Published online Nov 27, 2021. doi: 10.4254/wjh.v13.i11.1568
Peer-review started: March 10, 2021
First decision: May 2, 2021
Revised: May 12, 2021
Accepted: September 8, 2021
Article in press: September 8, 2021
Published online: November 27, 2021
Processing time: 259 Days and 0.5 Hours
Liver cancer is the sixth most commonly occurring cancer and costs millions of lives per year. The diagnosis of hepatocellular carcinoma (HCC) has relied on scanning techniques and serum-based markers such as α-fetoprotein. These measures have limitations due to their detection limits and asymptomatic conditions during the early stages, resulting in late-stage cancer diagnosis where targeted chemotherapy or systemic treatment with sorafenib is offered. However, the aid of conventional therapy for patients in the advanced stage of HCC has limited outcomes. Thus, it is essential to seek a new treatment strategy and improve the diagnostic techniques to manage the disease. Researchers have used the omics profile of HCC patients for sub-classification of tissues into different groups, which has helped us with prognosis. Despite these efforts, a promising target for treatment has not been identified. The hurdle in this situation is genetic and epigenetic variations in the tumor, leading to disparities in response to treatment. Understanding reversible epigenetic changes along with clinical traits help to define new markers for patient categorization and design personalized therapy. Many clinical trials of inhibitors of epigenetic modifiers (also known as epi-drugs) are in progress. Epi-drugs like azacytidine or belinostat are already approved for other cancer treatments. Furthermore, epigenetic changes have also been observed in drug-resistant HCC tumors. In such cases, combinatorial treatment of epi-drugs with systemic therapy or trans-arterial chemoembolization might re-sensitize resistant cells.
Core Tip: This review article focuses on the limitations of diagnosis and treatment of hepatocellular carcinoma (HCC). Furthermore, the use of omics technology with clinical attributes for categorizing HCC patients in order that personalized treatment can be designed to prolong survival is discussed. Finally, the potential of epi-drugs in targeting epigenetic changes in the disease and resistance has been proposed.
- Citation: Natu A, Singh A, Gupta S. Hepatocellular carcinoma: Understanding molecular mechanisms for defining potential clinical modalities. World J Hepatol 2021; 13(11): 1568-1583
- URL: https://www.wjgnet.com/1948-5182/full/v13/i11/1568.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i11.1568
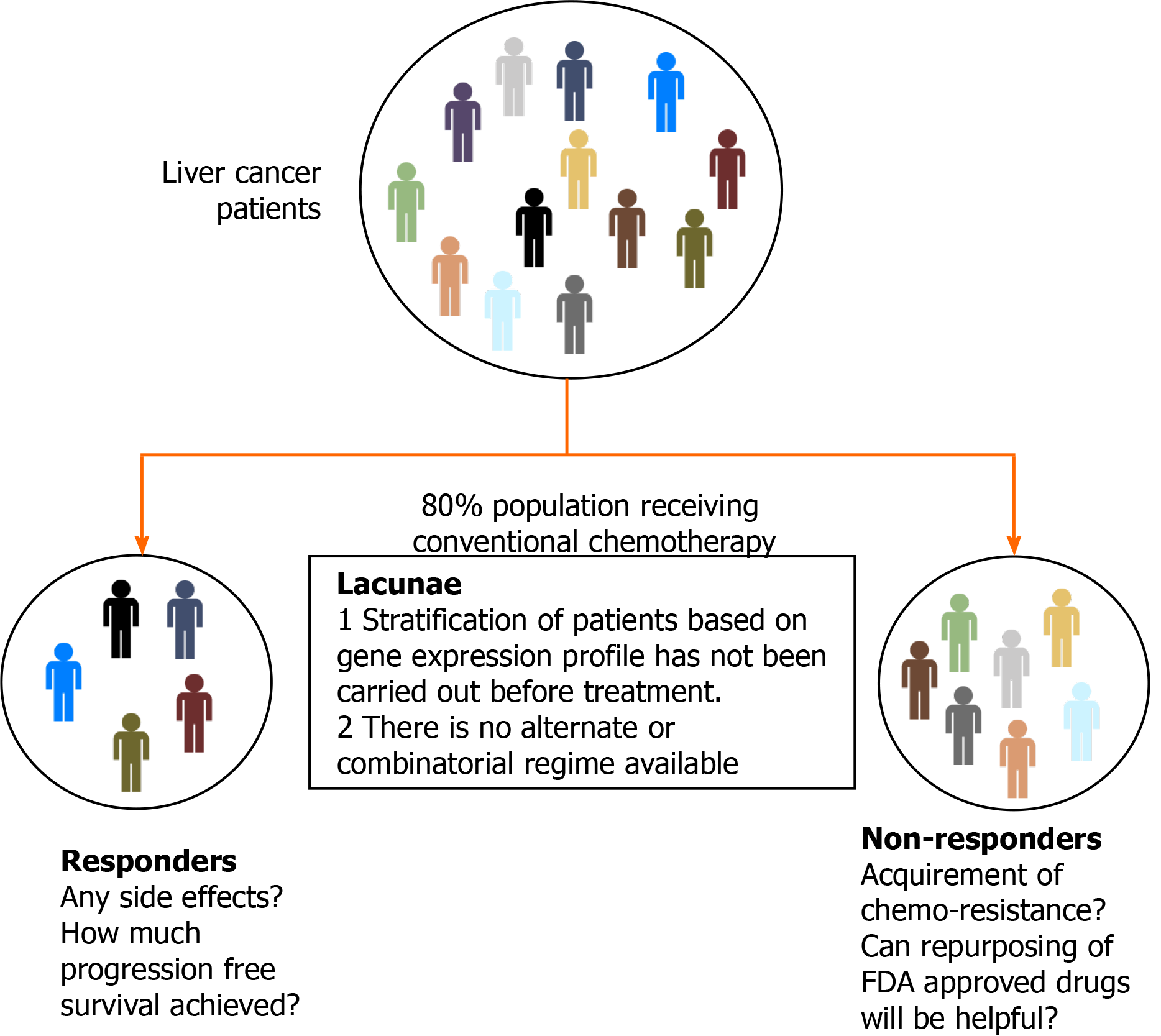
Liver cancer ranks sixth in cancer incidence globally and accounts for 8.2% of total cancer deaths. The different categories of primary liver cancer are intrahepatic cholangiocarcinoma, hepatocellular carcinoma (HCC), fibrolamellar carcinoma, and hepatoblastoma. These categories have distinct changes in their molecular, histological, and pathological features. HCC alone accounts for 85%-90% of liver cancer cases[1]. Almost 2/3 of the population affected by HCC is found in east Asian and south-east Asian countries, making this disease endemic to the region[2]. Globally, 5-year median survival is below 20% for HCC[3]. Major risk factors for HCC include chronic infection with hepatitis B virus and hepatitis C virus, excessive consumption of alcohol, exposure to aflatoxin, physiological state such as non-alcoholic fatty liver disease, and diabetes[4]. According to the Barcelona Clinic Cancer Liver Classification (BCLC) algorithm, curative care for HCC involves tumor resection, ablation, and liver transplantation[5]. However, this mode of treatment is offered to patients diagnosed in an early stage of the disease. Current research suggests that only 20% of patients are diagnosed in the early stage[6]. The lacunae in diagnosis are the unavailability of promising liquid-based biomarkers and detection limits of scanning techniques. Palliative care involving chemo/radiation-based treatment is given to patients with intermediate and advanced stage disease. Following this, 70% of patients come back with a relapse of disease and suffer treatment side effects[7,8].
A new approach should be considered to identify diagnostic markers and achieve better therapy response to overcome disease management challenges. Recent advances in the omics field shed light on the pathogenesis and molecular classification of HCC[9-11]. The omics approach can help to investigate new markers to improve the therapeutic outcome. Liver carcinogenesis involves both genetic and epigenetic changes. It is impossible to target all genetic variations due to tumor heterogeneity, but gene signature can be manipulated as epigenetic changes are reversible[12]. Therefore, epi-drug-based treatment may act as an alternate treatment strategy instead of targeting a single protein or molecular pathway. Epi-drugs can be beneficial not only for the treatment of HCC but also for dealing with cancer resistance[13,14].
This article focuses on the existing approach for diagnosis and treatment in the management of HCC. We also review transcriptomic-based signatures of HCC for patient sub-categorization and their potential implications for diagnosis and therapy. Finally, we propose an epi-drug based treatment strategy based on the epigenetic landscape of HCC.
Five standard WHO-approved guidelines include the European Association for the Study of Liver Disease (EASL)[15], American Association for the Study of Liver Diseases (AASLD)[16], Asia-Pacific Association Study of the Liver[17], EASL-EORTC Clinical Practice Guidelines[18], and the updated AASLD guidelines are used for diagnosis of liver cancer. The diagnosis is primarily based on imaging techniques such as ultrasound, computed tomography (CT) scan, and conventional magnetic resonance imaging (MRI)[19]. Invasive biopsies are not helpful for the diagnosis of liver tumors. The myriad risk factors involved in biopsy are the local spread of HCC along the needle track and different complications observed in individual patients[20]. The early-stage diagnosis of HCC continues to be crucial due to reduced sensitivity and specificity of the diagnostic methods, due to which an ample number of tumors are undetected. The complete list of diagnostic methods with detection limits is shown in Table 1. The various factors responsible for undetectable tumors involve a lack of specific markers and asymptomatic condition during the early stages of HCC[21]. Thus, the diagnosis of tumor occurs when it has spread and has reached an advanced stage.
| Diagnostic methods | Definition/concept | Diagnostic limit/range | Ref. |
| Contrast-enhanced ultrasound | Inexpensive, non-invasive, first choice for screening HCC; Real time dynamic of blood supply. | Small HCC less than 1 cm | [101] |
| Multi phasic enhanced computed tomography | 3 dimensional reconstructions, high sensitivity | 1-2 cm HCC lesion | [102] |
| Magnetic resonance imaging | High resolution anatomic details, pre-contrast and multi-phasic enhanced 3D; Diffusion weighted imaging-functional imaging | 2-3 cm HCC lesion | [103] |
| Positron emission tomography | Hepatocyte-specific PET tracer, 2-[18F] fluoro-2-deoxy-D-galactose, is used which accumulates in the liver compared with other tissues | Detection of small intrahepatic; HCC lesions | [104] |
| AFP | Elevated in HCC, non-specific | Range: > 500 ng/mL | [23] |
| α-L-fucosidase | Expressed in liver cirrhosis | Cut-off: 870 nmol/L | [105] |
| Des-γ-carboxy prothrombin | Sensitive; Not expressed in other liver disease | Cut-off: 40 mAU/mL | [105,106] |
| HSP90α + AFP +TKI | Combination of markers have improved diagnostic value | HSP90- (76.65-144.00); AFP- (5.33-2000.00); TK1- (0.57-2.30) | [26] |
| AFP, GPC3, and GP73 | Useful markers for early diagnosis and prognosis | Upregulated | [27,107] |
| microRNA: miR-21, miR-199, and miR-122, miR-23a | Specific for diagnosis of HCC; Extremely sensitive | Cut-off value of ≥ 210 | [108,109] |
The diagnostic marker used most frequently is serum α-fetoprotein (AFP)[22]. AFP level increases beyond 20 ng/mL in more than 70% of patients with HCC. However, AFP elevations are not explicitly associated with HCC as AFP levels from 10-500 ng/mL and even occasionally to 1000 ng/mL may be seen in patients with a high degree of necro-inflammatory activity such as chronic viral hepatitis[23]. Chan et al[24] in 2008 have shown that AFP could be better used as a prognostic marker to evaluate response to treatment and detection of recurrence instead of diagnosis[25]. Studies have shown that multiple combinations of markers provide more appropriate results in diagnosis than a single marker. A recent study investigated the use of HSP90α (heat shock protein 90) combined with AFP and thymidine kinase 1 to diagnose HCC with more efficiency[26]. A study from Beijing YouAn Hospital found that for early diagnosis of HBV-related HCC, a combination of AFP, GPC3, and GP73 had the highest diagnostic value[27]. Ghosh et al[28] have shown that the exosome encap
Another marker, α-L-fucosidase (AFU), is expressed in liver cirrhosis patients[29]. However, limited research is available regarding the utility of AFU in the diagnosis of HCC. In the liver and gallbladder, cell membrane protein 5’-nucleotidase (5’-NT) is released into the blood during hepatic injury or obstruction[30]. It has been observed that 5’-NT levels also increase with age and during pregnancy[31]. Other markers such as AFP-L3, glypican-3, and des-γ-carboxy prothrombin also show inconsistent data due to low sensitivity and specificity. Hence, the discovery of putative liquid biomarkers is required, which can associate with tumor progression, recurrence, and effectiveness of therapeutic programs.
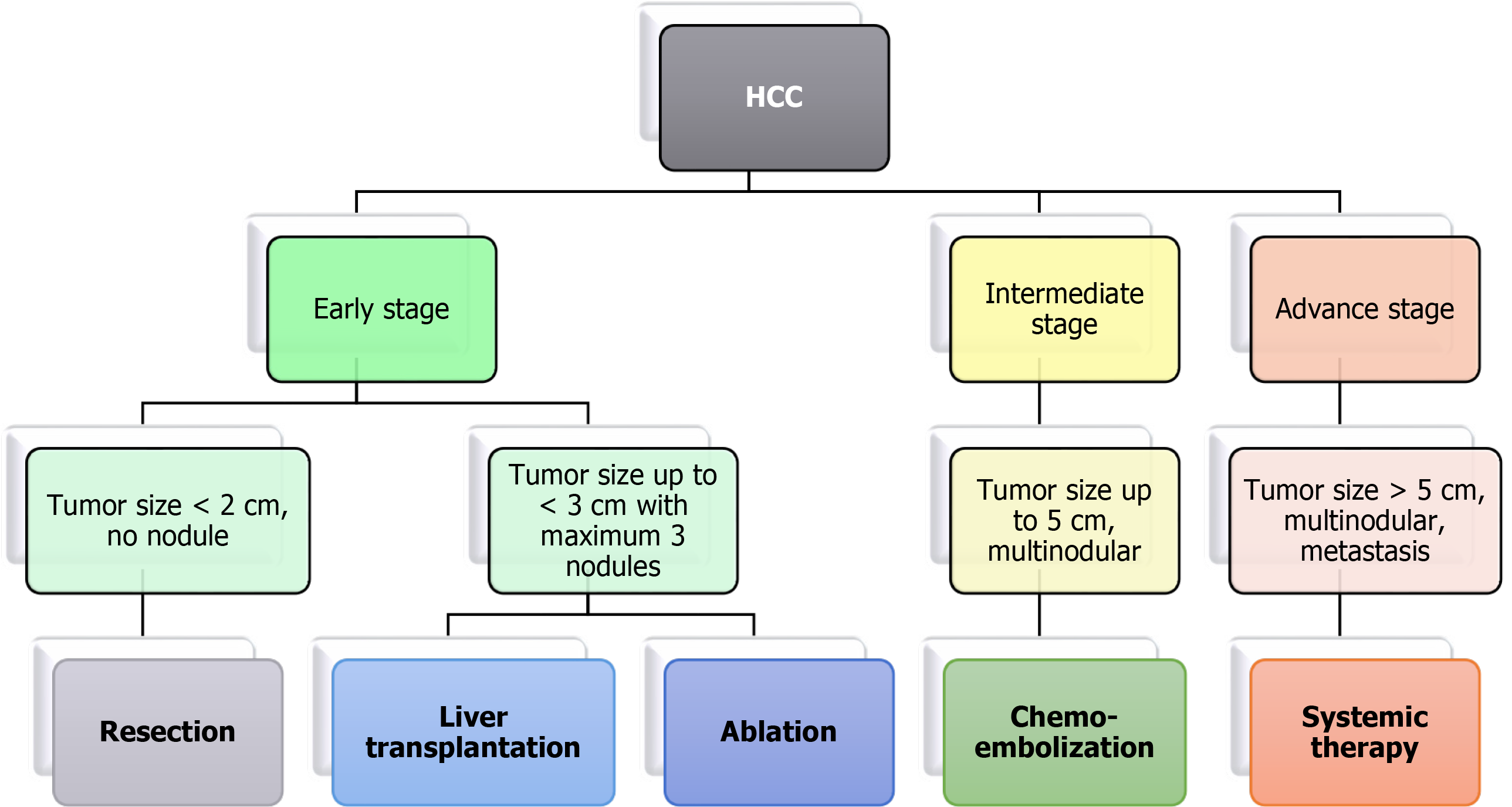
Treatment of HCC is decided based on different stages of tumor detection[32]. The BCLC algorithm is widely used for treatment as it considers tumor stage, liver function, performance status, and treatment impact (Figure 1). Early-stage cases are treated with surgery, ablation, or liver transplantation. The patients undergoing surgery showed 70% recurrence within five years[33]. The currently used methods for tumor ablation in HCC are percutaneous ethanol injection (PEI) and radiofrequency ablation (RFA). PEI consists of the direct injection of absolute ethanol into HCC nodules[34]. RFA is responsive in tumors > 4 cm in size. It involves necrosis of the tumor using a needle tip electrode that reaches temperatures up to 100°C[35]. Microwave ablation and irreversible electroporation have shown more promising results than tumor removal with PEI[36].
Patients with an intermediate stage having a tumor size greater than 5 cm or multinodular HCC with no vascular invasion are treated with trans-arterial chemoembolization (TACE). TACE is used to obstruct the nutrient supply to the tumor using the occlusion of arterial blood vessels[37]. Chemotherapeutic drugs such as doxorubicin or cisplatin are given during embolization, allowing prolonged exposure of the drug to tumor cells, resulting in tumor reduction. Yeo et al[38] showed that the overall response rate for doxorubicin-treated patients was 10.5%. Moreover, doxorubicin alone and combined with PIAF had no significant difference in response rate but showed treatment-associated toxicity in patients. Another study showed that combinatorial treatment of fluorouracil, leucovorin, and oxaliplatin failed to improve survival compared to doxorubicin[39]. In a multicohort study involving patients with unresectable tumors treated with TACE, overall survival (OS) was approximately 26-40 mo, with only 52% of patients achieving treatment benefits[40,41]. In some cases, selective internal radiation therapy is used in patients with intermediate-stage HCC. Intraarterial infusion of radioisotope labeled microspheres is carried out in this modality. Another radiation-based technique known as stereotactic body radiation is used for patients with > 3 cm of the tumor.
Systemic chemotherapy is given for advanced stages of HCC. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) have recommended sorafenib and lenvatinib as first-line systemic therapy for patients with unresectable HCC[42]. Brivanib, sunitinib, erlotinib, and regorafenib are other preferred drugs for late-stage HCC treatment. Kudo et al[43] observed that treatment with lenvatinib results in significantly higher OS than sorafenib and improvement in all secondary efficacy endpoints. This trial further results in FDA approval of lenvatinib as the first line of therapy for HCC[43]. Sorafenib and sunitinib are protein kinase inhibitors targeting VEGFR, PDGFR, and the Raf kinase pathway. However, a study suggested that sunitinib had an adverse effect in these patients and had no advantage over sorafenib[44]. Moreover, sorafenib has been extensively explored in the systemic treatment of advanced stage HCC and combination with TACE, but it provided contradictory results[45,46]. Brivanib is an inhibitor of FGF1 and VEGFR2. Phase II clinical trials of brivanib showed the ineffectiveness of the drug compared to sorafenib for improving OS[47,48]. The EGFR inhibitor erlotinib or cetuximab was administered in phase II clinical trials of advanced stages of HCC. However, the trial results did not show the anti-tumor effect of cetuximab in HCC patients[49]. Interestingly, erlotinib showed a positive response in treatment by increasing OS to 13 mo and a response rate of 59%[50].
As discussed earlier, ablation treatment is possible in less than 40% of patients due to late diagnosis, and only 20% are treated with TACE. For the patients with advanced stages of HCC, treatment modalities are limited to systemic therapy, and response rates are also significantly less due to resistance towards available chemotherapy. Multimodal treatment involving more than one therapeutic drug has also failed in different combinations due to cytotoxicity and poor trial outcomes. Despite the significant research in targeted therapy of HCC management, a promising drug is yet to be identified. Thus, the hunt for combinatorial treatment with different therapeutic agents continues (Figure 2).
Over the past years, HCC classification has mainly focused on histological analysis of tumor tissues. However, the molecular profile and clinical attributes have a significant impact on the prognosis of the disease, thereby redefining HCC into several subgroups. Boyault et al[51] published molecular classification systems for HCC composed of 6 groups. The groups were based on mutation profile, disease prognosis, and transcription landscape. The first group included patients with hepatitis B infection and low viral load, increased AFP levels, and high IGF2 expression, whereas the second group included patients with a high viral titer and associated microva
Tumor morphology-based classification has been proposed by Murakata et al[53]. The nodal status of the tumor was correlated with survival and recurrence of the disease. Moreover, the miRNA profile of HCC patients has been used to classify sorafenib responders[54]. c-myc signaling and EB-1 protein were functionally linked with HCC[55]. Similar findings were observed by Lee et al[56] in progenitor-like HCC, which correlated with poor prognosis. In another study, HCC progenitor-like signature consisting of CK-19, Ep-CAM, and CD133 was seen by Woo et al[57]. Morofuji et al[58] identified the gene signature of early recurrent HCC, including ERK1, PKG, Apaf1, and Bcl-X. Furthermore, ERK1 and Bcl-X were identified as genes associated with the poor prognosis of HCC[58]. However, these studies did not consider the survival status of an individual while proposing subtypes.
Jiang et al[59] showed that heterogeneity exists in proteomic profiling of paired early-stage HCC patients. The tumors were segregated into three subtypes: S-I, S-II, and S-III. S-I tumors had increased expression of liver-associated functional proteins. In contrast, S-II and S-III had a more proliferative nature due to overexpression of cell-cycle-related proteins. Furthermore, S-III were more aggressive and had a high expression of KRT19 and MMP9, associated with poor prognosis. Gao et al[60] sub-grouped 159 HBV infected patients based on survival, tumor thrombus, and multi-omics profile. These sub-groups were classified based on metabolic rewiring, alterations in the microenvironment, and cellular proliferation. Moreover, the study proposed two prognostic markers PYCR2 and ADH1A.
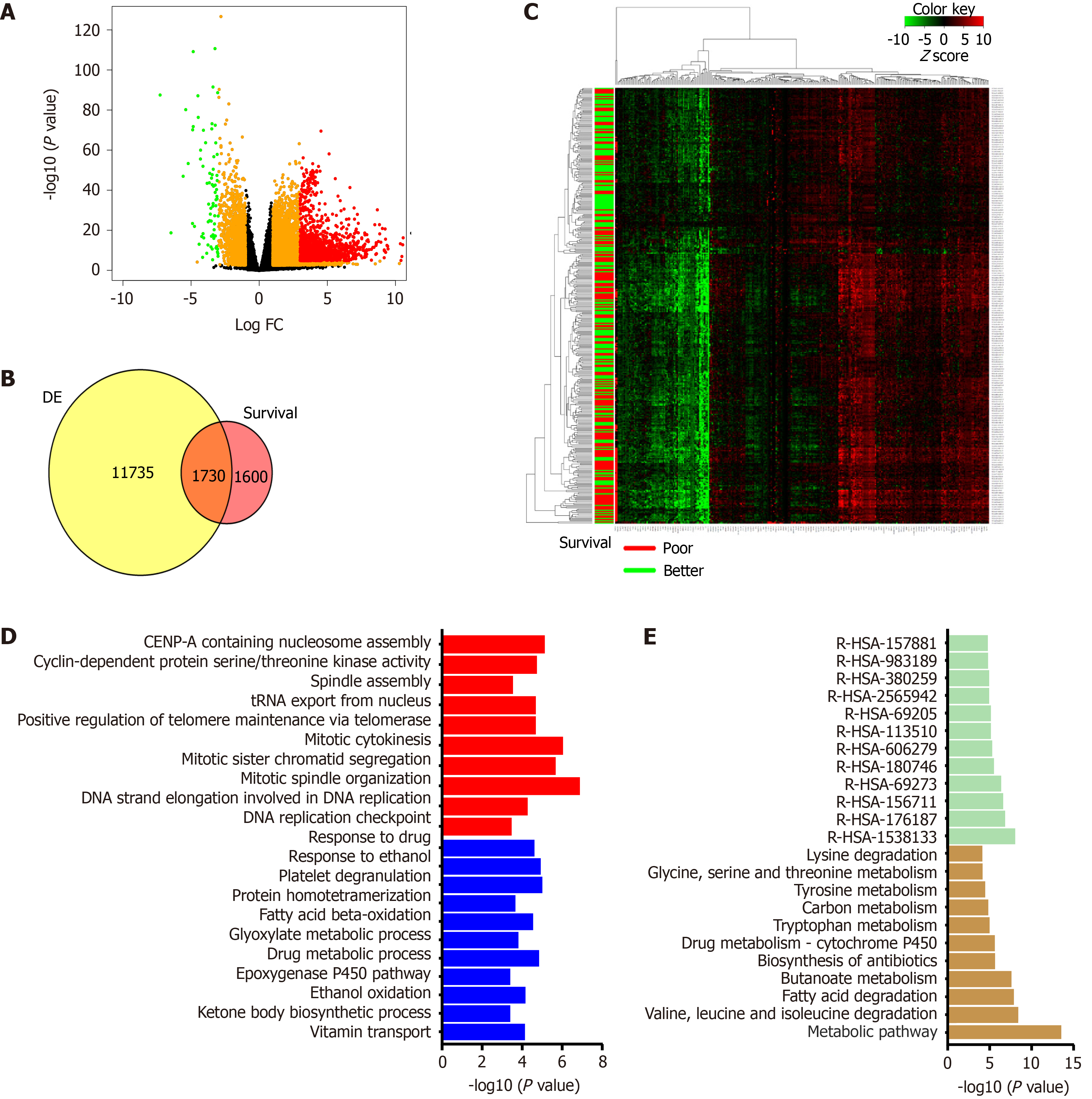
In the past decade, data generated under the TCGA consortium can be used to understand the gene expression profile of patients and obtain correlations with clinical attributes[9]. Machine learning algorithms are necessary to analyze such multivariate data. The molecular alterations obtained from the cancer genome atlas liver hepatocellular carcinoma (TCGA-LIHC) cohort (423 patients) can be explored to predict new targets and rationalize the combinatorial therapy. Transcriptome data generated from TCGA-LIHC identified over 13000 differentially expressed genes compared to cut-margin samples, and around 3330 genes correlated with poor survival (P value < 0.05). Furthermore, 1730 genes overlapped between the DE gene list and genes correlated with patient survival. The majority of overlapped genes showed more than 30% alteration compared to adjacent normal in this cohort and had a significant association with OS. Patients were categorized into different groups using clustering analysis of gene expression. It was observed that these genes belong to metabolism-related pathways and the cellular proliferation-related family (Figure 3). Deep learning computational framework on the TCGA-LIHC dataset suggested that aggressive subtype has TP53 inactivation with high expression of KRT10, EPCAM, and active AKT, WNT signaling[61]. Furthermore, drugs and small molecular compounds are available to target these genes. Schulze et al[62] reported that potential gene targets have FDA-approved drugs in 28% of liver tumors. Therefore, these genes can be used for prognosis of the disease, and targeting them may improve patient survival.
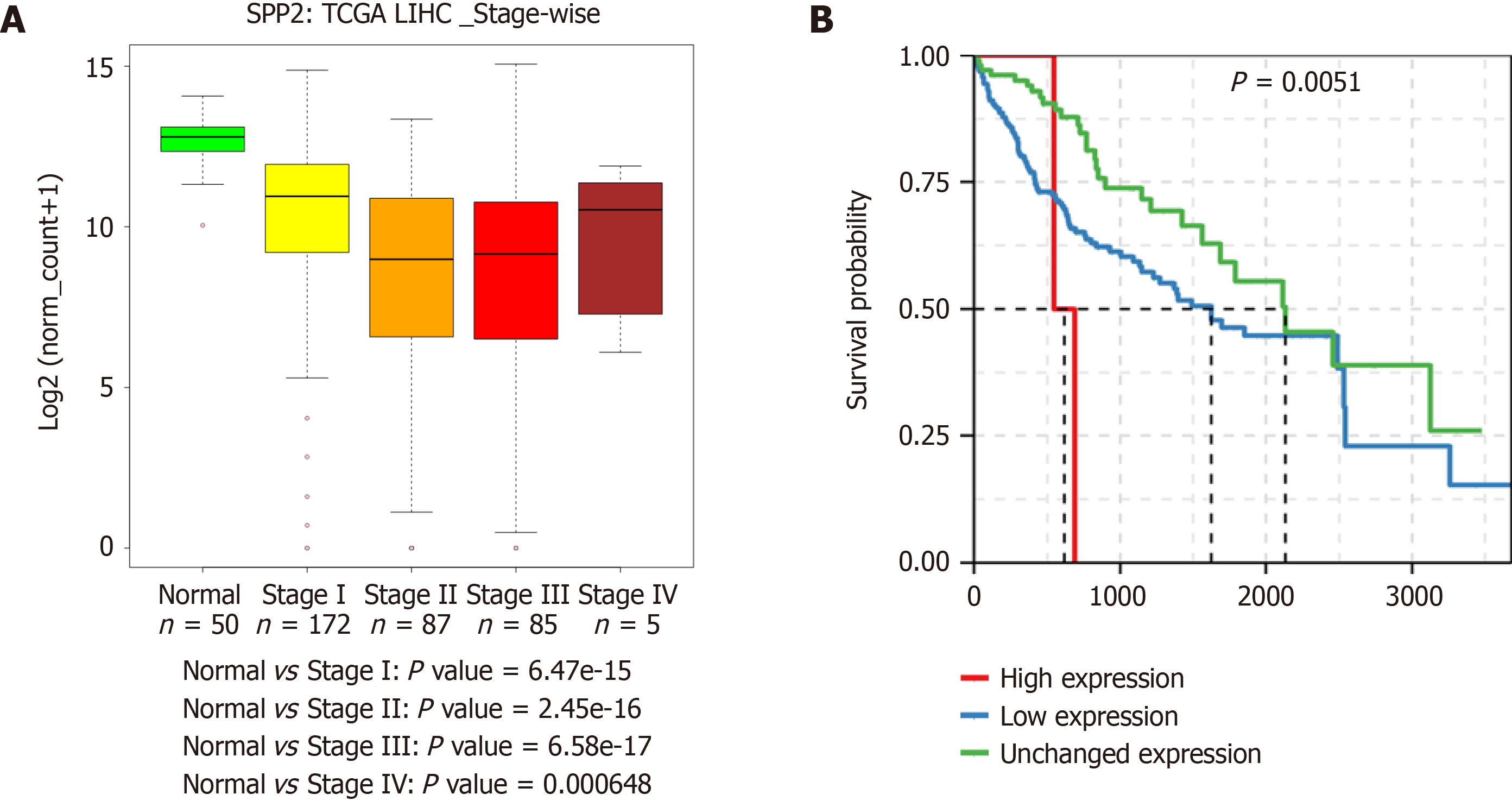
Gene expression analysis of liver cancer samples can also be utilized to identify new markers for diagnostic purposes. For example, SPP2 is downregulated at the transcript level in HCC. This gene is deregulated in multiple HCC cohorts. Moreover, a stage-wise decrease at the transcript level was observed in HCC TCGA data. Also, the downregulation of SPP2 leads to a significant decrease in patient survival (Figure 4). This observation indicates that SPP2 level is associated with normal liver function, and a change in levels can be a measure of liver carcinogenesis.
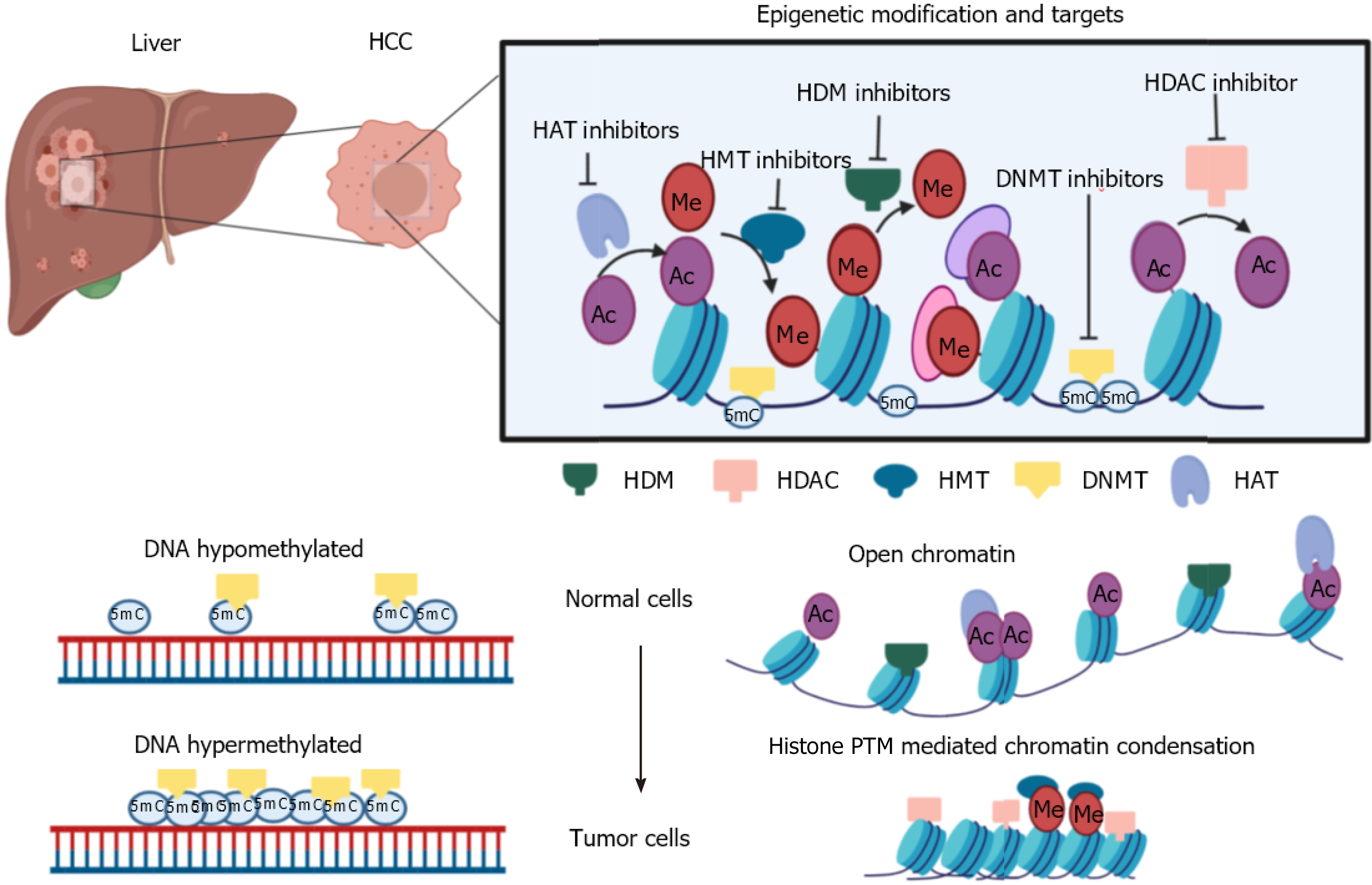
The lack of success in disease management can be explained by the multifactorial nature of carcinogenesis involving multiple mutations and global level epigenome alterations[63-65]. Epigenetic changes being reversible can be useful to understand the relationship between tumor biology and help in redefining therapeutic response[12]. Epigenetics deals with changes in gene expression without change in the DNA sequences[66]. Despite all cells having the same DNA sequence, the epigenome decides cell fate regarding differentiation, cell proliferation, and cell death[67,68]. The widely studied epigenetic marks are DNA methylation, histone post-translational modifications, and non-coding RNAs. DNA methylation is the most characterized heritable epigenetic mark. This is where a methyl group is transferred onto the cytosine of the CpG di-nucleotide-rich region in DNA by DNMT enzymes[69]. DNA methylation plays a vital role in gene inactivation, genomic imprinting, attaining tissue-specific gene expression, and X chromosome inactivation[69].
Similar to DNA modification, histone proteins also undergo post-translational modifications carried out by chromatin modifiers, namely writers, readers, and erasers[70]. The well-studied modifications include methylation, acetylation, phospho
Different research groups have extensively studied the epigenetic landscape of liver carcinogenesis. Moreover, in the past few years, researchers are investigating the epigenetic basis of chemoresistance in HCC. Lie et al[74] showed that lysine-specific demethylase 1 (LSD1) is upregulated in LGR5+ cells contributing to stemness and chemoresistance properties. Mechanistically, LSD1 removes the H3K4 methylation mark from the promoter of genes which inhibit Wnt-signaling. Thus, promoting pathway activation, which is essential for stemness and chemoresistance[74]. EpCAM+ liver cancer cells have high expression of chromodomain helicase DNA binding protein (CHD4), a DNA damage response protein. The abundance of CHD4 in liver cancer cells leads to epirubicin resistance[75]. Zinc-fingers and homeoboxes 2 (ZHX2) is one of the signature proteins which is downregulated in liver CSCs and is associated with tumor progression. It has been found that low expression of ZHX2 is correlated with epigenetic regulation of OCT4, SOX4, and NANOG by H3K36 methylation[76]. Oriana Lo Re et al[77] observed that low expression of MacroH2A1 leads to paracrine mediated chemoresistance and imparts CSCs properties to the tumor cells. Another study showed that the regulator of chromosome condensation 2 promotes metastasis and cisplatin resistance in HCC[78]. Ling et al[79] discovered that USP22 helps to attain chemoresistance by hypoxia-driven p53 mutant tumors. Hypoxia-induced expression of carbonyl reductase 1 leading to chemoresistance in HCC was observed by Tak et al[80]. H19 long non-coding (lnc)RNA has been shown to sensitize sorafenib or doxorubicin-resistant liver cancer cells[81]. The lncRNA CRNDE has been shown to interact with histone methyltransferase to enhance their effect on the inhibition of tumor suppressors and induce resistance in tumor cells[82].
Epigenetic alterations can be targeted by the class of small-molecule inhibitors that specifically inhibit or reverse the changes[83]. This class of inhibitors are referred to as epi-drugs. Different research groups have synthesized epi-drugs for all three prominent families of epigenetic modifiers- readers, writers, and erasers. Many epi-drugs have cleared pre-clinical trials, and initial phase trials have shown promising results. Few epi-drugs are clinically approved for the treatment of hematological malignancies. In some studies, treatment of solid tumors with an epi-drug helps in sensitizing tumor cells to chemotherapy[84,85]. These findings have promoted the research on inhibitors of HDAC, HAT, and DNMTs in combination with chemotherapeutic drugs. In HCC and gastric cancer, the inactive or suppressed state of tumor suppressor genes (TSGs) is mainly attributed to the overexpression of DNMTs and HDACs, leading to heterochromatinization. Reversion of the chromatin state using epi-drugs further leads to activation of TSGs and prevents tumor growth[86]. Ongoing pre-clinical trials have been carried out with HDAC and DNMT inhibitors in combination or in comparison with each other to study the anti-tumor effects of the drugs. Guadecitabine (SGI-110), a DNMT inhibitor with sorafenib and oxaliplatin, is in phase II clinical trials for HCC (NCT01752933). Multicenter phase I/II clinical trials using belinostat (HDAC inhibitor) in patients with unresectable HCC showed a tumor stabilization effect[87]. One study showed that the combination of panobinostat and sorafenib significantly decreased tumor volume by inducing apoptosis in the tumor[88]. A group of researchers observed that the DNMT inhibitor 5’-aza-2’ deoxycytidine and HDAC inhibitor SAHA down-regulated DNMT1, DNMT3a, DNMT3b, and HDAC1 and upregulated GSTP1 and SOCS1 gene expression, which further resulted in inhibition of cell viability and induced apoptosis[89]. A detailed list of potential epi-drugs is given in Table 2. These findings indicate the ability of epi-drugs, which can restructure the treatment strategy for HCC.
| Drugs | Classification | Approved year | Indicated disease | Reference/ clinical trial number |
| Azacytidine | DNMT inhibitor | 2004 | MDS | NCT01186939 |
| 2009 | AML | NCT00887068 | ||
| Decitabine | DNMT inhibitor | 2006 | MDS | NCT01751867 |
| 2011 | AML | NCT00260832 | ||
| Vorinostat | HDAC inhibitor | 2006 | CTCL | NCT00773747 |
| Romidepsin | HDAC inhibitor | 2009 | TCL | NCT02296398 |
| Belinostat | HDAC inhibitor | 2015 | PTCL | NCT01839097 |
| Panobinostat | HDAC inhibitor | 2015 | MM | NCT01023308 |
| 2016 | CML | NCT00451035 | ||
| 2017 | TCL | NCT00490776 |
The most effective way of controlling HCC is preventing the disease by spreading knowledge of etiological agents and hepatitis B vaccination. An increase in surveillance is one of the strategies to achieve better survival. This practice helps in the early diagnosis of HCC, monitors progression-free survival, and improves quality of life. Diagnosis of HCC at an early stage is crucial in order to start treatment at the right time and improve patient survival. Due to the reduced sensitivity of current diagnostic techniques, ultrasound scanning of high-risk individuals should be carried out every three months. Although ultrasound is cost-effective compared to MRI and CT scans, there is scope for developing more advanced MRI or CT versions to detect small lesions in the liver. Similarly, there is a need for an appropriate combination of liquid biomarkers used for the investigation of liver carcinogenesis. From a treatment perspective, upon early diagnosis, liver transplantation is preferred over surgical removal or ablation as it is has less than 15% chance of recurrence[90].
The primary cause of treatment failure in cancer is resistance to available chemotherapy, which results in relapse. From heterogeneous tumors, cells respond to treatment differently, and a rare small percentage of cells found in the quiescent G0 state of the cell cycle can escape treatment. These cells are inherently resistant to chemotherapy and involved in relapse. Studies have shown that tumor cells maintain the drug-tolerant state via chromatin-mediated changes after drug treatment[13]. The drug-tolerant persister (DTP) stage is reversible; however, prolonged exposure to chemotherapeutic drugs results in stable drug resistance properties[91-93]. DTP cells have non-random differential gene expressions, implicating chromatin-mediated changes leading to hetero-chromatinization of the transposable elements such as LINE1[94]. Recent findings suggest that ablation of the DTP cell population with FDA-approved epi-drugs impedes the development of resistance and relapse[13,94]. Hangauer et al[95] have shown DTP cells dependence on mesenchymal state and GPX4 (lipid hydroperoxide) for survival. Furthermore, inhibition of GPX4 triggers cell death of DTP cells via the ferroptosis pathway, indicating ferroptosis is required for the survival of DTP cells[95]. Thus, targeting inherently resistant residual cells could be helpful in reducing relapse in patients. However, more research on the identification and characterization of DTP cells is required to choose the appropriate drug combination for treatment purposes.
Targeted drug delivery is the critical factor in improving treatment outcomes and reducing the drug's side effects. Currently, researchers are investigating nanoparticle-mediated drug delivery. In addition, modified liposomal formulation showed a successful therapeutic response in HCC due to tumor-directed delivery and low drug load in the system[96]. Albumin is also a suitable drug-carrier molecule. An albumin-tagged drug has more potent effects compared to the drug alone[97]. Other materials such as dendrimers, micelles, polysaccharides, and silica are also used as carrier molecules[98-100]. Still, the hunt for an effective delivery system continues for targeted delivery.
Existing diagnostic methods are inadequate for the early detection of HCC. Similarly, implemented treatment modalities are unsuccessful in improving the survival of patients and result in cytotoxicity in normal cells. The use of credible biomarkers in the prognosis of HCC is essential to reduce mortality due to the disease. In the future, clinicians should focus on patient stratification based on molecular signatures and decide the treatment strategy to achieve maximum therapy outcome. The development of a combinatorial regime consisting of epi-drugs is urgently needed to treat the tumor mass.
Natu A and Singh A thank ACTREC-TMC for the research fellowship. We thank Gupta laboratory members for valuable discussions and inputs.
Provenance and peer review: Invited article; Externally peer reviewed.
Specialty type: Oncology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zeng YY S-Editor: Wu YXJ L-Editor: Webster JR P-Editor: Li JH
| 1. | Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology. 2017;152:745-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 839] [Article Influence: 104.9] [Reference Citation Analysis (2)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55820] [Article Influence: 7974.3] [Reference Citation Analysis (132)] |
| 3. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11573] [Cited by in RCA: 13166] [Article Influence: 1880.9] [Reference Citation Analysis (4)] |
| 4. | Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2066] [Cited by in RCA: 3170] [Article Influence: 528.3] [Reference Citation Analysis (37)] |
| 5. | Llovet JM, Fuster J, Bruix J; Barcelona-Clínic Liver Cancer Group. The Barcelona approach: diagnosis, staging, and treatment of hepatocellular carcinoma. Liver Transpl. 2004;10:S115-S120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 500] [Cited by in RCA: 514] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 6. | Farinati F, Sergio A, Baldan A, Giacomin A, Di Nolfo MA, Del Poggio P, Benvegnu L, Rapaccini G, Zoli M, Borzio F, Giannini EG, Caturelli E, Trevisani F. Early and very early hepatocellular carcinoma: when and how much do staging and choice of treatment really matter? BMC Cancer. 2009;9:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 7. | Bruix J, Sala M, Llovet JM. Chemoembolization for hepatocellular carcinoma. Gastroenterology. 2004;127:S179-S188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 396] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 8. | Launay-Vacher V, Deray G. Hypertension and proteinuria: a class-effect of antiangiogenic therapies. Anticancer Drugs. 2009;20:81-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 9. | Cancer Genome Atlas Research Network. Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169:1327-1341.e23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1578] [Cited by in RCA: 1729] [Article Influence: 216.1] [Reference Citation Analysis (1)] |
| 10. | Walther Z, Jain D. Molecular pathology of hepatic neoplasms: classification and clinical significance. Patholog Res Int. 2011;2011:403929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Marquardt JU, Galle PR, Teufel A. Molecular diagnosis and therapy of hepatocellular carcinoma (HCC): an emerging field for advanced technologies. J Hepatol. 2012;56:267-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 139] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 12. | Brien GL, Valerio DG, Armstrong SA. Exploiting the Epigenome to Control Cancer-Promoting Gene-Expression Programs. Cancer Cell. 2016;29:464-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 115] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 13. | Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69-80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2190] [Cited by in RCA: 1953] [Article Influence: 130.2] [Reference Citation Analysis (0)] |
| 14. | Dalvi MP, Wang L, Zhong R, Kollipara RK, Park H, Bayo J, Yenerall P, Zhou Y, Timmons BC, Rodriguez-Canales J, Behrens C, Mino B, Villalobos P, Parra ER, Suraokar M, Pataer A, Swisher SG, Kalhor N, Bhanu NV, Garcia BA, Heymach JV, Coombes K, Xie Y, Girard L, Gazdar AF, Kittler R, Wistuba II, Minna JD, Martinez ED. Taxane-Platin-Resistant Lung Cancers Co-develop Hypersensitivity to JumonjiC Demethylase Inhibitors. Cell Rep. 2017;19:1669-1684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 15. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5593] [Cited by in RCA: 6058] [Article Influence: 865.4] [Reference Citation Analysis (3)] |
| 16. | Merriman RB, Tran TT. AASLD practice guidelines: The past, the present, and the future. Hepatology. 2016;63:31-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 17. | Omata M, Cheng AL, Kokudo N, Kudo M, Lee JM, Jia J, Tateishi R, Han KH, Chawla YK, Shiina S, Jafri W, Payawal DA, Ohki T, Ogasawara S, Chen PJ, Lesmana CRA, Lesmana LA, Gani RA, Obi S, Dokmeci AK, Sarin SK. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int. 2017;11:317-370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1628] [Cited by in RCA: 1643] [Article Influence: 205.4] [Reference Citation Analysis (0)] |
| 18. | European Association for the Study of the Liver; European Organisation for Research and Treatment of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4059] [Cited by in RCA: 4520] [Article Influence: 347.7] [Reference Citation Analysis (2)] |
| 19. | Bargellini I, Battaglia V, Bozzi E, Lauretti DL, Lorenzoni G, Bartolozzi C. Radiological diagnosis of hepatocellular carcinoma. J Hepatocell Carcinoma. 2014;1:137-148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Di Tommaso L, Spadaccini M, Donadon M, Personeni N, Elamin A, Aghemo A, Lleo A. Role of liver biopsy in hepatocellular carcinoma. World J Gastroenterol. 2019;25:6041-6052. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 106] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (4)] |
| 21. | Dimitroulis D, Damaskos C, Valsami S, Davakis S, Garmpis N, Spartalis E, Athanasiou A, Moris D, Sakellariou S, Kykalos S, Tsourouflis G, Garmpi A, Delladetsima I, Kontzoglou K, Kouraklis G. From diagnosis to treatment of hepatocellular carcinoma: An epidemic problem for both developed and developing world. World J Gastroenterol. 2017;23:5282-5294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 228] [Cited by in RCA: 229] [Article Influence: 28.6] [Reference Citation Analysis (4)] |
| 22. | Galle PR, Foerster F, Kudo M, Chan SL, Llovet JM, Qin S, Schelman WR, Chintharlapalli S, Abada PB, Sherman M, Zhu AX. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019;39:2214-2229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 397] [Article Influence: 66.2] [Reference Citation Analysis (0)] |
| 23. | Johnson PJ. The role of serum alpha-fetoprotein estimation in the diagnosis and management of hepatocellular carcinoma. Clin Liver Dis. 2001;5:145-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 265] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 24. | Chan SL, Mo FK, Johnson PJ, Hui EP, Ma BB, Ho WM, Lam KC, Chan AT, Mok TS, Yeo W. New utility of an old marker: serial alpha-fetoprotein measurement in predicting radiologic response and survival of patients with hepatocellular carcinoma undergoing systemic chemotherapy. J Clin Oncol. 2009;27:446-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 214] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 25. | Merani S, Majno P, Kneteman NM, Berney T, Morel P, Mentha G, Toso C. The impact of waiting list alpha-fetoprotein changes on the outcome of liver transplant for hepatocellular carcinoma. J Hepatol. 2011;55:814-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 134] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 26. | Tang Y, Li K, Cai Z, Xie Y, Tan X, Su C, Li J. HSP90α combined with AFP and TK1 improved the diagnostic value for hepatocellular carcinoma. Biomark Med. 2020;14:869-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | Wu M, Liu Z, Li X, Zhang A, Li N. Dynamic Changes in Serum Markers and Their Utility in the Early Diagnosis of All Stages of Hepatitis B-Associated Hepatocellular Carcinoma. Onco Targets Ther. 2020;13:827-840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 28. | Ghosh S, Bhowmik S, Majumdar S, Goswami A, Chakraborty J, Gupta S, Aggarwal S, Ray S, Chatterjee R, Bhattacharyya S, Dutta M, Datta S, Chowdhury A, Dhali GK, Banerjee S. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int J Cancer. 2020;147:2934-2947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 92] [Article Influence: 18.4] [Reference Citation Analysis (1)] |
| 29. | Fawzy Montaser M, Amin Sakr M, Omar Khalifa M. Alpha-L-fucosidase as a tumour marker of hepatocellular carcinoma. Arab J Gastroenterol. 2012;13:9-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 30. | Hyder MA, Hasan M, Mohieldein A. Comparative Study of 5'-Nucleotidase Test in Various Liver Diseases. J Clin Diagn Res. 2016;10:BC01-BC03. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 31. | Mikolasevic I, Filipec-Kanizaj T, Jakopcic I, Majurec I, Brncic-Fischer A, Sobocan N, Hrstic I, Stimac T, Stimac D, Milic S. Liver Disease During Pregnancy: A Challenging Clinical Issue. Med Sci Monit. 2018;24:4080-4090. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 32. | Bruix J, Sherman M; Practice Guidelines Committee, American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4333] [Cited by in RCA: 4507] [Article Influence: 225.4] [Reference Citation Analysis (0)] |
| 33. | Ishizawa T, Hasegawa K, Aoki T, Takahashi M, Inoue Y, Sano K, Imamura H, Sugawara Y, Kokudo N, Makuuchi M. Neither multiple tumors nor portal hypertension are surgical contraindications for hepatocellular carcinoma. Gastroenterology. 2008;134:1908-1916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 582] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 34. | Zuo CJ, Wang PJ, Shao CW, Wang MJ, Tian JM, Xiao Y, Ren FY, Hao XY, Yuan M. CT-guided percutaneous ethanol injection with disposable curved needle for treatment of malignant liver neoplasms and their metastases in retroperitoneal lymph nodes. World J Gastroenterol. 2004;10:58-61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 35. | Sharma A, Moore WH, Lanuti M, Shepard JA. How I do it: radiofrequency ablation and cryoablation of lung tumors. J Thorac Imaging. 2011;26:162-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 36. | Yu H, Burke CT. Comparison of percutaneous ablation technologies in the treatment of malignant liver tumors. Semin Intervent Radiol. 2014;31:129-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 37. | Lencioni R, Petruzzi P, Crocetti L. Chemoembolization of hepatocellular carcinoma. Semin Intervent Radiol. 2013;30:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 150] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 38. | Yeo W, Mok TS, Zee B, Leung TW, Lai PB, Lau WY, Koh J, Mo FK, Yu SC, Chan AT, Hui P, Ma B, Lam KC, Ho WM, Wong HT, Tang A, Johnson PJ. A randomized phase III study of doxorubicin vs cisplatin/interferon alpha-2b/doxorubicin/fluorouracil (PIAF) combination chemotherapy for unresectable hepatocellular carcinoma. J Natl Cancer Inst. 2005;97:1532-1538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 455] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 39. | Qin S, Bai Y, Lim HY, Thongprasert S, Chao Y, Fan J, Yang TS, Bhudhisawasdi V, Kang WK, Zhou Y, Lee JH, Sun Y. Randomized, multicenter, open-label study of oxaliplatin plus fluorouracil/Leucovorin vs doxorubicin as palliative chemotherapy in patients with advanced hepatocellular carcinoma from Asia. J Clin Oncol. 2013;31:3501-3508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 375] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 40. | Lencioni R, de Baere T, Soulen MC, Rilling WS, Geschwind JF. Lipiodol transarterial chemoembolization for hepatocellular carcinoma: A systematic review of efficacy and safety data. Hepatology. 2016;64:106-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 521] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 41. | Kudo M, Han G, Finn RS, Poon RT, Blanc JF, Yan L, Yang J, Lu L, Tak WY, Yu X, Lee JH, Lin SM, Wu C, Tanwandee T, Shao G, Walters IB, Dela Cruz C, Poulart V, Wang JH. Brivanib as adjuvant therapy to transarterial chemoembolization in patients with hepatocellular carcinoma: A randomized phase III trial. Hepatology. 2014;60:1697-1707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 285] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 42. | Benson AB, D'Angelica MI, Abbott DE, Abrams TA, Alberts SR, Anaya DA, Anders R, Are C, Brown D, Chang DT, Cloyd J, Covey AM, Hawkins W, Iyer R, Jacob R, Karachristos A, Kelley RK, Kim R, Palta M, Park JO, Sahai V, Schefter T, Sicklick JK, Singh G, Sohal D, Stein S, Tian GG, Vauthey JN, Venook AP, Hammond LJ, Darlow SD. Guidelines Insights: Hepatobiliary Cancers, Version 2.2019. J Natl Compr Canc Netw. 2019;17:302-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 202] [Article Influence: 40.4] [Reference Citation Analysis (1)] |
| 43. | Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, Baron A, Park JW, Han G, Jassem J, Blanc JF, Vogel A, Komov D, Evans TRJ, Lopez C, Dutcus C, Guo M, Saito K, Kraljevic S, Tamai T, Ren M, Cheng AL. Lenvatinib vs sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391:1163-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3128] [Cited by in RCA: 3824] [Article Influence: 546.3] [Reference Citation Analysis (1)] |
| 44. | Cheng AL, Kang YK, Lin DY, Park JW, Kudo M, Qin S, Chung HC, Song X, Xu J, Poggi G, Omata M, Pitman Lowenthal S, Lanzalone S, Yang L, Lechuga MJ, Raymond E. Sunitinib vs sorafenib in advanced hepatocellular cancer: results of a randomized phase III trial. J Clin Oncol. 2013;31:4067-4075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 523] [Cited by in RCA: 593] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 45. | Park JW, Kim YJ, Kim DY, Bae SH, Paik SW, Lee YJ, Kim HY, Lee HC, Han SY, Cheong JY, Kwon OS, Yeon JE, Kim BH, Hwang J. Sorafenib with or without concurrent transarterial chemoembolization in patients with advanced hepatocellular carcinoma: The phase III STAH trial. J Hepatol. 2019;70:684-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 150] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 46. | Zhao Y, Wang WJ, Guan S, Li HL, Xu RC, Wu JB, Liu JS, Li HP, Bai W, Yin ZX, Fan DM, Zhang ZL, Han GH. Sorafenib combined with transarterial chemoembolization for the treatment of advanced hepatocellular carcinoma: a large-scale multicenter study of 222 patients. Ann Oncol. 2013;24:1786-1792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 47. | Llovet JM, Decaens T, Raoul JL, Boucher E, Kudo M, Chang C, Kang YK, Assenat E, Lim HY, Boige V, Mathurin P, Fartoux L, Lin DY, Bruix J, Poon RT, Sherman M, Blanc JF, Finn RS, Tak WY, Chao Y, Ezzeddine R, Liu D, Walters I, Park JW. Brivanib in patients with advanced hepatocellular carcinoma who were intolerant to sorafenib or for whom sorafenib failed: results from the randomized phase III BRISK-PS study. J Clin Oncol. 2013;31:3509-3516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 485] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 48. | Johnson PJ, Qin S, Park JW, Poon RT, Raoul JL, Philip PA, Hsu CH, Hu TH, Heo J, Xu J, Lu L, Chao Y, Boucher E, Han KH, Paik SW, Robles-Aviña J, Kudo M, Yan L, Sobhonslidsuk A, Komov D, Decaens T, Tak WY, Jeng LB, Liu D, Ezzeddine R, Walters I, Cheng AL. Brivanib vs sorafenib as first-line therapy in patients with unresectable, advanced hepatocellular carcinoma: results from the randomized phase III BRISK-FL study. J Clin Oncol. 2013;31:3517-3524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 596] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 49. | Zhu AX, Stuart K, Blaszkowsky LS, Muzikansky A, Reitberg DP, Clark JW, Enzinger PC, Bhargava P, Meyerhardt JA, Horgan K, Fuchs CS, Ryan DP. Phase 2 study of cetuximab in patients with advanced hepatocellular carcinoma. Cancer. 2007;110:581-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 181] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 50. | Philip PA, Mahoney MR, Allmer C, Thomas J, Pitot HC, Kim G, Donehower RC, Fitch T, Picus J, Erlichman C. Phase II study of Erlotinib (OSI-774) in patients with advanced hepatocellular cancer. J Clin Oncol. 2005;23:6657-6663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 344] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 51. | Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, Jeannot E, Hérault A, Saric J, Belghiti J, Franco D, Bioulac-Sage P, Laurent-Puig P, Zucman-Rossi J. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 827] [Cited by in RCA: 926] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 52. | Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, Watanabe G, Gabriel S, Friedman SL, Kumada H, Llovet JM, Golub TR. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385-7392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 956] [Cited by in RCA: 943] [Article Influence: 58.9] [Reference Citation Analysis (0)] |
| 53. | Murakata A, Tanaka S, Mogushi K, Yasen M, Noguchi N, Irie T, Kudo A, Nakamura N, Tanaka H, Arii S. Gene expression signature of the gross morphology in hepatocellular carcinoma. Ann Surg. 2011;253:94-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 54. | Nishida N, Arizumi T, Hagiwara S, Ida H, Sakurai T, Kudo M. MicroRNAs for the Prediction of Early Response to Sorafenib Treatment in Human Hepatocellular Carcinoma. Liver Cancer. 2017;6:113-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (1)] |
| 55. | Orimo T, Ojima H, Hiraoka N, Saito S, Kosuge T, Kakisaka T, Yokoo H, Nakanishi K, Kamiyama T, Todo S, Hirohashi S, Kondo T. Proteomic profiling reveals the prognostic value of adenomatous polyposis coli-end-binding protein 1 in hepatocellular carcinoma. Hepatology. 2008;48:1851-1863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 56. | Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, Roberts LR, Demetris AJ, Sun Z, Nevens F, Roskams T, Thorgeirsson SS. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 748] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 57. | Woo HG, Lee JH, Yoon JH, Kim CY, Lee HS, Jang JJ, Yi NJ, Suh KS, Lee KU, Park ES, Thorgeirsson SS, Kim YJ. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010;70:3034-3041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 159] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 58. | Morofuji N, Ojima H, Hiraoka N, Okusaka T, Esaki M, Nara S, Shimada K, Kishi Y, Kondo T. Antibody-based proteomics to identify an apoptosis signature for early recurrence of hepatocellular carcinoma. Clin Proteomics. 2016;13:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 59. | Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X, Xing B, Sun W, Ren L, Hu B, Li C, Zhang L, Qin G, Zhang M, Chen N, Huang Y, Zhou J, Liu M, Zhu X, Qiu Y, Sun Y, Huang C, Yan M, Wang M, Liu W, Tian F, Xu H, Wu Z, Shi T, Zhu W, Qin J, Xie L, Fan J, Qian X, He F; Chinese Human Proteome Project (CNHPP) Consortium. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567:257-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 623] [Article Influence: 103.8] [Reference Citation Analysis (0)] |
| 60. | Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z, Huang C, Li J, Dong X, Zhou Y, Liu Q, Ma L, Wang X, Zhou J, Liu Y, Boja E, Robles AI, Ma W, Wang P, Li Y, Ding L, Wen B, Zhang B, Rodriguez H, Gao D, Zhou H, Fan J. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell. 2019;179:561-577.e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 617] [Article Influence: 123.4] [Reference Citation Analysis (0)] |
| 61. | Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res. 2018;24:1248-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 588] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 62. | Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud AL, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc JF, Mazzaferro V, Calvo F, Villanueva A, Nault JC, Bioulac-Sage P, Stratton MR, Llovet JM, Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505-511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1006] [Cited by in RCA: 1342] [Article Influence: 134.2] [Reference Citation Analysis (0)] |
| 63. | Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1689] [Cited by in RCA: 1863] [Article Influence: 116.4] [Reference Citation Analysis (0)] |
| 64. | Herceg Z, Hainaut P. Genetic and epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis. Mol Oncol. 2007;1:26-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 167] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 65. | Shah MA, Denton EL, Arrowsmith CH, Lupien M, Schapira M. A global assessment of cancer genomic alterations in epigenetic mechanisms. Epigenetics Chromatin. 2014;7:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 66. | Weinhold B. Epigenetics: the science of change. Environ Health Perspect. 2006;114:A160-A167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 322] [Cited by in RCA: 313] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 67. | Kanherkar RR, Bhatia-Dey N, Csoka AB. Epigenetics across the human lifespan. Front Cell Dev Biol. 2014;2:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 229] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 68. | Srinageshwar B, Maiti P, Dunbar GL, Rossignol J. Role of Epigenetics in Stem Cell Proliferation and Differentiation: Implications for Treating Neurodegenerative Diseases. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 69. | Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2332] [Cited by in RCA: 2978] [Article Influence: 248.2] [Reference Citation Analysis (0)] |
| 70. | Gillette TG, Hill JA. Readers, writers, and erasers: chromatin as the whiteboard of heart disease. Circ Res. 2015;116:1245-1253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 165] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 71. | Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1676] [Cited by in RCA: 1642] [Article Influence: 126.3] [Reference Citation Analysis (0)] |
| 72. | Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3359] [Cited by in RCA: 4033] [Article Influence: 288.1] [Reference Citation Analysis (0)] |
| 73. | Shimada Y, Mohn F, Bühler M. The RNA-induced transcriptional silencing complex targets chromatin exclusively via interacting with nascent transcripts. Genes Dev. 2016;30:2571-2580. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 74. | Lei ZJ, Wang J, Xiao HL, Guo Y, Wang T, Li Q, Liu L, Luo X, Fan LL, Lin L, Mao CY, Wang SN, Wei YL, Lan CH, Jiang J, Yang XJ, Liu PD, Chen DF, Wang B. Lysine-specific demethylase 1 promotes the stemness and chemoresistance of Lgr5+ liver cancer initiating cells by suppressing negative regulators of β-catenin signaling. Oncogene. 2015;34:3214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 75. | Nio K, Yamashita T, Okada H, Kondo M, Hayashi T, Hara Y, Nomura Y, Zeng SS, Yoshida M, Sunagozaka H, Oishi N, Honda M, Kaneko S. Defeating EpCAM(+) liver cancer stem cells by targeting chromatin remodeling enzyme CHD4 in human hepatocellular carcinoma. J Hepatol. 2015;63:1164-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 76. | Lin Q, Wu Z, Yue X, Yu X, Wang Z, Song X, Xu L, He Y, Ge Y, Tan S, Wang T, Song H, Yuan D, Gong Y, Gao L, Liang X, Ma C. ZHX2 restricts hepatocellular carcinoma by suppressing stem cell-like traits through KDM2A-mediated H3K36 demethylation. EBioMedicine. 2020;53:102676. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 77. |
Lo Re O, Mazza T, Giallongo S, Sanna P, Rappa F, Vinh Luong T, Li Volti G, Drovakova A, Roskams T, Van Haele M, Tsochatzis E, Vinciguerra M. Loss of histone macroH2A1 in hepatocellular carcinoma cells promotes paracrine-mediated chemoresistance and |
| 78. | Chen Q, Jiang P, Jia B, Liu Y, Zhang Z. RCC2 contributes to tumor invasion and chemoresistance to cisplatin in hepatocellular carcinoma. Hum Cell. 2020;33:709-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 79. | Ling S, Shan Q, Zhan Q, Ye Q, Liu P, Xu S, He X, Ma J, Xiang J, Jiang G, Wen X, Feng Z, Wu Y, Feng T, Xu L, Chen K, Zhang X, Wei R, Zhang C, Cen B, Xie H, Song P, Liu J, Zheng S, Xu X. USP22 promotes hypoxia-induced hepatocellular carcinoma stemness by a HIF1α/USP22 positive feedback loop upon TP53 inactivation. Gut. 2020;69:1322-1334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 149] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 80. | Tak E, Lee S, Lee J, Rashid MA, Kim YW, Park JH, Park WS, Shokat KM, Ha J, Kim SS. Human carbonyl reductase 1 upregulated by hypoxia renders resistance to apoptosis in hepatocellular carcinoma cells. J Hepatol. 2011;54:328-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 81. | Schultheiss CS, Laggai S, Czepukojc B, Hussein UK, List M, Barghash A, Tierling S, Hosseini K, Golob-Schwarzl N, Pokorny J, Hachenthal N, Schulz M, Helms V, Walter J, Zimmer V, Lammert F, Bohle RM, Dandolo L, Haybaeck J, Kiemer AK, Kessler SM. The long non-coding RNA H19 suppresses carcinogenesis and chemoresistance in hepatocellular carcinoma. Cell Stress. 2017;1:37-54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 82. | Xie SC, Zhang JQ, Jiang XL, Hua YY, Xie SW, Qin YA, Yang YJ. LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death Dis. 2020;11:676. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 83. | Berdasco M, Esteller M. Clinical epigenetics: seizing opportunities for translation. Nat Rev Genet. 2019;20:109-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 332] [Article Influence: 55.3] [Reference Citation Analysis (0)] |
| 84. | Italiano A, Soria JC, Toulmonde M, Michot JM, Lucchesi C, Varga A, Coindre JM, Blakemore SJ, Clawson A, Suttle B, McDonald AA, Woodruff M, Ribich S, Hedrick E, Keilhack H, Thomson B, Owa T, Copeland RA, Ho PTC, Ribrag V. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 2018;19:649-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 464] [Article Influence: 66.3] [Reference Citation Analysis (0)] |
| 85. | Li J, Hao D, Wang L, Wang H, Wang Y, Zhao Z, Li P, Deng C, Di LJ. Epigenetic targeting drugs potentiate chemotherapeutic effects in solid tumor therapy. Sci Rep. 2017;7:4035. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 86. | Amnekar RV, Khan SA, Rashid M, Khade B, Thorat R, Gera P, Shrikhande SV, Smoot DT, Ashktorab H, Gupta S. Histone deacetylase inhibitor pre-treatment enhances the efficacy of DNA-interacting chemotherapeutic drugs in gastric cancer. World J Gastroenterol. 2020;26:598-613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 14] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (1)] |
| 87. | Yeo W, Chung HC, Chan SL, Wang LZ, Lim R, Picus J, Boyer M, Mo FK, Koh J, Rha SY, Hui EP, Jeung HC, Roh JK, Yu SC, To KF, Tao Q, Ma BB, Chan AW, Tong JH, Erlichman C, Chan AT, Goh BC. Epigenetic therapy using belinostat for patients with unresectable hepatocellular carcinoma: a multicenter phase I/II study with biomarker and pharmacokinetic analysis of tumors from patients in the Mayo Phase II Consortium and the Cancer Therapeutics Research Group. J Clin Oncol. 2012;30:3361-3367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 160] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 88. | Lachenmayer A, Toffanin S, Cabellos L, Alsinet C, Hoshida Y, Villanueva A, Minguez B, Tsai HW, Ward SC, Thung S, Friedman SL, Llovet JM. Combination therapy for hepatocellular carcinoma: additive preclinical efficacy of the HDAC inhibitor panobinostat with sorafenib. J Hepatol. 2012;56:1343-1350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 169] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 89. | Sanaei M, Kavoosi F, Esmi Z. The Effect of 5-Aza-2'-Deoxycytidine in Combination to and in Comparison with Vorinostat on DNA Methyltransferases, Histone Deacetylase 1, Glutathione S-Transferase 1 and Suppressor of Cytokine Signaling 1 Genes Expression, Cell Growth Inhibition and Apoptotic Induction in Hepatocellular LCL-PI 11 Cell Line. Int J Hematol Oncol Stem Cell Res. 2020;14:45-55. [PubMed] |
| 90. | Mehta N, Heimbach J, Harnois DM, Sapisochin G, Dodge JL, Lee D, Burns JM, Sanchez W, Greig PD, Grant DR, Roberts JP, Yao FY. Validation of a Risk Estimation of Tumor Recurrence After Transplant (RETREAT) Score for Hepatocellular Carcinoma Recurrence After Liver Transplant. JAMA Oncol. 2017;3:493-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 291] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 91. | Ravindran Menon D, Das S, Krepler C, Vultur A, Rinner B, Schauer S, Kashofer K, Wagner K, Zhang G, Bonyadi Rad E, Haass NK, Soyer HP, Gabrielli B, Somasundaram R, Hoefler G, Herlyn M, Schaider H. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene. 2015;34:4448-4459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 100] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 92. | Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, Maruvka YE, Ji F, Bhang HE, Krishnamurthy Radhakrishna V, Siravegna G, Hu H, Raoof S, Lockerman E, Kalsy A, Lee D, Keating CL, Ruddy DA, Damon LJ, Crystal AS, Costa C, Piotrowska Z, Bardelli A, Iafrate AJ, Sadreyev RI, Stegmeier F, Getz G, Sequist LV, Faber AC, Engelman JA. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22:262-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 584] [Cited by in RCA: 732] [Article Influence: 81.3] [Reference Citation Analysis (0)] |
| 93. | Ramirez M, Rajaram S, Steininger RJ, Osipchuk D, Roth MA, Morinishi LS, Evans L, Ji W, Hsu CH, Thurley K, Wei S, Zhou A, Koduru PR, Posner BA, Wu LF, Altschuler SJ. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun. 2016;7:10690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 313] [Cited by in RCA: 407] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 94. | Guler GD, Tindell CA, Pitti R, Wilson C, Nichols K, KaiWai Cheung T, Kim HJ, Wongchenko M, Yan Y, Haley B, Cuellar T, Webster J, Alag N, Hegde G, Jackson E, Nance TL, Giresi PG, Chen KB, Liu J, Jhunjhunwala S, Settleman J, Stephan JP, Arnott D, Classon M. Repression of Stress-Induced LINE-1 Expression Protects Cancer Cell Subpopulations from Lethal Drug Exposure. Cancer Cell. 2017;32:221-237.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 176] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 95. | Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, Galeas J, Dhruv HD, Berens ME, Schreiber SL, McCormick F, McManus MT. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247-250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1046] [Cited by in RCA: 1169] [Article Influence: 146.1] [Reference Citation Analysis (0)] |
| 96. | Shah SM, Goel PN, Jain AS, Pathak PO, Padhye SG, Govindarajan S, Ghosh SS, Chaudhari PR, Gude RP, Gopal V, Nagarsenker MS. Liposomes for targeting hepatocellular carcinoma: use of conjugated arabinogalactan as targeting ligand. Int J Pharm. 2014;477:128-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 97. | Zhou Q, Ching AK, Leung WK, Szeto CY, Ho SM, Chan PK, Yuan YF, Lai PB, Yeo W, Wong N. Novel therapeutic potential in targeting microtubules by nanoparticle albumin-bound paclitaxel in hepatocellular carcinoma. Int J Oncol. 2011;38:721-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 98. | Madaan K, Kumar S, Poonia N, Lather V, Pandita D. Dendrimers in drug delivery and targeting: Drug-dendrimer interactions and toxicity issues. J Pharm Bioallied Sci. 2014;6:139-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 395] [Cited by in RCA: 364] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 99. | Zhang N, Wardwell PR, Bader RA. Polysaccharide-based micelles for drug delivery. Pharmaceutics. 2013;5:329-352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 100. | Bharti C, Nagaich U, Pal AK, Gulati N. Mesoporous silica nanoparticles in target drug delivery system: A review. Int J Pharm Investig. 2015;5:124-133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 399] [Cited by in RCA: 411] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 101. | Maida M, Orlando E, Cammà C, Cabibbo G. Staging systems of hepatocellular carcinoma: a review of literature. World J Gastroenterol. 2014;20:4141-4150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 95] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 102. | Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5972] [Cited by in RCA: 6573] [Article Influence: 469.5] [Reference Citation Analysis (1)] |
| 103. | Chou R, Cuevas C, Fu R, Devine B, Wasson N, Ginsburg A, Zakher B, Pappas M, Graham E, Sullivan SD. Imaging Techniques for the Diagnosis of Hepatocellular Carcinoma: A Systematic Review and Meta-analysis. Ann Intern Med. 2015;162:697-711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 135] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 104. | Sørensen M, Frisch K, Bender D, Keiding S. The potential use of 2-[¹⁸F]fluoro-2-deoxy-D-galactose as a PET/CT tracer for detection of hepatocellular carcinoma. Eur J Nucl Med Mol Imaging. 2011;38:1723-1731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 105. | Bialecki ES, Di Bisceglie AM. Diagnosis of hepatocellular carcinoma. HPB (Oxford). 2005;7:26-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 205] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 106. | Lok AS, Sterling RK, Everhart JE, Wright EC, Hoefs JC, Di Bisceglie AM, Morgan TR, Kim HY, Lee WM, Bonkovsky HL, Dienstag JL; HALT-C Trial Group. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology. 2010;138:493-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 450] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 107. |
El-Saadany S, El-Demerdash T, Helmy A, Mayah WW, El-Sayed Hussein B, Hassanien M, Elmashad N, Fouad MA, Basha EA.
Diagnostic Value of Glypican-3 for Hepatocellular Carcinomas |
| 108. | Elhendawy M, Abdul-Baki EA, Abd-Elsalam S, Hagras MM, Zidan AA, Abdel-Naby AY, Watny M, Elkabash IA, Salem ML, Elshanshoury M, Soliman S, Abdou S. MicroRNA signature in hepatocellular carcinoma patients: identification of potential markers. Mol Biol Rep. 2020;47:4945-4953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 109. | Abdalla MA, Haj-Ahmad Y. Promising Candidate Urinary MicroRNA Biomarkers for the Early Detection of Hepatocellular Carcinoma among High-Risk Hepatitis C Virus Egyptian Patients. J Cancer. 2012;3:19-31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |