Published online Sep 26, 2017. doi: 10.4252/wjsc.v9.i9.159
Peer-review started: May 10, 2017
First decision: June 16, 2017
Revised: June 29, 2017
Accepted: July 14, 2017
Article in press: July 16, 2017
Published online: September 26, 2017
Processing time: 134 Days and 9.7 Hours
To establish a model to enrich and characterize stem-like cells from murine normal liver and hepatocellular carcinoma (HCC) cell lines and to further investigate stem-like cell association with epithelial-to-mesenchymal transition (EMT).
In this study, we utilized a stem cell conditioned serum-free medium to enrich stem-like cells from mouse HCC and normal liver cell lines, Hepa 1-6 and AML12, respectively. We isolated the 3-dimensional spheres and assessed their stemness characteristics by evaluating the RNA levels of stemness genes and a cell surface stem cell marker by quantitative reverse transcriptase-PCR (qRT-PCR). Next, we examined the relationship between stem cells and EMT using qRT-PCR.
Three-dimensional spheres were enriched by culturing murine HCC and normal hepatocyte cell lines in stem cell conditioned serum-free medium supplemented with epidermal growth factor, basic fibroblast growth factor and heparin sulfate. The 3-dimensional spheres had enhanced stemness markers such as Klf4 and Bmi1 and hepatic cancer stem cell (CSC) marker Cd44 compared to parental cells grown as adherent cultures. We report that epithelial markers E-cadherin and ZO-1 were downregulated, while mesenchymal markers Vimentin and Fibronectin were upregulated in 3-dimensional spheres. The 3-dimensional spheres also exhibited changes in expression of Snai, Zeb and Twist family of EMT transcription factors.
Our novel method successfully enriched stem-like cells which possessed an EMT phenotype. The isolation and characterization of murine hepatic CSCs could establish a precise target for the development of more effective therapies for HCC.
Core tip: Although existing therapies can initially eliminate the bulk population of a tumor, the stem cell properties of cancer stem cells (CSCs) enable them to survive and repopulate the tumor, resulting in disease relapse. Therefore, elimination of CSCs has the potential to improve patient outcomes and survival. Isolation and characterization of liver CSCs is essential for the selective targeting of this crucial population of cells. We report that the sphere culture method is a more precise and reliable tool for the enrichment of murine stem-like cells which relies on their functional property of anchorage-independent growth.
- Citation: Jayachandran A, Shrestha R, Dhungel B, Huang IT, Vasconcelos MYK, Morrison BJ, Ramlogan-Steel CA, Steel JC. Murine hepatocellular carcinoma derived stem cells reveal epithelial-to-mesenchymal plasticity. World J Stem Cells 2017; 9(9): 159-168
- URL: https://www.wjgnet.com/1948-0210/full/v9/i9/159.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v9.i9.159
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide affecting one million individuals annually[1]. HCC is associated with high mortality rates largely due to the development of resistance to chemotherapy or radiotherapy, recurrence after surgery or intra-hepatic metastases[2]. While treatments such as surgical liver resection and liver transplantation have had a significant impact in early-stage HCC, these treatments have limited efficacy in most patients with advanced stage HCC[3]. Moreover, Sorafenib, the only available drug for advanced stage HCC has limited efficacy[4]. A better understanding of the biology of HCC would have a major impact on the management of this disease.
According to the stem cell model of carcinogenesis cancers are initiated and maintained by a rare fraction of cells called cancer stem cells (CSCs) or cancer initiating cells (CICs)[5,6]. The presence of CSCs with biological properties such as multipotency and self-renewal, similar to those of normal stem cells, was first reported in leukemia and subsequently in diverse malignancies including breast cancer, glioblastoma, prostate cancer, colon cancer and liver carcinoma[7-13]. CSCs have proven to play a central role in the development, maintenance, metastasis, and recurrence of HCC[14-16]. Therefore the prospective identification and isolation of CSCs in HCC could generate a better understanding of hepatocarcinogenesis and facilitate the identification of novel druggable targets for development of more efficient therapeutic strategies.
Recent evidence indicates that CSCs may be generated with the reactivation of the developmental epithelial-to-mesenchymal transition (EMT) program, which impacts tumor metastatic potential[17-19]. EMT describes a reprogramming of epithelial cells that leads to a phenotype switch from an epithelial to a mesenchymal cellular state. This cellular plasticity occurs during normal development as part of processes such as gastrulation and neural crest cell migration. During cancer progression, this phenotype is associated with metastatic dissemination, acquisition of drug resistance and acquisition of CSC state[20,21]. Whereas the role of EMT in HCC metastasis is well documented, its role in HCC CSC generation is only just emerging[22].
Although a number of cell surface markers have been identified for the enrichment of HCC derived CSCs, there is no general consensus on the best CSC markers for HCC[23,24]. We used an alternate method for the enrichment of HCC CSCs based on functional aspect of CSCs. CSCs exhibit anchorage-independent growth and form spheres that possess the capacity for self-renewal and tumorigenicity, when grown in a stem cell conditioned serum-free medium[25]. Sphere formation assay thus represent a more precise tool for the enrichment of CSCs. This study therefore aimed to enrich stem-like cells from mouse HCC and normal liver cell lines with the goal to better characterize the 3-dimensional spheres. We also sought to examine the relationship between CSCs and EMT.
Murine HCC cell line Hepa 1-6 and normal liver cell line AML12 were procured from American Type Culture Collection (ATCC) and maintained as per ATCC protocols. The cell lines Hepa 1-6 and AML12 were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Lonza, Australia) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Assay Matrix, Australia) and 1% penicillin/streptomycin (P/S) (Thermofischer Scientific, Australia) and incubated at 37 °C under a humidified atmosphere with 5% CO2 in air.
Cells were cultured as 3-dimensional spheres using a stem cell conditioned serum-free medium which is based on a neural stem cell medium[25]. Stem cell conditioned serum-free medium was prepared by adding 1:1 mixture of DMEM and HAM’s F12 medium (Lonza, Australia) supplemented with 4 μg/mL heparin sulfate (Sigma-Aldrich, United States), 1% penicillin/streptomycin (P/S) (ThermoFischer Scientific, Australia), 2% bovine serum albumin (BSA) (Sigma-Aldrich, United States), 20 ng/mL recombinant human epidermal growth factor (rhEGF) (Lonza, Australia) and 10 ng/mL recombinant human basic fibroblast growth factor (rhbFGF) (Lonza, Australia). Briefly, adherent cells were detached and collected following Trypsin-EDTA (ThermoFisher Scientific, Australia) treatment. Cells were washed three times with 50 mL 1 × PBS to remove serum. Cells were counted and seeded at 5000 cells/ml in a T-25 ultra-low-attachment flask (Corning Incorporated, United States) and cultured with stem cell medium at 37 °C in a humidified atmosphere of 5% CO2 in air.
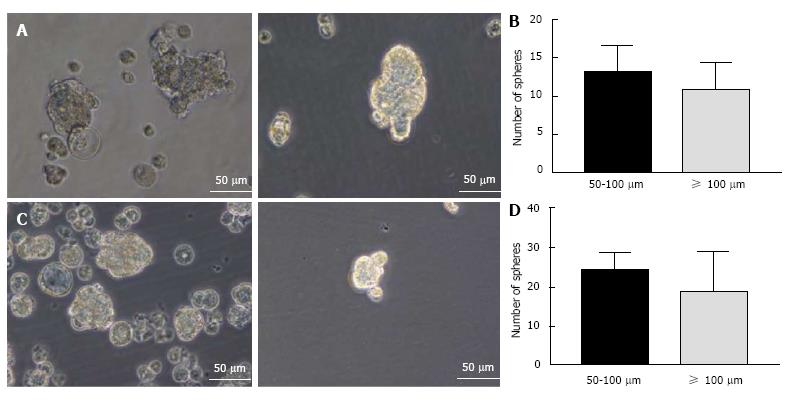
Cells were seeded at 2000 cells/well in a 6-well ultra-low-attachment plates (Corning Incorporated, United States) and cultured with stem cell medium. Diameter of 3-dimensional spheroids and number of spheres per culture well were counted on day 5 using an inverted microscope equipped with a digital camera (Olympus DP21, Japan).
The parental cells were plated at the same density as the sphere cells and on day 5 total cellular RNA was extracted using the Isolate II Bioline RNA synthesis kit (Bioline, Australia) as per the manufacturer’s protocol. We performed on column DNAase digestion using RNase-Free DNase at room temperature (20 °C-30 °C) for 15 min in accordance to Bioline RNA synthesis kit instructions. Spectrophotometric quantification using the Nanodrop 2000 c (ThermoFisher, United States) confirmed purity of RNA and absence of DNA in our samples. One micrograms of the extracted RNA was reverse transcribed using the Bioline SensiFAST cDNA synthesis kit (Bioline, Australia).
Following reverse transcription, quantitative reverse transcriptase-PCR (qRT-PCR) was performed using Lo-ROX SYBR Green (Bioline, Australia). Reactions were run in 384-well plates on a ViiA7 Applied Biosystems Real-Time PCR system. Amplification was performed according to a three-step cycle procedure consisting of 40 cycles of denaturation at 95 °C for 5 s, annealing at 60 °C for 10 s and extension at 75 °C for 15 s. E-cadherin expression was evaluated using QuantiFast SYBR Green PCR Kit (Qiagen, United States) following the manufacturer’s instructions. Amplification was performed according to a two-step cycling procedure consisting of 40 cycles of denaturation at 95 °C for 10 s and combined annealing/extension at 60 °C for 30 s. Beta-Actin (ActB) was used as an internal control. The primers used are listed in Table 1. Expression levels were normalized to ActB and are presented as copies of target gene per 10000 copies of ActB, calculated using the formula: 2 (CTActB-CTtarget) × 10000. The copy number values were calculated from a minimum of three independent biological replicates.
| Primer | Sequence (5’-3’) |
| ActB forward | ATGGAGGGGAATACAGCCC |
| ActB reverse | TTCTTTGCAGCTCCTTCGTT |
| Klf4 forward | CAGTGGTAAGGTTTCTCGCC |
| Klf4 reverse | GCCACCCACACTTGTGACTA |
| Bmi1 forward | TGGTTGTTCGATGCATTTCT |
| Bmi1 reverse | CTTTCATTGTCTTTTCCGCC |
| Cd44 forward | AGCGGCAGGTTACATTCAAA |
| Cd44 reverse | CAAGTTTTGGTGGCACACAG |
| E-Cadherin forward | AAAAGAAGGCTGTCCTTGGC |
| E-Cadherin reverse | GAGGTCTACACCTTCCCGGT |
| ZO-1 forward | CCTGTGAAGCGTCACTGTGT |
| ZO-1 reverse | CGCGGAGAGAGACAAGATGT |
| Vimentin forward | AGAGAGAGGAAGCCGAAAGC |
| Vimentin reverse | TCCACTTTCCGTTCAAGGTC |
| Fibronectin forward | ACTGGATGGGGTGGGAAT |
| Fibronectin reverse | GGAGTGGCACTGTCAACCTC |
| Snai1 forward | AGTGGGAGCAGGAGAATGG |
| Snai1 reverse | CTTGTGTCTGCACGACCTGT |
| Snai2 forward | GATGTGCCCTCAGGTTTGAT |
| Snai2 reverse | GGCTGCTTCAAGGACACATT |
| Zeb1 forward | TCATCGGAATCTGAATTTGC |
| Zeb1 reverse | CCAGGTGTAAGCGCAGAAAG |
| Zeb2 forward | TGCGTCCACTACGTTGTCAT |
| Zeb2 reverse | TCTTATCAATGAAGCAGCCG |
| Twist1 forward | CATGTCCGCGTCCCACTA |
| Twist1 reverse | TCCATTTTCTCCTTCTCTGGA |
| Twist2 forward | GCCTGAGATGTGCAGGTG |
| Twist2 reverse | GTCTCAGCTACGCCTTCTCC |
All experiments were repeated at least three times and representative results are presented. All statistical comparisons of data sets were performed using Student’s two-tailed t-test in GraphPad Prism software version 7.00 (GraphPad Software Inc). Statistical significance was set at aP <0.05, bP <0.01 and eP <0.001.
Mouse HCC cell line Hepa 1-6 and normal mouse liver cell line AML12 were used for induction of spheres. Both cell lines could form anchorage-independent, non-adherent 3-dimensional spheres when grown in conditioned serum-free culture medium supplemented with rhEGF, rhbFGF and heparin sulfate (Figure 1A and C). Both cell lines formed floating small spheres which eventually form 3-dimensional structures by day 5. No adherent cells were detected. The number of spheres were counted and appeared to be similar in both the cell types (Figure 1B and D).
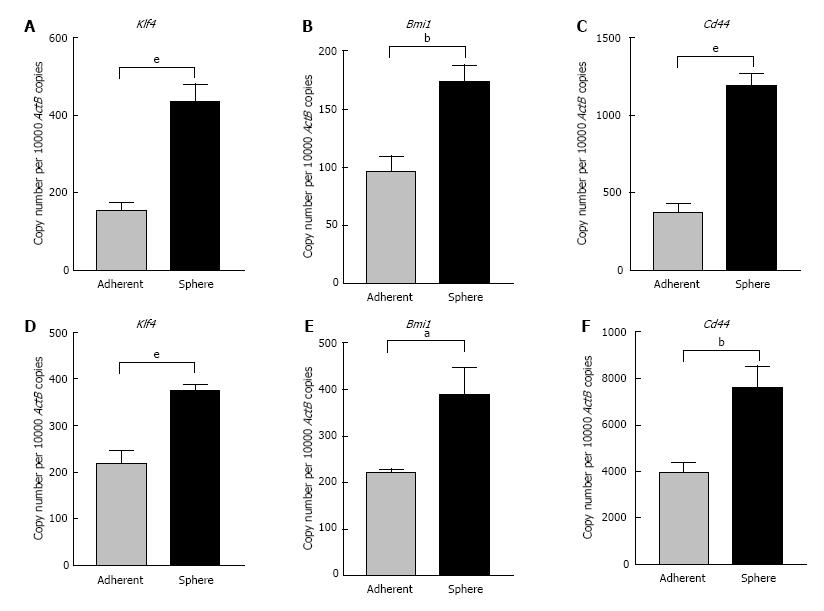
With the goal of better characterizing the cells enriched by sphere culture, we examined the expression levels of some stem cell-associated genes important for the proliferation, self-renewal and differentiation of stem cells. As controls, the parental cells were plated as adherent cultures at the same density as the spheres. On day 5 RNA was extracted from 3-dimensional sphere cultures and adherent cultures. qRT-PCR analysis revealed markedly elevated expression of embryonic stem cell-associated genes Kruppel like factor 4 (Klf4) and Bmi1 polycomb ring finger oncogene (Bmi1) in Hepa 1-6 spheres compared with parental cells (Figure 2A and B). Cd44, a cell surface adhesion molecule which has been used as a CSC marker in HCC showed significantly increased expression in Hepa 1-6 spheres compared with adherent parental cells (Figure 2C). Similarly, AML12 derived 3-dimensional spheres also expressed significantly higher mRNA levels of Klf4 and Bmi1 compared with the adherent AML12 population (Figure 2D and E). Higher expression of Cd44 was detected in spheres from AML12 compared with the parental cells (Figure 2F). These results indicate that the conditioned stem cell serum-free medium is a precise tool for the selective enrichment of hepatic mouse stem-like cells.
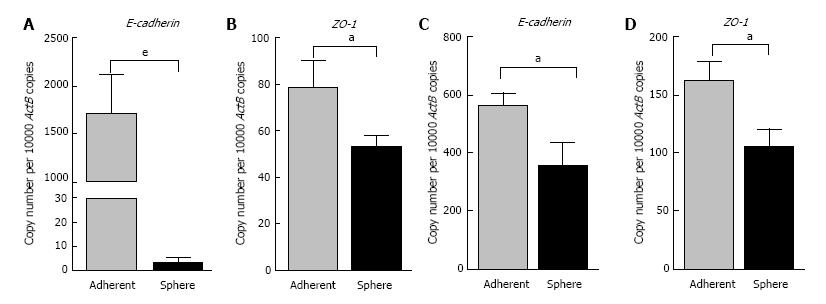
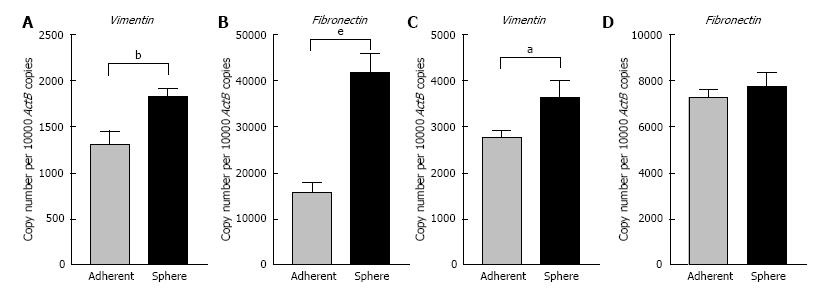
To elucidate whether there were connections between the spheres and EMT phenotype, we assessed the EMT characteristics of the 3-dimensional spheres from Hepa 1-6 and AML12. At the molecular level, EMT is characterized by a series of coordinated changes including down-regulation of the adherens junction molecule E-cadherin and tight junction molecule Zonula occludens-1 (ZO-1) and upregulation of Vimentin, an intermediate filament and Fibronectin, a key molecule of extracellular matrix. We observed that the expression of classical epithelial marker genes, E-cadherin and ZO-1 were significantly downregulated in 3-dimensional spheres from both Hepa 1-6 and AML12 compared with parental cells (Figure 3). These 3-dimensional spheres also exhibited the characteristic features of a mesenchymal phenotype with high expression of Vimentin and Fibronectin (Figure 4). These findings suggest that the stem cell phenotype is closely linked with an EMT phenotype.
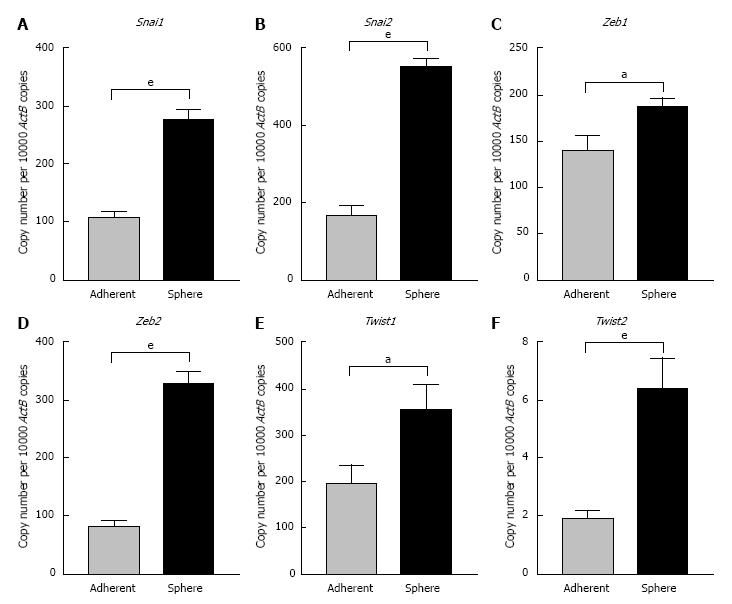
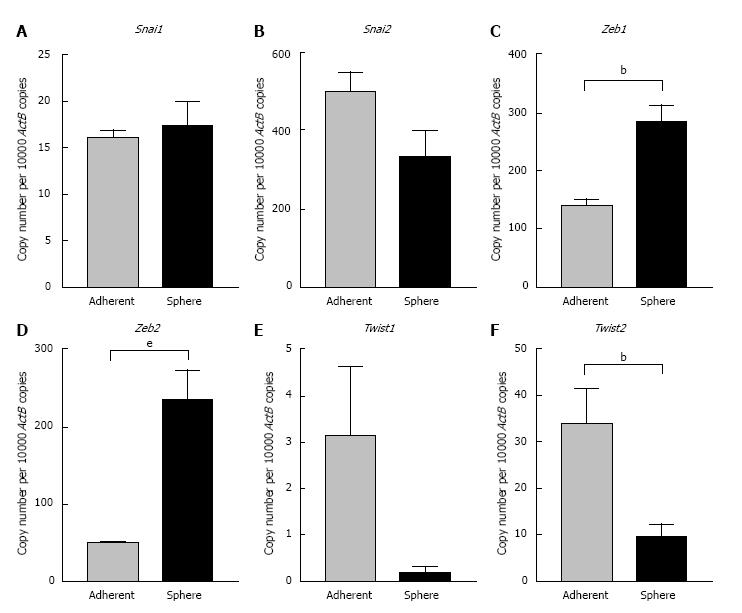
To further confirm the occurrence of EMT process in CSCs we examined the expression levels of core EMT transcription factors that govern cellular plasticity. In Hepa 1-6 spheres we observed significant upregulation of Snai family of transcription factors (Snai1 and 2), Zinc-finger E-box-binding homeobox family of transcription factors (Zeb1 and 2) and helix-loop-helix Twist family of transcription factors (Twist1 and 2) compared with adherent parental cells (Figure 5). We observed a significant increase in the Zeb family of transcription factors in AML12 3-dimensional spheres compared with parental cells. AML12 3-dimensional spheres showed downregulation of Snai2, Twist1 and Twist2 RNA levels (Figure 6). This raises the possibility that distinct family of transcription factor may enable maintenance of CSC cellular plasticity in different cell types. Together, these features of EMT strongly suggest a possible relationship of EMT with the hepatic stem-like cell phenotype.
Worldwide, HCC, a primary liver cancer is one of the most common malignancies with a poor outcome[2]. Non-resectable advanced stage HCC remains an incurable disease for which novel therapies are urgently needed. Accumulating evidence suggests that CSCs play an important role in HCC tumorigenicity and the reactivation of EMT process has been implicated in the generation of CSCs[22]. The CSC field has experienced rapid advances in the past decade and a number of strategies have been applied to identify and harvest them[12-14,26-28]. Several markers have been proposed for the identification of CSCs in HCC, but not all are uniformly expressed in all CSC populations and single markers have been deemed insufficient to represent the real CSC phenotype[24]. Alternately, the sphere culture method, which is not dependent on markers, has been increasingly utilized in various tumors, including HCC for isolating, enriching, maintaining or expanding the potential CSC subpopulations[25,29-32]. To our knowledge, this is the first time that murine HCC and normal hepatocyte cell lines have been examined for sphere forming capacity, enrichment of stem-like cells and occurrence of epithelial-mesenchymal plasticity.
Enrichment and characterization of murine derived CSCs provides a better understanding of how these CSCs interact with the CSC niche environment and host immune system in order to form a tumor and are indispensable for the development of new therapies for the elimination of CSCs. In HCC, the majority of studies of CSCs have utilized patient-derived material or established human tumor cell lines inoculated into immunocompromised mice[29,31,32]. The immunocompromised mouse microenvironments do not recapitulate the microenvironment in a human patient with naturally occurring cancer and have limited value in assessing therapies targeting CSCs. Moreover, the ability of cells to grow in immunocompromised mice does not distinguish CSCs from non-CSCs, as it demonstrates selection for cells that can best adapt to growth in murine tissue, and therefore might not represent a true approximation of CSCs[25]. We have previously demonstrated that immunocompetent syngeneic models allow for interactions of the recipient mouse host immune system with CSCs, a situation that more closely models cancer in humans[25]. Future studies are needed to address whether mouse HCC derived CSCs are able to initiate tumors in syngeneic immunocompetent mice compared with the parental counterparts.
Our stem cell enrichment medium comprised of serum free media supplemented with rhEGF, rhbFGF and heparin sulfate, while others have previously used media supplements such as B27, leukemia inhibitory factor, N-acetyl-L-cysteine and neural survival factor for enriching human HCC CSCs[29,30]. Our finding that murine 3-dimensional spheres had enhanced expression of stem cell markers namely, Klf4, Bmi1 and Cd44 lends credence to the use of the sphere culture model for CSC enrichment. Positive expression of KLF4 was correlated with tumor relapse and a poor prognosis in patients with HCC[33]. CD44 expression was highly correlated with decreased overall survival in HCC patients[34] while high BMI1 expression was associated with a poor prognosis in HCC patients[35].
Finally we demonstrate a striking association between the expression of CSC and EMT markers. The biologic link between EMT phenotypes and CSCs has recently been evidenced in many types of cancer, including HCC[3,22]. E-cadherin functions as a key gatekeeper of the epithelial state. Loss or downregulation of E-cadherin has been considered to be a hallmark of EMT[20,21]. In our study, 3-dimensional spheres demonstrated down-regulation of E-cadherin and ZO-1. We also found that the 3-dimensional spheres exhibited high Vimentin and Fibronectin, the phenotypes of mesenchymal cells that have more aggressive biological behaviour. Most notably, we found elevation of core EMT transcription factors in 3-dimensional spheres. Downregulation of E-cadherin is often mediated by core EMT-controlling transcription factors of Snai, Zeb and Twist families which have recently been molecularly linked to self-renewal programs[36]. AML12 have yielded mixed results for EMT transcription factors in spheres with downregulation of Twists and Snai2. This indicates apparent cell type-specific differences and the cause for this variance in transcription factor expression remains elusive and warrants further investigation. Taken together, our findings indicate that EMT transcription factors such as Snai1, Zeb1 and 2 may provide opportunities for therapeutic targeting of CSC via blocking EMT. An in-depth investigation of crosstalk of stemness with EMT is essential for a better understanding of tumor progression in HCC. It is clear that further studies of CSC characterization will be critical to better understand plasticity and the mediators of phenotype switching as contributors to HCC initiation, progression, treatment failure and disease relapse. As a central player in these processes, EMT transcription factors may well serve as druggable targets in strategies to better treat HCC.
We would like to acknowledge the Gallipoli Medical Research Foundation and the Cyril Gilbert Foundation for funding of this project. We would like to thank Dr Prashanth Prithviraj from the Fiona Elsey Cancer Research Institute for reviewing the statistics in this manuscript.
Cancer stem cells (CSCs) have proven to play a central role in the development, maintenance, metastasis, and recurrence of hepatocellular carcinoma (HCC). Therefore the prospective identification and isolation of CSCs in HCC could generate a better understanding of hepatocarcinogenesis and facilitate the identification of novel druggable targets for development of more efficient therapeutic strategies.
Although a number of cell surface markers have been identified for the enrichment of HCC derived CSCs, there is no general consensus on the best CSC markers for HCC. The authors used an alternate method for the enrichment of HCC CSCs based on functional aspect of CSCs.
To the knowledge, this is the first time that murine HCC and normal hepatocyte cell lines have been examined for sphere forming capacity, enrichment of stem-like cells and occurrence of epithelial-mesenchymal plasticity.
The authors’ findings indicate that EMT transcription factors such as Snai1, Zeb1 and 2 may provide opportunities for therapeutic targeting of CSC via blocking EMT. An in-depth investigation of crosstalk of stemness with EMT is essential for a better understanding of tumor progression in HCC. It is clear that further studies of CSC characterization will be critical to better understand plasticity and the mediators of phenotype switching as contributors to HCC initiation, progression, treatment failure and disease relapse.
CSC is cancer stem cells which have biological properties such as multipotency and self-renewal, similar to those of normal stem cells. EMT describes epithelial-to-mesenchymal transition, a reprogramming of epithelial cells that leads to a phenotype switch from an epithelial to a mesenchymal cellular state.
This manuscript is interesting, presenting a feasible method for concentrating a stem-like population from hepatic cancer cells by extending their previously reported technique for enriching a cancer-initiating population from lung cancer cell lines.
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country of origin: Australia
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Cao T, Kiselev SL, Saeki K S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1097] [Cited by in RCA: 1102] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 2. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4265] [Article Influence: 236.9] [Reference Citation Analysis (2)] |
| 3. | Mir N, Jayachandran A, Dhungel B, Shrestha R, Steel JC. Epithelial-to-Mesenchymal Transition: a Mediator of Sorafenib Resistance in Advanced Hepatocellular Carcinoma. Curr Cancer Drug Targets. 2017;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 4. | Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9016] [Cited by in RCA: 10265] [Article Influence: 603.8] [Reference Citation Analysis (2)] |
| 5. | Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6844] [Cited by in RCA: 6914] [Article Influence: 288.1] [Reference Citation Analysis (0)] |
| 6. | Morrison BJ, Morris JC, Steel JC. Lung cancer-initiating cells: a novel target for cancer therapy. Target Oncol. 2013;8:159-172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 7. | Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4851] [Cited by in RCA: 4861] [Article Influence: 173.6] [Reference Citation Analysis (1)] |
| 8. | Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983-3988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7830] [Cited by in RCA: 7727] [Article Influence: 351.2] [Reference Citation Analysis (0)] |
| 9. | Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821-5828. [PubMed] |
| 10. | Patrawala L, Calhoun-Davis T, Schneider-Broussard R, Tang DG. Hierarchical organization of prostate cancer cells in xenograft tumors: the CD44+alpha2beta1+ cell population is enriched in tumor-initiating cells. Cancer Res. 2007;67:6796-6805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 267] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 11. | O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2977] [Cited by in RCA: 3051] [Article Influence: 160.6] [Reference Citation Analysis (0)] |
| 12. | Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, Nakauchi H, Taniguchi H. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 494] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 13. | Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, Zheng BJ, Guan XY. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542-2556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 899] [Cited by in RCA: 922] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 14. | Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu PW, Lam CT, Poon RT, Fan ST. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 883] [Cited by in RCA: 927] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 15. | Ma S, Lee TK, Zheng BJ, Chan KW, Guan XY. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749-1758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 605] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 16. | Song W, Li H, Tao K, Li R, Song Z, Zhao Q, Zhang F, Dou K. Expression and clinical significance of the stem cell marker CD133 in hepatocellular carcinoma. Int J Clin Pract. 2008;62:1212-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 182] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 17. | Kawai T, Yasuchika K, Ishii T, Katayama H, Yoshitoshi EY, Ogiso S, Kita S, Yasuda K, Fukumitsu K, Mizumoto M. Keratin 19, a Cancer Stem Cell Marker in Human Hepatocellular Carcinoma. Clin Cancer Res. 2015;21:3081-3091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 135] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 18. | Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704-715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6972] [Cited by in RCA: 6806] [Article Influence: 400.4] [Reference Citation Analysis (0)] |
| 19. | Morel AP, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1116] [Cited by in RCA: 1240] [Article Influence: 72.9] [Reference Citation Analysis (1)] |
| 20. | Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 754] [Article Influence: 62.8] [Reference Citation Analysis (0)] |
| 21. | Jayachandran A, Anaka M, Prithviraj P, Hudson C, McKeown SJ, Lo PH, Vella LJ, Goding CR, Cebon J, Behren A. Thrombospondin 1 promotes an aggressive phenotype through epithelial-to-mesenchymal transition in human melanoma. Oncotarget. 2014;5:5782-5797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 104] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 22. | Tögel L, Nightingale R, Chueh AC, Jayachandran A, Tran H, Phesse T, Wu R, Sieber OM, Arango D, Dhillon AS. Dual Targeting of Bromodomain and Extraterminal Domain Proteins, and WNT or MAPK Signaling, Inhibits c-MYC Expression and Proliferation of Colorectal Cancer Cells. Mol Cancer Ther. 2016;15:1217-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 23. | Liu LL, Fu D, Ma Y, Shen XZ. The power and the promise of liver cancer stem cell markers. Stem Cells Dev. 2011;20:2023-2030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 24. | Wilson GS, Hu Z, Duan W, Tian A, Wang XM, McLeod D, Lam V, George J, Qiao L. Efficacy of using cancer stem cell markers in isolating and characterizing liver cancer stem cells. Stem Cells Dev. 2013;22:2655-2664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 25. | Morrison BJ, Steel JC, Morris JC. Sphere culture of murine lung cancer cell lines are enriched with cancer initiating cells. PLoS One. 2012;7:e49752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 26. | Deleyrolle LP, Ericksson G, Morrison BJ, Lopez JA, Burrage K, Burrage P, Vescovi A, Rietze RL, Reynolds BA. Determination of somatic and cancer stem cell self-renewing symmetric division rate using sphere assays. PLoS One. 2011;6:e15844. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 27. | Ma S, Chan KW, Lee TK, Tang KH, Wo JY, Zheng BJ, Guan XY. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res. 2008;6:1146-1153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 367] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 28. | Zhu Z, Hao X, Yan M, Yao M, Ge C, Gu J, Li J. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int J Cancer. 2010;126:2067-2078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 225] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 29. | Cao L, Zhou Y, Zhai B, Liao J, Xu W, Zhang R, Li J, Zhang Y, Chen L, Qian H. Sphere-forming cell subpopulations with cancer stem cell properties in human hepatoma cell lines. BMC Gastroenterol. 2011;11:71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 215] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 30. | Hashimoto N, Tsunedomi R, Yoshimura K, Watanabe Y, Hazama S, Oka M. Cancer stem-like sphere cells induced from de-differentiated hepatocellular carcinoma-derived cell lines possess the resistance to anti-cancer drugs. BMC Cancer. 2014;14:722. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 31. | Li J, Yu Y, Wang J, Yan Z, Liu H, Wang Y, Ding M, Cui L, Wu M, Jiang X. Establishment of a novel system for the culture and expansion of hepatic stem-like cancer cells. Cancer Lett. 2015;360:177-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 32. | Kwon YC, Bose SK, Steele R, Meyer K, Di Bisceglie AM, Ray RB, Ray R. Promotion of Cancer Stem-Like Cell Properties in Hepatitis C Virus-Infected Hepatocytes. J Virol. 2015;89:11549-11556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 33. | Yin X, Li YW, Jin JJ, Zhou Y, Ren ZG, Qiu SJ, Zhang BH. The clinical and prognostic implications of pluripotent stem cell gene expression in hepatocellular carcinoma. Oncol Lett. 2013;5:1155-1162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 34. | Luo Y, Tan Y. Prognostic value of CD44 expression in patients with hepatocellular carcinoma: meta-analysis. Cancer Cell Int. 2016;16:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 35. | Zhai R, Tang F, Gong J, Zhang J, Lei B, Li B, Wei Y, Liang X, Tang B, He S. The relationship between the expression of USP22, BMI1, and EZH2 in hepatocellular carcinoma and their impacts on prognosis. Onco Targets Ther. 2016;9:6987-6998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 36. | Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 640] [Cited by in RCA: 815] [Article Influence: 74.1] [Reference Citation Analysis (0)] |