Published online Nov 27, 2021. doi: 10.4254/wjh.v13.i11.1494
Peer-review started: February 24, 2021
First decision: May 3, 2021
Revised: June 6, 2021
Accepted: October 14, 2021
Article in press: October 14, 2021
Published online: November 27, 2021
Processing time: 272 Days and 21.4 Hours
Fatty liver has been present in the lives of patients and physicians for almost two centuries. Vast knowledge has been generated regarding its etiology and consequences, although a long path seeking novel and innovative diagnostic biomarkers for nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH) is still envisioned. On the one hand, proteomics and lipidomics have emerged as potential noninvasive resources for NAFLD diagnosis. In contrast, metabolomics has been able to distinguish between NAFLD and NASH, even detecting degrees of fibrosis. On the other hand, genetic and epigenetic markers have been useful in monitoring disease progression, eventually functioning as target therapies. Other markers involved in immune dysregulation, oxidative stress, and inflammation are involved in the instauration and evolution of the disease. Finally, the fascinating gut microbiome is significantly involved in NAFLD and NASH. This review presents state-of-the-art biomarkers related to NAFLD and NASH and new promises that could eventually be positioned as diagnostic resources for this disease. As is evident, despite great advances in studying these biomarkers, there is still a long path before they translate into clinical benefits.
Core Tip: Nonalcoholic fatty liver disease is increasing in prevalence worldwide. Liver biopsy is considered the gold standard for diagnosis, but it has several limitations. Given the burden on the healthcare system caused by liver fibrosis in a population with metabolic syndrome, there is a priority for noninvasive and accurate diagnostic biomarkers that differentiate patients with steatosis from those with nonalcoholic steatohepatitis, stage fibrosis, predict progression, and monitor treatment response. These biomarkers could assist clinicians in early interventions, avoiding complications and improving prognosis. Here, we summarize the current evidence and future directions.
- Citation: Castillo-Castro C, Martagón-Rosado AJ, Ortiz-Lopez R, Garrido-Treviño LF, Villegas-Albo M, Bosques-Padilla FJ. Promising diagnostic biomarkers of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: From clinical proteomics to microbiome. World J Hepatol 2021; 13(11): 1494-1511
- URL: https://www.wjgnet.com/1948-5182/full/v13/i11/1494.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i11.1494
Thomas Addison first described “fatty liver” in 1836 in England; however, it was not until 1885 when Bartholow made an association between obesity and fatty liver. In 1938, Charles Connor demonstrated a link between fatty liver and progression to cirrhosis in diabetic patients. Throughout the 1950s and up to the 1970s, pathologists reported similarities between alcoholic liver disease and hepatic histological changes in obese and diabetic patients. In 1980, Jurgen Ludwig[1] described patients who denied excessive alcohol consumption yet still had chronic liver disease and histo
As reported in the most recent guidelines, NAFLD is defined as the presence of steatosis in > 5% of hepatocytes in the absence of significant ongoing or recent alcohol consumption and other known causes of liver disease. While in 2005 it had a global prevalence of 15%, a rapid increase in sedentarism and excessive calorie intake independent of diet has pushed it to 24%, with the highest rates in South America (31%) and the Middle East (32%), followed by Asia (27%), the United States (24%), and Europe (23%)[2]. In persons with obesity or type 2 diabetes, it increases up to 70%-90%[3]. Although there is a significant difference between ethnicities within these populations, the exact explanation remains unknown[2].
NAFLD is a necessary and opportune diagnosis, given that 59% progress to NASH. From this stage, 41% continue to fibrosis, with 40% ending with cirrhosis, increasing their risk of a liver transplant, cardiovascular disease, and mortality if there are no interventions[4]. In our country, the Mexican population has several risk factors for the disease because there is a high incidence of overweight and obesity[5], making the NAFLD prevalence likely to surpass 50%. Up to 82% of obese patients who have undergone bariatric surgery present NAFLD, alongside 36% of women with obesity[6].
An international panel has now proposed to rename the disease metabolic dysfunction-associated fatty liver disease to represent the hepatic manifestation of a multisystemic disorder. Until now, the diagnosis was reached by the exclusion of other liver diseases; however, as the pathogenesis is better understood, it is now perceived as a distinct disease and requires a positive diagnosis, which is why it is proposed that the criteria be based on histological, imaging, or blood biomarker evidence of fat accumulation in the liver in addition to one of the following three: Overweight/ obesity, type 2 diabetes mellitus, or evidence of metabolic dysregulation (at least two metabolic risk abnormalities)[1].
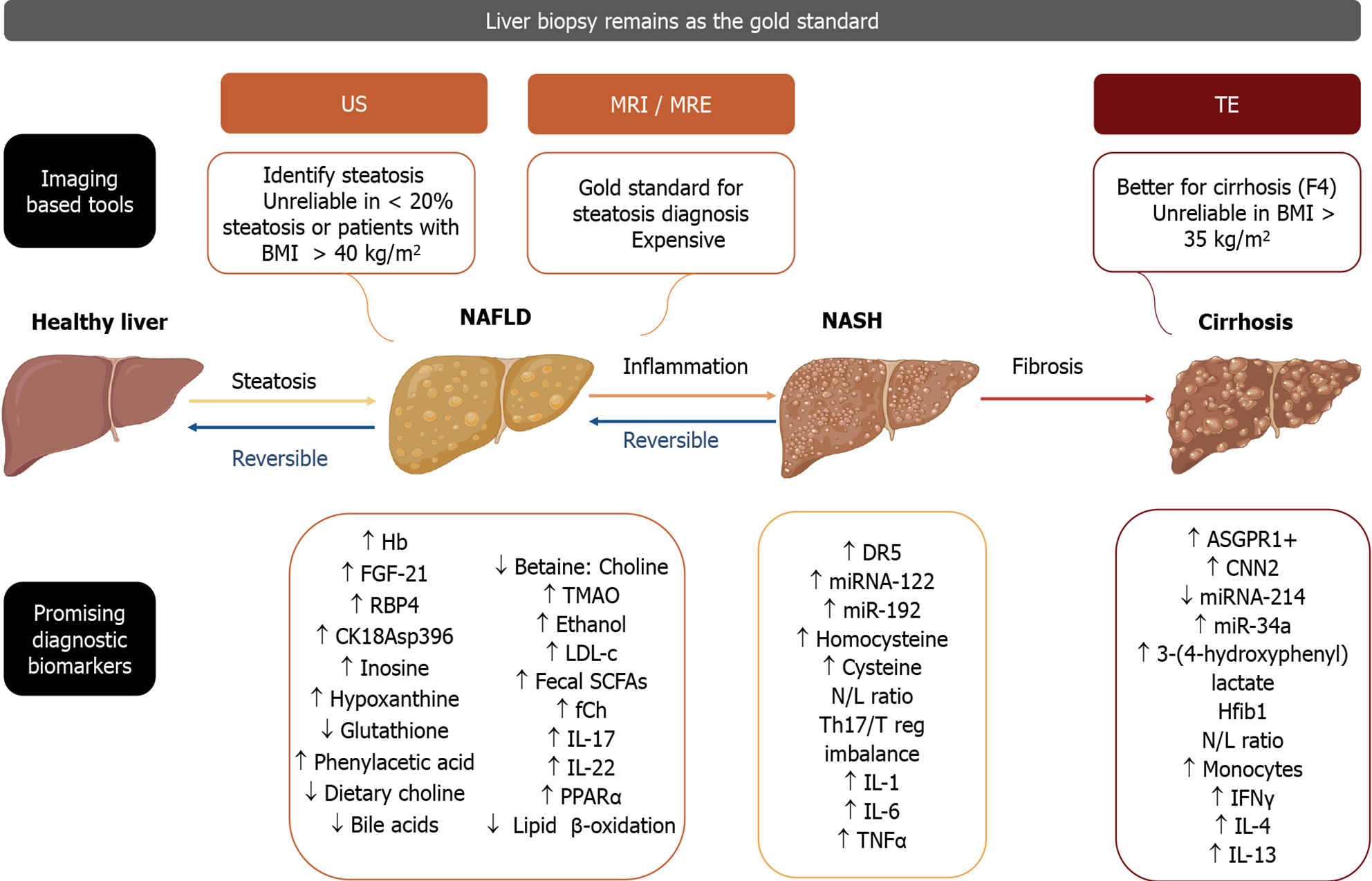
Today, the liver biopsy remains the gold standard for diagnosing and monitoring liver disease, with the disadvantage of being a costly and invasive procedure[7], which is why it is important to look into possible new noninvasive diagnostic tools, such as biomarkers, use of transcriptomics, proteomics, metabolomics, and now “glycomics”[8]. These should aid in predicting liver disease severity, progression, and response to lifestyle changes and pharmacological treatment[9]. The objective of this article is to review concisely and present the potential diagnostic biomarkers for NAFLD and NASH (Figure 1).
The concentrations of several plasma components are determined in routine clinical practice, including electrolytes, molecules, and proteins. Plasma proteins, which constitute the plasma proteome, are released as a result of inflammation, apoptosis, and oxidative stress (OS)[10]. Mass spectrometry-based proteomics[9] and two-dimensional electrophoresis are powerful tools for studying differences[11] in the plasma proteome. There are differences in protein expression among patients with NAFLD and healthy controls. Proteomics technologies have gained relevance as potential non-invasive diagnostic methods for NAFLD.
Plasma proteomics may be secreted by the liver or as a result of the response of the host to steatosis. Hemoglobin is currently the most replicated proteomic biomarker in NAFLD[12]. Studies have found that higher hemoglobin levels are associated with a higher incidence of NAFLD[12]. Circulating aminotransferase [aspartate aminotransferase (AST) and alanine aminotransferase (ALT)] levels are markers of several liver diseases, including NASH. Changes in these enzymes are one of the most commonly observed abnormalities[10].
Fibroblast growth factor 21 is another protein secreted in response to peroxisome proliferator-activated receptor (PPAR)-α activation, and several studies support its potential use as a biomarker for NAFLD[13,14]. The elevation of retinol-binding protein 4 has also been associated with liver fat accumulation[15]. Some glycoproteins like serum fucosylated haptoglobin and Mac-2 binding protein are predictors of hepatocyte ballooning and liver fibrosis[16].
Cytokeratin-18 fragments, such as CK18Asp396, are other proteins that have been extensively studied. These are produced during apoptosis (M30) or cell death (M65). CK18 is the most reviewed biomarker to evaluate liver inflammation[15], but current knowledge does not support its use in clinical practice[17] because of its modest accuracy[8].
Increased cytokeratin-18 levels have good predictive value for NASH vs normal livers but do not differentiate NASH vs simple steatosis[18,19]. Cytokeratin-18 serum levels decrease parallel with histological improvement, but its predictive value is not better than ALT in identifying histological responders[20].
Circulating concentrations of cytokeratin-18 fragments were proposed as the most reliable predictors of NASH in patients with NAFLD[21].
Another important plasma component includes circulating extracellular vesicles (EVs), which are small cell-derived membrane-surrounded structures enclosed by a phospholipid bilayer, with a specific cargo of bioactive molecules of cell origin. There are three types according to their size: Exosomes (40-100 nm), microvesicles or microparticles (0.1-1 μm), and apoptotic bodies (1-4 μm)[22].
They can be detected in several body fluids and can serve several functions by delivering a variety of bioactive molecules, including non-coding RNAs, proteins, lipids, and nucleic acids[23]. Recent studies have provided insight on the bioavailability of circulating EVs in various fluids and, as a consequence, on their potential use as biomarkers for various diseases such as cancer[20,24,25], cardiovascular disease[26], renal disease[27], and liver disease[28,29].
Some authors consider them noninvasive “liquid biopsies” for NASH diagnosis, and studies suggest they can assess disease severity[30]. Serum levels of total and hepatocyte-derived EVs correlate with NASH clinical characteristics, and disease severity in experimental models of NASH, liver and blood levels of EVs are increased and correlate positively with changes in liver histology[31].
Povero et al[30] performed a study isolating EVs from controls with histologically confirmed NASH without cirrhosis and patients with cirrhotic NASH[30]. After the characterization of EV structural features, they found that differences in the quantity and protein components of circulating EVs could be potentially useful for differentiating patients with NASH from controls and patients with pre-cirrhotic NASH from patients with cirrhotic NASH[30].
Notably, asialoglycoprotein receptor 1-positive hepatocyte-specific EVs may represent a surrogate noninvasive biomarker of portal hypertension in patients with cirrhotic NASH. If confirmed, these findings may support the clinical utility of asialoglycoprotein receptor 1-positive EVs (hepatocyte-specific EVs) as a potential alternative to an invasive hepatic venous pressure gradient[30].
Patients with NAFLD or NASH secrete increased levels of microvesicles derived from macrophages/monocytes [CD14(+)] and natural killer (NK) T cells; these levels correlate with NASH severity based on histology[28]. Hirsova et al[32] have demonstrated that lipids that stimulate death receptor 5 on hepatocytes also induce the release of hepatocyte EVs that activate an inflammatory phenotype in macro
However, a major problem in translating this research into clinically useful information is a lack of reproducibility and rigorous criteria for reporting these biomarkers. Proteomics analysis of EVs from patients with advanced NASH is currently limited.
Exosomes are a type of EVs secreted in most cells[22]. These nanovesicles of endocytic origin are present in nearly all-human fluids. Exosomes have several bioactive molecules, including proteins, lipids, and genetic materials[33]. They are conduits for intracellular transfer, and their signals can induce fibrosis, macrophage activation, cytokine secretion, and remodeling extracellular matrix (ECM) production and inactivate hepatic stellate cells (HSC)[34]. Hepatocytes are exosome-secreting cells that are also regulated by hepatic and extrahepatic exosomes[33].
Koeck et al[35] found that exosomes from visceral adipose tissue were involved in the progression of NAFLD by inducing dysregulation of the transforming growth factor-beta (TGF-β) pathway in hepatocytes and HSCs in vitro[35]. Another study by Seo et al[36] detected that during liver injury, damaged hepatocytes produce exosomes that activate toll-like receptor 3, which exacerbates liver fibrosis by enhancing interleukin-17A (IL-17A) production by γδ T cells[36].
Liver fibrogenic pathways are primarily controlled by HSC, which produces and responds to fibrotic mediators such as connective tissue growth factor (CCN2)[37]. Tadokoro et al[29] found that CCN2 upregulation in fibrotic or steatotic livers is associated with the downregulation of microRNA-214 (miRNA-214). miR-214 levels increased in quiescent HSC-secreted exosomes compared with active HSC-released exosomes[29]. On the other hand, exosomal CCN2 may amplify fibrogenic signaling and might be useful for assessing hepatic fibrosis[37].
Chen et al[38] found that the miR-214 promoter binds to the basic helix-loop-helix transcription factor (Twist1), which drives miR-214 expression and results in CNN2 suppression. Twist 1 expression was suppressed during HSC activation. The amounts of Twist1, miR-214, or CCN2 in circulating exosomes from fibrotic mice reflected fibrosis-induced changes in the liver[38]. These findings suggest that during liver fibrosis, exosomes contain specific types of biomarkers, which could be helpful in the diagnosis and progression of liver diseases.
Circulating microRNAs (miRNA) are RNA molecules that do not encode proteins but regulate gene expression in the body, binding to target mRNAs and interfering with their translation[22]. They are expressed in several liver cell types and may offer a biologically stable blood-based biomarker tool for the detection and stratification of liver disease[29].
Tadokoro et al[29] have suggested that serum/plasma miR-122 correlates with liver damage. They have also identified that miR-155 might serve as a liver inflammation biomarker. The one limitation found is that this miRNA cannot differentiate different liver damage etiologies[29].
Another study reported that miRNA-122 and miR-192 levels are dynamic and increase over time, closely correlating with the histopathological severity of NASH[31]. The miR-29 family (miR-29a, miR-29b, miR-29c) mediates the regulation of liver fibrosis through several cellular signaling pathways such as the nuclear transcription factor-kappa B pathway, TGF, and phosphatidylinositol 3-kinase/AKT signaling in HSC with upregulation of ECM genes for the progression of liver fibrosis[39].
Members of the miR-34 family (miR-34a, miR-34b, miR-34c) have pleiotropic roles in the cell cycle and promote the progression of hepatic fibrosis by activation of HSC[39]. miR-34a appears to have an important role in liver fibrosis by regulating the deposition of ECM[40]. miR-30c and miR-193 are also involved in fibrotic remodeling processes that modify the TGF-β-dependent regulation of ECM-related genes in HSCs[41].
The miR-15 family mainly regulates the TGF-β pathway. The activation of HSCs relates to miR15a and miR15b, and they are thought to be essential for apoptosis by targeting Bcl-2 and the caspase signaling pathway[42]. The miR-378 family (specially miR-378a-3p) suppresses the activation of HSCs by directly targeting Gli3[43]. miR-571 closely correlates with the liver cirrhosis stage, and it is upregulated in human hepatocytes and HSC[44]. miR-503 also acts on HSC activation and hepatic fibrosis through the TGF-β/SMAD pathway[45].
The miR-199 family and miR-200 family are responsible for ECM deposition and the release of profibrotic cytokines, which might play profibrotic or anti-fibrotic roles[39]. HSCs also have anti-fibrotic miRNAs, and these include miR-19b, miR-29, miR-30, miR-101, miR-122, miR-133a, miR-144, miR-146a, miR-150-5p, miR-155, miR-195, miR-200a, miR-214, miR-335, miR-370, miR-454, miR-483, etc. The latter are responsible for the maintenance of the quiescent phenotype of normal HSCs[46]. Thus, these studies evidence the role of microRNAs as potential biomarkers of liver damage in NAFLD.
Technological advances in metabolomic analyses on feces, serum, plasma, urine, or liver biopsies led to identifying different metabolites in patients with NAFLD or NASH[47]. Recent studies have found that the severity of fibrosis is associated with serum metabolite changes[48-50].
Remarkably, some metabolites come from the host or the diet, but most need the participation of gut microbes. Notably, inosine and hypoxanthine are enriched in serum samples from patients with mild or moderate NAFLD[47]. Another study found that liver steatosis correlates with phenylacetic acid levels in humans[51]. Glutathione plasma concentration is significantly lower in subjects with liver steatosis, while in subjects with NASH, homocysteine and cysteine concentrations in plasma are higher[52].
Choline, betaine, and circulating methylamines: Choline is an essential component of phosphatidylcholine (a precursor of acetylcholine), mostly obtained from the diet[53]. It is known that a reduction in dietary choline is related to an increase in liver fat. Mice fed with a choline deficient diet are identified as a characteristic model of NAFLD[54]. Choline can be oxidized to betaine, and it has been found that patients with increasing severity of NAFLD have a decreased betaine to choline ratio[55]. The gut microbiota metabolizes choline into trimethylamine (TMA), which is further metabolized into trimethylamine-N-oxide (TMAO) in the liver[56]. Studies suggest that NAFLD severity is associated with increased urinary levels of TMA and TMAO, while TMAO seems to be associated with NAFLD severity[47].
TMAO and bile acids: Gut microbiota regulates secondary bile acid metabolism and inhibits the liver synthesis of lipids by alleviating farnesoid X-activated receptor inhibition[57]. TMAO is a gut-dependent metabolite of choline. A decreased level of bile acids could be associated with TMAO production and NAFLD since it induces a decrease in the bile acid pool by inhibiting two key enzymes of bile acid metabolism: Cytochrome P450 (CYP)7A1 and CYP27A1[55]. Some studies have found adverse associations between the circulating TMAO levels and the presence and severity of NAFLD and a favorable betaine-NAFLD relationship in participants[55].
Three-(4-hydroxyphenyl) lactate: Three-(4-hydroxyphenyl) lactate is a derived product of amino acid metabolism. It was consistently associated with increased liver fibrosis severity in a test and validation cohort[48].
Ethanol: Gut microbiota leads to endogenous ethanol production, which might be a liver toxin involved in NAFLD and NASH development[47]. A study showed that Klebsiella pneumoniae can produce ethanol from glucose in the absence of alcohol consumption, and it might be associated with NAFLD[58].
Human serum and plasma are composed of lipids that play important roles in energy storage, metabolic regulation, signaling, etc.[10]. Technological advances have made possible the identification of specific alterations in lipids and metabolites in the feces, serum, plasma, urine, and liver of patients with NAFLD[47].
Choline is a dietary component metabolized in the liver, necessary for cell function. Epidemiological studies suggest that increased free choline levels are related to the degree of hepatic steatosis fibrosis[59].
Kalhan et al[60] have shown that plasma levels of triglycerides[60] and low-density lipoprotein cholesterol are higher in patients with NAFLD[52]; however, differences in this lipidomic profile are also observed in obesity. Therefore, this lack of specificity remains a limitation for their use. Barr et al[61] described a lipidomic signature associated with NAFLD progression to distinguish NASH from steatosis, depending on the body mass index in a large cohort of samples[61].
Gorden et al[62] described a panel of 20 lipids that differentiate patients with NASH and liver steatosis[62]. Later, Kimberly et al[63] identified the association between anandamide (endocannabinoid derived from arachidonic acid metabolism) and NAFLD severity[63]. Tokushige et al[64] reported 28 metabolites associated with liver fibrosis, showing a decrease of dehydroepiandrosterone sulfate and etiocholanolone-S with the progression of fibrosis[64].
Puri et al[65] analyzed plasma lipids and eicosanoid metabolites in NAFLD and NASH patients. They reported increased plasma monounsaturated fatty acids and primary palmitoleic and oleic acids and decreased linoleic acid. Plasmalogen levels were significantly decreased in NASH, and 11-HETE (a nonenzymatic product of arachidonic acid) was increased in NASH[65]. Loomba et al[66] assessed the lipidomic profile in NAFLD and NASH patients and reported that 11,12-dihydroxy- eicosatrie
Short-chain fatty acids (SCFAs) are comprised of butyrate, acetate, and propionate. They are produced in the colon through microbial fermentation of dietary fiber and are a substrate that increases liver triglyceride levels[67]. They are also involved in fatty acid synthesis and gluconeogenesis[68]. Human studies have observed an increased fecal concentration of SCFAs in patients with NAFLD and/or NASH[69].
In NAFLD, lipid metabolism is disrupted, and lipotoxicity is a key mechanism for NAFLD progression. Lipidomic profiling might provide a novel biomarker for the noninvasive prediction of NASH.
The role of genetic and epigenetic factors in the progression of liver fibrosis is well documented. It is known that key regulatory genes partially control the cell phenotype. Several genes are involved in the pathogenesis and histological stage of liver fibrosis, although the mechanisms underlying gene regulation are highly complex and need additional research[70].
Chromosome 15, designated Hfib1 (hepatic fibrogenic gene 1), affects the stage of liver fibrosis[71]. The core of risk genes that control fibrosis progression has been defined by quantitative trait locus analysis in mouse strains by genome-wide interval mapping, which identified several genomic loci related to fibrosis phenotypes on chromosomes 4, 5, 7, 12, and 17[72].
Bruschi et al[73] reported that PLPNA3 quantification correlates with the liver fibrosis stage. Expression of PLPNA3 in biopsies from NASH patients is increased during progression from mild to severe liver fibrosis. Carriers of the I148M single-nucleotide polymorphism (C>G) had higher PLPNA3 and serum liver enzyme (ALT/AST) levels, along with steatosis grade inflammation ballooning and NAFLD activity score, compared with non-polymorphism carriers[73]. On the other hand, Sharma et al[74] stated that neurocan is associated with NASH and liver fibrosis in patients of European ancestry. Another study found that patients of Indian descent with neurocan variations had higher ALT levels[74].
Epigenetics describes reversible gene expression changes that do not imply changes in the DNA sequence and are entirely cell type-specific. Epigenetic mechanisms initiate and sustain chromatin modifications by facilitating gene transcription, cell phenotype, and consequently, organ function. These mechanisms include DNA methylation, histone modifications, and noncoding RNAs mediating gene silencing[75].
Aberrant DNA methylation is associated with fibrosis. Komatsu et al[76] suggested that DNA hypomethylation in fibrogenic genes is crucial for the onset and progression of liver fibrosis[76]. Mann et al[77] confirmed this functional association of DNA methylation with liver fibrosis. The transdifferentiation of HSC to profibrogenic myofibroblast phenotype was suppressed in vitro by the DNMT inhibitor 5’-aza-deoxycytidine[77]. The development of fibrosis is also related to changes in the expression of enzymes that regulate DNA methylation and hydroxymethylation[78].
Epigenetic modulation on the PPAR-γ gene promoter is involved in HSC differentiation. Aberrant expression of a series of chemokines in HSCs aggravate inflammation and OS[79].
Small non-coding RNAs contribute to various pathologic states of liver disease, but miRNA has been previously reviewed. The detection of genetic and epigenetic markers may be helpful in the recognition and monitoring of disease evolution and can eventually be applied for targeted therapies.
NASH pathology encompasses an intricate network of mechanisms. OS activates Kupffer cells (KC), and KC activation triggers an innate and adaptative immune response, including the release of cytokines and chemokines that activate NK T (NKT) cells and HSCs[80]. Besides, there is augmented infiltration of different immune cells, such as monocytes, T lymphocytes, and neutrophils, in the activation and in situ expansion of liver cells, like KC or stellate cells. Activated KC and NKT cells promote additional fat accumulation in the liver. KC, neutrophils, NKT cells, and inflammatory T cells [T helper (Th)1, Th17, CD8+ T cells] enhance liver inflammation and contribute to the development of fibrosis[81].
The neutrophil to lymphocyte ratio (N:L ratio) has been proposed as a novel noninvasive marker to predict NASH and advanced fibrosis in patients with NAFLD[82]. In patients with cirrhosis, these cells are functionally deficient, with impaired chemotaxis, phagocytosis, and intracellular killing. Their function correlates with 90-d survival[83].
On the other hand, monocytes are myeloid-derived cells that migrate to inflammation sites, phagocytose microbes, and secrete cytotoxins. They are spontaneously activated in patients with liver fibrosis. Cirrhotic patients have an increased peripheral frequency of monocytes, impaired phagocytosis, and reduced responses to stimulation[84].
Studies have reported that NK cells are dysregulated in liver diseases. One study found that IL-17- and IL-22- secreting iNKT cells are dominant at the beginning of liver steatosis, and IFNγ/IL-4/IL-13-secreting iNKT cells are prevalent at the most advanced course of the disease[85].
Notably, CD4+ T cells are reduced in patients with liver fibrosis. This finding could explain the increased risk of spontaneous bacterial peritonitis in these patients[86]. CD8+ T cells isolated from mice hepatic cells expressed an increased cytotoxic IL-10 phenotype and CD8+ T cell depletion[87].
Th17 cells and T regulatory cells (Treg) originate from naïve T cell precursors. Th17 cells are important for pathogen clearance and inflammation. Treg cells in patients with liver fibrosis are significant[88]. There is a Th17/Treg imbalance that positively correlates with NASH histological progression[89].
Innate lymphoid cells are lymphocytes that secrete cytokines and chemokines in response to pathogenic tissue damage. They have a role in inflammation and fibro
Detoxification is a crucial hepatic activity. It is vulnerable to OS and inflammation. An increase in free fatty acids is critical for the elevation of reactive oxygen species (ROS). A balance between the ROS and antioxidant systems is necessary for adequate cell function[80]. OS causes liver damage by altering DNA molecules, proteins, and lipids and modulating pathways associated with gene transcription, protein expression, cell apoptosis, and HSC activation. Inflammation is manifested as inflammatory cell infiltration in the liver to fight pathogen invasion. When the stimuli are persistent, it can lead to cell injury and lipid accumulation associated with an increased risk of severe liver disease, including steatohepatitis and fibrosis[91].
In NASH, ROS are generated in several ways that can alter signaling pathways, such as cell kinases, phosphatases, and transcription factors, which impact cell proliferation, differentiation, and apoptosis. They can lead to cirrhosis via the rebuilding of stellate cells and ECM within the liver. Substantial hepatic ROS is produced by excessive angiotensin II and activated CYP2E1, resulting in impaired beta-oxidation and eventually fatty liver[91].
Lipotoxicity in NAFLD causes OS and induces organelle damage due to decreased antioxidant systems, mitochondrial dysfunction, and an increase in unfolded protein response by endoplasmic reticulum stress[80]. On the other hand, there is an impairment of α-oxidation due to a decrease in PPARα activity, which upturns hepatic lipid levels. Fatty acid overload is the major source of reducing equivalents responsible for increased ROS production. Also, TNF-α and lipid peroxidation products could induce mitochondrial dysfunction. Mitochondrial damage will result in secondary lipid α-oxidation inhibition and a further increase in the degree of steatosis[80].
Furthermore, inflammatory cytokines such as IL-1-β, TNF-α, and IL-17/20/33, chemokines, like monocyte chemoattractant protein-1 and C-X-C chemokine ligand 10, and the toll-like receptor pathway are intensively involved in the regulation of hepatic fibrogenesis[91]. Macrophage activation and influx in the liver are important for the progression of NAFLD since hepatic macrophages promote NASH development via cytokines IL-1, IL-6, and TNF-α[92]. Liver failure causes an increase of TNF-α, IL-6, and angiotensin II[80].
The intestinal barrier is composed of chemical, physical, and immunological barriers. Maintaining a healthy barrier is essential to prevent microbial translocation and keep the liver safe to prevent systemic inflammation[93].
Differences in the taxonomic composition of the intestinal microbiome in NAFLD (an increased proportion of Firmicutes and a reduced proportion of Bacteriodetes) change metabolic function. The availability of bile acids, endogenous alcohols, and voltaic organic compounds increases. When these changes are combined with reduced SCFAs and choline, the integrity of the intestinal barrier is reduced[93].
Gut barrier disruption is recognized in patients with cirrhosis. The epithelial layers show structural abnormalities related to increased intestinal permeability or bacterial translocation[94]. Permeability can be measured by the urinary excretion of radiolabeled 51chromium-ethylenediamine tetraacetic acid or by measuring volatile organic compounds formed by the fermentation of some dietary polysaccharides[95].
Cardiotonic steroids (CTS) are part of a group of specific ligands of Na+, K+-ATPase, a ubiquitously expressed enzyme responsible for the maintenance of electrochemical gradients across the cell membrane through active transport[96] that provokes a variety of cell signals[70]. In the last decades, studies have revealed the role of Na+, K+-ATPase and its signaling in various diseases, including inflammation and fibrosis[97].
CTS increase cholesterol synthesis in liver HepG2 cells, which augments the activity and expression of 3-hydroxy-3-methylglutaryl-coenzyme A reductase[98]. Disturbed cholesterol balance underlies cardiovascular disease and an increasing number of other diseases, such as neurodegenerative diseases, cancers, and liver disease[99].
Elevated CTS might encourage increased cholesterol levels in the liver and worsen liver fibrosis by activating HSCs[100] and other redox-inflammatory pathways[101]. This increase in cholesterol levels could precipitate hepatocyte injury and macrophage activation that could lead to liver fibrosis progression. However, even CTS seem to have an important role in hepatocyte lipotoxicity and fibrosis; to our knowledge, they have not been studied as biomarkers for liver disease progression.
A large community of viruses, bacteria, archaea, and fungi live in the gastrointestinal tract and composes the gut microbiota[102]. It has critical roles in digestion, immunity, and metabolism[103]. Recently, the characterization of gut microbiota has evolved rapidly due to the advances in sequencing technology, permitting the creation of a gut microbiota gene catalogue[102]. The collective genetic material of the microbiota is often referred to as the “gut microbiome”. It encodes pathways that produce small bioactive molecules derived from dietary or metabolic precursors and may alter human health[104].
Thus, knowledge of microbiome characteristics in different metabolic diseases has increased in the past years. There has been great interest in dysbiosis (alterations in the composition and balance of microbiota[104]). Microbiota alterations are being studied as possible diagnostic biomarkers to improve personalized care. Animal studies have demonstrated a potential causal role of gut microbiota in NAFLD development[105]. However, extrapolating mouse model experimental information to humans has several limitations[106]. Consequently, signatures specific to liver alterations would be useful as NAFLD diagnostic biomarkers. However, discrepant microbiome signatures might be linked to the heterogeneity of diet, drugs, infections, environmental exposures, among others[104].
Alterations in the gut microbiome have been associated with the progression and severity of NAFLD[107]. Proteobacteria are enriched in steatosis[103,108,109]. Patients with NAFLD, compared with healthy individuals, also have significant changes at the phylum (increased Enterobacteriaceae[109] and decreased Rikenellaceae and Ruminococcaceae[109]) and genera level (increased Escherichia[109], Dorea, and Peptoniphilus and decreased Anaerosporobacter, Coprococcus, Faecalibacterium, and Prevotella)[103].
When comparing people with NASH vs healthy controls, some patterns are observed that also overlap with the NAFLD microbiome: Phylum (increased Proteobacteria[50,109-111]), family (increased Enterobacteriaceae[109,110] and decreased Ruminococcaceae[110-113] and Rikenellaceae[110]), and genera (increased Dorea[111] and decreased Faecalibacterium[110,113,114], Coprococcus[110,112,113], and Anaeros
Few projects have studied microbial composition as a function of fibrosis progression. Bacteroides vulgatus and Escherichia coli are the most abundant species in advanced fibrosis (F3–F4)[50]. Models have been proposed to use the microbiome as a reservoir for diagnostic signatures of NAFLD fibrosis[50], but further confirmation in independent cohorts and across geographical regions is necessary to assess their clinical relevance.
Microbial signatures of liver fibrosis are related to a severe shift in taxa conformation, leading to a growth in pathogenic taxa and a decline in metabolically beneficial taxa[115]. However, the evaluation of gut microbiota contribution to liver disease progression (from steatosis to NASH and NASH cirrhosis) is limited and bacterial markers are frequently identified in a given study yet not confirmed in independent cohorts.
Although some studies consider gut bacterial groups as promising markers of different stages of liver disease, if the microbiota is a causal factor and how it interacts with the complex pathophysiological processes driving disease progression from mild fibrosis to severe fibrosis is still under investigation[50,109].
Dense and complex populations of intestinal viruses reside in the gut and interact with other microorganisms and the human host[116,117]. Most intestinal viruses are bacteriophages (phages), viruses that can specifically infect bacteria[118]. Phages may serve as important microbiota genetic diversity reservoirs by acting as vehicles for the horizontal transfer of virulence, antibiotic resistance, and metabolic determinants among bacteria[119].
Lang et al[120] studied the fecal viromes from NAFLD patients and controls. They found associated histologic markers of NAFLD severity with significant decreases in viral diversity and proportion of bacteriophages[120]. The intestinal virome is specific for every individual, and viral diversity measures were the third and fifth most important variables following a higher AST and higher age. The most important viral species belonged to Lactococcus phages, and several Lactococcus phages were less present in patients with NAFLD and NASH.
Fungi and archaea are important components of the human microbiota. Recent findings have revealed that mycobiome (commensal fungi at barrier surfaces) can influence host immunity and the development and progression of human inflammatory diseases[121]. The human gut mycobiome is dominated by Saccharomyces, Malassezia, Candida, and Cladosporium and are an important modulator for local and peripheral immune responses. Patients with liver fibrosis have decreased fungal diversity and increased Candida[122]. Gut mycobiota disturbance might produce metabolites called mycotoxins (trichothecenes, zearalenone, fumonisins, ochratoxins, aflatoxins) that can alter gut health by compromising intestinal epithelia[123,124].
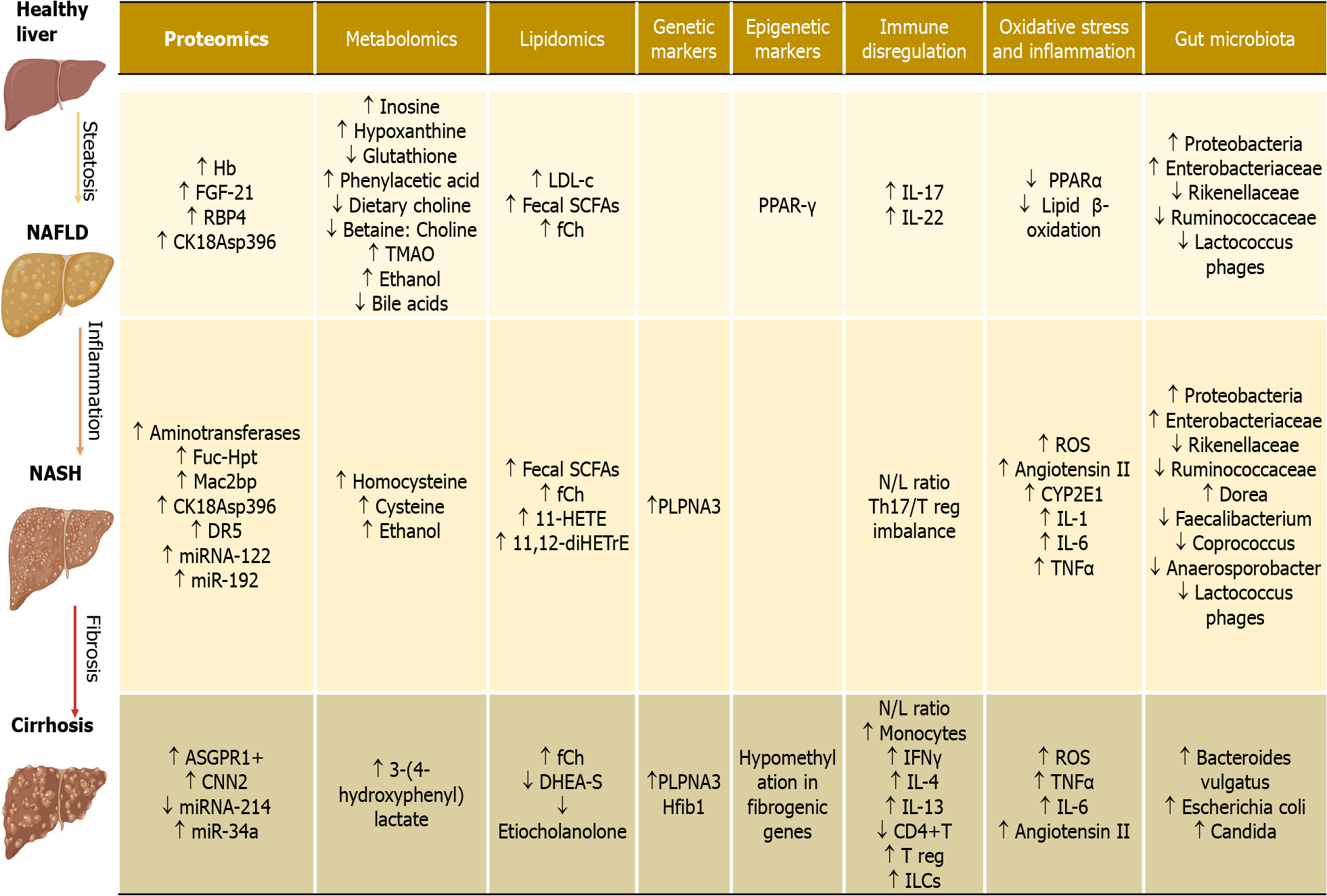
The increasing burden of NAFLD worldwide has encouraged the search for novel biomarkers to detect liver diseases. Liver biopsy is currently the gold standard for diagnosis and staging, but it has several limitations, including sampling errors, invasiveness, inter-observer variability, and related procedure risks. Researchers have faced the challenge of developing novel biomarkers in past decades, and significant advances have been made. A promising biomarker should be liver-specific, accessible and accurate, replicable, and available in clinical laboratories. As summarized in this article, most studies have focused on proteomics, metabolomics, genome-wide association studies, microbiome, and inflammation markers. Still, some may be more specific for NAFLD while others for NASH, although the challenge for determining the etiology and staging the degree of severity remains a limitation (Figure 2).
The evaluation of future biomarkers for the assessment of liver fibrosis could greatly impact the health system. There is a priority for non-invasive diagnostic tools to fulfil medical needs, differentiate patients with steatosis from those with NASH and fibrosis, predict disease progression, and monitor patients to evaluate the therapeutic response. In the following years, it would be expected that a physician who faces a hepatic patient could suspect hepatic disease, perform imaging studies, and from there have a set of potential biomarkers that they may request to have a concrete and specific diagnosis. Some of these biomarkers have strong diagnostic performance, but current evidence shows a lack of reproducibility. Besides, the analytical, clinical validity of the methodology is lacking. Validity is necessary to translate basic research into real clinical application. Even if we perform this validation, it is unlikely that a single biomarker could fulfil this necessity. A combination of these biomarkers could soon be used to create a diagnostic panel. This panel, combined with the patient´s clinical history and clinical data, could certainly lead to a medical decision that results in an accurate diagnosis and treatment. This result must be the goal in the following years.
Through this review, we have shown that despite a wide range of potential biomarkers for the different stages of hepatic steatosis and fibrosis, there is still a long path to the translation of these resources. We provide evidence of the current absence of an efficient, non-invasive, and widely accessible test for NAFLD and NASH detection. Biomarkers are still in early stages. Rigorous, well-designed comprehensive studies are required to determine the actual benefit these may pose for determining the risk, diagnosis, and progression of the hepatic patient. In conclusion, our review compiles significant efforts to find new promising biomarkers for liver disease, still leaving great challenges. There is still a need to define normal reference levels in healthy individuals and the different stages of the disease and to determine the clinical sensitivity and specificity of biomarkers to develop a clinical diagnostic panel.
Provenance and peer review: Invited article; Externally peer reviewed.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Mexico
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sporea I S-Editor: Fan JR L-Editor: Filipodia P-Editor: Fan JR
| 1. | Fouad Y, Waked I, Bollipo S, Gomaa A, Ajlouni Y, Attia D. What's in a name? Liver Int. 2020;40:1254-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 212] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 2. | Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4054] [Cited by in RCA: 3800] [Article Influence: 542.9] [Reference Citation Analysis (2)] |
| 3. | Byrne CD, Patel J, Scorletti E, Targher G. Tests for diagnosing and monitoring non-alcoholic fatty liver disease in adults. BMJ. 2018;362:k2734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 4. | Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5322] [Cited by in RCA: 7542] [Article Influence: 838.0] [Reference Citation Analysis (0)] |
| 5. | Méndez-Sánchez N, Villa AR, Chávez-Tapia NC, Ponciano-Rodriguez G, Almeda-Valdés P, González D, Uribe M. Trends in liver disease prevalence in Mexico from 2005 to 2050 through mortality data. Ann Hepatol. 2005;4:52-55. [PubMed] |
| 6. | Bernal-Reyes R, Castro-Narro G, Malé-Velázquez R, Carmona-Sánchez R, González-Huezo MS, García-Juárez I, Chávez-Tapia N, Aguilar-Salinas C, Aiza-Haddad I, Ballesteros-Amozurrutia MA, Bosques-Padilla F, Castillo-Barradas M, Chávez-Barrera JA, Cisneros-Garza L, Flores-Calderón J, García-Compeán D, Gutiérrez-Grobe Y, Higuera de la Tijera MF, Kershenobich-Stalnikowitz D, Ladrón de Guevara-Cetina L, Lizardi-Cervera J, López-Cossio JA, Martínez-Vázquez S, Márquez-Guillén E, Méndez-Sánchez N, Moreno-Alcantar R, Poo-Ramírez JL, Ramos-Martínez P, Rodríguez-Hernández H, Sánchez-Ávila JF, Stoopen-Rometti M, Torre-Delgadillo A, Torres-Villalobos G, Trejo-Estrada R, Uribe-Esquivel M, Velarde-Ruiz Velasco JA. The Mexican consensus on nonalcoholic fatty liver disease. Rev Gastroenterol Mex (Engl Ed). 2019;84:69-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 7. | Long MT, Gandhi S, Loomba R. Advances in non-invasive biomarkers for the diagnosis and monitoring of non-alcoholic fatty liver disease. Metabolism. 2020;111S:154259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 8. | Giraudi PJ, Stephenson AM, Tiribelli C, Rosso N. Novel high-throughput applications for NAFLD diagnostics and biomarker dis-covery. Hepatoma Res. 2021;2021. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 9. | Geyer PE, Holdt LM, Teupser D, Mann M. Revisiting biomarker discovery by plasma proteomics. Mol Syst Biol. 2017;13:942. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 557] [Cited by in RCA: 601] [Article Influence: 75.1] [Reference Citation Analysis (0)] |
| 10. | Iruarrizaga-Lejarreta M, Bril F, Noureddin M, Ortiz P, Lu SC, Mato JM. Emerging circulating biomarkers for the diagnosis and assess-ment of treatment responses in patients with hepatic fat accumula-tion, nash and liver fibrosis. Translational Research Methods in Diabetes, Obesity, and Nonalcoholic Fatty Liver Disease: A Focus on Early Phase Clinical Drug Development. Gainesville: Springer, 2019: 423–448. |
| 11. | Rodriguez-Suarez E, Mato JM, Elortza F. Proteomics analysis of human nonalcoholic fatty liver. Methods Mol Biol. 2012;909:241-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | Yilmaz Y. Serum proteomics for biomarker discovery in nonalcoholic fatty liver disease. Clin Chim Acta. 2012;413:1190-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Li H, Dong K, Fang Q, Hou X, Zhou M, Bao Y, Xiang K, Xu A, Jia W. High serum level of fibroblast growth factor 21 is an independent predictor of non-alcoholic fatty liver disease: a 3-year prospective study in China. J Hepatol. 2013;58:557-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 14. | Seo JA, Kim NH, Park SY, Kim HY, Ryu OH, Lee KW, Lee J, Kim DL, Choi KM, Baik SH, Choi DS, Kim SG. Serum retinol-binding protein 4 levels are elevated in non-alcoholic fatty liver disease. Clin Endocrinol (Oxf). 2008;68:555-560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 77] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 15. | Leoni S, Tovoli F, Napoli L, Serio I, Ferri S, Bolondi L. Current guidelines for the management of non-alcoholic fatty liver disease: A systematic review with comparative analysis. World J Gastroenterol. 2018;24:3361-3373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 412] [Cited by in RCA: 393] [Article Influence: 56.1] [Reference Citation Analysis (7)] |
| 16. | Kamada Y, Akita M, Takeda Y, Yamada S, Fujii H, Sawai Y, Doi Y, Asazawa H, Nakayama K, Mizutani K, Yakushijin T, Miyazaki M, Ezaki H, Hiramatsu N, Yoshida Y, Kiso S, Imai Y, Kawada N, Takehara T, Miyoshi E. Serum Fucosylated Haptoglobin as a Novel Diagnostic Biomarker for Predicting Hepatocyte Ballooning and Nonalcoholic Steatohepatitis. PLoS One. 2013;8:e66328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 17. | European Association for the Study of the Liver (EASL); European Association for the Study of Diabetes (EASD); European Association for the Study of Obesity (EASO). EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. Diabetologia. 2016;59:1121-1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 506] [Article Influence: 56.2] [Reference Citation Analysis (2)] |
| 18. | Shen J, Chan HL, Wong GL, Chan AW, Choi PC, Chan HY, Chim AM, Yeung DK, Yu J, Chu WC, Wong VW. Assessment of non-alcoholic fatty liver disease using serum total cell death and apoptosis markers. Aliment Pharmacol Ther. 2012;36:1057-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 19. | Estep M, Mehta R, Bratthauer G, Alaparthi L, Monge F, Ali S, Abdelatif D, Younoszai Z, Stepanova M, Goodman ZD, Younossi ZM. Hepatic sonic hedgehog protein expression measured by computer assisted morphometry significantly correlates with features of non-alcoholic steatohepatitis. BMC Gastroenterol. 2019;19:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 20. | Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470-1476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4100] [Cited by in RCA: 3944] [Article Influence: 232.0] [Reference Citation Analysis (0)] |
| 21. | Stefan N, Häring HU, Cusi K. Non-alcoholic fatty liver disease: causes, diagnosis, cardiometabolic consequences, and treatment strategies. Lancet Diabetes Endocrinol. 2019;7:313-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 586] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 22. | Shah R, Patel T, Freedman JE. Circulating Extracellular Vesicles in Human Disease. N Engl J Med. 2018;379:958-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 526] [Article Influence: 75.1] [Reference Citation Analysis (0)] |
| 23. | Maas SLN, Breakefield XO, Weaver AM. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017;27:172-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 977] [Cited by in RCA: 1083] [Article Influence: 135.4] [Reference Citation Analysis (0)] |
| 24. | Juan T, Fürthauer M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin Cell Dev Biol. 2018;74:66-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 329] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 25. | Hornick NI, Huan J, Doron B, Goloviznina NA, Lapidus J, Chang BH, Kurre P. Serum Exosome MicroRNA as a Minimally-Invasive Early Biomarker of AML. Sci Rep. 2015;5:11295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 208] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 26. | Dickhout A, Koenen RR. Extracellular Vesicles as Biomarkers in Cardiovascular Disease; Chances and Risks. Front Cardiovasc Med. 2018;5:113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 27. | Erdbrügger U, Le TH. Extracellular Vesicles in Renal Diseases: More than Novel Biomarkers? J Am Soc Nephrol. 2016;27:12-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 159] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 28. | Kornek M, Lynch M, Mehta SH, Lai M, Exley M, Afdhal NH, Schuppan D. Circulating microparticles as disease-specific biomarkers of severity of inflammation in patients with hepatitis C or nonalcoholic steatohepatitis. Gastroenterology. 2012;143:448-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 164] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 29. | Tadokoro T, Morishita A, Masaki T. Diagnosis and Therapeutic Management of Liver Fibrosis by MicroRNA. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 30. | Povero D, Yamashita H, Ren W, Subramanian MG, Myers RP, Eguchi A, Simonetto DA, Goodman ZD, Harrison SA, Sanyal AJ, Bosch J, Feldstein AE. Characterization and Proteome of Circulating Extracellular Vesicles as Potential Biomarkers for NASH. Hepatol Commun. 2020;4:1263-1278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 31. | Povero D, Eguchi A, Li H, Johnson CD, Papouchado BG, Wree A, Messer K, Feldstein AE. Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS One. 2014;9:e113651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 218] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 32. | Hirsova P, Ibrahim SH, Krishnan A, Verma VK, Bronk SF, Werneburg NW, Charlton MR, Shah VH, Malhi H, Gores GJ. Lipid-Induced Signaling Causes Release of Inflammatory Extracellular Vesicles From Hepatocytes. Gastroenterology. 2016;150:956-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 392] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 33. | Cai S, Cheng X, Pan X, Li J. Emerging role of exosomes in liver physiology and pathology. Hepatol Res. 2017;47:194-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (1)] |
| 34. | Chen L, Brenner DA, Kisseleva T. Combatting Fibrosis: Exosome-Based Therapies in the Regression of Liver Fibrosis. Hepatol Commun. 2019;3:180-192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 35. | Koeck ES, Iordanskaia T, Sevilla S, Ferrante SC, Hubal MJ, Freishtat RJ, Nadler EP. Adipocyte exosomes induce transforming growth factor beta pathway dysregulation in hepatocytes: a novel paradigm for obesity-related liver disease. J Surg Res. 2014;192:268-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 159] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 36. | Seo W, Eun HS, Kim SY, Yi HS, Lee YS, Park SH, Jang MJ, Jo E, Kim SC, Han YM, Park KG, Jeong WI. Exosome-mediated activation of toll-like receptor 3 in stellate cells stimulates interleukin-17 production by γδ T cells in liver fibrosis. Hepatology. 2016;64:616-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 154] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 37. | Charrier A, Chen R, Chen L, Kemper S, Hattori T, Takigawa M, Brigstock DR. Exosomes mediate intercellular transfer of pro-fibrogenic connective tissue growth factor (CCN2) between hepatic stellate cells, the principal fibrotic cells in the liver. Surgery. 2014;156:548-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 105] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 38. | Chen L, Chen R, Kemper S, Charrier A, Brigstock DR. Suppression of fibrogenic signaling in hepatic stellate cells by Twist1-dependent microRNA-214 expression: Role of exosomes in horizontal transfer of Twist1. Am J Physiol Gastrointest Liver Physiol. 2015;309:G491-G499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 39. | Jiang XP, Ai WB, Wan LY, Zhang YQ, Wu JF. The roles of microRNA families in hepatic fibrosis. Cell Biosci. 2017;7:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 40. | Yan G, Li B, Xin X, Xu M, Ji G, Yu H. MicroRNA-34a Promotes Hepatic Stellate Cell Activation via Targeting ACSL1. Med Sci Monit. 2015;21:3008-3015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 41. | Roy S, Benz F, Vargas Cardenas D, Vucur M, Gautheron J, Schneider A, Hellerbrand C, Pottier N, Alder J, Tacke F, Trautwein C, Roderburg C, Luedde T. miR-30c and miR-193 are a part of the TGF-β-dependent regulatory network controlling extracellular matrix genes in liver fibrosis. J Dig Dis. 2015;16:513-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 42. | Guo CJ, Pan Q, Li DG, Sun H, Liu BW. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J Hepatol. 2009;50:766-778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 241] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 43. | Hyun J, Wang S, Kim J, Rao KM, Park SY, Chung I, Ha CS, Kim SW, Yun YH, Jung Y. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun. 2016;7:10993. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 185] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 44. | Roderburg C, Mollnow T, Bongaerts B, Elfimova N, Vargas Cardenas D, Berger K, Zimmermann H, Koch A, Vucur M, Luedde M, Hellerbrand C, Odenthal M, Trautwein C, Tacke F, Luedde T. Micro-RNA profiling in human serum reveals compartment-specific roles of miR-571 and miR-652 in liver cirrhosis. PLoS One. 2012;7:e32999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 45. | Xie X, Dou CY, Zhou Y, Zhou Q, Tang HB. MicroRNA-503 Targets Mothers Against Decapentaplegic Homolog 7 Enhancing Hepatic Stellate Cell Activation and Hepatic Fibrosis. Dig Dis Sci. 2021;66:1928-1939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 46. | Ezhilarasan D. MicroRNA interplay between hepatic stellate cell quiescence and activation. Eur J Pharmacol. 2020;885:173507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 47. | Chu H, Duan Y, Yang L, Schnabl B. Small metabolites, possible big changes: a microbiota-centered view of non-alcoholic fatty liver disease. Gut. 2019;68:359-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 240] [Article Influence: 40.0] [Reference Citation Analysis (1)] |
| 48. | Caussy C, Loomba R. Gut microbiome, microbial metabolites and the development of NAFLD. Nat Rev Gastroenterol Hepatol. 2018;15:719-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 49. | Dong S, Zhan ZY, Cao HY, Wu C, Bian YQ, Li JY, Cheng GH, Liu P, Sun MY. Urinary metabolomics analysis identifies key biomarkers of different stages of nonalcoholic fatty liver disease. World J Gastroenterol. 2017;23:2771-2784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 59] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 50. | Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, Dulai PS, Caussy C, Bettencourt R, Highlander SK, Jones MB, Sirlin CB, Schnabl B, Brinkac L, Schork N, Chen CH, Brenner DA, Biggs W, Yooseph S, Venter JC, Nelson KE. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017;25:1054-1062.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 551] [Cited by in RCA: 754] [Article Influence: 94.3] [Reference Citation Analysis (0)] |
| 51. | Hoyles L, Fernández-Real JM, Federici M, Serino M, Abbott J, Charpentier J, Heymes C, Luque JL, Anthony E, Barton RH, Chilloux J, Myridakis A, Martinez-Gili L, Moreno-Navarrete JM, Benhamed F, Azalbert V, Blasco-Baque V, Puig J, Xifra G, Ricart W, Tomlinson C, Woodbridge M, Cardellini M, Davato F, Cardolini I, Porzio O, Gentileschi P, Lopez F, Foufelle F, Butcher SA, Holmes E, Nicholson JK, Postic C, Burcelin R, Dumas ME. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med. 2018;24:1070-1080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 303] [Cited by in RCA: 492] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 52. | Engin A. Non-Alcoholic Fatty Liver Disease. Adv Exp Med Biol. 2017;960:443-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 186] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 53. | Sherriff JL, O'Sullivan TA, Properzi C, Oddo JL, Adams LA. Choline, Its Potential Role in Nonalcoholic Fatty Liver Disease, and the Case for Human and Bacterial Genes. Adv Nutr. 2016;7:5-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 141] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 54. | Sergeeva KV. [Blood serum and liver phospholipids in rats with chronic choline-protein deficiency]. Biull Eksp Biol Med. 1975;79:47-49. [PubMed] |
| 55. | Chen YM, Liu Y, Zhou RF, Chen XL, Wang C, Tan XY, Wang LJ, Zheng RD, Zhang HW, Ling WH, Zhu HL. Associations of gut-flora-dependent metabolite trimethylamine-N-oxide, betaine and choline with non-alcoholic fatty liver disease in adults. Sci Rep. 2016;6:19076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 254] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 56. | Zeisel SH, daCosta KA, Youssef M, Hensey S. Conversion of dietary choline to trimethylamine and dimethylamine in rats: dose-response relationship. J Nutr. 1989;119:800-804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 110] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 57. | Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1367] [Cited by in RCA: 1684] [Article Influence: 140.3] [Reference Citation Analysis (0)] |
| 58. | Yuan J, Chen C, Cui J, Lu J, Yan C, Wei X, Zhao X, Li N, Li S, Xue G, Cheng W, Li B, Li H, Lin W, Tian C, Zhao J, Han J, An D, Zhang Q, Wei H, Zheng M, Ma X, Li W, Chen X, Zhang Z, Zeng H, Ying S, Wu J, Yang R, Liu D. Fatty Liver Disease Caused by High-Alcohol-Producing Klebsiella pneumoniae. Cell Metab. 2019;30:675-688.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 323] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 59. | Imajo K, Fujita K, Yoneda M, Shinohara Y, Suzuki K, Mawatari H, Takahashi J, Nozaki Y, Sumida Y, Kirikoshi H, Saito S, Nakamuta M, Matsuhashi N, Wada K, Nakajima A. Plasma free choline is a novel non-invasive biomarker for early-stage non-alcoholic steatohepatitis: A multi-center validation study. Hepatol Res. 2012;42:757-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 60. | Kalhan SC, Guo L, Edmison J, Dasarathy S, McCullough AJ, Hanson RW, Milburn M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism. 2011;60:404-413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 457] [Cited by in RCA: 430] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 61. | Barr J, Caballería J, Martínez-Arranz I, Domínguez-Díez A, Alonso C, Muntané J, Pérez-Cormenzana M, García-Monzón C, Mayo R, Martín-Duce A, Romero-Gómez M, Lo Iacono O, Tordjman J, Andrade RJ, Pérez-Carreras M, Le Marchand-Brustel Y, Tran A, Fernández-Escalante C, Arévalo E, García-Unzueta M, Clement K, Crespo J, Gual P, Gómez-Fleitas M, Martínez-Chantar ML, Castro A, Lu SC, Vázquez-Chantada M, Mato JM. Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J Proteome Res. 2012;11:2521-2532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 185] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 62. | Gorden DL, Myers DS, Ivanova PT, Fahy E, Maurya MR, Gupta S, Min J, Spann NJ, McDonald JG, Kelly SL, Duan J, Sullards MC, Leiker TJ, Barkley RM, Quehenberger O, Armando AM, Milne SB, Mathews TP, Armstrong MD, Li C, Melvin WV, Clements RH, Washington MK, Mendonsa AM, Witztum JL, Guan Z, Glass CK, Murphy RC, Dennis EA, Merrill AH Jr, Russell DW, Subramaniam S, Brown HA. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J Lipid Res. 2015;56:722-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 287] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 63. | Kimberly WT, O'Sullivan JF, Nath AK, Keyes M, Shi X, Larson MG, Yang Q, Long MT, Vasan R, Peterson RT, Wang TJ, Corey KE, Gerszten RE. Metabolite profiling identifies anandamide as a biomarker of nonalcoholic steatohepatitis. JCI Insight. 2017;2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 64. | Tokushige K, Hashimoto E, Kodama K, Tobari M, Matsushita N, Kogiso T, Taniai M, Torii N, Shiratori K, Nishizaki Y, Ohga T, Ohashi Y, Sato T. Serum metabolomic profile and potential biomarkers for severity of fibrosis in nonalcoholic fatty liver disease. J Gastroenterol. 2013;48:1392-1400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 65. | Puri P, Wiest MM, Cheung O, Mirshahi F, Sargeant C, Min HK, Contos MJ, Sterling RK, Fuchs M, Zhou H, Watkins SM, Sanyal AJ. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 2009;50:1827-1838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 521] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 66. | Loomba R, Quehenberger O, Armando A, Dennis EA. Polyunsaturated fatty acid metabolites as novel lipidomic biomarkers for noninvasive diagnosis of nonalcoholic steatohepatitis. J Lipid Res. 2015;56:185-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 163] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 67. | Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, Gordon JI. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. 2008;105:16767-16772. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1270] [Cited by in RCA: 1151] [Article Influence: 67.7] [Reference Citation Analysis (0)] |
| 68. | den Besten G, Lange K, Havinga R, van Dijk TH, Gerding A, van Eunen K, Müller M, Groen AK, Hooiveld GJ, Bakker BM, Reijngoud DJ. Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am J Physiol Gastrointest Liver Physiol. 2013;305:G900-G910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 413] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 69. | Rau M, Rehman A, Dittrich M, Groen AK, Hermanns HM, Seyfried F, Beyersdorf N, Dandekar T, Rosenstiel P, Geier A. Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United European Gastroenterol J. 2018;6:1496-1507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 246] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 70. | Zehra M, Curry JC, Pillai SS, Lakhani HV, Edwards CE, Sodhi K. Elucidating Potential Profibrotic Mechanisms of Emerging Biomarkers for Early Prognosis of Hepatic Fibrosis. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 71. | Haston CK, Zhou X, Gumbiner-Russo L, Irani R, Dejournett R, Gu X, Weil M, Amos CI, Travis EL. Universal and radiation-specific loci influence murine susceptibility to radiation-induced pulmonary fibrosis. Cancer Res. 2002;62:3782-3788. [PubMed] |
| 72. | Liebe R, Hall RA, Williams RW, Dooley S, Lammert F. Systems genetics of hepatocellular damage in vivo and in vitro: identification of a critical network on chromosome 11 in mouse. Physiol Genomics. 2013;45:931-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 73. | Bruschi FV, Tardelli M, Herac M, Claudel T, Trauner M. Metabolic regulation of hepatic PNPLA3 expression and severity of liver fibrosis in patients with NASH. Liver Int. 2020;40:1098-1110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 74. | Sharma M, Mitnala S, Vishnubhotla RK, Mukherjee R, Reddy DN, Rao PN. The Riddle of Nonalcoholic Fatty Liver Disease: Progression From Nonalcoholic Fatty Liver to Nonalcoholic Steatohepatitis. J Clin Exp Hepatol. 2015;5:147-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 101] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 75. | Moran-Salvador E, Mann J. Epigenetics and Liver Fibrosis. Cell Mol Gastroenterol Hepatol. 2017;4:125-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 76. | Komatsu Y, Waku T, Iwasaki N, Ono W, Yamaguchi C, Yanagisawa J. Global analysis of DNA methylation in early-stage liver fibrosis. BMC Med Genomics. 2012;5:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 77. | Mann J, Oakley F, Akiboye F, Elsharkawy A, Thorne AW, Mann DA. Regulation of myofibroblast transdifferentiation by DNA methylation and MeCP2: implications for wound healing and fibrogenesis. Cell Death Differ. 2007;14:275-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 200] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 78. | You H, Sun YM. [Cellular mechanism in the fibrogenesis of liver fibrosis]. Zhonghua Gan Zang Bing Za Zhi. 2012;20:563-564. [PubMed] |
| 79. | Zhao Q, Qin CY, Zhao ZH, Fan YC, Wang K. Epigenetic modifications in hepatic stellate cells contribute to liver fibrosis. Tohoku J Exp Med. 2013;229:35-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 80. | Gonzalez A, Huerta-Salgado C, Orozco-Aguilar J, Aguirre F, Tacchi F, Simon F, Cabello-Verrugio C. Role of Oxidative Stress in Hepatic and Extrahepatic Dysfunctions during Nonalcoholic Fatty Liver Disease (NAFLD). Oxid Med Cell Longev. 2020;2020:1617805. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (2)] |
| 81. | Nati M, Haddad D, Birkenfeld AL, Koch CA, Chavakis T, Chatzigeorgiou A. The role of immune cells in metabolism-related liver inflammation and development of non-alcoholic steatohepatitis (NASH). Rev Endocr Metab Disord. 2016;17:29-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 108] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 82. | Alkhouri N, Morris-Stiff G, Campbell C, Lopez R, Tamimi TA, Yerian L, Zein NN, Feldstein AE. Neutrophil to lymphocyte ratio: a new marker for predicting steatohepatitis and fibrosis in patients with nonalcoholic fatty liver disease. Liver Int. 2012;32:297-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 201] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 83. | Irvine KM, Ratnasekera I, Powell EE, Hume DA. Causes and Consequences of Innate Immune Dysfunction in Cirrhosis. Front Immunol. 2019;10:293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 125] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 84. | Sargenti K, Johansson Å, Bertilsson S, Mattsby-Baltzer I, Klintman D, Kalaitzakis E. Dysfunction of Circulating Polymorphonuclear Leukocytes and Monocytes in Ambulatory Cirrhotics Predicts Patient Outcome. Dig Dis Sci. 2016;61:2294-2302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 85. | Maricic I, Marrero I, Eguchi A, Nakamura R, Johnson CD, Dasgupta S, Hernandez CD, Nguyen PS, Swafford AD, Knight R, Feldstein AE, Loomba R, Kumar V. Differential Activation of Hepatic Invariant NKT Cell Subsets Plays a Key Role in Progression of Nonalcoholic Steatohepatitis. J Immunol. 2018;201:3017-3035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 86. | Confer BD, Lopez R, Zein NN. CD4+ T-Cell function and the risk for developing spontaneous bacterial peritonitis in patients with cirrhosis. J Clin Gastroenterol. 2013;47:807-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 87. | Breuer DA, Pacheco MC, Washington MK, Montgomery SA, Hasty AH, Kennedy AJ. CD8+ T cells regulate liver injury in obesity-related nonalcoholic fatty liver disease. Am J Physiol Gastrointest Liver Physiol. 2020;318:G211-G224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 88. | Huang CH, Jeng WJ, Ho YP, Teng W, Chen WT, Chen YC, Lin SM, Chiu CT, Sheen IS, Lin CY. Increased regulatory T cells in patients with liver cirrhosis correlated with hyperbilirubinemia and predict bacterial complications. J Gastroenterol Hepatol. 2015;30:775-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 89. | Chackelevicius CM, Gambaro SE, Tiribelli C, Rosso N. Th17 involvement in nonalcoholic fatty liver disease progression to non-alcoholic steatohepatitis. World J Gastroenterol. 2016;22:9096-9103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (1)] |
| 90. | Panda SK, Colonna M. Innate Lymphoid Cells in Mucosal Immunity. Front Immunol. 2019;10:861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 200] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 91. | Li S, Hong M, Tan HY, Wang N, Feng Y. Insights into the Role and Interdependence of Oxidative Stress and Inflammation in Liver Diseases. Oxid Med Cell Longev. 2016;2016:4234061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 252] [Cited by in RCA: 236] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 92. | Shabangu CS, Huang JF, Hsiao HH, Yu ML, Chuang WL, Wang SC. Liquid Biopsy for the Diagnosis of Viral Hepatitis, Fatty Liver Steatosis, and Alcoholic Liver Diseases. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 93. | Tranah TH, Edwards LA, Schnabl B, Shawcross DL. Targeting the gut-liver-immune axis to treat cirrhosis. Gut. 2021;70:982-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 117] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 94. | Such J, Guardiola JV, de Juan J, Casellas JA, Pascual S, Aparicio JR, Solá-Vera J, Pérez-Mateo M. Ultrastructural characteristics of distal duodenum mucosa in patients with cirrhosis. Eur J Gastroenterol Hepatol. 2002;14:371-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 59] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 95. | Cho MS, Kim SY, Suk KT, Kim BY. Modulation of gut microbiome in nonalcoholic fatty liver disease: pro-, pre-, syn-, and antibiotics. J Microbiol. 2018;56:855-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 96. | Orellana AM, Kinoshita PF, Leite JA, Kawamoto EM, Scavone C. Cardiotonic Steroids as Modulators of Neuroinflammation. Front Endocrinol (Lausanne). 2016;7:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 97. | Xu Y, Marck P, Huang M, Xie JX, Wang T, Shapiro JI, Cai L, Feng F, Xie Z. Biased Effect of Cardiotonic Steroids on Na/K-ATPase-Mediated Signal Transduction. Mol Pharmacol. 2021;99:217-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 98. | Campia I, Gazzano E, Pescarmona G, Ghigo D, Bosia A, Riganti C. Digoxin and ouabain increase the synthesis of cholesterol in human liver cells. Cell Mol Life Sci. 2009;66:1580-1594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 99. | Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21:225-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 1141] [Article Influence: 190.2] [Reference Citation Analysis (0)] |
| 100. | Arguello G, Balboa E, Arrese M, Zanlungo S. Recent insights on the role of cholesterol in non-alcoholic fatty liver disease. Biochim Biophys Acta. 2015;1852:1765-1778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 230] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 101. | Liu J, Lilly MN, Shapiro JI. Targeting Na/K-ATPase Signaling: A New Approach to Control Oxidative Stress. Curr Pharm Des. 2018;24:359-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 102. | Sharpton SR, Ajmera V, Loomba R. Emerging Role of the Gut Microbiome in Nonalcoholic Fatty Liver Disease: From Composition to Function. Clin Gastroenterol Hepatol. 2019;17:296-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 125] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 103. | Aron-Wisnewsky J, Vigliotti C, Witjes J, Le P, Holleboom AG, Verheij J, Nieuwdorp M, Clément K. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat Rev Gastroenterol Hepatol. 2020;17:279-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 692] [Article Influence: 138.4] [Reference Citation Analysis (0)] |
| 104. | Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med. 2016;375:2369-2379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1826] [Cited by in RCA: 2326] [Article Influence: 258.4] [Reference Citation Analysis (0)] |
| 105. | Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, Camporez JP, Shulman GI, Gordon JI, Hoffman HM, Flavell RA. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179-185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1620] [Cited by in RCA: 1882] [Article Influence: 144.8] [Reference Citation Analysis (0)] |
| 106. | Van Herck MA, Vonghia L, Francque SM. Animal Models of Nonalcoholic Fatty Liver Disease-A Starter's Guide. Nutrients. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 263] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 107. | Brandl K, Schnabl B. Intestinal microbiota and nonalcoholic steatohepatitis. Curr Opin Gastroenterol. 2017;33:128-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 134] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 108. | Grabherr F, Grander C, Effenberger M, Adolph TE, Tilg H. Gut Dysfunction and Non-alcoholic Fatty Liver Disease. Front Endocrinol (Lausanne). 2019;10:611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 109. | Shen F, Zheng RD, Sun XQ, Ding WJ, Wang XY, Fan JG. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int. 2017;16:375-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 412] [Article Influence: 51.5] [Reference Citation Analysis (1)] |
| 110. | Safari Z, Gérard P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD). Cell Mol Life Sci. 2019;76:1541-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 340] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 111. | Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, Bailey J, Myers RP, Rioux KP. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11:868-75.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 451] [Cited by in RCA: 522] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 112. | Wang B, Jiang X, Cao M, Ge J, Bao Q, Tang L, Chen Y, Li L. Altered Fecal Microbiota Correlates with Liver Biochemistry in Nonobese Patients with Non-alcoholic Fatty Liver Disease. Sci Rep. 2016;6:32002. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 167] [Cited by in RCA: 274] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 113. | de Faria Ghetti F, Oliveira DG, de Oliveira JM, de Castro Ferreira LEVV, Cesar DE, Moreira APB. Influence of gut microbiota on the development and progression of nonalcoholic steatohepatitis. Eur J Nutr. 2018;57:861-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 114. | Tsai MC, Liu YY, Lin CC, Wang CC, Wu YJ, Yong CC, Chen KD, Chuah SK, Yao CC, Huang PY, Chen CH, Hu TH, Chen CL. Gut Microbiota Dysbiosis in Patients with Biopsy-Proven Nonalcoholic Fatty Liver Disease: A Cross-Sectional Study in Taiwan. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 115. | Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR, Sikaroodi M, Gillevet PM. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 659] [Cited by in RCA: 837] [Article Influence: 76.1] [Reference Citation Analysis (0)] |
| 116. | Lima M, Andrade A, Oliveira G, Nicoli J, Martins F, Kroon E, Abrahão J. Virus and microbiota relationships in humans and other mammals: An evolutionary view. Hum Micro J. 2019;11:100050. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 117. | Carding SR, Davis N, Hoyles L. Review article: the human intestinal virome in health and disease. Aliment Pharmacol Ther. 2017;46:800-815. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 167] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 118. | Schmelcher M, Loessner MJ. Bacteriophage endolysins: applications for food safety. Curr Opin Biotechnol. 2016;37:76-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 144] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 119. | Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. Phage as agents of lateral gene transfer. Curr Opin Microbiol. 2003;6:417-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 334] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 120. | Lang S, Demir M, Martin A, Jiang L, Zhang X, Duan Y, Gao B, Wisplinghoff H, Kasper P, Roderburg C, Tacke F, Steffen HM, Goeser T, Abraldes JG, Tu XM, Loomba R, Stärkel P, Pride D, Fouts DE, Schnabl B. Intestinal Virome Signature Associated With Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology. 2020;159:1839-1852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 134] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 121. | Li XV, Leonardi I, Iliev ID. Gut Mycobiota in Immunity and Inflammatory Disease. Immunity. 2019;50:1365-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 170] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 122. | Jiang L, Stärkel P, Fan JG, Fouts DE, Bacher P, Schnabl B. The gut mycobiome: a novel player in chronic liver diseases. J Gastroenterol. 2021;56:1-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 123. | Liew WP, Mohd-Redzwan S. Mycotoxin: Its Impact on Gut Health and Microbiota. Front Cell Infect Microbiol. 2018;8:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 237] [Cited by in RCA: 249] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 124. | BioRender. Adapted from "Nonalcoholic Fatty Liver Disease (NAFLD) Spec-trum. [cited 10 January 2021]. Available from: https://app.biorender.com/biorender-templates. |