Published online Dec 27, 2020. doi: 10.4254/wjh.v12.i12.1367
Peer-review started: July 31, 2020
First decision: September 21, 2020
Revised: September 29, 2020
Accepted: October 19, 2020
Article in press: October 19, 2020
Published online: December 27, 2020
Processing time: 139 Days and 14.3 Hours
The majority of hepatocellular carcinoma (HCC) cases are associated with the hepatitis B virus (HBV) infection. Autophagy related protein 9A (ATG9A) is a transmembrane protein required for autophagosome formation. In order to investigate the role of ATG9A in HBV-associated HCC, ATG9A protein expression was determined in tumor liver tissues and compared with adjacent nontumor tissues from HCC patients with or without HBV infection. In HBV-associated HCC tissues, ATG9A protein level was increased in tumor liver tissues, but not in cases of non-HBV HCC. Our findings suggested that ATG9A might be involved in HBV and cancer cell survival. Therefore, we aimed to analyze the function of ATG9A in HBV replication using RNA interference to evaluate the HBV DNA level using real-time PCR. In the present study, there were no significant differences between shATG9A-transfected HepG2.2.15 cells and the mock control. However, we found that silencing ATG9A affected apoptosis in HepG2.2.15 and HepG2 cell lines. Our results indicated that ATG9A might be partly involved in the survival of HCC. Thus, the inhibition of ATG9A together with other targets might be a potential drug target for HCC treatment.
Core Tip: Autophagy related protein 9A (ATG9A) protein expression was increased in tumor liver tissues compared to adjacent nontumor tissues from hepatocellular carcinoma (HCC) patients with hepatitis B virus infection. We showed that silencing ATG9A increased cell apoptosis of HepG2.2.15 and HepG2 cells. These results suggested that ATG9A protein is involved in the survival of HCC. The inhibition of ATG9A combined with other targets might be a potential drug target for HCC treatment.
- Citation: Kimkong I, Kunanopparat A. Autophagy related protein 9A increase in hepatitis B virus-associated hepatocellular carcinoma and the role in apoptosis. World J Hepatol 2020; 12(12): 1367-1371
- URL: https://www.wjgnet.com/1948-5182/full/v12/i12/1367.htm
- DOI: https://dx.doi.org/10.4254/wjh.v12.i12.1367
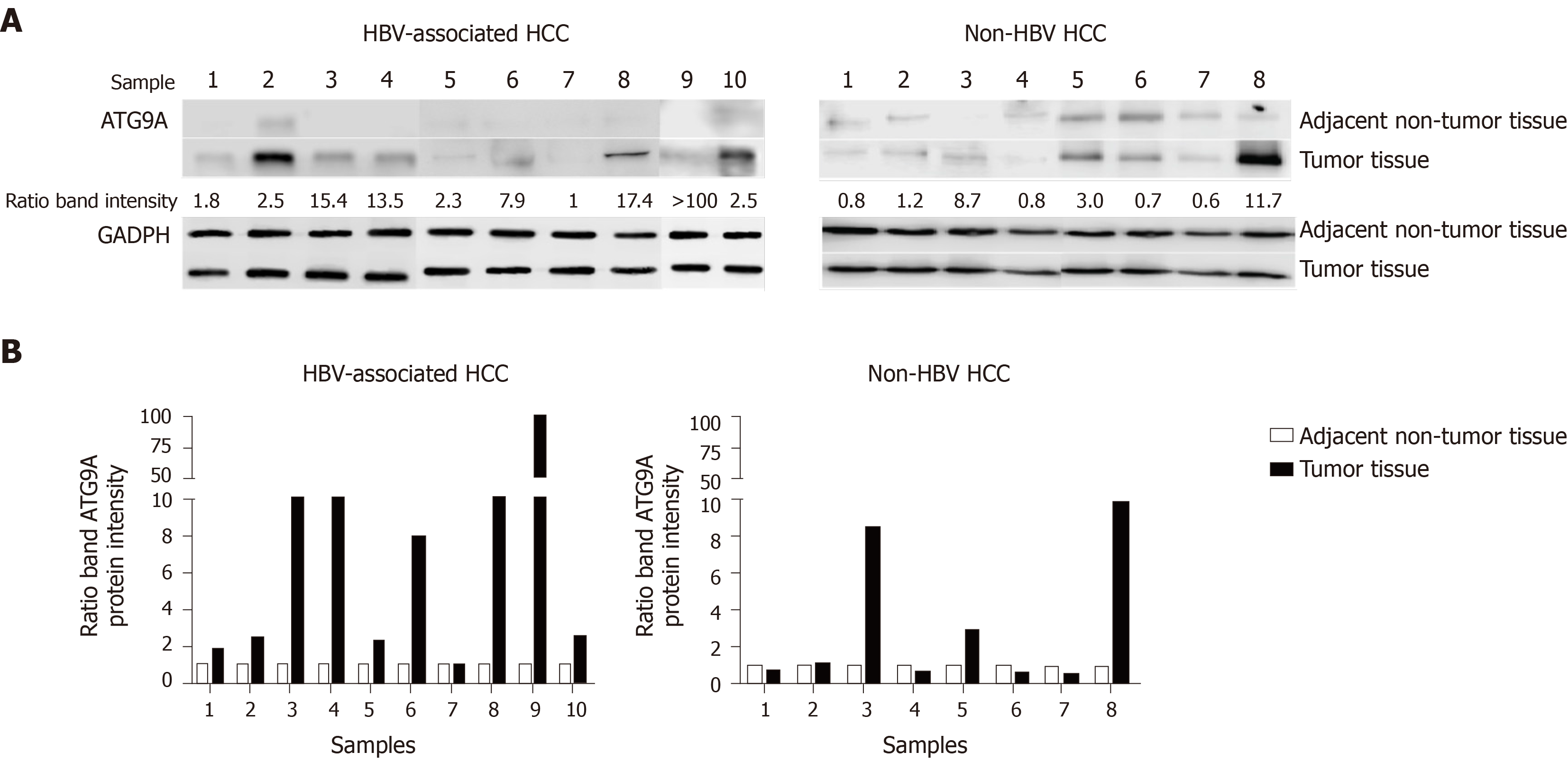
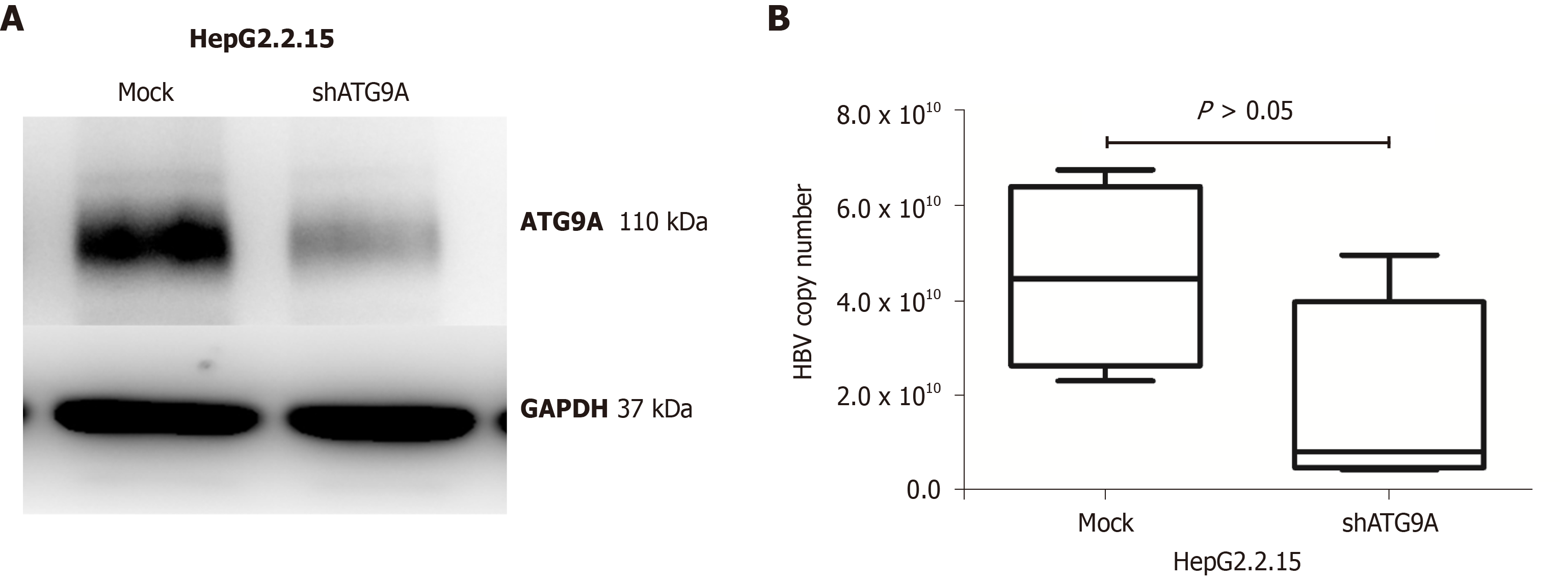
Autophagy related protein 9A (ATG9A) is a transporter membrane molecule required for initial autophagosome formation in the autophagy pathway[1]. ATG9A has been identified as having the function of a stimulator of interferon (IFN) genes (STING)inhibition. A loss of ATG9A results in enhanced assembly of STING/TANK-binding kinase 1 complexes in response to dsDNA, leading to an increase in innate immune responses[2]. Silencing of ATG9A in macrophages increases STING-mediated IFN-β production and promotes cell viability[3]. Our previous study reported that gene and protein expressions of ATG9A were upregulated in HepG2 and HepG2.2.15 cells compared with a THLE-2 hepatic cell line[4]. Thus, in this study we investigated the role of ATG9A in hepatitis B virus (HBV)-associated hepatocellular carcinoma (HCC) tissues. We found that ATG9A protein levels were highly increased in tumor liver tissues in HBV-associated HCC (9 of the 10 sample pairs). In the case of non-HBV HCC, ATG9A protein levels were decreased or slightly increased in tumor liver tissues (Figure 1). Therefore, we hypothesized that HBV induces the upregulation of ATG9A to benefit its replication. To determine the effect of ATG9A on HBV replication, HBV DNA was quantified from shATG9A-transfected cells and compared to mock cells. We observed no significant difference in shATG9A transfected cells (Figure 2).
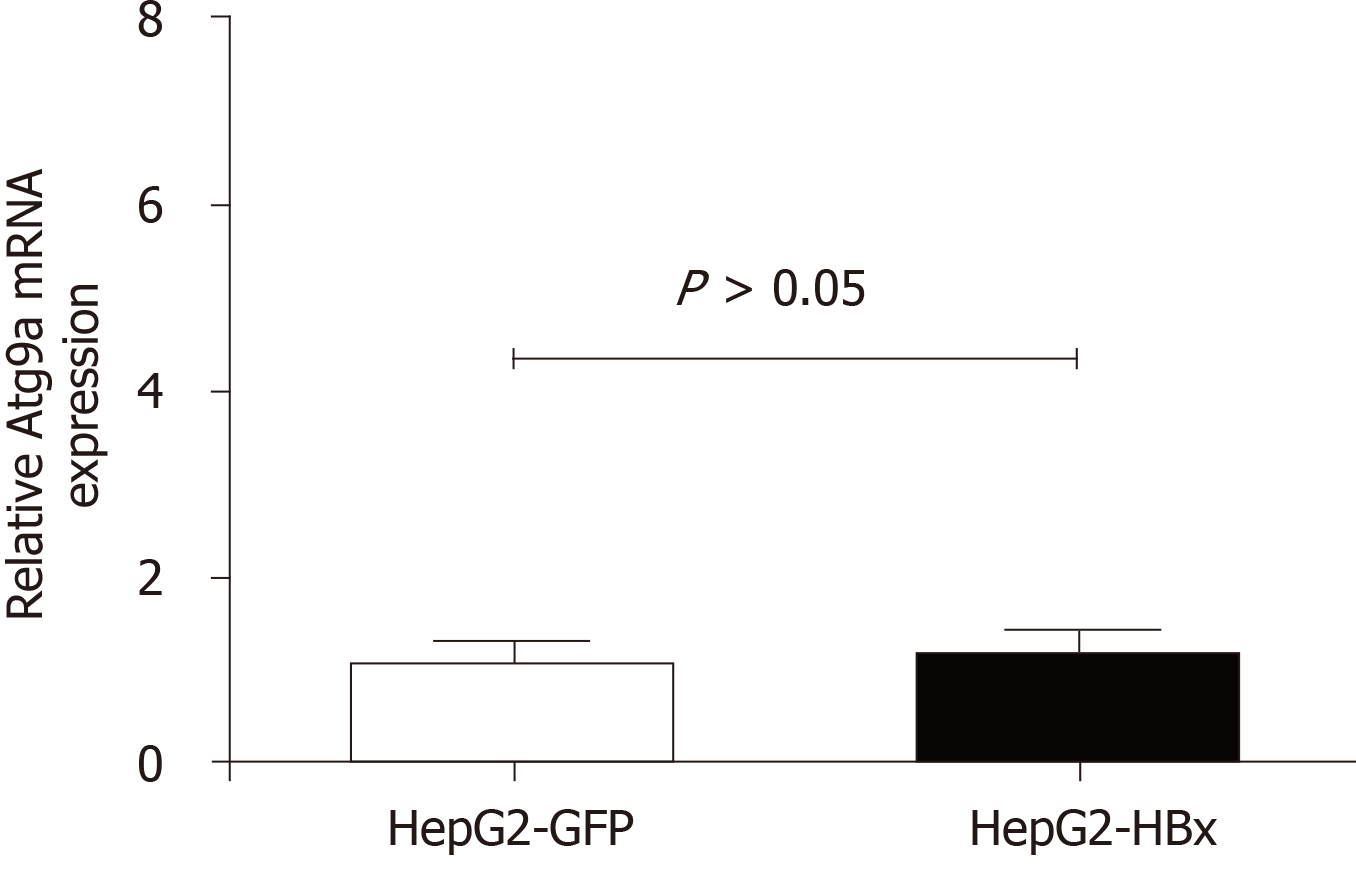
HBV induces autophagy via the HBx protein and is directly involved in starvation-induced autophagy via upregulation of Beclin-1 expression[5]. HBx also binds and activates phosphatidylinositol 3-kinase class III for autophagy induction[6]. Our study showed that overexpression of HBx did not affect ATG9A expression (Figure 3), suggesting that the function of ATG9A may not involve HBV replication or viral clearance.
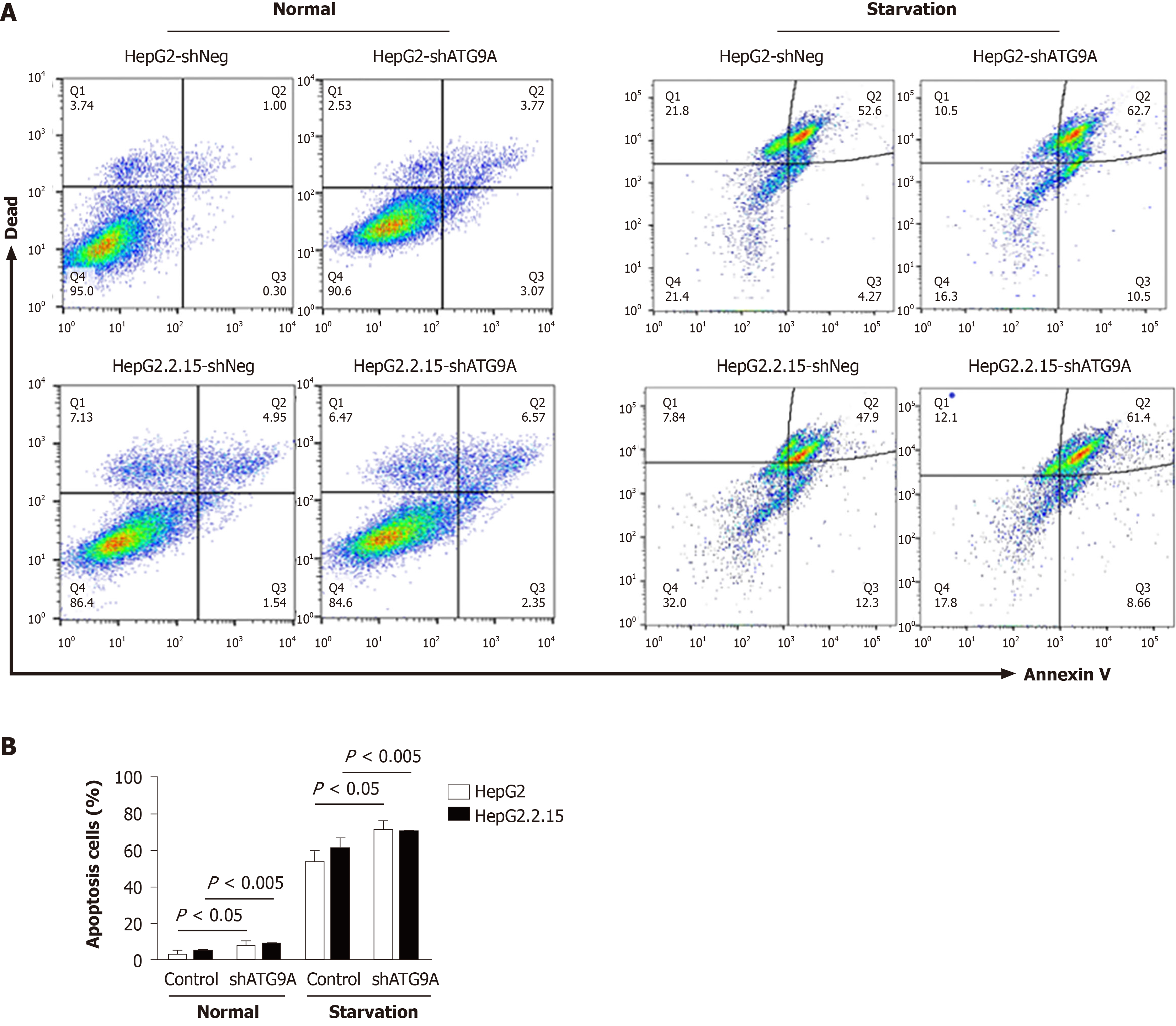
Autophagy is involved in tumor progression and tumor suppression. Several studies have shown that HBV induces autophagy for cell survival in an unsuitable environment[7]. In order to search for the effect of ATG9A on apoptosis, we performed flow cytometry in HepG2.2.15 cells and compared against HepG2 cells after ATG9A silencing. We found that silencing ATG9A increased apoptosis in both cell lines (Figure 4), suggesting that ATG9A is involved in cell apoptosis related to HCC.
In conclusion, we provide information that ATG9A is highly expressed in HBV-associated HCC tissue samples and plays a role in cell apoptosis. Further studies are needed to investigate the mechanism of ATG9A-mediated inhibition of apoptosis in HCC.
We are grateful to all the patients who participated in this study. We would like to thank Tangkijvanich P and Sirichindakul B for tissue biopsy collection and Hirankarn N for valuable advice.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Thailand
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chung YH, Fan X S-Editor: Gao CC L-Editor: Filipodia P-Editor: Xing YX
| 1. | Zhuang X, Chung KP, Cui Y, Lin W, Gao C, Kang BH, Jiang L. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci USA. 2017;114:E426-E435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 191] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 2. | Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842-20846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 563] [Cited by in RCA: 672] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 3. | Mitzel DN, Lowry V, Shirali AC, Liu Y, Stout-Delgado HW. Age-enhanced endoplasmic reticulum stress contributes to increased Atg9A inhibition of STING-mediated IFN-β production during Streptococcus pneumoniae infection. J Immunol. 2014;192:4273-4283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 4. | Kunanopparat A, Kimkong I, Palaga T, Tangkijvanich P, Sirichindakul B, Hirankarn N. Increased ATG5-ATG12 in hepatitis B virus-associated hepatocellular carcinoma and their role in apoptosis. World J Gastroenterol. 2016;22:8361-8374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 13] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 5. | Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, Li F, Wang Y, Tiollais P, Li T, Zhao M. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 201] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 6. | Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci USA. 2010;107:4383-4388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 253] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 7. | Ávalos Y, Canales J, Bravo-Sagua R, Criollo A, Lavandero S, Quest AF. Tumor suppression and promotion by autophagy. Biomed Res Int. 2014;2014:603980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 138] [Article Influence: 12.5] [Reference Citation Analysis (0)] |