Published online Oct 26, 2017. doi: 10.4252/wjsc.v9.i10.179
Peer-review started: April 26, 2017
First decision: June 6, 2017
Revised: August 28, 2017
Accepted: September 12, 2017
Article in press: September 13, 2017
Published online: October 26, 2017
Processing time: 182 Days and 18.8 Hours
To identify and characterize functionally distinct subpopulation of adipose-derived stem cells (ADSCs).
ADSCs cultured from mouse subcutaneous adipose tissue were sorted fluorescence-activated cell sorter based on aldehyde dehydrogenase (ALDH) activity, a widely used stem cell marker. Differentiation potentials were analyzed by utilizing immunocytofluorescece and its quantitative analysis.
Approximately 15% of bulk ADSCs showed high ALDH activity in flow cytometric analysis. Although significant difference was not seen in proliferation capacity, the adipogenic and osteogenic differentiation capacity was higher in ALDHHi subpopulations than in ALDHLo. Gene set enrichment analysis revealed that ribosome-related gene sets were enriched in the ALDHHi subpopulation.
High ALDH activity is a useful marker for identifying functionally different subpopulations in murine ADSCs. Additionally, we suggested the importance of ribosome for differentiation of ADSCs by gene set enrichment analysis.
Core tip: Aldehyde dehydrogenase (ALDH) activity is widely used as a stem cell marker in several types of normal or malignant tissues. However, there was no report of ALDH activity in murine adipose-derived stem cells (ADSCs). Here, our study demonstrated a subpopulation defined by high ALDH activity within murine ADSCs. The subpopulation with high ALDH activity (ALDHHi) showed enhanced differentiation potentials into adipocyte and osteocyte. Furthermore, gene set enrichment analysis revealed that ribosome-related gene sets were enriched in ALDHHi of murine ADSCs. We showed relationship between ALDHHi and ribosome biosynthesis, providing a novel insight of mesenchymal stem cell biology.
- Citation: Itoh H, Nishikawa S, Haraguchi T, Arikawa Y, Eto S, Hiyama M, Iseri T, Itoh Y, Nakaichi M, Sakai Y, Tani K, Taura Y, Itamoto K. Aldehyde dehydrogenase activity helps identify a subpopulation of murine adipose-derived stem cells with enhanced adipogenic and osteogenic differentiation potential. World J Stem Cells 2017; 9(10): 179-186
- URL: https://www.wjgnet.com/1948-0210/full/v9/i10/179.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v9.i10.179
Stem cells can self-renew and differentiate into specialized cells of various tissues[1]. Therefore, these cells, for example, embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), hematopoietic stem cells, and mesenchymal stem cells (MSCs), have been the object of basic research and clinical applications. Among these types of stem cells, MSCs, as represented by adipose-derived stem/stromal cells (ADSCs) and bone marrow-derived stem/stromal cells (BMSCs), have been recognized as useful material for cell-based therapy[2]. MSCs have been isolated from various tissues, including adipose tissue, the bone marrow, peripheral blood, cord blood, the liver, dental pulp, and fetal tissue; of these, adipose tissue is one of the most abundant source of MSCs[3]. ADSCs possess multipotency and have the potential to differentiate into cell types such as adipocytes, osteocytes, chondrocytes, neurons, vascular endothelial cells, cardiomyocytes, myoblasts, and islet β-cells under appropriate conditions[4].
The researches have suggested that ADSCs are heterogeneous and comprise phenotypically and/or functionally different subpopulations[5-7]. For example, the cluster of differentiation (CD)73+ subpopulation of murine ADSCs possesses increased potential for cardiomyocyte differentiation[6]. The CD90+ subpopulation of murine ADSCs has higher tube-forming ability than the CD90– subpopulation, which has high adipogenic potential[8]. The CD90+ subpopulation also exhibits higher efficiency of iPSC induction than the CD90– subpopulation[5]. Human ADSCs also include the CD105Lo subpopulation, which has high osteogenic potential[7]. Some studies have identified different subpopulations in ADSCs on the basis of surface antigen markers[5-7]. However, it is unclear how these markers (e.g., CD90 and CD105) are functionally related to cell differentiation.
In mice, aldehyde dehydrogenase (ALDH) is a superfamily comprising 20 intracellular enzymes and is responsible for the oxidization of various aldehydes[9]. High ALDH activity has been shown in normal hematopoietic stem cells, neural stem cells, and cancer stem cells in various types of neoplastic diseases[10]. Therefore, high ALDH activity is considered to be a common marker for normal and malignant stem cells. In human ADSCs, however, only one study has been performed on the ALDHHi subpopulation, whose significance in differentiation potential is unclear[11]. Moreover, to our knowledge, the existence of the ALDHHi subpopulation within murine ADSCs has not yet been reported.
In the current study, the ALDHHi and ALDHLo subpopulations of murine ADSCs were sorted using flow cytometry. The differentiation potential and proliferation of the sorted ALDHHi and ALDHLo subpopulations were analyzed. Furthermore, we analyzed the transcriptional profiles of the ALDHHi and ALDHLo subpopulations by utilizing gene set enrichment analysis (GSEA).
C57BL/6J mice were purchased from Kyudo Co., Ltd (Saga, Japan). All animal experiments were carried out in accordance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978) and the institutional guidelines of Yamaguchi University. The animal experiments were approved by the institutional animal experiment ethics committee of Yamaguchi University.
Murine ADSCs were isolated from twenty of C57BL/6J female mice of 4- to 6-wk-old, as previously described[8]. Briefly, the subcutaneous adipose tissue was resected, washed with Dulbecco’s phosphate-buffered saline (DPBS; Wako, Osaka, Japan), and cut into small pieces. The adipose tissue pieces were digested in high glucose Dulbecco’s modified Eagle’s medium (DMEM; Wako, Osaka, Japan) containing 1.0 mg/mL collagenase type I (Sigma-Aldrich, St. Louis, MO, United States), 10% fetal bovine serum (FBS; Sigma-Aldrich), and antibiotic-antimycotic agents (PSM; penicillin: 100 U/mL, streptomycin: 100 μg/mL, and amphotericin B: 0.25 μg/mL, final concentrations; Nacalai Tesque, Kyoto, Japan), using a shaking incubator at 37.5 °C and 250 rpm for 1 h. The digested tissue was filtered through a sterile ø100-μm nylon mesh (EASYstrainer, 100 μm; Greiner Bio-One Japan, Tokyo, Japan), followed by centrifugation at 400 × g for 5 min in DPBS supplemented with 1% FBS and 1 mmol/L EDTA∙3Na (Wako, Osaka, Japan). The pellet was resuspended in DMEM supplemented with 10% FBS and antibacterial/antimycotic agent and was cultured at 37.0 °C in a 5% CO2 atmosphere, using ø10 cm dish (Corning, NY, United States). When the cultures reached 80%-90% confluence, the ADSCs were dissociated from the dish by using Accutase solution (Innovative Cell Technologies, San Diego, United States), and seeded into new dishes.
Adherent ADSCs from passage 4 were dissociated using Accutase solution; 1 × 106 cells were resuspended and incubated for 5 min on ice with 2 μL of anti-mouse CD16/32 rat monoclonal antibody (BioLegend, San Diego, CA, United States). Cells were stained with 1 μL viability probe (Zombie NIR, Biolegend) for 20 min at room temperature to stain dead cells. ALDH activity was assessed by utilizing the ALDEFLUOR kit (Stemcell Technologies, Vancouver, Canada) according to the manufacturer’s instructions. Briefly, 1 × 106 cells were resuspended in 1 mL assay buffer and 5 μL ALDEFLUOR reagent was added after thorough mixing; then, 0.5 mL of the cell suspension was transferred to a new tube with 5 μL diethylaminobenzaldehyde (DEAB) reagent (ALDH inhibitor) for negative control of ALDH activity. Flow cytometric analysis and cell sorting were performed using Accuri C6 (BD Bioscience, San Jose, CA, United States) and the SH800 cell sorter (Sony, Tokyo, Japan). Flow cytometric data were analyzed with the FlowJo (Tree Star, Ashland, OR, United States) software.
To assess the viability of the ADSC subpopulations, we used a cell WST-8 assay (Cell Counting Kit-8; Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. Briefly, sorted ALDHHi or ALDHLo murine ADSCs were seeded in 96-well plates at a density of 3 × 103 cells/well. After 12, 24, 48, and 72 h, 100 μL fresh medium containing 10 μL CCK-8 solution was added to each well, followed by incubation at 37 °C for 1 h. The absorbance of each well at 450 nm was measured using an Epoch microplate spectrophotometer (BioTek Instruments, Winooski, VT, United States). Six replicates were prepared for each group.
The adipogenic and osteogenic differentiations of ADSCs were characterized using a Mouse Mesenchymal Stem Cell Functional Identification Kit (R and D Systems, Minneapolis, MN, United States) according to the manufacturer’s instructions. Briefly, for adipogenic differentiation, cells (3 × 103/well) were cultured at 37 °C in a 5% CO2 atmosphere in a 96-well plate in 100 μL adipogenic differentiation medium composed of α-minimal essential medium (αMEM) supplemented with 10% FBS, 1% PSM, L-glutamine, and 50 μL adipogenic supplement containing hydrocortisone, isobutylmethylxanthine, and indomethacin for 15 d in 37 °C and a 5% CO2 atmosphere.
For osteogenic differentiation, cells were cultured in osteogenic differentiation medium composed of 5 mL α-MEM basal medium and 250 μL osteogenic supplement containing ascorbate-phosphate, β-glycerolphosphate, and recombinant human bone morphogenetic protein-2 for 15 d in 37 °C and a 5% CO2 atmosphere. The medium was replaced every 2-3 d.
To assess adipogenic and osteogenic differentiation by immunocytochemistry, cultured cells were fixed in 4% paraformaldehyde phosphate buffer solution (Wako, Osaka, Japan) for 20 min. After the cells were washed with DPBS, they were permeabilized and blocked with DPBS supplemented with 0.3% Triton X-100 (Sigma-Aldrich), and 10% FBS for 45 min. The cells were subsequently incubated for 1 h in DPBS containing 10 μg/mL goat anti-mouse fatty acid binding protein (FABP) 4 polyclonal antibody to label adipocytes or were incubated with 10 μg/mL goat anti-mouse osteopontin polyclonal antibody to label osteocytes. They were then washed with DPBS and incubated for 1 h in DPBS containing phycoerythrin (PE)-conjugated rabbit anti-goat IgG antibody [rabbit F(ab’)2 anti-goat IgG-H and L (PE), pre-adsorbed, Abcam, Cambridge, United Kingdom]. Nuclei were stained with 5 μg/mL Hoechst 33342 (Dojindo Laboratories, Kumamoto, Japan). Photographs were obtained and analyzed using a fluorescence microscope (BZ-9000; Keyence, Osaka, Japan) and its analysis software.
Gene expression array analysis and GSEA were performed on the published gene expression profile of C57BL/6 mice divided by ALDHhi and ALDHlo subpopulations of ADSCs. About 3 × 106 cells from each subpopulation were lysed and total RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Cyanine-3 (Cy3)-labeled cRNA was prepared from 0.1 μg total RNA by using the Low Input Quick Amp Labeling Kit (Agilent Technologies, Santa Clara, CA, United States) according to the manufacturer’s instructions; this was followed by RNeasy column purification (Qiagen). Dye incorporation and cRNA yield were checked with the NanoDrop ND-2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). Cy3-labelled cRNA (0.6 μg) was fragmented at 60 °C for 30 min in a reaction volume of 25 μL containing 1 × Agilent fragmentation buffer and 2 × Agilent blocking agent following the manufacturer’s instructions. On completion of the fragmentation reaction, 25 μL of 2 × Agilent hybridization buffer was added to the fragmentation mixture and hybridized to SurePrint G3 Mouse GE 8 × 60 K Ver1.0 (Agilent Technologies) for 17 h at 65 °C in a rotating Agilent hybridization oven. After hybridization, the microarrays were washed for 1 min at room temperature with GE Wash Buffer 1 (Agilent Technologies) and 1 min with 37 °C GE Wash buffer 2 (Agilent Technologies). The slides were scanned immediately after washing on the Agilent SureScan Microarray Scanner (G2600D), using one color scan setting for 8 × 60 k array slides (scan area, 61 × 21.6 mm; scan resolution, 3 μm; dye channel set for Green PMT was set to 100%). The scanned images were analyzed with Feature Extraction Software 11.5.1.1 (Agilent Technologies), using default parameters to obtain the subtracted background and spatially detrended Processed Signal intensities.
Statistical analysis was performed using GraphPad Prism version 6.01 for Windows (GraphPad Software, La Jolla, CA, United States). The results have been expressed in terms of mean ± SE. Comparisons of two groups were performed with the independent t-test. Multiple comparisons were performed with one-way analysis of variance. Data were considered statistically significant when the P value was ≤ 0.05.
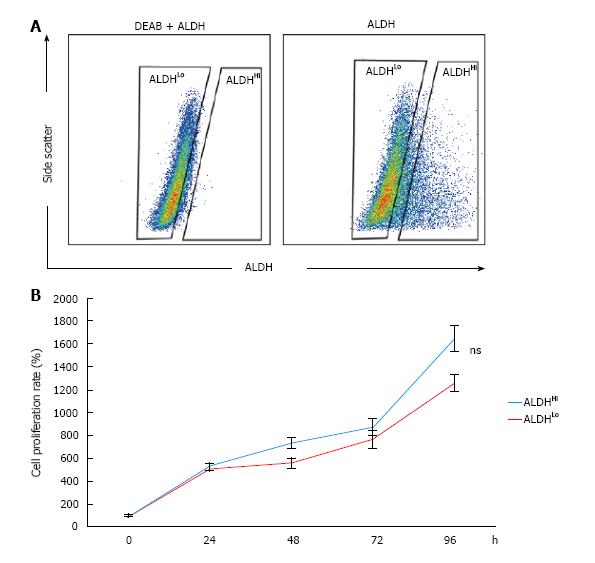
To identify the subpopulation defined by ALDH activity in murine ADSCs, single-cell suspensions of cultured murine ADSCs were stained using the ALDEFLUOR kit and analyzed with flow cytometry. A small subpopulation with distinctively high ALDH activity (ALDHHi cells) was detected within the bulk populations of ADSCs (Figure 1A). The percentage of ALDHHi cells was approximately 15% of the bulk murine ADSC population (Figure 1A). However, on adding the ALDH inhibitor N,N-diethylaminobenzaldehyde (DEAB), a distinct ALDHHi subpopulation was not detected (Figure 1A). To assess the difference in the proliferation potentials of the ALDHHi and ALDHLo subpopulations, we measured the proliferation rate of each subpopulation by using the WST assay. The proliferation potential of the ALDHHi subpopulation of ADSCs was not significantly different compared to the ALDHLo subpopulation (Figure 1B).
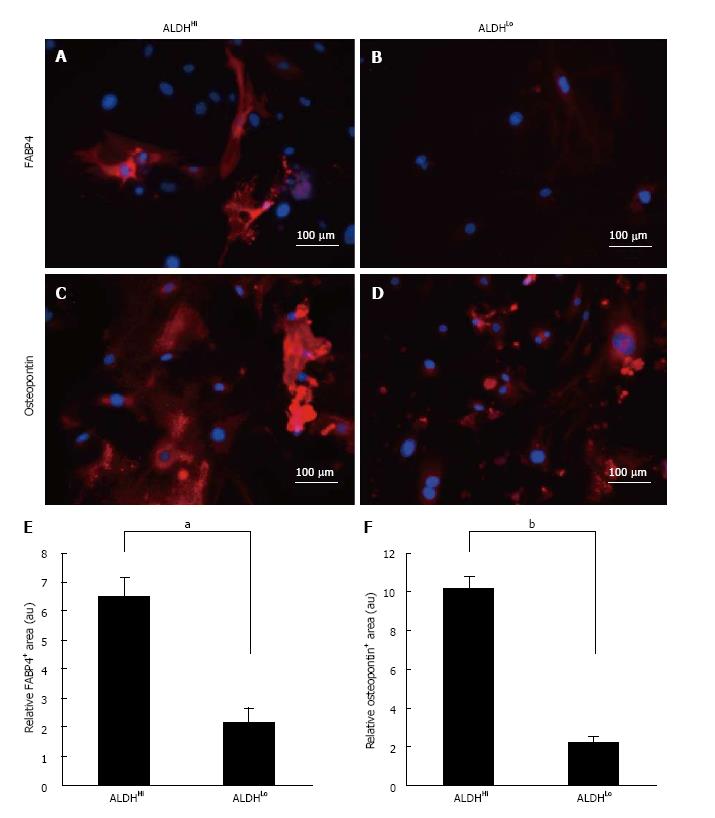
To assess the adipogenic and osteogenic differentiation potential of the two subpopulations, sorted ALDHHi and ALDHLo cells were cultured under adipogenic or osteogenic differentiation conditions. After in vitro differentiation, immunofluorescence staining for FABP4 (marker of adipocytes) and immunofluorescence staining for osteopontin (marker of osteocytes) were performed (Figure 2A-D). ADSCs that differentiated into adipocytes appeared as accumulated lipid droplets in the cytosol in each ALDHHi and ALDHLo subpopulation (Figure 2A and B). Furthermore, immunofluorescence staining for osteopontin revealed that ADSCs that differentiated into osteocytes appeared as accumulated granules in the cytosol in each ALDHHi and ALDHLo subpopulation (Figure 2C and D).
Adipogenic and osteogenic differentiation of each ALDHHi and ALDHLo subpopulation was quantitatively assessed using the BZ-9000 microscope and its analysis software. Ten visual fields were taken randomly for every 3 wells, and the immunofluorescence-staining positive-areas in 30 visual fields were analyzed. Subsequently, the immunofluorescence-staining-positive area was divided by the Hoechst 33342-positive area for each of the 30 visual fields. The ALDHHi subpopulation was found to have significantly more adipogenic and osteogenic relative-differentiation-marker-positive areas than the ALDHLo subpopulation (Figure 2E and F).
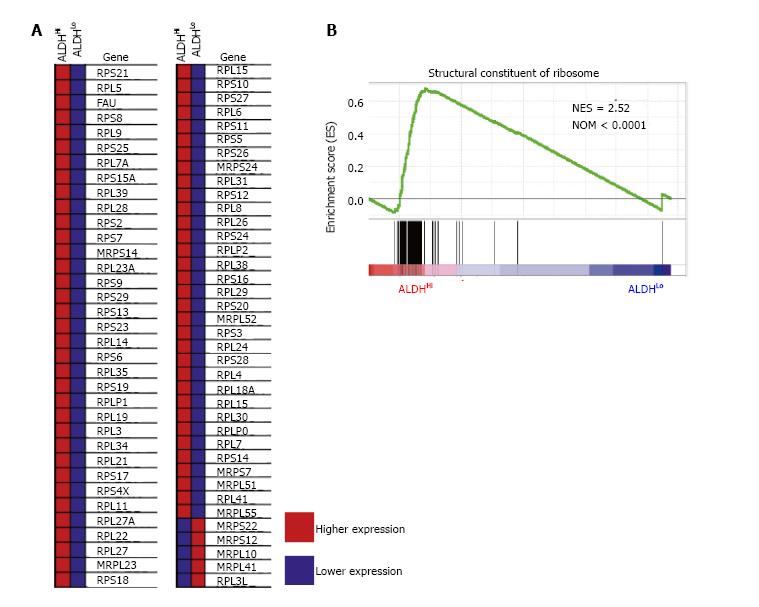
To identify the sets of gene that were up- or down-regulated in the ALDHHi subpopulations, we performed GSEA for the published gene expression profile of C57BL/6 mice divided by the ALDHHi and ALDHLo subpopulations of ADSCs. Intriguingly, high gene set enrichment scores were obtained for the structural constituents of ribosomes (Figure 3).
MSCs are reported to commonly express CD29, CD73, CD90, and CD105 and to be negative for markers such as CD45 and CD56[12,13]. There have been many studies on cell surface antigen markers of ADSCs, such as CD34 and CD44[12,14,15]. Recently, however, studies have shown that some markers such as CD90 or CD105 are not expressed homogenously in bulk ADSC populations but are expressed in small ADSC subpopulations, suggesting that ADSCs are phenotypically heterogeneous[5,7,8]. In our current study, we detected ALDH activity as a stem cell marker in murine ADSCs. High ALDH activity has been reported as a marker for cells such as hematopoietic stem cells and cancer stem cells[10]. However, not many studies have been performed on ALDH activity and cell differentiation potential in MSCs. In one of these studies, Estes et al[11] showed the presence of a subpopulation with high ALDH activity in human ADSCs; however, no difference was found in terms of differentiation potential. In our present study, the cultured murine bulk ADSC population contained approximately 15% of the ALDHHi subpopulation. Additionally, in the induction experiment for adipogenic and osteogenic differentiation for each sorted ALDHHi and ALDH Lo subpopulation, significantly higher adipogenic and osteogenic potentials were found in the ALDHHi subpopulation. The ALDHHi subpopulation had higher cell differentiation potential than the ALDHLo subpopulation. To the best of our knowledge, this is the first report on the functionally distinguishable subpopulation defined by ALDH activity within murine ADSCs.
Relationships between ribosome biogenesis and stem cells have been described only recently. For example, it was reported that the transition from self-renewal to differentiation depends on the enhancement of ribosome biogenesis accompanied by increased protein synthesis in female Drosophila germline stem cells[16]. Slow growth, low biosynthesis and markedly reduced ribosome biogenesis were observed in hematopoietic stem cells that lacked RUNX1, which is known to promote the transcription of essential ribosome-related proteins[17]. We have few reports about relationship between ribosome biogenesis and MSCs. One of these reports presented one of core proteins of 60S ribosome is necessary for differentiation of osteocyte from MSCs[18]. In our current study, GSEA revealed the significant enrichment of ribosome-related genes in the ALDHHi subpopulation compared to that in the ALDHLo subpopulation, suggesting that ribosome biogenesis is part of the mechanism underlying the higher differentiation potential of the ALDHHi subpopulation.
ADSCs can be obtained in a less invasive manner from adipose tissue. Therefore, ADSCs are considered to be a promising source of cell-based therapy in the clinical setting. ADSCs have already been used in clinical studies for cardiovascular disease, breast reconstruction after mastectomy, spinal cord injury, cirrhosis, renal insufficiency, skin fistula after surgery, and skin fistula with Crohn’s disease[4,19,20]. Some of those trials reported the therapy to be safe and effective; however, there is obvious room for improvement. For instance, in a phase 3 trial for therapy with allogeneic expanded ADSCs for treatment-refractory complex perianal fistulas in patients with Crohn’s disease (ADMIRE-CD trial), approximately 50% of patients who received ADSC-therapy experienced remissions[21]. Although this is a significant achievement, further research and development are required in relation to the patients who did not respond to this trial.
Purification of specific subpopulations and engineering of ADSCs into cells that are highly efficient in differentiating into specific tissues might help obtain basic knowledge for cell-based therapy, which is more specific to individual disease conditions of each organ for which ADSCs are used. Further investigation is required to identify the underlying mechanisms that regulate ribosome biogenesis and differentiation in ALDHHi ADSCs.
In conclusion, we demonstrated that murine ADSCs have a distinct subpopulation defined by ALDH activity. Furthermore, the ALDHHi subpopulation had higher osteogenic and adipogenic differentiation potential than the ALDHLo subpopulation. Ribosome biosynthesis is suggested to be a remarkable difference between ALDHHi and ALDHLo subpopulations.
Adipose-derived stem cells (ADSCs) are recognized as useful materials for regenerative therapy. Recent study revealed the existence of subpopulations in ADSCs by surface antigen markers. However, functions of these markers remain elusive. Aldehyde dehydrogenase (ALDH) activity is commonly used as functional marker to identify human and mouse hematopoietic stem cell, though there has been no report about identification of a subpopulation(s) in murine ADSCs using ALDH.
Several surface antigen markers are reported to be capable of prospectively identifying distinct ADSCs subpopulations in human and murine. However, the function(s) of those reported markers are poorly understood. ALDH has its known function, such as a protective effect to hematopoietic stem cells through acetaldehyde detoxification, although it is not known in ADSCs.
The authors suggest a novel area of research consisted of ALDH, stem cell, and ribosome biosynthesis, by reporting here ALDHHi murine ADSCs are highly capable of differentiation, and have enriched ribosome-related gene sets.
The authors current findings of ALDHHi subpopulation of ADSCs might provide future application for enrichment of more useful cells which is applicable to an efficient cell-based therapy. Moreover, by elucidating mechanisms of the higher differentiation potentials shown in ALDHHi subpopulation of ADSCs might provide knowledge of a key regulator(s) of differentiation, and links between ribosome biosynthesis.
ADSCs: Adipose-derived stem cells can be obtained from adipose tissues and induced to differentiate into adipocytes, osteocytes and chondrocytes; ALDH: ALDH is a superfamily comprising 20 intracellular enzymes and is responsible for the oxidization of various aldehydes. Some reports identified ALDH is a marker that detect hematopoietic stem cells and cancer stem cells; GSEA: Gene set enrichment analysis is a comprehensive analysis of gene expression by a computational method.
This is a very interesting and well executed piece of work, with suitable controls.
The authors thank the staff of Yamaguchi University Animal Medical Center for their support.
Manuscript source: Unsolicited manuscript
Specialty type: Cell and tissue engineering
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C, C, C, C
Grade D (Fair): D, D
Grade E (Poor): 0
P- Reviewer: Goebel WS, Kan L, Kiselev SL, Lee Y, Ramírez M, Shawcross SG, Tanabe S, Wakao H S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Li L, Xie T. Stem cell niche: structure and function. Annu Rev Cell Dev Biol. 2005;21:605-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 854] [Cited by in RCA: 814] [Article Influence: 42.8] [Reference Citation Analysis (0)] |
| 2. | Keating A. Mesenchymal stromal cells: new directions. Cell Stem Cell. 2012;10:709-716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 560] [Cited by in RCA: 607] [Article Influence: 46.7] [Reference Citation Analysis (0)] |
| 3. | Levi B, Longaker MT. Concise review: adipose-derived stromal cells for skeletal regenerative medicine. Stem Cells. 2011;29:576-582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 155] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 4. | Konno M, Hamabe A, Hasegawa S, Ogawa H, Fukusumi T, Nishikawa S, Ohta K, Kano Y, Ozaki M, Noguchi Y. Adipose-derived mesenchymal stem cells and regenerative medicine. Dev Growth Differ. 2013;55:309-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 5. | Kawamoto K, Konno M, Nagano H, Nishikawa S, Tomimaru Y, Akita H, Hama N, Wada H, Kobayashi S, Eguchi H. CD90- (Thy-1-) high selection enhances reprogramming capacity of murine adipose-derived mesenchymal stem cells. Dis Markers. 2013;35:573-579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | Li Q, Qi LJ, Guo ZK, Li H, Zuo HB, Li NN. CD73+ adipose-derived mesenchymal stem cells possess higher potential to differentiate into cardiomyocytes in vitro. J Mol Histol. 2013;44:411-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 7. | Yamamoto M, Nakata H, Hao J, Chou J, Kasugai S, Kuroda S. Osteogenic Potential of Mouse Adipose-Derived Stem Cells Sorted for CD90 and CD105 In Vitro. Stem Cells Int. 2014;2014:576358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Takahashi H, Haraguchi N, Nishikawa S, Miyazaki S, Suzuki Y, Mizushima T, Nishimura J, Takemasa I, Yamamoto H, Mimori K. Biological and clinical availability of adipose-derived stem cells for pelvic dead space repair. Stem Cells Transl Med. 2012;1:803-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 9. | Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697-720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 658] [Cited by in RCA: 604] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 10. | Balber AE. Concise review: aldehyde dehydrogenase bright stem and progenitor cell populations from normal tissues: characteristics, activities, and emerging uses in regenerative medicine. Stem Cells. 2011;29:570-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 11. | Estes BT, Wu AW, Storms RW, Guilak F. Extended passaging, but not aldehyde dehydrogenase activity, increases the chondrogenic potential of human adipose-derived adult stem cells. J Cell Physiol. 2006;209:987-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 92] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 12. | Troyer DL, Weiss ML. Wharton’s jelly-derived cells are a primitive stromal cell population. Stem Cells. 2008;26:591-599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 596] [Cited by in RCA: 550] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 13. | Jones RJ, Barber JP, Vala MS, Collector MI, Kaufmann SH, Ludeman SM, Colvin OM, Hilton J. Assessment of aldehyde dehydrogenase in viable cells. Blood. 1995;85:2742-2746. [PubMed] |
| 14. | Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143-147. [PubMed] |
| 15. | Kuroda Y, Dezawa M. Mesenchymal stem cells and their subpopulation, pluripotent muse cells, in basic research and regenerative medicine. Anat Rec (Hoboken). 2014;297:98-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 16. | Sanchez CG, Teixeira FK, Czech B, Preall JB, Zamparini AL, Seifert JR, Malone CD, Hannon GJ, Lehmann R. Regulation of Ribosome Biogenesis and Protein Synthesis Controls Germline Stem Cell Differentiation. Cell Stem Cell. 2016;18:276-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 177] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 17. | Cai X, Gao L, Teng L, Ge J, Oo ZM, Kumar AR, Gilliland DG, Mason PJ, Tan K, Speck NA. Runx1 Deficiency Decreases Ribosome Biogenesis and Confers Stress Resistance to Hematopoietic Stem and Progenitor Cells. Cell Stem Cell. 2015;17:165-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 178] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 18. | Lin YM, Wu CC, Chang YC, Wu CH, Ho HL, Hu JW, Chang RC, Wang CT, Ouyang P. Target disruption of ribosomal protein pNO40 accelerates aging and impairs osteogenic differentiation of mesenchymal stem cells. Biochem Biophys Res Commun. 2016;469:903-910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 19. | Illouz Y-Gr, Sterodimas A. Adipose stem cells and regenerative medicine. Heidelberg; New York: Springer 2011; . |
| 20. | Hong SJ, Traktuev DO, March KL. Therapeutic potential of adipose-derived stem cells in vascular growth and tissue repair. Curr Opin Organ Transplant. 2010;15:86-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 118] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 21. | Panés J, García-Olmo D, Van Assche G, Colombel JF, Reinisch W, Baumgart DC, Dignass A, Nachury M, Ferrante M, Kazemi-Shirazi L. Expanded allogeneic adipose-derived mesenchymal stem cells (Cx601) for complex perianal fistulas in Crohn’s disease: a phase 3 randomised, double-blind controlled trial. Lancet. 2016;388:1281-1290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 628] [Cited by in RCA: 730] [Article Influence: 81.1] [Reference Citation Analysis (1)] |