control the proliferation and differentiation of cells, such as TEAD family members[74]. The Hippo/Yap signaling pathway is likely an integrator of a large number of alternative growth factor signaling pathways and regulates liver size by balancing negative and positive regulatory signals[72]. The Hippo signaling pathway does not have any specific receptors and is regulated by molecules that control cellular polarity and morphology, intercellular adhesion and other processes. The activity of this pathway is modulated in response to mechanical deformation and intercellular adhesion defects and cell adhesion to the intercellular matrix. Consequently, Hippo signaling senses cellular and tissue integrity[75]. Intranuclear Yap is located in periportal hepatocytes and BECs, whereas pericentral hepatocytes contain few current molecules, which is exactly the opposite of the constitutive Wnt ligand content; therefore, current pathways inhibit one another[76]. Table 1 summarizes the main molecular factors in liver regeneration.
| Factor of regeneration | Influence on LR |
| TNF-α | Induction of CDK-1 |
| IL-6 | Activation of the JAK/STAT, MAPK, and PI3K/AKT signaling pathways |
| Hh signaling pathway | ECM remodulation; induction of progenitor cell and liver epithelial cell expansion; induction of glutaminolysis; inhibition of hepatocyte, BEC, Ito cell and progenitor cell apoptosis |
| ALR | lfALR: Enhancement of the hepatocyte response to IL-6 and STAT3 phosphorylation induction. MAPK signaling pathway activation; NK cells inhibition; increase in IL-6, TNFα and iNOS production by Kupffer cells, sfALR: Inhibition of proapoptotic stimuli |
| NRF2 | Regulation of M phase entry, hepatocyte proliferation, maintenance of newly formed hepatocytes in a differentiated state |
| Growth factors (HGF, TGF-α, EGF, HB-EGF) | Stimulation of DNA synthesis and cell proliferation via Ras-MAPK and PI3K/AKT signaling pathway activation |
| BAs | Activation of CDK2, cell cycle, regulation of termination phase and terminate liver size, decrease in the inflammatory cytokine production, enhancement of BA excretion and HCO3ˉ, Clˉ secretion, control of BA polarity |
| Wnt-β-catenin | Hepatocyte proliferation induction |
| Notch signaling pathway | Modulation of HPC differentiation toward BECs, regulation of hepatocyte proliferation, mitotic rhythms, cyclin E1, A2 and B1 |
| IL-1 | DNA synthesis inhibitor |
| SOCSs | c-MET and JAK-STAT signaling pathway inhibition |
| TGF-β1, activin A, BMPs | Induction of apoptosis to correct excessive liver mass |
| HNF4 | Regulation of hepatocyte differentiation, initiation of the termination phase, antagonism YAP and TGF-β/SMAD3, prevention of excessive connective tissue synthesis, inhibition of HPC proliferation and migration |
| Hippo/YAP signaling pathway | Terminal liver size control |
The hepatic acinus is the structural and functional unit of the liver. It consists of three zones. The hepatocytes in the first zone have a periportal location and are specialized in gluconeogenesis and the beta-oxidation of fatty acids; conversely, hepatocytes in the third zone lie pericentral and perform glycolysis, lipogenesis and detoxification. Therefore, hepatocytes are functionally heterogeneous and express various genes depending on their localization[77]. Experiments performed on mice have mainly investigated diploid hepatocyte populations within the third zone, which express the early progenitor cell markers Tbx3 and Axin2 and can proliferate twice as fast as other hepatocytes. This ability depends on Wnt ligand expression in nearby SECs[78]. Sun et al[79] reported that Axin2+ pericentral hepatocytes are not confined to the liver stem cell compartment and do not have an enhanced capacity for proliferation. Cells of every zone participate in cellular homeostasis. The authors further controverted the opinion that Axin2+ pericentral hepatocytes translocate to the periportal zone. Axin2+ induction was identified in every zone of the acinus during regeneration[79].
Hybrid hepatocytes, which account for 5% of all hepatocytes, were found in the first zone of acinus. These cells express the hepatic transcription factors Hnf4a and sulfur oxide (Sox) 9, which are active in BECs. Hybrid hepatocytes are capable of differentiation into BECs and hepatocytes and help recover liver mass after various chronic diseases[80].
In contrast with the regenerative pool of the third zone, cells of the first zone do not proliferate in the absence of functional damage. The specific marker of periportal hepatocytes is the major facilitator superfamily domain-containing 2a[9]. Liver regeneration in the homeostatic state was performed via hepatocyte self-renewal within each acinus zone. Pu et al[81] elicited the capacity of major facilitator superfamily domain-containing 2a+ hepatocytes to proliferate more actively after PH and to completely replace pericentral hepatocytes during CCl4ˉ induced chronic injury. Therefore, depending on the type and duration of damage, liver regeneration occurs via different pools of cells[81].
Immune cells, including Kupffer cells, circulating monocytes and lymphocytes, play an important role in liver regeneration. Kupffer cells secrete mediators of proliferation, such as HGF, IL-6, TNF-α and Wnts, stimulating angiogenesis via VEGF-A secretion. Circulating monocytes play an important role in the first hours of liver regeneration, as indicated by the significantly increased number of adhesion molecules on the SEC surface. Monocytes play a role because it takes time for Kupffer cells to reach the sinusoids from the space of Disse[82]. Organ damage initiates the release of chemoattractants, such as osteopontin, monocyte chemoattractant protein-1, and intercellular adhesion molecule 1, resulting in macrophage recruitment to the liver where lipopolysaccharide and the C3a and C5a components of the complement system activate the NF-κB signaling pathway and the synthesis of IL-6 and TNF-α[83]. Apart from macrophage activation, components of the complement system directly influence liver regeneration. C5a binds to C5aR1, whose expression on hepatocyte surfaces significantly increases during regeneration, inducing cellular growth. C3a and C3b might facilitate liver regeneration since organ recovery within C3-deficient mice (C3–/–) was disturbed in a previous experiment[84].
NK and natural killer T cells inhibit regenerative processes via interferon (IFN)-γ secretion, which stimulates the synthesis of antiproliferative proteins, such as STAT1, IRF-1, and p21CIP1/WAF1, by hepatocytes. The influence of natural killer T cells on liver regeneration is significantly lower[83,85]. The medium limitation of NK cell activation is required for normal liver regeneration, which provides the increased expression of the coinhibitory receptor T cell Ig and ITIM domain (TIGIT) on these cell surfaces. TIGIT binds to the poliovirus receptor (PVR), which is located on hepatocyte and Kupffer cell surfaces and results in the inhibition of INF-γ secretion by NK cells. Hepatocyte expression of PVR significantly increased because the current protein is not only a ligand of NK cell receptors but is also a mediator of cellular growth, adhesion, migration and immunomodulation. However, the main role of PVR in liver regeneration seems to be interaction with TIGIT[86]. Eosinophils are key cells that secrete IL-4, which stimulates the G1 phase entrance of hepatocytes by binding to IL4Rα[87].
Hepatic stellate cells (HSCs, Ito cells) are located in the space of Disse and function in retinoid storage during the inactive stage[88,89]. Ito cells have regenerative potential and, in addition to growth factor secretion, can exhibit stem cell properties. Thus, stellate cells have the capacity to differentiate into HPCs, hepatocytes and BECs based on the influence of certain cytokines[90,91]. Ito cells demonstrate this capacity, as described above, in the case of chronic liver diseases, including cirrhosis[92]. Swiderska-Syn et al[93] demonstrated that hepatocytes require modulation of the epithelial-mesenchymal transition in multipotent progenitors derived from HSCs. A crucial role in this process is canonical Hh signaling. Although Ito cells have characteristics of multipotent cells, they improve the supportive role of each progenitor pool rather than nullify the importance of other liver progenitor populations[93]. HPC expansion and infiltration are correlated with ECM remodeling. HSCs engage in the degradation of collagen, forming an HPC niche that is rich in laminin, hyaluronic acid (HA) and collagen III, which are necessary for the development of the undifferentiated HPC phenotype. Collagen type I and fibronectin promote cell cycle arrest and HPC differentiation into hepatocytes and BECs[94]. Several studies have contradicted the capacity of HSCs to give rise to an epithelial pool of liver cells in various models of liver injury and in isolated cell cultures. The sources of hepatocytes and BECs are mature hepatocytes and bipotential liver progenitor cells[95,96]. Kordes et al[97] showed that the pancreatic stellate cells of rats express stem cell markers, such as CD133 and nestin, and have the possibility to display the β-catenin-dependent Wnt and Notch signaling pathways, which are required for stem cell maintenance and expansion. Transplantation of these cells after the surgical removal of 70% of the liver mass and the inhibition of hepatocyte proliferation (2AAF/PHX) led to the transdifferentiation of current cells into Hnf4α+ hepatocytes and panCK+ BECs[97]. Further studies in the given field are required because the role of HSCs in liver regeneration is significant. Research by Mabuchi et al[98] defined the importance of HSC and hepatocyte interactions in the early phases of liver regeneration, resulting in HSC activation[98]. Activated stellate cells transdifferentiate into myofibroblasts, secreting ECM components and cytokines, which drive the proliferation and differentiation of liver cells[92]. Among the cytokines secreted by Ito cells, HGF, lymphotoxin-beta, FGF, IL-6, NOTCH, delta-like noncanonical Notch ligand 1 and TGF-β1 play important roles[92,99]. HSCs regulate HPC proliferation via the antiproliferative effect of TGF-β1, which controls the termination phase of liver regeneration. Ito cells regulate the cytokine profile, affecting various phases of liver mass restoration[100]. Konishi et al[73] demonstrated the intensification of hepatocyte proliferation via HSC activation after ischemia-reperfusion injury (IRI). Herein, activated YAP and TAZ served as the inductors of HSC proliferation in the postischemic liver[73].
In addition to playing a role in thrombogenesis, platelets are involved in the development of inflammation and several syndromes; they also lead to the metastasis of some tumors and are required for liver regeneration. Previous studies have elicited impaired regeneration after PH under conditions of thrombocytopenia, whereas an elevated level of platelets was associated with enhanced regeneration since platelets produce HGF, Platelet-derived growth factor and TGF-β[101]. Partial resection or chronic liver injury leads to platelet accumulation in the sinusoids and space of Disse, likely via von Willebrand factor (vWF) secretion by SECs[102]. vWF plays a crucial role in the early stages of liver regeneration by promoting platelet adhesion, which is significantly decreased when anti-vWF antibodies are present. After the initiation of regeneration, the secretion of vWF antigens increases. The postoperative level of vWF antigens may be used to predict the survival prognosis[103]. Platelets secrete various growth factors that positively influence liver regeneration. The most important secreted cytokines are HGF, IGF and serotonin, which promote hepatocyte proliferation[104]. Human platelets do not secrete a considerable amount of HGF; therefore, the primary platelet mediator of liver regeneration is IGF-1. Apart from hepatocytes, platelets also interact with SECs and Kupffer cells and thus positively affect liver regeneration. Sphingosine-1-phosphate is secreted by platelets and stimulates SEC proliferation and IL-6 secretion, which drives DNA synthesis within hepatocytes. The interaction between platelets and Kupffer cells leads to the activation of both cells[105]. Platelets enhance the Kupffer cell secretion of mediators, i.e., TNF-α and IL-6, that are required for liver regeneration[101]. Platelets can either activate angiogenesis or inhibit it, depending on the mediator secreted from α-granules. Thus, thrombospondin 1 is an antiangiogenic mediator, whereas VEGF has a proangiogenic function. As long as platelets secrete both of these mediators, the PH outcome depends on the pattern of α-granule secretion[106]. Since platelets secrete many mitogens, the transfusion of blood enriched with platelets promotes liver regeneration after PH; however, it may lead to complications, including fatality[3].
A general scheme of the molecular processes involved in the various phases of typical liver mass regeneration is shown in Figure 1.
The mechanisms described above are specific for healthy livers and occur among living liver donors. However, in most cases, liver resection occurs within patients with impaired liver function, and subsequent regeneration proceeds in a nonstandard way, which can lead to hepatic failure and death[107].
Acute liver failure caused by intoxication, viral hepatitis A, B or E, autoimmune liver disease, etc., is often followed by widespread necrotic and apoptotic zones, and adequate liver regeneration becomes impossible[107]. During acute liver failure, the main regenerative role is given to HPCs, as indicated by the increased level of alpha-fetoprotein (AFP). Therefore, a high AFP level is correlated with a positive prognosis after acetaminophen-induced liver damage[12]. The immune system regulates liver regeneration via necrotic cell phagocytosis and controls inflammatory reactions in response to injury. The number of proliferative macrophages in the liver significantly increases after organ damage, and monocytes are recruited from the bloodstream and differentiate into macrophages in response to increasing the colony-stimulating factor 1 levels. Colony-stimulating factor 1 injection promotes liver regeneration after PH; conversely, a low level of the current factor is correlated with a negative patient prognosis[11].
Liver steatosis is associated with an impaired regenerative function, in which GADD34 plays an important role since its increased expression promotes liver regeneration within mice. IRI often complicates the posttransplantation period and impairs typical liver regeneration. The current complication is followed by increased receptor for advanced glycation end product levels, which might be a therapeutic target. Thus, receptor for advanced glycation end product inhibitor injection leads to a reduction in organ damage and the induction of liver regeneration. The excessive synthesis of ECM components by activated HSCs inhibits hepatocyte proliferation, and if macrophage MMPs do not promote connective tissue restitution, the angioarchitecture of hepatic lobules is impaired, resulting in cirrhosis[107]. In the liver, damage due to cirrhosis and hepatitis B or C often reveals hepatocytes with BEC markers, such as epithelial cell adhesion molecule (EpCAM), on their surface. The presence of these intermediate hepatobiliary cells is thought to be explained by their origin from biliary compartment progenitors[108].
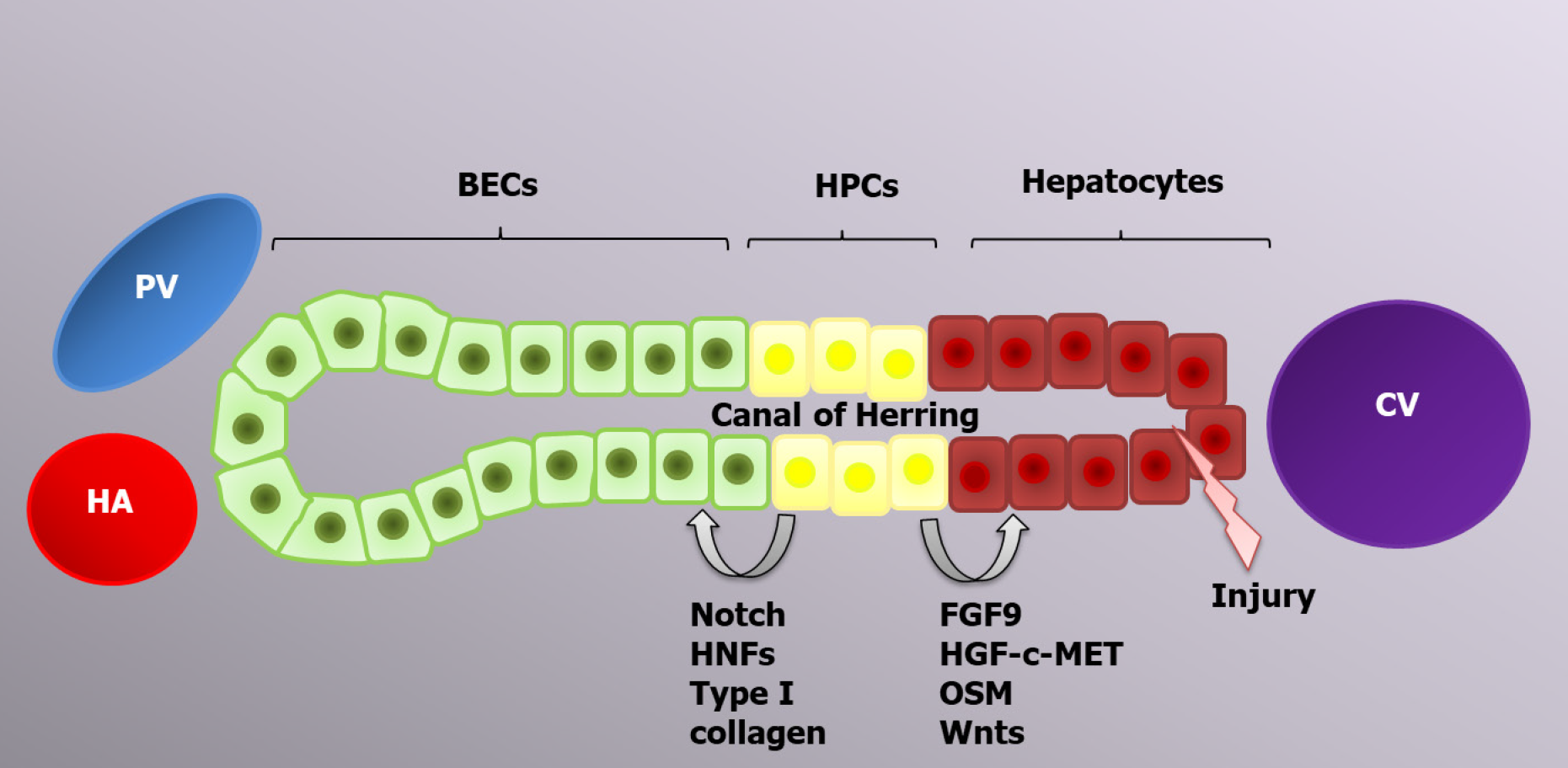
Hepatocytes are the main cells driving typical liver regeneration, whereas alternative liver regeneration is performed by HPCs[92]. The process of progenitor-dependent liver regeneration is shown in Figure 2.
Therefore, the liver regenerative capacity is significantly impaired during chronic liver diseases due to the accumulation of senescent hepatocytes[109,110]. In this case, liver mass restoration is performed by HPCs[111]. HPCs are located in the canals of Hering and have a bipotential nature; in other words, they can differentiate into both hepatocytes and BECs, the choice of which is determined by the activation of certain genes[12,108]. The canals of Hering connect the hepatocyte canalicular system and the biliary tree, and such a location of HPCs is consistent with their bipotential features[94]. Transplantation of current cells leads to liver regeneration enhancement via HPC proliferation and differentiation, which can be applicable for the treatment of certain liver diseases[112-115]. CK19, EpCAM and CD133 are markers common to both HPCs and BECs. Trop2 (Tacstd2) is a transmembrane molecule that is present on the HPC surface and absent on BECs; therefore, it can play a role as a specific marker, similar to Foxl1[116].
The origin of HPCs is still being researched. Many scientists think that HPCs arise from mature differentiated BECs due to the presence of similar markers and cell localization. The expression of hepatocyte markers, such as albumin, AFP and HNF4α, appears earlier than HPC expansion. Newly formed HPCs have various markers on their surface, including the BEC markers HNF1b and CK19, which are maintained until the HPCs differentiate into mature hepatocytes[117]. Hepatocytes and BECs are formed from common cells, called hepatoblasts, during the second trimester of embryonic development. Consequently, the possibility of hepatocyte to BEC transdifferentiation and vice versa is genetically feasible and might be programmed to form a facultative pool of progenitors[12].
HPC compartment activation in the human liver is called ductular reaction because of the role of ductular epithelium activation. In the niche, HPCs are surrounded by epithelial and nonparenchymal cells, immune cells, and the components of the ECM, which transport activating signals[118]. As long as HPCs drive the regeneration of massive or chronic damage facilitated by immune cells, inflammatory cytokines, such as TNF-α, lymphotoxin-β, interferon-γ and IL-6, will play a crucial role in HPC activation. TNF-like weak inducer of apoptosis (TWEAK) is a TNF superfamily member and the main inducer of HPC activation[119]. Macrophages and NK cells are primary sources of TWEAK ligands. The interaction with target cells is realized by FGF-inducible 14 receptors. The TWEAK/ FGF-inducible 14 interaction leads to ductular reaction initiation via activation of the NF-κB signaling pathway[120]. HPC regulation is also performed by free oxygen radicals, which act as second messengers, realizing the balance between self-renewal and the differentiation of current cells. Low reactive oxygen species levels promote HPC proliferation via extracellular-signal-regulated kinase 1/2, Jun 1/2, Wnt and NF-κB signaling[121].
HPC differentiation into hepatocytes and BECs is regulated by a variety of signaling pathways. Thus, FGF9, the HGF-с-MET signaling pathway[122] and oncostatin M activate AKT and STAT3, which are required for HPC differentiation into hepatocytes, whereas HNF-6, HNF-1β and NOTCH signaling lead to BEC development[123,124]. All-trans retinoic acid is a significant active metabolite of vitamin A that is involved in HPC differentiation by increasing miR-200a expression, which regulates cell autophagy[125]. Ma et al[126] demonstrated the regulatory function of autophagy in HPC differentiation into hepatocytes via activation of the Wnt/β-catenin signaling pathway. Autophagy can also regulate HPC differentiation into BECs since it inhibits the Notch1 signaling pathway, which is required for the development of biliary duct cells. Therefore, autophagy is decreased during the early stages of liver regeneration[127].
Recently, a new pool of multipotential biliary progenitor cells, which can differentiate into hepatocytes, BECs and the islets of Langerhans cells, was identified in peribiliary glands, which are epithelial invaginations of extrahepatic and large intrahepatic biliary ducts[108]. This pool was named biliary tree stem/progenitor cells (BTSCs). BTSCs express stem cell markers such as Sox17, Pdx1, Sox9, EpCAM, Sall4 and Lgr5 on their surface. BTSCs are primarily involved in biliary epithelium regeneration in chronic diseases such as primary sclerosing cholangitis, cholangiocarcinoma, nonanastomotic strictures and biliary atresia[128].
Micro ribonucleic acids (MiRNAs) are short molecules of 19–25 nucleotides in length that regulate the posttranscriptional silencing of target genes. One miRNA molecule can regulate hundreds of mRNAs, thus controlling the expression of various genes[129,130]. After PH, miRNA expression is primarily decreased (miR-16, miR-22, miR-23, miR-24, miR-26a, miR-29, miR-30, miR-31, miR-33, miR-122a, miR-126, miR-127, miR-145, miR-150 and miR-378); however, the expression of certain miRNAs increases (miR-21, miR-26b, miR-192, miR-194, MiR34a, miR-122, miR-203 and miR-221), thus affecting the hepatocyte cell cycle[131]. Table 2 summarizes the significant miRNAs in liver regeneration after PH.
| miRNA | Expression change | Target genes | Influence on LR |
| miR-21 | Increased | Rhob, Sox7, Crebl2, Bcl-2, Btg2, Timp3, Reck, Pdcd4, Tgfbi, Smad7, PTEN | Induction |
| miR -19a | Increased | PTEN | Induction |
| miR-214 | Increased | PTEN | Induction |
| miR-203 | Increased | SOCS3 | Induction |
| miR-27a/b | Increased | Tmub1 | Induction |
| miR-503 | Decreased | Cyclin D1, Cyclin E2, CDC25A, CDKN1B, CHK1 | Suppression |
| miR-23a | Decreased | TNF-α, c-Myc CCNL2, HNF4G MET | Suppression |
| miR-150 | Decreased | TNF-α, survivin, FoxP1, c-Myb | Suppression |
| miR-663 | Decreased | TGF-β1, AP-1, Jun-B, Jun-D | Suppression |
| miR-378 | Decreased | Odc1 | Suppression |
| miR-34a | Decreased | INHBB | Suppression |
| miR-33 | Decreased | CDK6, EEF1A1, RAP2A | Suppression |
| miR-26a | Decreased | MAP3K2, MXI1, SENP5, CCND2, CCNE2 | Suppression |
Castro et al[132] demonstrated the crucial role of miRNAs in liver regeneration after PH. Thus, it was demonstrated that the expression of 26 different miRNAs changes during regeneration, notably in both increasing and decreasing ways. The expressions of miR-19a, -21, and-214 were significantly increased. MiR-21 transcription is activated by activator protein 1 (AP-1), which is also required for the activation of the important Stat3 and TGF-β signaling pathways[132]. Ng et al[133] pointed out the regulatory role of miR-21 in hepatocyte cell cycle events preceding the S phase via the indirect induction of cyclin D1 translation, which occurs due to a reduction in cell cycle inhibitor expression. MiR-21 has a binding site on Ras homolog gene family member B, whose expression leads to the suppression of Akt1 activation, thus regulating cyclin D1 expression via mTORC1[133]. Additionally, miR-21 plays a significant role in decreasing phosphatase and tensin homolog expression, resulting in increased Akt and mTOR activities[134]. MiR-203 induces liver regeneration via IL-6/STAT3 signaling enhancement and SOCS3 expression inhibition[135]. MiR-27a/b regulates hepatocyte proliferation during regeneration because it suppresses Tmub1 expression[136], which suppresses the IL-6/STAT3 signaling pathway[137].
The decreased expression occurs within molecules such as miRs-503, -23a, -150, -663, -654 and is associated with their negative influence on liver regeneration. Thus, miR-150 inhibits TNF-α expression, which is essential for liver regeneration[138]. Increased miR-503 expression leads to the enhancement of essential cell cycle gene expression, including that of cyclin D1, E1, E2, F, Wee1, CDC25A and CHK1[139]. The AP-1 transcription factors, including the Jun and Fos family members, are the target genes of miR-663[140]. The c-Jun/AP-1 signaling pathway controls hepatocyte proliferation and has antiapoptotic activity via p-53-dependent pathway suppression[141]. An important negative regulator of hepatocyte epithelial-mesenchymal transition is miR-378, whose expression is decreased by Smo during liver regeneration, resulting in Hh-pathway activation and the transdifferentiation of hepatocytes and BECs into myofibroblasts[142]. MiR-34a expression is significantly decreased during the first days after PH, whereas the expression of its target genes (Notch1, Notch 4 and Hes1) is increased, leading to hepatocyte differentiation and growth enhancement[143]. MiRs inhibiting liver regeneration are also important because they prevent excessive regeneration. Among these molecules, for example, miR-33 suppresses CDK6 and CCND1[144], and miR-26a targets CCND2 and CCNE2[145].
A further understanding of the miRNAs involved in normal and progenitor- dependent liver regeneration can improve the use of miRNAs for the diagnosis of different liver diseases, control the adequacy of liver regeneration and act as a potential therapy for insufficient liver regeneration.
Therapeutic methods for insufficient liver regeneration treatment are lacking, although many studies have focused on the efficiency of various molecules in promoting liver regeneration. Shi et al[146] determined that baicalin can stimulate liver regeneration after acetaminophen-induced acute liver injury in mice via inducing hepatocyte proliferating cell nuclear antigen, increasing cyclin D1 expression and Nrf2 cytosolic accumulation, and enhancing IL-18 Levels, leading to the upregulation of hepatocyte proliferation. So et al[147] showed the promotion of liver regeneration after the inhibition of EGFR or MEK/extracellular signal-regulated kinase (ERK) and the genetic suppression of the EGFR-ERK-SOX9 axis via inducing HPC-to-hepatocyte differentiation in zebrafish. The research of Xiang et al[148] noted the therapeutic effect of IL-22Fc in inducing liver regeneration in acute-on-chronic liver failure patients due to the shift from anti-regenerative IFN-γ/STAT1 to the pro-regenerative IL-6/STAT3 pathway. Li et al[149] reported that aldose reductase (AR) is a new potential therapeutic target for enhancing normal and fatty liver regeneration after surgery and IRI because the knockout of AR leads to enhanced oxisome proliferator activated receptor-α and oxisome proliferator activated receptor-γ expression, thus improving energy metabolism in the liver. The research of Loforese et al[150] revealed that the inhibition of MST1 and MST2 with si-RNA resulted in improved hepatocyte proliferation in aged mice after PH; therefore, Ste20-like kinases 1/2 may be a potential therapeutic target. Many other molecules and molecular pathways have been shown to enhance liver regeneration in experimental models. Further studies would help implicate the potential therapy in the clinic and improve the survival of patients with different liver diseases in the near future.
Mesenchymal stem cells (MSCs) have a self-renewal capacity and are derived from the bone marrow, adipose tissue, umbilical cord, etc. They are the subject of focus in regenerative medicine and serve as a potential therapy for different liver diseases[151,152]. MSCs were shown to improve liver regeneration in patients with cirrhosis by elevating anti-apoptotic factors, such as HGF and IGF-1, and angiogenic and mitogenic factors. In acute liver failure animal models, MSCs have been shown to promote liver regeneration mostly by suppressing the oxidative stress and inflammation via reducing TNF-α, IFN-γ and IL-4 Levels and stimulating liver regeneration with various released factors such as PGE2 and delta-like 4[153,154]. MSCs can also stimulate liver regeneration after PH by upregulating hepatic cell proliferation and downregulating fat accumulation and HGF, IL-6, IL-10 and TNF-α serum levels[155].
Further studies in this field can help determine how to prevent hepatic failure after surgical interventions and acute and chronic injuries via improving liver regeneration.
Liver regeneration is driven by multiple molecular processes. Biomolecular factors permit the possibility of targeted therapy to prevent serious complications, such as liver failure due to a decreased cellular regenerative potential.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Russia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Bhat M, Kaul R, Youness RA S-Editor: Zhang L L-Editor: A P-Editor: Wang LL
| 1. | van Gulik TM, van Gulik MM, Koning HH. [Prometheus and liver regeneration: the dissection of a myth]. Ned Tijdschr Geneeskd. 2018;162. [PubMed] |
| 2. | Chen KY, Shen X, Diehl AM. Prometheus revisited. J Clin Invest. 2018;128:2192-2193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 3. | Tao Y, Wang M, Chen E, Tang H. Liver Regeneration: Analysis of the Main Relevant Signaling Molecules. Mediators Inflamm. 2017;2017:4256352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 4. | Manco R, Leclercq IA, Clerbaux LA. Liver Regeneration: Different Sub-Populations of Parenchymal Cells at Play Choreographed by an Injury-Specific Microenvironment. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 5. | Michalopoulos GK. Principles of liver regeneration and growth homeostasis. Compr Physiol. 2013;3:485-513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 174] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 6. | Duncan AW, Soto-Gutierrez A. Liver repopulation and regeneration: new approaches to old questions. Curr Opin Organ Transplant. 2013;18:197-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 7. | Mao SA, Glorioso JM, Nyberg SL. Liver regeneration. Transl Res. 2014;163:352-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 8. | Abu Rmilah A, Zhou W, Nelson E, Lin L, Amiot B, Nyberg SL. Understanding the marvels behind liver regeneration. Wiley Interdiscip Rev Dev Biol. 2019;8:e340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 9. | Gilgenkrantz H, Collin de l'Hortet A. Understanding Liver Regeneration: From Mechanisms to Regenerative Medicine. Am J Pathol. 2018;188:1316-1327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 113] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 10. | López-Luque J, Fabregat I. Revisiting the liver: from development to regeneration - what we ought to know! Int J Dev Biol. 2018;62:441-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Wirth KM, Kizy S, Steer CJ. Liver Regeneration in the Acute Liver Failure Patient. Clin Liver Dis. 2018;22:269-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Kholodenko IV, Yarygin KN. Cellular Mechanisms of Liver Regeneration and Cell-Based Therapies of Liver Diseases. Biomed Res Int. 2017;2017:8910821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 13. | Bhat M, Pasini E, Baciu C, Angeli M, Humar A, Macparland S, Feld J, McGilvray I. The basis of liver regeneration: A systems biology approach. Ann Hepatol. 2019;18:422-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 14. | Drixler TA, Vogten JM, Gebbink MF, Carmeliet P, Voest EE, Borel Rinkes IH. Plasminogen mediates liver regeneration and angiogenesis after experimental partial hepatectomy. Br J Surg. 2003;90:1384-1390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176:2-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 578] [Cited by in RCA: 569] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 16. | Min JS, DeAngelis RA, Reis ES, Gupta S, Maurya MR, Evans C, Das A, Burant C, Lambris JD, Subramaniam S. Systems Analysis of the Complement-Induced Priming Phase of Liver Regeneration. J Immunol. 2016;197:2500-2508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 17. | Schmidt-Arras D, Rose-John S. IL-6 pathway in the liver: From physiopathology to therapy. J Hepatol. 2016;64:1403-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 460] [Cited by in RCA: 651] [Article Influence: 72.3] [Reference Citation Analysis (0)] |
| 18. | Fazel Modares N, Polz R, Haghighi F, Lamertz L, Behnke K, Zhuang Y, Kordes C, Häussinger D, Sorg UR, Pfeffer K, Floss DM, Moll JM, Piekorz RP, Ahmadian MR, Lang PA, Scheller J. IL-6 Trans-signaling Controls Liver Regeneration After Partial Hepatectomy. Hepatology. 2019;70:2075-2091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 85] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 19. | Gupta P, Venugopal SK. Augmenter of liver regeneration: A key protein in liver regeneration and pathophysiology. Hepatol Res. 2018;48:587-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 20. | Ibrahim S, Weiss TS. Augmenter of liver regeneration: Essential for growth and beyond. Cytokine Growth Factor Rev. 2019;45:65-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 21. | Nalesnik MA, Gandhi CR, Starzl TE. Augmenter of liver regeneration: A fundamental life protein. Hepatology. 2017;66:266-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 22. | Araújo TG, de Oliveira AG, Tobar N, Saad MJ, Moreira LR, Reis ER, Nicola EM, de Jorge GL, dos Tártaro RR, Boin IF, Teixeira AR. Liver regeneration following partial hepatectomy is improved by enhancing the HGF/Met axis and Akt and Erk pathways after low-power laser irradiation in rats. Lasers Med Sci. 2013;28:1511-1517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 23. | Fajardo-Puerta AB, Mato Prado M, Frampton AE, Jiao LR. Gene of the month: HGF. J Clin Pathol. 2016;69:575-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 24. | Berasain C, Avila MA. The EGFR signalling system in the liver: from hepatoprotection to hepatocarcinogenesis. J Gastroenterol. 2014;49:9-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 130] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 25. | Natarajan A, Wagner B, Sibilia M. The EGF receptor is required for efficient liver regeneration. Proc Natl Acad Sci. 2007;104:17081-17086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 245] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 26. | Morales-González Á, Bautista M, Madrigal-Santillán E, Posadas-Mondragón A, Anguiano-Robledo L, Madrigal-Bujaidar E, Álvarez-González I, Fregoso-Aguilar T, Gayosso-Islas E, Sánchez-Moreno C, Morales-González JA. Nrf2 modulates cell proliferation and antioxidants defenses during liver regeneration induced by partial hepatectomy. Int J Clin Exp Pathol. 2017;10:7801-7811. [PubMed] |
| 27. | Zou Y, Hu M, Lee J, Nambiar SM, Garcia V, Bao Q, Chan JY, Dai G. Nrf2 is essential for timely M phase entry of replicating hepatocytes during liver regeneration. Am J Physiol Gastrointest Liver Physiol. 2015;308:G262-G268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 28. | Zou Y, Lee J, Nambiar SM, Hu M, Rui W, Bao Q, Chan JY, Dai G. Nrf2 is involved in maintaining hepatocyte identity during liver regeneration. PLoS One. 2014;9:e107423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | Liu HX, Keane R, Sheng L, Wan YJ. Implications of microbiota and bile acid in liver injury and regeneration. J Hepatol. 2015;63:1502-1510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 30. | van de Laarschot LF, Jansen PL, Schaap FG, Olde Damink SW. The role of bile salts in liver regeneration. Hepatol Int. 2016;10:733-740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 31. | de Haan L, van der Lely SJ, Warps AK, Hofsink Q, Olthof PB, de Keijzer MJ, Lionarons DA, Mendes-Dias L, Bruinsma BG, Uygun K, Jaeschke H, Farrell GC, Teoh N, van Golen RF, Li T, Heger M. Post-hepatectomy liver regeneration in the context of bile acid homeostasis and the gut-liver signaling axis. J Clin Transl Res. 2018;4:1-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 32. | Alvarez-Sola G, Uriarte I, Latasa MU, Jimenez M, Barcena-Varela M, Santamaría E, Urtasun R, Rodriguez-Ortigosa C, Prieto J, Berraondo P, Fernandez-Barrena MG, Berasain C, Avila MA. Bile acids, FGF15/19 and liver regeneration: From mechanisms to clinical applications. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1326-1334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 33. | Kong B, Sun R, Huang M, Chow MD, Zhong XB, Xie W, Lee YH, Guo GL. Fibroblast Growth Factor 15-Dependent and Bile Acid-Independent Promotion of Liver Regeneration in Mice. Hepatology. 2018;68:1961-1976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 34. | Péan N, Doignon I, Garcin I, Besnard A, Julien B, Liu B, Branchereau S, Spraul A, Guettier C, Humbert L, Schoonjans K, Rainteau D, Tordjmann T. The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology. 2013;58:1451-1460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 35. | Keitel V, Häussinger D. Role of TGR5 (GPBAR1) in Liver Disease. Semin Liver Dis. 2018;38:333-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 36. | Merlen G, Ursic-Bedoya J, Jourdainne V, Kahale N, Glenisson M, Doignon I, Rainteau D, Tordjmann T. Bile acids and their receptors during liver regeneration: "Dangerous protectors". Mol Aspects Med. 2017;56:25-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 37. | Valizadeh A, Majidinia M, Samadi-Kafil H, Yousefi M, Yousefi B. The roles of signaling pathways in liver repair and regeneration. J Cell Physiol. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 38. | Russell JO, Monga SP. Wnt/β-Catenin Signaling in Liver Development, Homeostasis, and Pathobiology. Annu Rev Pathol. 2018;13:351-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 341] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 39. | Preziosi M, Okabe H, Poddar M, Singh S, Monga SP. Endothelial Wnts regulate β-catenin signaling in murine liver zonation and regeneration: A sequel to the Wnt-Wnt situation. Hepatol Commun. 2018;2:845-860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 108] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 40. | Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1167] [Cited by in RCA: 1352] [Article Influence: 112.7] [Reference Citation Analysis (0)] |
| 41. | Chapouly C, Guimbal S, Hollier PL, Renault MA. Role of Hedgehog Signaling in Vasculature Development, Differentiation, and Maintenance. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 42. | Qi X, Schmiege P, Coutavas E, Wang J, Li X. Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature. 2018;560:128-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 149] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 43. | Jiang K, Liu Y, Fan J, Zhang J, Li XA, Evers BM, Zhu H, Jia J. PI(4)P Promotes Phosphorylation and Conformational Change of Smoothened through Interaction with Its C-terminal Tail. PLoS Biol. 2016;14:e1002375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 44. | Hu A, Song BL. The interplay of Patched, Smoothened and cholesterol in Hedgehog signaling. Curr Opin Cell Biol. 2019;61:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 45. | Gao L, Zhang Z, Zhang P, Yu M, Yang T. Role of canonical Hedgehog signaling pathway in liver. Int J Biol Sci. 2018;14:1636-1644. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 46. | Ozretić P, Trnski D, Musani V, Maurac I, Kalafatić D, Orešković S, Levanat S, Sabol M. Non-canonical Hedgehog signaling activation in ovarian borderline tumors and ovarian carcinomas. Int J Oncol. 2017;51:1869-1877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 47. | Ho Wei L, Arastoo M, Georgiou I, Manning DR, Riobo-Del Galdo NA. Activation of the Gi protein-RHOA axis by non-canonical Hedgehog signaling is independent of primary cilia. PLoS One. 2018;13:e0203170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 48. | Ochoa B, Syn WK, Delgado I, Karaca GF, Jung Y, Wang J, Zubiaga AM, Fresnedo O, Omenetti A, Zdanowicz M, Choi SS, Diehl AM. Hedgehog signaling is critical for normal liver regeneration after partial hepatectomy in mice. Hepatology. 2010;51:1712-1723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 144] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 49. | Machado MV, Diehl AM. Hedgehog signalling in liver pathophysiology. J Hepatol. 2018;68:550-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 107] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 50. | Papa S, Bubici C. Feeding the Hedgehog: A new meaning for JNK signalling in liver regeneration. J Hepatol. 2018;69:572-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 51. | Langiewicz M, Schlegel A, Saponara E, Linecker M, Borger P, Graf R, Humar B, Clavien PA. Hedgehog pathway mediates early acceleration of liver regeneration induced by a novel two-staged hepatectomy in mice. J Hepatol. 2017;66:560-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 52. | Jalan-Sakrikar N, De Assuncao TM, Lu J, Almada LL, Lomberk G, Fernandez-Zapico ME, Urrutia R, Huebert RC. Hedgehog Signaling Overcomes an EZH2-Dependent Epigenetic Barrier to Promote Cholangiocyte Expansion. PLoS One. 2016;11:e0168266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 53. | Swiderska-Syn M, Xie G, Michelotti GA, Jewell ML, Premont RT, Syn WK, Diehl AM. Hedgehog regulates yes-associated protein 1 in regenerating mouse liver. Hepatology. 2016;64:232-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 99] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 54. | Du K, Hyun J, Premont RT, Choi SS, Michelotti GA, Swiderska-Syn M, Dalton GD, Thelen E, Rizi BS, Jung Y, Diehl AM. Hedgehog-YAP Signaling Pathway Regulates Glutaminolysis to Control Activation of Hepatic Stellate Cells. Gastroenterology 2018; 154: 1465-1479. e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 251] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 55. | Omenetti A, Choi S, Michelotti G, Diehl AM. Hedgehog signaling in the liver. J Hepatol. 2011;54:366-373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 232] [Cited by in RCA: 220] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 56. | Siebel C, Lendahl U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol Rev. 2017;97:1235-1294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 451] [Cited by in RCA: 704] [Article Influence: 88.0] [Reference Citation Analysis (0)] |
| 57. | Morell CM, Fiorotto R, Fabris L, Strazzabosco M. Notch signalling beyond liver development: emerging concepts in liver repair and oncogenesis. Clin Res Hepatol Gastroenterol. 2013;37:447-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 58. | Adams JM, Jafar-Nejad H. The Roles of Notch Signaling in Liver Development and Disease. Biomolecules. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 59. | Lu J, Zhou Y, Hu T, Zhang H, Shen M, Cheng P, Dai W, Wang F, Chen K, Zhang Y, Wang C, Li J, Zheng Y, Yang J, Zhu R, Wang J, Lu W, Xia Y, De Assuncao TM, Jalan-Sakrikar N, Huebert RC, Bin Zhou, Guo C. Notch Signaling Coordinates Progenitor Cell-Mediated Biliary Regeneration Following Partial Hepatectomy. Sci Rep. 2016;6:22754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 60. | Morell CM, Strazzabosco M. Notch signaling and new therapeutic options in liver disease. J Hepatol. 2014;60:885-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 61. | Ortica S, Tarantino N, Aulner N, Israël A, Gupta-Rossi N. The 4 Notch receptors play distinct and antagonistic roles in the proliferation and hepatocytic differentiation of liver progenitors. FASEB J. 2014;28:603-614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 62. | Zhang F, Zhang J, Li X, Li B, Tao K, Yue S. Notch signaling pathway regulates cell cycle in proliferating hepatocytes involved in liver regeneration. J Gastroenterol Hepatol. 2018;33:1538-1547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 63. | Yang X, He C, Zhu L, Zhao W, Li S, Xia C, Xu C. Comparative Analysis of Regulatory Role of Notch Signaling Pathway in 8 Types Liver Cell During Liver Regeneration. Biochem Genet. 2019;57:1-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 64. | Liu M, Chen P. Proliferationinhibiting pathways in liver regeneration (Review). Mol Med Rep. 2017;16:23-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 65. | Khan MGM, Ghosh A, Variya B, Santharam MA, Kandhi R, Ramanathan S, Ilangumaran S. Hepatocyte growth control by SOCS1 and SOCS3. Cytokine. 2019;121:154733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 66. | Pascale RM, Feo F, Calvisi DF. The complex role of bone morphogenetic protein 9 in liver damage and regeneration: New evidence from in vivo and in vitro studies. Liver Int. 2018;38:1547-1549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 67. | Addante A, Roncero C, Almalé L, Lazcanoiturburu N, García-Álvaro M, Fernández M, Sanz J, Hammad S, Nwosu ZC, Lee SJ, Fabregat I, Dooley S, Ten Dijke P, Herrera B, Sánchez A. Bone morphogenetic protein 9 as a key regulator of liver progenitor cells in DDC-induced cholestatic liver injury. Liver Int. 2018;38:1664-1675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 68. | Marí M, Morales A. Bone morphogenetic protein-9/activin-like kinase 1 axis a new target for hepatic regeneration and fibrosis treatment in liver injury. Hepatobiliary Surg Nutr. 2017;6:414-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 69. | Huck I, Gunewardena S, Espanol-Suner R, Willenbring H, Apte U. Hepatocyte Nuclear Factor 4 Alpha Activation Is Essential for Termination of Liver Regeneration in Mice. Hepatology. 2019;70:666-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 70. | Zhou J, Sun X, Yang L, Wang L, Ran G, Wang J, Cao Q, Wu L, Bryant A, Ling C, Pi L. Hepatocyte nuclear factor 4α negatively regulates connective tissue growth factor during liver regeneration. FASEB J. 2020;34:4970-4983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 71. | Wang P, Cong M, Liu T, Xu H, Wang L, Sun G, Yang A, Zhang D, Huang J, Sun Y, Zhao W, Ma H, Jia J, You H. Inhibitory effects of HNF4α on migration/maltransformation of hepatic progenitors: HNF4α-overexpressing hepatic progenitors for liver repopulation. Stem Cell Res Ther. 2017;8:183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 72. | Michalopoulos GK. Hepatostat: Liver regeneration and normal liver tissue maintenance. Hepatology. 2017;65:1384-1392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 318] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 73. | Konishi T, Schuster RM, Lentsch AB. Proliferation of hepatic stellate cells, mediated by YAP and TAZ, contributes to liver repair and regeneration after liver ischemia-reperfusion injury. Am J Physiol Gastrointest Liver Physiol. 2018;314:G471-G482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 74. | Lu L, Finegold MJ, Johnson RL. Hippo pathway coactivators Yap and Taz are required to coordinate mammalian liver regeneration. Exp Mol Med. 2018;50:e423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 127] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 75. | Moya IM, Halder G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat Rev Mol Cell Biol. 2019;20:211-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 664] [Article Influence: 110.7] [Reference Citation Analysis (0)] |
| 76. | Patel SH, Camargo FD, Yimlamai D. Hippo Signaling in the Liver Regulates Organ Size, Cell Fate, and Carcinogenesis. Gastroenterology. 2017;152:533-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 230] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 77. | Kietzmann T. Metabolic zonation of the liver: The oxygen gradient revisited. Redox Biol. 2017;11:622-630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 322] [Cited by in RCA: 349] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 78. | Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015;524:180-185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 600] [Cited by in RCA: 540] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 79. | Sun T, Pikiolek M, Orsini V, Bergling S, Holwerda S, Morelli L, Hoppe PS, Planas-Paz L, Yang Y, Ruffner H, Bouwmeester T, Lohmann F, Terracciano LM, Roma G, Cong F, Tchorz JS. AXIN2+ Pericentral Hepatocytes Have Limited Contributions to Liver Homeostasis and Regeneration. Cell Stem Cell 2020; 26: 97-107. e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 116] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 80. | Alison MR, Lin WR. Diverse routes to liver regeneration. J Pathol. 2016;238:371-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 81. | Pu W, Zhang H, Huang X, Tian X, He L, Wang Y, Zhang L, Liu Q, Li Y, Zhao H, Liu K, Lu J, Zhou Y, Huang P, Nie Y, Yan Y, Hui L, Lui KO, Zhou B. Mfsd2a+ hepatocytes repopulate the liver during injury and regeneration. Nat Commun. 2016;7:13369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 91] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 82. | Preziosi ME, Monga SP. Update on the Mechanisms of Liver Regeneration. Semin Liver Dis. 2017;37:141-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 83. | Li N, Hua J. Immune cells in liver regeneration. Oncotarget. 2017;8:3628-3639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 84. | Thorgersen EB, Barratt-Due A, Haugaa H, Harboe M, Pischke SE, Nilsson PH, Mollnes TE. The Role of Complement in Liver Injury, Regeneration, and Transplantation. Hepatology. 2019;70:725-736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 164] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 85. | Ridgway WM, Gershwin ME. Prometheus unbound: NKT cells inhibit hepatic regeneration. Hepatology. 2014;60:1133-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 86. | Bi J, Zheng X, Chen Y, Wei H, Sun R, Tian Z. TIGIT safeguards liver regeneration through regulating natural killer cell-hepatocyte crosstalk. Hepatology. 2014;60:1389-1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 87. | Goh YP, Henderson NC, Heredia JE, Red Eagle A, Odegaard JI, Lehwald N, Nguyen KD, Sheppard D, Mukundan L, Locksley RM, Chawla A. Eosinophils secrete IL-4 to facilitate liver regeneration. Proc Natl Acad Sci USA. 2013;110:9914-9919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 212] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 88. | Lua I, Asahina K. The Role of Mesothelial Cells in Liver Development, Injury, and Regeneration. Gut Liver. 2016;10:166-176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 89. | Senoo H, Mezaki Y, Fujiwara M. The stellate cell system (vitamin A-storing cell system). Anat Sci Int. 2017;92:387-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 74] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 90. | Yin C, Evason KJ, Asahina K, Stainier DY. Hepatic stellate cells in liver development, regeneration, and cancer. J Clin Invest. 2013;123:1902-1910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 439] [Cited by in RCA: 571] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 91. | Tsolaki E, Yannaki E. Stem cell-based regenerative opportunities for the liver: State of the art and beyond. World J Gastroenterol. 2015;21:12334-12350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 50] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 92. | Shang H, Wang Z, Song Y. Liver progenitor cells-mediated liver regeneration in liver cirrhosis. Hepatol Int. 2016;10:440-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 93. | Swiderska-Syn M, Syn WK, Xie G, Krüger L, Machado MV, Karaca G, Michelotti GA, Choi SS, Premont RT, Diehl AM. Myofibroblastic cells function as progenitors to regenerate murine livers after partial hepatectomy. Gut. 2014;63:1333-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 93] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 94. | Zhu C, Coombe DR, Zheng MH, Yeoh GC, Li L. Liver progenitor cell interactions with the extracellular matrix. J Tissue Eng Regen Med. 2013;7:757-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 95. | Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere JP, Schwabe RF. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 750] [Cited by in RCA: 1052] [Article Influence: 95.6] [Reference Citation Analysis (0)] |
| 96. | Schaub JR, Malato Y, Gormond C, Willenbring H. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 2014;8:933-939. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 218] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 97. | Kordes C, Sawitza I, Götze S, Häussinger D. Stellate cells from rat pancreas are stem cells and can contribute to liver regeneration. PLoS One. 2012;7:e51878. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 98. | Mabuchi A, Mullaney I, Sheard PW, Hessian PA, Mallard BL, Tawadrous MN, Zimmermann A, Senoo H, Wheatley AM. Role of hepatic stellate cell/hepatocyte interaction and activation of hepatic stellate cells in the early phase of liver regeneration in the rat. J Hepatol. 2004;40:910-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 99. | Aimaiti Y, Jin X, Shao Y, Wang W, Li D. Hepatic stellate cells regulate hepatic progenitor cells differentiation via the TGF-β1/Jagged1 signaling axis. J Cell Physiol. 2019;234:9283-9296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 100. | Saito Y, Morine Y, Shimada M. Mechanism of impairment on liver regeneration in elderly patients: Role of hepatic stellate cell function. Hepatol Res. 2017;47:505-513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 101. | Takahashi K, Liang C, Oda T, Ohkohchi N. Platelet and liver regeneration after liver surgery. Surg Today. 2020;50:974-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (1)] |
| 102. | Meyer J, Lejmi E, Fontana P, Morel P, Gonelle-Gispert C, Bühler L. A focus on the role of platelets in liver regeneration: Do platelet-endothelial cell interactions initiate the regenerative process? J Hepatol. 2015;63:1263-1271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 103. | Starlinger P, Pereyra D, Haegele S, Braeuer P, Oehlberger L, Primavesi F, Kohler A, Offensperger F, Reiberger T, Ferlitsch A, Messner B, Beldi G, Staettner S, Brostjan C, Gruenberger T. Perioperative von Willebrand factor dynamics are associated with liver regeneration and predict outcome after liver resection. Hepatology. 2018;67:1516-1530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 104. | Lisman T, Porte RJ. Mechanisms of platelet-mediated liver regeneration. Blood. 2016;128:625-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 105. | Kurokawa T, Ohkohchi N. Platelets in liver disease, cancer and regeneration. World J Gastroenterol. 2017;23:3228-3239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 62] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 106. | Aryal B, Yamakuchi M, Shimizu T, Kadono J, Furoi A, Gejima K, Komokata T, Hashiguchi T, Imoto Y. Therapeutic implication of platelets in liver regeneration -hopes and hues. Expert Rev Gastroenterol Hepatol. 2018;12:1219-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 107. | Forbes SJ, Newsome PN. Liver regeneration - mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol. 2016;13:473-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 294] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 108. | Itoh T. Stem/progenitor cells in liver regeneration. Hepatology. 2016;64:663-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 109. | Van Haele M, Roskams T. Hepatic Progenitor Cells: An Update. Gastroenterol Clin North Am. 2017;46:409-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 110. | Dan YY. Chasing the facultative liver progenitor cell. Hepatology. 2016;64:297-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 111. | Raven A, Forbes SJ. Hepatic progenitors in liver regeneration. J Hepatol. 2018;69:1394-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 112. | Lu WY, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T, Guest RV, Wojtacha D, Man TY, Mackinnon A, Ridgway RA, Kendall T, Williams MJ, Jamieson T, Raven A, Hay DC, Iredale JP, Clarke AR, Sansom OJ, Forbes SJ. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol. 2015;17:971-983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 356] [Cited by in RCA: 372] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 113. | Alwahsh SM, Rashidi H, Hay DC. Liver cell therapy: is this the end of the beginning? Cell Mol Life Sci. 2018;75:1307-1324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 114. | Habeeb MA, Vishwakarma SK, Bardia A, Khan AA. Hepatic stem cells: A viable approach for the treatment of liver cirrhosis. World J Stem Cells. 2015;7:859-865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 115. | Kakinuma S, Nakauchi H, Watanabe M. Hepatic stem/progenitor cells and stem-cell transplantation for the treatment of liver disease. J Gastroenterol. 2009;44:167-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 116. | Miyajima A, Tanaka M, Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell. 2014;14:561-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 412] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 117. | Michalopoulos GK, Khan Z. Liver Stem Cells: Experimental Findings and Implications for Human Liver Disease. Gastroenterology. 2015;149:876-882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 118. | Best J, Manka P, Syn WK, Dollé L, van Grunsven LA, Canbay A. Role of liver progenitors in liver regeneration. Hepatobiliary Surg Nutr. 2015;4:48-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 119. | Tanaka M, Itoh T, Tanimizu N, Miyajima A. Liver stem/progenitor cells: their characteristics and regulatory mechanisms. J Biochem. 2011;149:231-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 120. | Dwyer BJ, Olynyk JK, Ramm GA, Tirnitz-Parker JE. TWEAK and LTβ Signaling during Chronic Liver Disease. Front Immunol. 2014;5:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 121. | di Bello G, Vendemiale G, Bellanti F. Redox cell signaling and hepatic progenitor cells. Eur J Cell Biol. 2018;97:546-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 122. | Liu W, Wang Y, Sun Y, Wu Y, Ma Q, Shi Y, He R, Zhang T, Ma Y, Zuo W, Wu Z. Clonal expansion of hepatic progenitor cells and differentiation into hepatocyte-like cells. Dev Growth Differ. 2019;61:203-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 123. | Kitade M, Kaji K, Yoshiji H. Relationship between hepatic progenitor cell-mediated liver regeneration and non-parenchymal cells. Hepatol Res. 2016;46:1187-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 124. | Mao Y, Tang S, Yang L, Li K. Inhibition of the Notch Signaling Pathway Reduces the Differentiation of Hepatic Progenitor Cells into Cholangiocytes in Biliary Atresia. Cell Physiol Biochem. 2018;49:1074-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 125. | Hu C, Liang X, Fang S, Xu L, Gong M, Wang Y, Bi Y, Hong S, He Y. ATRA induces the differentiation of hepatic progenitor cells by upregulating microRNA-200a. In Vitro Cell Dev Biol Anim. 2019;55:713-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 126. | Ma Z, Li F, Chen L, Gu T, Zhang Q, Qu Y, Xu M, Cai X, Lu L. Autophagy promotes hepatic differentiation of hepatic progenitor cells by regulating the Wnt/β-catenin signaling pathway. J Mol Histol. 2019;50:75-90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 127. | Zeng J, Jing Y, Shi R, Pan X, Lai F, Liu W, Li R, Gao L, Hou X, Wu M, Wei L. Autophagy regulates biliary differentiation of hepatic progenitor cells through Notch1 signaling pathway. Cell Cycle. 2016;15:1602-1610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 128. | Overi D, Carpino G, Cardinale V, Franchitto A, Safarikia S, Onori P, Alvaro D, Gaudio E. Contribution of Resident Stem Cells to Liver and Biliary Tree Regeneration in Human Diseases. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 129. | Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 853] [Cited by in RCA: 1814] [Article Influence: 226.8] [Reference Citation Analysis (0)] |
| 130. | Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 640] [Cited by in RCA: 852] [Article Influence: 85.2] [Reference Citation Analysis (0)] |
| 131. | Chen X, Zhao Y, Wang F, Bei Y, Xiao J, Yang C. MicroRNAs in Liver Regeneration. Cell Physiol Biochem. 2015;37:615-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 132. | Castro RE, Ferreira DM, Zhang X, Borralho PM, Sarver AL, Zeng Y, Steer CJ, Kren BT, Rodrigues CM. Identification of microRNAs during rat liver regeneration after partial hepatectomy and modulation by ursodeoxycholic acid. Am J Physiol Gastrointest Liver Physiol. 2010;299:G887-G897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 80] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 133. | Ng R, Song G, Roll GR, Frandsen NM, Willenbring H. A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J Clin Invest. 2012;122:1097-1108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 130] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 134. | Chen X, Song M, Chen W, Dimitrova-Shumkovska J, Zhao Y, Cao Y, Song Y, Yang W, Wang F, Xiang Y, Yang C. MicroRNA-21 Contributes to Liver Regeneration by Targeting PTEN. Med Sci Monit. 2016;22:83-91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 135. | Chen XB, Zheng XB, Cai ZX, Lin XJ, Xu MQ. MicroRNA-203 promotes liver regeneration after partial hepatectomy in cirrhotic rats. J Surg Res. 2017;211:53-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 136. | Lan X, Li G, Liu H, Fu H, Chen P, Liu M. MiR-27a/b Regulates Liver Regeneration by Posttranscriptional Modification of Tmub1. Dig Dis Sci. 2018;63:2362-2372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 137. | Fu H, Dong R, Zhang Y, Xu J, Liu M, Chen P. Tmub1 negatively regulates liver regeneration via inhibiting STAT3 phosphorylation. Cell Signal. 2019;55:65-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 138. | Yi PS, Zhang M, Xu MQ. Role of microRNA in liver regeneration. Hepatobiliary Pancreat Dis Int. 2016;15:141-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 139. | Salehi S, Brereton HC, Arno MJ, Darling D, Quaglia A, O'Grady J, Heaton N, Aluvihare VR. Human liver regeneration is characterized by the coordinated expression of distinct microRNA governing cell cycle fate. Am J Transplant. 2013;13:1282-1295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 140. | Tili E, Michaille JJ, Adair B, Alder H, Limagne E, Taccioli C, Ferracin M, Delmas D, Latruffe N, Croce CM. Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting JunB and JunD. Carcinogenesis. 2010;31:1561-1566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 199] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 141. | Stepniak E, Ricci R, Eferl R, Sumara G, Sumara I, Rath M, Hui L, Wagner EF. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev. 2006;20:2306-2314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 186] [Cited by in RCA: 184] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 142. | Kim J, Hyun J, Wang S, Lee C, Jung Y. MicroRNA-378 is involved in hedgehog-driven epithelial-to-mesenchymal transition in hepatocytes of regenerating liver. Cell Death Dis. 2018;9:721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 143. | Wang XP, Zhou J, Han M, Chen CB, Zheng YT, He XS, Yuan XP. MicroRNA-34a regulates liver regeneration and the development of liver cancer in rats by targeting Notch signaling pathway. Oncotarget. 2017;8:13264-13276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 144. | Cirera-Salinas D, Pauta M, Allen RM, Salerno AG, Ramírez CM, Chamorro-Jorganes A, Wanschel AC, Lasuncion MA, Morales-Ruiz M, Suarez Y, Baldan Á, Esplugues E, Fernández-Hernando C. Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle. 2012;11:922-933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 141] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 145. | Zhou J, Ju W, Wang D, Wu L, Zhu X, Guo Z, He X. Down-regulation of microRNA-26a promotes mouse hepatocyte proliferation during liver regeneration. PLoS One. 2012;7:e33577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 146. | Shi L, Zhang S, Huang Z, Hu F, Zhang T, Wei M, Bai Q, Lu B, Ji L. Baicalin promotes liver regeneration after acetaminophen-induced liver injury by inducing NLRP3 inflammasome activation. Free Radic Biol Med. 2020;160:163-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 147. | So J, Kim M, Lee SH, Ko S, Lee DA, Park H, Azuma M, Parsons MJ, Prober D, Shin D. Attenuating the EGFR-ERK-SOX9 axis promotes liver progenitor cell-mediated liver regeneration in zebrafish. Hepatology. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 148. | Xiang X, Feng D, Hwang S, Ren T, Wang X, Trojnar E, Matyas C, Mo R, Shang D, He Y, Seo W, Shah VH, Pacher P, Xie Q, Gao B. Interleukin-22 ameliorates acute-on-chronic liver failure by reprogramming impaired regeneration pathways in mice. J Hepatol. 2020;72:736-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 141] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 149. | Li CX, Wang HW, Jiang WJ, Li GC, Zhang YD, Luo CH, Li XC. The Inhibition of Aldose Reductase Accelerates Liver Regeneration through Regulating Energy Metabolism. Oxid Med Cell Longev. 2020;2020:3076131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 150. | Loforese G, Malinka T, Keogh A, Baier F, Simillion C, Montani M, Halazonetis TD, Candinas D, Stroka D. Impaired liver regeneration in aged mice can be rescued by silencing Hippo core kinases MST1 and MST2. EMBO Mol Med. 2017;9:46-60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 151. | Varderidou-Minasian S, Lorenowicz MJ. Mesenchymal stromal/stem cell-derived extracellular vesicles in tissue repair: challenges and opportunities. Theranostics. 2020;10:5979-5997. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 191] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 152. | Hu C, Zhao L, Zhang L, Bao Q, Li L. Mesenchymal stem cell-based cell-free strategies: safe and effective treatments for liver injury. Stem Cell Res Ther. 2020;11:377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 153. | Zhang S, Yang Y, Fan L, Zhang F, Li L. The clinical application of mesenchymal stem cells in liver disease: the current situation and potential future. Ann Transl Med. 2020;8:565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 154. | Hu C, Zhao L, Wu Z, Li L. Transplantation of mesenchymal stem cells and their derivatives effectively promotes liver regeneration to attenuate acetaminophen-induced liver injury. Stem Cell Res Ther. 2020;11:88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 155. | Hu C, Wu Z, Li L. Mesenchymal stromal cells promote liver regeneration through regulation of immune cells. Int J Biol Sci. 2020;16:893-903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 104] [Article Influence: 20.8] [Reference Citation Analysis (0)] |