Published online Apr 26, 2016. doi: 10.4252/wjsc.v8.i4.170
Peer-review started: November 3, 2015
First decision: December 4, 2015
Revised: December 30, 2015
Accepted: February 14, 2016
Article in press: February 16, 2016
Published online: April 26, 2016
Processing time: 168 Days and 6.7 Hours
Carcinogenic transformation of somatic cells resembles nuclear reprogramming toward the generation of pluripotent stem cells. These events share eternal escape from cellular senescence, continuous self-renewal in limited but certain population of cells, and refractoriness to terminal differentiation while maintaining the potential to differentiate into cells of one or multiple lineages. As represented by several oncogenes those appeared to be first keys to pluripotency, carcinogenesis and nuclear reprogramming seem to share a number of core mechanisms. The retinoblastoma tumor suppressor product retinoblastoma (RB) seems to be critically involved in both events in highly complicated manners. However, disentangling such complicated interactions has enabled us to better understand how stem cell strategies are shared by cancer cells. This review covers recent findings on RB functions related to stem cells and stem cell-like behaviors of cancer cells.
Core tip: Carcinogenic transformation of somatic cells resembles nuclear reprogramming toward the generation of pluripotent stem cells. The retinoblastoma tumor suppressor product retinoblastoma (RB) seems to be critically involved in both events in highly complicated manners. This review covers recent findings on RB functions related to stem cells and stem cell-like behaviors of cancer cells.
- Citation: Kohno S, Kitajima S, Sasaki N, Takahashi C. Retinoblastoma tumor suppressor functions shared by stem cell and cancer cell strategies. World J Stem Cells 2016; 8(4): 170-184
- URL: https://www.wjgnet.com/1948-0210/full/v8/i4/170.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v8.i4.170
The cell-of-origin in various types of cancers has been one of the most important areas of research in modern cancer biology, because deep understanding of this can help design of future cancer therapies[1-4]. Many studies have indicated that distinct cells-of-origin give rise to distinct features of cancers, and can often predict the prognosis of patients[5,6]. However, the debate on cell-of-origin of each cancer is often controversial, because cancer phenotypes do not stereotypically reflect phenotypes of their true cells-of-origin. This is at least partially due to the high developmental plasticity that is acquired after tumor initiation and during tumor progression.
The cancer stem cell hypothesis proposes a model in which like in normal tissues, cancer cells obey the hierarchy of development where stem cell-like cancer cells are placed at the top. This hypothesis does not always explain the cell-of-origin of a specific cancer; however, it has led us to the idea that cancer cells prefer to employ stem cell strategies in order to maintain tumor-initiating clones[7,8]. This theory has been reinforced by many findings, including the switchable cell fates of cancer cells, the inseparable relationship between pluripotency and teratogenicity, the requirement for oncogenic elements for the generation of induced pluripotent stem (iPS) cells, and the oncogenic activities of many embryonic genes[9,10].
Sophisticated gene-engineered and tumor-grafted mouse models have been used to trace the cells-of-origin for specific cancers, for instance in case of prostate adenocarcinomas, to basal or luminal cells[2]. During these investigations, the existence of unexpectedly high levels of developmental plasticity became apparent, when comparing the cell-of-origin to its resultant tumor. In addition, researchers found switchable cell fates in cultured cancer cell lines that were induced by artificially altering the status of genes, such as tumor suppressor genes.
Although the significance of epigenetic alterations is still unclear, carcinogenesis results from the step-wise accumulation of readable lesions in the genome[11]. Given the gain of developmental plasticity, even if it is transient, is essential for carcinogenesis, then the next question should be as follows: Which genes are mechanistically involved in this gain of plasticity?
This question is not satisfactorily answered yet, but a part of answer may be informed by the switchable cell fate of cancer cells. The retinoblastoma tumor suppressor gene (RB) is closely implicated in the change in developmental phenotypes of many types of cancers, including lung cancer, breast cancer, prostate cancer, osteosarcoma, and soft tissue sarcoma. This phenomenon has been attributed to the physical or genetic interaction between the RB gene product (RB) and tissue-specific transcription factors[12]. However, emerging evidence indicates that inactivation of RB in particular genetic backgrounds or in certain contexts can lead cells to an undifferentiated state that resembles that of most immature cells, such as embryonic stem cells[13-15].
We also know that the targeted inactivation of RB, in combination with p53, provides strong experimental tools to determine the cell-of-origin of various types of cancers[16-18]. Indeed, these two tumor suppressor pathways are the most commonly inactivated in human cancers, and simultaneous inactivation is sufficient to induce cancers from various types of somatic cells[19]. Therefore, one of the optimal ways to understand RB function in the context of full carcinogenesis would be to determine RB functions in a p53-deficient genetic background.
This review briefly summarizes the well-established functions of RB in mammalian cells, presents cross-species evidence for the possible link between RB function and the control of stem cell activities, and describes findings that may explain the molecular mechanisms underlying this link. The RB locus was identified more than a quarter century ago; however, researchers are still providing new wineskins to new wines.
The RB gene was first identified as a tumor suppressor in the childhood malignancies retinoblastoma and osteosarcoma[20]. Somatic RB loss typically causes unilateral retinoblastoma with no obvious risk for other types of malignancies. However, germline RB mutation often results in bilateral retinoblastoma, and carriers are at very high risk of various types of cancer over their lifetimes[21]. Therefore, researchers proposed that RB might be involved in the core mechanisms of tumorigenesis. Indeed, unveiling the functions of RB in controlling cell cycle progression provided a big breakthrough to the field of cancer research[22].
A primary RB function in cell cycle control is exerted at the G1/S transition. RB undergoes dephosphorylation at the end of the M phase with the aid of protein phosphatases (PPs) and resumes its phosphorylated state during the G1 phase by the action of cyclin D/cyclin-dependent kinase (CDK) 4 or 6 complexes[23]. Most of cellular mitogenic signals converge on the transcriptional upregulation of D-type cyclins. This could be one reason that cells in the G1 phase are most vulnerable to extracellular growth stimuli[23,24].
Phosphorylation of RB alters its three dimensional (3D) structure. This results primarily in the loss of binding affinity to E2F family transcription factors[25,26]. Among nine identified E2F family members (E2F1, 2, 3A, 3B, 4-8), RB was shown to bind to at least E2F1, 2, and 3A. Each of these three family members is able to positively transactivate genes, including cyclin E[27]. Upregulation of cyclin E in cooperation with CDK2 further promotes RB phosphorylation. This enables cells to cross the boundary between G1 and S. Further, with the aid of cyclin A, RB attains the maximal level of phosphorylation before cells enter the M phase[23]. In addition, when bound to hypophosphorylated RB, E2Fs form a transcriptional repressor complex that recruits histone deacetylase (HDAC) to epigenetically silence gene transcription[28]. Therefore, the phosphorylation status of RB dramatically changes the expression of E2F-targeted genes. The function of RB in restricting the G1/S transition is also mediated by its binding to SKP2, which destabilizes p27KIP1 by enhancing the ubiquitin-proteasome system when freed from phosphorylated RB[29,30]. This represents one of E2F-independent functions of RB in the control of cell cycle progression.
RB plays pivotal roles also in M phase, which is most typically represented by the impact of RB inactivation on the chromosomal instability (CIN). E2Fs target a number of M phase genes including MAD2 which functions by inhibiting the anaphase promoting complex/cyclosome (APC/C)-cell division cycle 20 (CDC20) complex. This complex regulates spindle assembly[31]. RB also controls the M phase by directly binding to cohesin and condensin II, two critical regulators of centromeric functions[32].
“How many total RB functions are cell cycle-dependent?” is an intriguing question. RB mutants found in partially penetrant retinoblastomas (low grade retinoblastomas with limited genetic inheritance) or retinomas that failed to inhibit the cell cycle but retained the ability to promote terminal differentiation suggested that RB functions in cell cycle control and differentiation might be distinct[33]. In addition, phenotypic analyses of Rb-deficient mice simultaneously lacking an E2F family member allowed at least partial discrimination of the E2F-dependent function from the E2F-independent function[34]. However, since E2Fs target both cell cycle-related and cell cycle-unrelated genes, discrimination of cell cycle-dependent functions from cell cycle-independent functions of RB based on the E2F-dependency is difficult.
Artificial and acute alteration of RB status in a wild type genetic background often greatly affects the cell cycle. For instance, it induces cell cycle exit (quiescence or cellular senescence), or inversely cell cycle re-entry[35,36]. This change in the cell cycle control is highly drastic, thus can mask cell cycle-independent phenotypes associated with altered RB activity. However, based on the experience of analyzing Rb-deficient mice, our group discovered several genetic backgrounds that allow mice or cells to exhibit cell cycle-independent phenotypes following RB inactivation[14,15,37,38]. The cell cycle-independent phenotypes include gain of undifferentiated phenotypes and altered chemo-resistance[15]. Therefore, we thought that control of the undifferentiated state of cells might represent at least a part of the cell cycle-independent functions of RB.
Whole genome sequencing studies have revealed that the RB loci undergoes the fourth most frequent loss-of-heterozygosity (LOH) found in whole human tumors, following CDKA2A, PTEN, and SMAD4. Thus, RB mutations definitely can be “driver mutations”[39]. However, the type of tumors in which RB mutations occur at their initiation is highly limited, including only retinoblastomas, small cell lung cancers (SCLC), osteosarcomas, and familial melanoma. In other types, in the vast majority of tumors, RB functions are largely maintained during initiation but these functions typically collapse while tumors undergo malignant progression[40].
The question of why RB mutation is rare in the majority of cancers has yielded many interesting answers. Lack of RB in many cell types has already been linked to apoptosis through E2Fs, ARF, and p53[41]. In addition, RB residing in mitochondria directly interacts with Bax, and thus regulates apoptosis in a completely E2F-independent and cell cycle-independent manner[42,43]. These pathways represent a disadvantage in carcinogenesis upon RB inactivation when it occurs at early steps. The 3T3 cells lacking Rb are less susceptible to Ras-transformation, indicating that preservation of RB functions in the interaction of mitogenic signals and the cell cycle might be important for tumor initiation[44].
Our group demonstrated that Rb-heterozygous mice generate adenomas or low-grade adenocarcinomas derived from calcitonin-producing cells (C cells) of neuro
endocrine origin that exhibit whole evidence of DNA damage response and cellular senescence. However, the genetic background of a homozygous lack of N-ras allowed these Rb-deficient C cell tumors to progress to highly invasive and metastatic adenocarcinomas[14,37]. RB appeared to regulate isoprenylation of the N-Ras protein. Isoprenylation (farnesyl moiety-transfer and geranylgeranyl moiety-transfer) is the chemical reaction that is essential for the initial maturation of this protein. RB loss causes intermediate level upregulation of N-Ras under particular culture conditions, which induces a DNA damage response and subsequently cellular senescence, thus antagonizing full carcinogenesis in a manner similar to oncogene-induced senescence (OIS). The mechanism whereby RB controls N-Ras isoprenylation involves E2F-dependent regulation of sterol regulatory element binding protein (SREBP) transcription factors and direct drive by E2Fs in some of mevalonate (MVA) pathway genes[14]. This study indicated a case that RB influences intra-cellular signaling primarily by controlling metabolic pathways. Many reports, including ours, have directly implicated RB in the control of cellular metabolism[36,45,46]. This may indicate that RB is simultaneously involved in the control of the cell cycle and metabolic regulation. This may further explain why cell cycle progression and cellular metabolism are tightly coupled. Cells never diminish their volume after rounds of cell division. This is because cells very strictly double biomass up to the time of mitosis, just as they strictly conserve genome size. From this point of view, cellular metabolism for biomass synthesis would not be passively controlled by the demands of cell cycle progression; rather, they are both actively and presumably simultaneously regulated by a common mechanism.
The outcomes of RB inactivation during malignant progression entail not only facilitated G1/S transition, but also many events critical for malignant cell behaviors. These include increased cell motility, angiogenesis, inflammatory response, metabolic rewiring, gain of undifferentiated developmental features, lineage change and altered drug resistance[36,45]. Since acquisition of these abilities is not always associated with cell cycle progression, many of these, except G1/S control, may more or less represent cell cycle-independent functions of RB. Particularly, gain of undifferentiated developmental features following RB inactivation and lineage changes presumably related to the developmental plasticity of tumors are central interests of this review article.
We need to be very careful in defining what is cell cycle-independent function of RB and what is not. E2F targets contain not only cell cycle-related genes. In addition, many LxCxE proteins that bind to RB are chromatin modifiers; hence, their roles in control of the cell cycle and differentiation are barely discernible. We later discuss the experimental system by which we addressed cell cycle-independent functions of RB in the context of regulation of undifferentiated cell states. In the next paragraph, we introduce accumulating biological evidence implicating RB in multiple stem cell systems. Later, we discuss possible cell cycle-dependent and independent mechanisms underlying RB functions in stem cells.
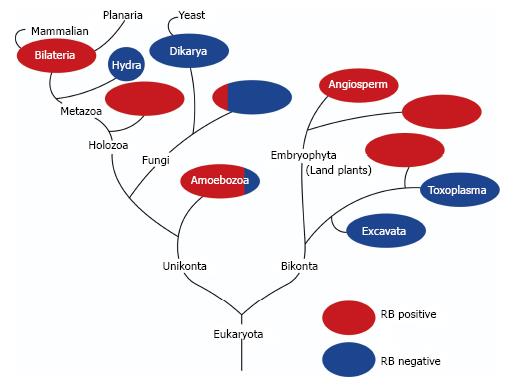
Figure 1 shows a phylogenic tree of genes in the RB family (Figure 1). RB gene orthologues do not exist in the genome of many of unicellular organisms, including yeasts, but appear in almost all multicellular organisms[47]. In addition, the component including RB alone and/or E2F transcription factors and DP protein(s) are well conserved from plants to animals[48]. This indicates that RB might be involved not only in cell-autonomous proliferation control but also in some machineries unique to multi-cellular organisms. Stem cell conservation attained by asymmetric cell division is apparently unique to multi-cellular organisms. Therefore, we present cross-species evidence that may link RB to stem cells.
Analysis of RB functions in embryonic development and embryonic stem cells provided a substantial amount of information regarding its potential roles in stem cells. The role of RB in various adult tissue stem cells was once thoroughly summarized by Sage[19]. Our current review focuses on the cells-of-origins in which RB inactivation likely gives the first cue for clonal expansion of stem or progenitor cells during carcinogenesis.
Retinoblastoma develops from the retina composed of multiple lineages of cells. The cell-of-origin of retinoblastoma is quite controversial because of the complexity of retinal development and tumor phenotype. More in concrete, findings that contribute to the controversy include the reported appearance of differentiated and undifferentiated developmental markers of mixed lineages within the same tumor derived from retinoblastoma patients and Rb-deficient mice[49]. Human retinoblastomas show typically differentiated features[50], and post-mitotic retinal cells can be the cell-of-origin in mouse models[51]. These findings suggest that promoting dedifferentiation and increasing flexibility of fate determination are initially attained by RB inactivation in retinal cells. In other words, RB inactivation in retinal somatic cells endows them with the ability to differentiate to multiple types of cells. The dispute over cell-of-origin might be resolved, since cone precursors have been shown to be specifically vulnerable to Rb-deficiency-induced clonal expansion[52]. However, given that the cell-of-origin has been truly traced to one lineage in retinoblastoma development, the findings on mixed and normally inconsistent developmental features co-existing in retinoblastoma tumor cells imply that RB-deficiency not only initiates tumors of a particular cell-of-origin, but also increases plasticity in lineage specification and probably induces dedifferentiation so that tumor cells employ multiple stem or progenitor cell strategies to fit to the retinal microenvironment.
There are several other types of cancers in which RB loss occurs prevalently at initiation: Osteosarcoma and SCLC. In these malignancies, discussions of the cell-of-origin and the role of RB in tumor initiation are inseparable similarly as in case of retinoblastoma. Mesenchymal stem cells or osteo-progenitors can be the cell-of-origin of osteosarcoma. The direct interaction between RB and osteoblast transcription factor Runx2 might at least partially explain the ability of RB to suppress tumor initiation in a lineage-specific manner. Rb deficiency in these cells often cooperates with p53 loss-of-function to generate osteosarcomas[53]. p53-deficiency is in some case sufficient to induce osteosarcoma, and p53-deficient osteosarcomas can be converted to brown fat tumors (hibernomas) with subsequent RB inactivation[12]. This has been attributed to increased expression of PPARγ which is governed by E2Fs. However, this fate change might also be mechanistically supported by function of RB to influence on cell fate decision that may occur prior to the decision of commitment to terminal differentiation. Hence, in osteosarcoma, like in retinoblastoma, RB might function in mesenchymal stem cells or osteo-progenitors to contribute to suppress the plasticity in lineage specification.
There are also debates on the cell-of-origin of SCLC. Again, simultaneous inactivation of RB and p53 is sufficient to induce SCLC in mouse lungs[3]. A study of cell type-specific deletion of RB and p53 indicated that neuroendocrine cells more often gave rise to SCLC than alveolar type II cells[54,55]. From this and other evidence, neuroendocrine cells are believed to be the predominant cell-of-origin of SCLC. The differential roles of RB and p53 in the formation of SCLC will be discussed later (see below).
The role of RB in controlling pluripotency was first addressed by testing whether tumor suppressor depletion facilitates iPS induction[56]. This study identified p53 but not RB to be an influential molecule in iPS induction. However, two later studies indicated that RB suppresses iPS induction from fibroblasts. A screen for short hairpin RNAs (shRNAs) that enhance iPS induction efficiency identified RB[57]. Another study indicated that RB cleavage by caspase 3/8 is critical for iPS induction[58]. More recently, RB was directly implicated in the transcriptional control of Oct4 and Sox2[59]. These findings indicate that RB inactivation leads to a state favorable for iPS induction by facilitating the induction of embryonic genes that induce a pluripotent state. This function of pRB may partly explain how stem cell functions can be shared by cancer cells. We indeed often observed increased expression of Oct4 and Sox2 in spheres induced by RB inactivation[59]. The effect of RB inactivation on the cell cycle control is also contributable to iPS cell induction. We will discuss later many of the signals required for iPS induction are possibly controlled by pRB (see below).
MEFs lacking all members of the RB family can form embryoid body-like 3D structures in suspended culture conditions that express higher levels of embryonic genes and can form teratoma-like tumors when inoculated into immune-deficient mice[13]. Since mostly tumor-derived spheres express higher levels of embryonic genes than when cultured under 2D conditions, this observation might result from carcinogenic change in MEFs.
Kareta et al[59] (our study) demonstrated that RB inactivation in p53-/- MEFs give rise to sphere formation without carcinogenic conversion. Sphere formation in the absence of serum and in the presence of limited growth factors (bFGF and EGF) is thought to represent increased self-renewal/symmetric cell division. Preceding this observation, we analyzed C cell adenocarcinomas developed in Rb-heterozygous mice that simultaneously lacked p53, Ink4a, Arf or Cdkn1a (p21). Rb-heterozygous mice typically develop low-grade C cell adenocarcinomas. Simultaneous lack of an additional gene allowed Rb-deficient C cells to develop full brown tumors at seemingly similar levels. However, as compared to other genotypes of C cell tumors that were all calcitonin-positive, thyroid tumors that developed in Rb+/-p53-/- mice showed virtually no expression of calcitonin. However, earlier neuroendocrine lineage markers, including synaptophysin, were expressed. These findings indicated that simultaneous inactivation of Rb and p53 induced a highly undifferentiated status in neuroendocrine cells that were originally destined to develop into C cells. We then analyzed Rb-/-; p53-/- MEFs in comparison with Rb+/+; p53-/- MEFs, They showed insignificant differences in cell cycle progression; however, Rb-/-; p53-/- MEFs showed significantly higher self-renewal activity and increased expression of embryonic genes. This may represent a cell cycle-independent function of RB in controlling stem cell-like features. Lastly, we screened an FDA-approved drug library and found that some drugs reported to be effective as cancer stem cell therapies were also effective in cells lacking Rb and p53[15].
Here, we focus on four tumor types (SCLC, breast cancer, prostate cancer, and soft tissue sarcoma) in which RB inactivation during tumor progression presumably contributes to the increased developmental plasticity of tumor cells.
p53 is the primary gene mutated in SCLC (75%-90%)[60]. p53 mutations are found also in normal bronchioles of patients, suggesting that this mutation most likely occurs at tumor initiation[61]. The second most frequent mutation occurs at RB loci. Recent comprehensive genomic profiling of SCLC revealed that bi-allelic losses at p53 and RB loci were 100% and 93%, respectively[62]. This signifies that the simultaneous inactivation in p53 and RB occurs in almost 90% of all cases. This study showed a previously unexpected high frequency of RB inactivation in SCLC. In terms of determining RB function in SCLC development, other gene mutations relevant to RB loss in RB-intact SCLC (7% of p53-mutated cases) should be noted. Semenova et al[3] discussed possible RB functions in p53-mutated SCLC cells. They pointed out that pluripotency genes such as OCT4 and SOX2 are frequently amplified in SCLC[63] and that enhancers of zesta 2 (EZH2), which is implicated in the neural stem cell maintenance, are very often upregulated in SCLC following RB inactivation[64,65]. Finally, they concluded that “RB loss (in SCLC) is associated with an increase in cell plasticity”[3]. Although more evidence is needed to finally accept this notion, it is definitely an attractive hypothesis.
Recent publication from Engelman group demonstrated that gain of resistance to tyrosine kinase inhibitors in non-small-cell lung cancers (NSCLCs) harboring epidermal growth factor receptor (EGFR) mutation is associated with fundamental histological transformation from NSCLC to SCLC at a certain frequency (5%-15%). Surprisingly, in such subset of resistant cancers, RB was lost at 100% frequency[66]. This report indicates that the developmental plasticity enhanced by RB inactivation is coupled to gain of drug resistance. The mechanism of drug resistance associated with NSCLC-to-SCLC conversion is currently unknown, but it would be definitely attractive to investigate RB functions in this context.
Breast cancers are frequently characterized by RB pathway inactivation, and low RB expression is a hallmark of basal-like breast cancers[67-69]. RB inactivation in luminal type breast cancer induces tamoxifen resistance[70], possibly owing to a gain-of-undifferentiated status following RB inactivation. Simultaneous inactivation of p53 and RB is prevalent in basal-like cancers[71]. Basal-like and luminal type breast cancers have been recently suggested to stem from common luminal progenitor cells[72]. Because p53 mutations are common in most breast cancer types[73], RB status might be one of the determinants of basal-like or luminal type cancers. If so, RB could be implicated in determining the fate of breast cancers.
RB was also implicated in epithelial-to-mesenchymal transition (EMT) in breast cancer cells. Taya group demonstrated that RB depletion in a luminal type MCF-7 breast cancer cells induced EMT and overexpression of RB inhibited the EMT in MCF10A non-tumorigenic breast mammary epithelial cells[74]. They also demonstrated that RB controls transcription of SLUG and ZEB-1 in cooperation with the transcription factor activator protein 2α.
RB is deeply implicated in prostate cancer development, especially during its progression. Although RB inactivation is observed in only 5% of primary prostate cancers, its rate rises to 40% in metastatic tumors. Furthermore, the RB signaling pathway is altered in 34% and 74% of primary tumors and metastatic loci, respectively[75]. One study demonstrated a high rate of RB loci deletion and DNA methylation in the RB promoter in metastatic castration-resistant prostate cancers (CRPC)[76]. RB depletion in hormone-dependent human prostate cancer cells induces androgen-independent cell growth through upregulation of androgen receptor (AR) in an E2F1-dependent manner[77]. RB and p130 are involved in the regulation of EZH2 transcription in prostate cancer cells, whose upregulation is often observed during prostate cancer progression[78]. Loss of RB in prostate cancer cells increases the expression of nucleolar and spindle-associated protein 1 (NUSAP1), which is associated with a poor prognosis in prostate cancer[79]. Recently, we observed that RB depletion in androgen-dependent prostate cancer cells induces several lipid metabolism-related genes and some typical malignant features, including tumor spheroid formation. These observations indicate that inactivation of RB strongly promotes prostate cancer progression.
Simultaneous mutations in RB and p53 are frequently found in human soft tissue sarcomas[80]. Conditional inactivation of both tumor suppressors by subcutaneous injection of AdCMVC into p53flox/flox; Rbflox/flox mice induced undifferentiated high-grade pleomorphic type sarcomas from locally resident cells. Inactivation of p53 but not RB is sufficient to induce well-differentiated sarcomas, such as rhabdomyosarcoma and leiomyosarcoma, but typically not sufficient to induce undifferentiated types of tumors[81]. These findings indicate that RB inactivation does not directly contribute to the initiation of sarcoma development, but rather does contribute to converting well-differentiated types of tumors to undifferentiated types in a p53-deficient background. We recapitulated this finding in an in vitro culture system. We first developed poorly spherogenic p53-null soft tissue sarcoma cell lines from soft tissue sarcomas that subcutaneously developed in approximately 10% of p53-/- mice with a C57BL/6 background. Additional depletion of Rb successfully induced less differentiated highly spherogenic and less differentiated sarcomas from “poorly spherogenic”p53-null soft tissue sarcoma cell lines. These findings indicate a possibility that in soft tissue sarcomas, RB directs both the self-renewal and plasticity of developmental features.
Relying on a large population of pluripotent adult stem cells, planarians exhibit extraordinary high regenerative capacities. Zhu and Pearson presented a comprehensive study on the RB system in these organisms[82]. Planarians possess unexpectedly few RB system components: A single Rb family member, single E2F (E2F4-1), and single DP. They are primarily expressed in planarian stem cells, and knockdown of any of these components significantly phenocopied the stem cell loss induced by irradiation or RNAi against stem cell-specific genes. The RB system was found to be indispensable for planarian stem cell self-renewal and survival; however, it was dispensable for late differentiation. Interestingly, planarians have 20 homologs to cyclin genes and none of them is homologous to cyclin E. An HDAC1 and a cyclin D homolog are expressed specifically in planarian stem cells, and knockdown of either of them induced deficiencies in stem cell functions. The simplicity of the RB system and ease of visualizing stem cell behaviors in planarians make this a valuable system for the RB research field, especially with regard to stem cell functions. Additionally, the molecular mechanism whereby the RB system contributes to the extraordinarily high regenerative capacity of this creature is great of interest.
RB family proteins and their binding partners had existed before multicellular organisms appeared on the earth, and are shared by plants and animals[82]. Arabidopsis thaliana has one ortholog of an RB family protein, which was named retinoblastoma-related protein (RBR)[83]. Inactivation of RBR led to the expansion of root stem cells without affecting the ability of progenitor cells (descendants) to self-renew and differentiate[48]. The mechanism whereby RBR controls maintenance of root stem cells seems to involve two transcription factors shortroot (SHR) and scarecrow (SCR). SCR interacts with RBR through an LxCxE motif. Thus, surprisingly, the role of LxCxE motifs is well conserved between animals and plants. More surprisingly, the RBR status affects the function of the SHR/SCR complex to spatiotemporally control the expression of a plant homolog of D-type cyclin (CYC D6;1). CYC D6;1, in cooperation with its corresponding cyclin-dependent kinase (CDK), promotes phosphorylation of RBR[83]. These findings indicate that in plants, the RB ortholog exerts the function of controlling stem cells through the regulation of well-conserved cell cycle machinery. Further investigation in this field might unveil many unexpected aspects of RB functions in the control of stem cells.
Analysis of the cell cycle status in embryonic stem (ES) cells provided valuable information on how its alteration might contribute to the acquisition of increased self-renewal and pluripotency. Conklin and Sage[84] provided a concise perspective on the possible roles of RB in maintaining the ES cell functions. ES cells have a rapid cell cycle. Because of the prolonged or continuous expression of cyclin E and A family members and lower or no expression of many CDK inhibitors, human and mouse ES cells maintain high levels of activity in multiple CDKs. pRB is consequently hyperphosphorylated for longer periods in ES cells than in normally cycling cells. No obvious cell cycle phenotype in mouse ES cells following inactivation of all RB family members might be consistent with this view[85,86].
An extraordinarily shorter G1 phase as compared to the relatively prolonged S phase in ES cells would be beneficial to lessen the susceptibility to differentiate upon receiving stimuli to lender cells to do so. As G1 as well is the period that is most vulnerable to mitogenic signals that directs cells to decide to proliferate, arrest or senesce, this phase could be the most critical one in the decision to differentiate upon various stimuli. Keeping the G1 phase shorter could be the primary role of RB hyperphosphorylation in ES cells. Consistent with this view, overexpression of the constitutively active (non-phosphorylatable) form of RB (RB7LP) in human ES cells induced cell cycle arrest, followed by spontaneous differentiation and p53-dependent cell death[87].
The difference in undifferentiated behaviors between Rb+/+; p53-/- and Rb-/-; p53-/- MEFs predicted that there could be cell cycle-independent functions of RB in controlling stemness. p53-/- MEFs exhibited similar proliferation phenotypes regardless of Rb genotype; however, Rb-/-; p53-/- showed increased sphere forming activity relative to Rb+/+; p53-/- MEFs[15]. The same phenomenon was observed in mouse soft tissue sarcoma cells and mouse mammary gland epithelial cells. Given that RB-deficiency has no or very little impact on cell proliferation in a p53-null genetic background, the mechanism whereby RB-deficiency effects stem cell-like features of multiple types of cells can be separated from that governs cell proliferation.
In contrast to mouse ES cells, inactivation of all RB family members in human ES cells exhibited abnormal quiescence, featured by G2/M arrest and cell death[87]. Cell death depended on the p53-p21 module, similar to RB overexpression. Thus, in human ES cells, both hypo- and hyper-activation of RB are counteracted by cell cycle arrest and p53 pathway activation, indicating a critical role for RB in the homeostatic control of ES cell activities. p53 is typically expressed in human ES cells at a low level[88]. This might be beneficial for ES cells so that they are not too sensitive to alterations in RB activity status. Mouse hematopoietic stem cells lacking all RB family proteins exhibit impaired quiescence control and apoptosis in lymphoid progenitor cells[89]. In MEFs, the absence of p53 endowed RB-deficient cells significant increase in self-renewal activity when cultured in the presence of limited growth factors[15]. However, in a p53-/- background, the RB status did not impact cell proliferation of MEFs under regular culture conditions. It should be noted again that in mouse ES cells, lack of RB family proteins generated no difference in the cell cycle[85,86]. These findings indicate that RB may control stemness beyond its role in cell cycle control.
Cellular senescence is believed to be strongly inhibited in stem cells, otherwise irreversible growth arrest could lead to total elimination of a stem cell pool from tissues. RB plays pivotal roles in inducing and maintaining cellular senescence, not only by controlling required transcription of genes, but also by being involved in senescence-associated heterochromatic foci (SAHF)[90]. Therefore, hyperactivation of RB can be harmful to stem cells, as was shown in a human ES cell study[87].
Surprisingly, loss of RB function, as well, can induce cellular senescence. As mentioned above, thyroid tumors that developed in Rb-heterozygous mice were typically low-grade adenocarcinomas or adenomas of C cell origin; however, the genetic background lacking N-ras[37] or either Ink4a, Arf, Suvh39[14], ATM[38] or p53[15] allow these tumors to develop into highly invasive and metastatic type medullary adenocarcinomas. The immunohistochemical observation of Rb-deficient C cell tumors lacking no other genes revealed whole evidence of cellular senescence, including increased expression of p16Ink4a and HP-1, and positive β-galactosidase staining[14]. Since a simultaneous lack of genes mediating DNA damage response (ATM) and cellular senescence (Ink4a) allowed malignant progression of C cell tumors, we concluded that these cellular responses prevented Rb-deficient premalignant cells from developing into malignant cells. Analysis of Rb-N-ras DKO mice revealed that the mechanism whereby RB loss induces a DNA damage response involved p130[14]. Another system in which RB loss possibly induces cellular senescence under particular culture conditions is MEFs. Rb loss alone does not allow MEFs to escape senescence when cultured at low density. However, simultaneous loss of N-ras or Ink4a allowed MEFs to escape cellular senescence upon low cell density plating[14]. The reason why N-ras loci are associated with susceptibility to senescence was explained by the E2F-dependent control of Ras isoprenylation[14].
There are three other tumor suppressors whose loss of function can induce cellular senescence in particular contexts. These are PTEN, VHL and NF1[90-93]. Somatic cells are protected from carcinogenesis when these tumor suppressor genes are inactivated at early steps of carcinogenesis. This could be the major reason that RB mutations are detected in only limited types of cancer at their initiation.
ES cells, iPS cells, and tissue stem cells seem to confer lower RB activity in order to accelerate self-renewal and to keep undifferentiated state. We do not know whether cellular senescence machineries are simultaneously suppressed during the period that RB function is suppressed in stem cells. Unlike Rb-/-; p53-/- MEFs, Rb-/-, Ink4a-/- MEFs did not form spheres[15]. However, regarding iPS induction, the Ink4a/Arf locus appeared to be a barrier to reprogramming[94]. Therefore, RB functions in controlling cellular senescence could be intimately involved in the regulation of stemness.
Chromosomal instability (CIN) might be one of the events that are seemingly not shared by normal stem cells and cancer cells. In tumor cells, RB inactivation, especially when combined with p53 mutation, led cells to accumulate chromosomal aberrations[95]. Inactivation of all RB family members in human ES cells causes CIN; however, clonal expansion of these cells was blocked by G2/M arrest and cell death[87]. Presumably, then, normal stem cells are much less tolerant of CIN than cancer cells. RB status could influence CIN through its effects on Mad2 transcription and its direct interaction with cohesion and condensin II[96]. The rapid cell cycle in ES cells might increase the risk of accumulating DNA damage due to hyper-replication and nucleotide deficiency[97]. How ES cells are protected from these risks and why cancer cells tolerate CIN needs to be clarified.
Through LxCxE motifs, RB interacts with numerous chromatin modifiers, including DNMT1, SUV39H1, Suv4-20H1, BRN1, BRG1, HDAC, and KDM5A/JARID1A/RBP2[36]. Among these, KDM5A might mediate RB function to control stemness. KDM5A demethylates tri- and di-methylated lysine 4 in histone H3. ES cells lacking KDM5A failed to maintain OCT4 and NANOG expression upon stimulation to promote differentiation[98]. KDM5A controls RB-dependent myogenic differentiation, at least partially, through the regulation of mitochondrial function[99]. Importantly, loss of KDM5A in Rb-heterozygous mice attenuated pituitary tumorigenesis[98]. KDM5A was first identified in a screen to find proteins that bind to pRB mutants unable to bind to E2Fs[100]. pRB mutants unable to bind to KDM5A failed to control differentiation. The mechanism whereby KDM5A controls mitochondrial function involves PGC-1/PPARγc1a[99]. It is of great interest to determine whether other genes under the influence of RB-KDM5A axis control stemness based on the effect on their epigenetic status.
Our experience with the stem cell-like behaviors exhibited by RB-p53 double deficient cells indicated that some of stem cell-like features are not reversed by RB reconstitution. This suggests that the effect of RB deficiency on stem cell-like behaviors might depend on its role in epigenetic control. Many chromatin modifiers carrying LxCxE motifs may be involved in the epigenetic function of pRB; however, so far, only some of them have been characterized regarding their role in controlling stem cell functions. We demonstrated that Rb in mouse cells exerts its influence on the epigenetic control of Ink4a, Shc, and FoxO6 through DNMT1; however, its significance in the control of stemness has not yet been elucidated[38].
RB plays critical roles in the terminal differentiation of cells owing to its genetic and physical interaction with tissue-specific transcription factors, including MYOD, C/EBP, GR, GATA-1, PU-1, CBFA-1, PDX1, RUNX2, and NF-IL6[36]. It is totally unknown whether the interaction between RB and these factors has any role in tissue stem cells. Myoblast regeneration induced by RB and ARF depletion in post-mitotic muscle cells[101] may involve the elimination of MYOD functions because there is a physical interaction between RB and MYOD. Additionally, the physical interaction of RB and RUNX2 may explain dedifferentiation in osteoblasts that would occur upon development of osteosarcoma.
The interaction of RB with ID2, KDM5A, and EID1 may govern differentiation in a less tissue-specific manner. ID2, when overexpressed in Nestin-expressing cells, induced precocious neural stem cell depletion[102]. RB is to some extent dispensable in brain development during the embryonic stage since Rb-deficient embryonic brains exhibited almost normal development, except for ectopic cell cycle entry and cell death in cortical neurons[103,104]. However, the role of RB in adult neural stem cells has not been sufficiently addressed yet.
There are reports that NANOG and SOX2 induce hyperphosphorylation of pRB through regulation of CDC25A and CDK6[84]. OCT4, in cooperation with miR-335, induces hyperphosphorylation of RB by suppressing PP1 through NIP1 and CCNF[105]. Following RB hyperphosphorylation, freed E2Fs transactivate OCT4-targeted genes. These findings indicate the general function of RB in orchestrating the embryonic gene network. The most surprising finding regarding the role of RB in the embryonic gene network was recently made by the Wernig and Sage laboratory[59]. They discovered that RB is directly involved in the transcriptional control of OCT4 and SOX2. They also demonstrated that Sox2 deletion attenuated pituitary tumorigenesis in Rb-heterozygous mice. These findings clearly revealed the strong influence of RB on the embryonic gene network.
These findings also explain the upregulation of a series of embryonic genes in MEFs lacking all RB family members[13] and also in those lacking Rb and p53[15] when forming embryoid body or spheres under nutrition-restricted and floating conditions. However, there was no evidence of upregulation of these genes when cells were cultured under 2D culture conditions in the presence of serum. This implies that regulation of OCT4 and SOX2 by RB is influenced by the environment where cells are placed and other genes that play a critical role in allowing RB to control embryonic genes. It would be of great interest to survey such genes.
There are significant similarities between stem cell metabolism and cancer cell metabolic reprogramming (rewiring)[106,107]. RB functions in various metabolic pathways have attracted attention from cancer researchers[36,45,46,108]. Given that RB is the central molecule in cell cycle control, it is reasonable that RB also responses to demands for increasing biomass that is coupled to the cell cycle. It is well known that RB controls thymidine kinase 1 (TK1) and dihydrofolate reductase (DHFR), both of which are required for nucleotide synthesis[109]. However, the biomass needed for doubling the cell size during the S phase contains not only nucleotides but also amino acids, lipids, and many other carbon metabolites. pRB undergoes post-translational modification by various nutrient signals involving SIRT1 and AMPK and by cyclin/CDK complexes stimulated by various mitogenic signals[110,111]. RB regulates the transcription or the activity of wide range of enzymes, signaling molecules, and transcription factors, including OXPHOS genes, MVA pathway genes, UCP-1, SOD2, ASCT2, GLS1, PKA, AKT, mTOR, PDK4, PGC-1, ERR, FOXc2, HIF-1, BNIP3, SREBP-1,2, PPAR and KDM5A[12,14,112-124]. Genes encoding many of these factors are supposed to be driven by E2F transcription factors. Myc is also a well-established downstream molecule of RB, and is implicated in the transcriptional control of GLUT1, HK2, PKM2, and LDH-A[45].
OXPHOS activity is significantly correlated to the efficacy of iPS induction[125]. Besides its interaction with OCT4 and SOX2, RB might be implicated in nuclear reprogamming through its influence on OXPHOS.
HIF-1α activation explains why hypoxia facilitates iPS induction and self-renewal of tumor cells[126,127]. Additionally, this molecule was implicated in the long-term maintenance of hematopoietic stem cell (LT-HSC) populations[128]. Thus, RB status presumably has a big impact on glycolysis and TCA activity through the functional interaction with HIF-1. We have recently identified an HIF-1-independent target of RB in the glycolytic pathway. It is of interest to address how RB and HIF-1 cooperate in the control of the glycolytic pathway.
Recent studies from two independent research groups highlighted the link between RB status and glutaminolysis in Drosophila and mammalian cells[116,129]. Activation of the glutamine pathway not only fuels the TCA cycle, but also contributes to control of the cellular reactive oxygen species (ROS) level by regulating glutathione synthesis[130]. This may give both stem cells and cancer cells an advantage in terms of the maintenance of stemness. Additionally, ASCT2 in cooperation with GLUT1 appears to regulate human hematopoietic stem cell lineage specification[131]. Increased dependency on glutamine metabolism is frequently observed in cancers, and has been linked to the emergence of drug resistance[132,133]. Glutamine metabolism could be an important link between stem cells and cancer cells.
While addressing the question of why Ras proteins are activated after RB loss, we discovered that RB controls a number of enzymes involved in the MVA pathway. This study has been extended to another study that addresses the role of the MVA pathway in controlling stem cell-like activity in prostate cancer cells. The mechanism that links RB to Ras include SREBP-1,2. The characterization of Rb-Srebf-1 DKO mice revealed that RB status has a big impact on the control of fatty acid quality. We also identified molecules that explain differential self-renewal activity between p53-/- cells and Rb-deficient p53-/- cells (see above). These include enzymes involved in the glycolytic pathway.
RB status has been linked to pro-inflammatory phenotypes in breast cancer cells[68]. This study highlighted COX2 as a target of the E2F transcription factor. However, our reassessment of these data revealed that many pro-inflammatory cytokines and chemokines, including IL-6, CCL2, and CCL5, are upregulated when breast cancers express lower levels of RB. A similar observation was made in mice lacking Rb in the back skin of p21-/- mice[134]. The tumors derived from these mice exhibited high levels of pro-inflammatory cytokines and evidence of infiltration by immune cells. Indeed, many cytokines and chemokines, and even their receptors, are often regulated by E2F transcription factors[135,136].
Among cytokines and chemokines, IL-6 and CCL2 are drawing attention as strong inducers of iPS cells. A study demonstrated that IL-6 is more potent than c-Myc in terms of their abilities to induce iPS cells[137]. Another study demonstrated that CCL2 plays pivotal role in the maintenance of pluripotency in ES and iPS cells[138]. There are many reports indicating that pro-inflammatory status is critical for cancer stem cells to evolve[139-142]. Many of pro-inflammatory factors stimulates JAK/STAT3 pathway, thus contribute to enhance self-renewal of stem cells and possibly cancer stem cells. This could be one of core mechanisms that are shared by stem cells and cancer cells. Our recent efforts revealed that the RB status in soft tissue sarcoma, breast and prostate cancer cells significantly alter the pro-inflammatory status of these cells, and this significantly enhances self-renewal activity and chemo-resistance.
RB may control innate immunity as well. The RB-E2F1 complex appears to regulate toll-like receptor 3 (TLR3)[143]. In Drosophila, RB, in cooperation with dCAP-D3, upregulates innate immunity[144]. Several reports demonstrated that RB-deficiency in tumors attenuates the innate immune response, thereby promoting tumor development[145]. There is also a report that showed a positive role for innate immunity in inducing iPS cells[146].
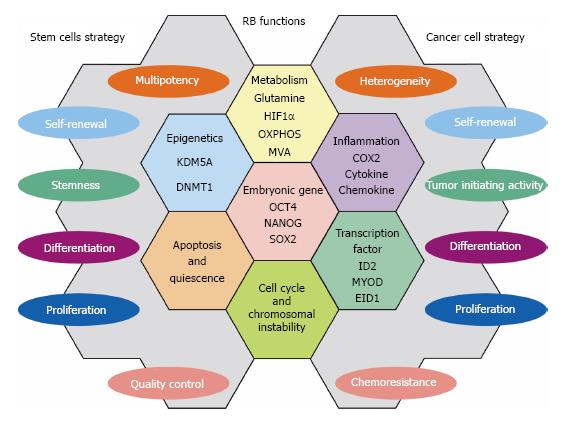
In this manuscript, we described a part of numerous RB functions that are employed commonly as strategies to control stem cells and suppress cancer cells. These findings may help readers to understand how stem cell strategies are shared by cancer (stem) cells. Figure 2 demonstrates RB functions that may be shared by stem cells and cancer cells. Although RB is one of tumor suppressors those have been characterized for long time and by big number of cancer researchers, our curiosity on its hidden roles in various biological events never fades away. In the near future, more number of stem cell and regenerative medicine researchers may stand up by this molecule. Although beyond the scope of this article, RB is implicated in the regeneration of many tissues/organs[19]. Discovery of its target in the context of the undifferentiated state of cancer cells or in drug resistance may lead us to develop powerful tools in both cancer therapy and regenerative medicine.
P- Reviewer: Gunther T, Syed V, Tomizawa M S- Editor: Qiu S L- Editor: A E- Editor: Lu YJ
| 1. | Visvader JE. Cells of origin in cancer. Nature. 2011;469:314-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1008] [Cited by in RCA: 1087] [Article Influence: 77.6] [Reference Citation Analysis (0)] |
| 2. | Blanpain C. Tracing the cellular origin of cancer. Nat Cell Biol. 2013;15:126-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 196] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 3. | Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 2015;29:1447-1462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 178] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 4. | Wang DJ, Ratnam NM, Byrd JC, Guttridge DC. NF-κB functions in tumor initiation by suppressing the surveillance of both innate and adaptive immune cells. Cell Rep. 2014;9:90-103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 5. | Wang ZA, Mitrofanova A, Bergren SK, Abate-Shen C, Cardiff RD, Califano A, Shen MM. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat Cell Biol. 2013;15:274-283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 253] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 6. | Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LC, Lannon WA, Grotzinger C, Del Rio M. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 758] [Article Influence: 63.2] [Reference Citation Analysis (0)] |
| 7. | Huels DJ, Sansom OJ. Stem vs non-stem cell origin of colorectal cancer. Br J Cancer. 2015;113:1-5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 8. | Skibinski A, Kuperwasser C. The origin of breast tumor heterogeneity. Oncogene. 2015;34:5309-5316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 114] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 9. | Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238-1242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 772] [Cited by in RCA: 747] [Article Influence: 46.7] [Reference Citation Analysis (0)] |
| 10. | Jeter CR, Yang T, Wang J, Chao HP, Tang DG. Concise Review: NANOG in Cancer Stem Cells and Tumor Development: An Update and Outstanding Questions. Stem Cells. 2015;33:2381-2390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 176] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 11. | Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719-724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2798] [Cited by in RCA: 2397] [Article Influence: 149.8] [Reference Citation Analysis (0)] |
| 12. | Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010;466:1110-1114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 262] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 13. | Liu Y, Clem B, Zuba-Surma EK, El-Naggar S, Telang S, Jenson AB, Wang Y, Shao H, Ratajczak MZ, Chesney J. Mouse fibroblasts lacking RB1 function form spheres and undergo reprogramming to a cancer stem cell phenotype. Cell Stem Cell. 2009;4:336-347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 14. | Shamma A, Takegami Y, Miki T, Kitajima S, Noda M, Obara T, Okamoto T, Takahashi C. Rb Regulates DNA damage response and cellular senescence through E2F-dependent suppression of N-ras isoprenylation. Cancer Cell. 2009;15:255-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 99] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 15. | Kitajima S, Kohno S, Kondoh A, Sasaki N, Nishimoto Y, Li F, Abdallah Mohammed MS, Muranaka H, Nagatani N, Suzuki M. Undifferentiated State Induced by Rb-p53 Double Inactivation in Mouse Thyroid Neuroendocrine Cells and Embryonic Fibroblasts. Stem Cells. 2015;33:1657-1669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Kwon MC, Berns A. Mouse models for lung cancer. Mol Oncol. 2013;7:165-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 111] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 17. | Sutherland KD, Berns A. Cell of origin of lung cancer. Mol Oncol. 2010;4:397-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 135] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 18. | Rubio R, Gutierrez-Aranda I, Sáez-Castillo AI, Labarga A, Rosu-Myles M, Gonzalez-Garcia S, Toribio ML, Menendez P, Rodriguez R. The differentiation stage of p53-Rb-deficient bone marrow mesenchymal stem cells imposes the phenotype of in vivo sarcoma development. Oncogene. 2013;32:4970-4980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 19. | Sage J. The retinoblastoma tumor suppressor and stem cell biology. Genes Dev. 2012;26:1409-1420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 20. | Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1992] [Cited by in RCA: 1881] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 21. | Kleinerman RA, Tucker MA, Tarone RE, Abramson DH, Seddon JM, Stovall M, Li FP, Fraumeni JF. Risk of new cancers after radiotherapy in long-term survivors of retinoblastoma: an extended follow-up. J Clin Oncol. 2005;23:2272-2279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 306] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 22. | Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3548] [Cited by in RCA: 3489] [Article Influence: 116.3] [Reference Citation Analysis (0)] |
| 23. | Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008;8:714-724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 297] [Cited by in RCA: 292] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 24. | Marshall C. How do small GTPase signal transduction pathways regulate cell cycle entry? Curr Opin Cell Biol. 1999;11:732-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 128] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 25. | Macdonald JI, Dick FA. Posttranslational modifications of the retinoblastoma tumor suppressor protein as determinants of function. Genes Cancer. 2012;3:619-633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Rubin SM. Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem Sci. 2013;38:12-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 193] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 27. | Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14:297-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 433] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 28. | Harbour JW, Dean DC. Chromatin remodeling and Rb activity. Curr Opin Cell Biol. 2000;12:685-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 93] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 29. | Ji P, Jiang H, Rekhtman K, Bloom J, Ichetovkin M, Pagano M, Zhu L. An Rb-Skp2-p27 pathway mediates acute cell cycle inhibition by Rb and is retained in a partial-penetrance Rb mutant. Mol Cell. 2004;16:47-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 133] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 30. | Wang H, Bauzon F, Ji P, Xu X, Sun D, Locker J, Sellers RS, Nakayama K, Nakayama KI, Cobrinik D. Skp2 is required for survival of aberrantly proliferating Rb1-deficient cells and for tumorigenesis in Rb1+/- mice. Nat Genet. 2010;42:83-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 102] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 31. | Manning AL, Dyson NJ. RB: mitotic implications of a tumour suppressor. Nat Rev Cancer. 2012;12:220-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 121] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 32. | Sage J, Straight AF. RB’s original CIN? Genes Dev. 2010;24:1329-1333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 33. | Sellers WR, Novitch BG, Miyake S, Heith A, Otterson GA, Kaye FJ, Lassar AB, Kaelin WG. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 275] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 34. | Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998;2:293-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 277] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 35. | Sage J, Miller AL, Pérez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 429] [Cited by in RCA: 420] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 36. | Takahashi C, Sasaki N, Kitajima S. Twists in views on RB functions in cellular signaling, metabolism and stem cells. Cancer Sci. 2012;103:1182-1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 37. | Takahashi C, Contreras B, Iwanaga T, Takegami Y, Bakker A, Bronson RT, Noda M, Loda M, Hunt JL, Ewen ME. Nras loss induces metastatic conversion of Rb1-deficient neuroendocrine thyroid tumor. Nat Genet. 2006;38:118-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 38. | Shamma A, Suzuki M, Hayashi N, Kobayashi M, Sasaki N, Nishiuchi T, Doki Y, Okamoto T, Kohno S, Muranaka H. ATM mediates pRB function to control DNMT1 protein stability and DNA methylation. Mol Cell Biol. 2013;33:3113-3124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 39. | Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893-898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 611] [Cited by in RCA: 547] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 40. | Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 712] [Cited by in RCA: 695] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 41. | Sharpless NE, DePinho RA. Telomeres, stem cells, senescence, and cancer. J Clin Invest. 2004;113:160-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 308] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 42. | Hilgendorf KI, Leshchiner ES, Nedelcu S, Maynard MA, Calo E, Ianari A, Walensky LD, Lees JA. The retinoblastoma protein induces apoptosis directly at the mitochondria. Genes Dev. 2013;27:1003-1015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 43. | Attardi LD, Sage J. RB goes mitochondrial. Genes Dev. 2013;27:975-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 44. | Williams JP, Stewart T, Li B, Mulloy R, Dimova D, Classon M. The retinoblastoma protein is required for Ras-induced oncogenic transformation. Mol Cell Biol. 2006;26:1170-1182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 45. | Clem BF, Chesney J. Molecular pathways: regulation of metabolism by RB. Clin Cancer Res. 2012;18:6096-6100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 46. | Nicolay BN, Dyson NJ. The multiple connections between pRB and cell metabolism. Curr Opin Cell Biol. 2013;25:735-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 47. | Desvoyes B, Fernández-Marcos M, Sequeira-Mendes J, Otero S, Vergara Z, Gutierrez C. Looking at plant cell cycle from the chromatin window. Front Plant Sci. 2014;5:369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 48. | Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005;123:1337-1349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 260] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 49. | McEvoy J, Flores-Otero J, Zhang J, Nemeth K, Brennan R, Bradley C, Krafcik F, Rodriguez-Galindo C, Wilson M, Xiong S. Coexpression of normally incompatible developmental pathways in retinoblastoma genesis. Cancer Cell. 2011;20:260-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 50. | Xu XL, Fang Y, Lee TC, Forrest D, Gregory-Evans C, Almeida D, Liu A, Jhanwar SC, Abramson DH, Cobrinik D. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell. 2009;137:1018-1031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 187] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 51. | Ajioka I, Martins RA, Bayazitov IT, Donovan S, Johnson DA, Frase S, Cicero SA, Boyd K, Zakharenko SS, Dyer MA. Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell. 2007;131:378-390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 150] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 52. | Xu XL, Singh HP, Wang L, Qi DL, Poulos BK, Abramson DH, Jhanwar SC, Cobrinik D. Rb suppresses human cone-precursor-derived retinoblastoma tumours. Nature. 2014;514:385-388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 171] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 53. | Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 282] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 54. | Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 384] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 55. | Song H, Yao E, Lin C, Gacayan R, Chen MH, Chuang PT. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc Natl Acad Sci USA. 2012;109:17531-17536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 215] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 56. | Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132-1135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1078] [Cited by in RCA: 1015] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 57. | Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 792] [Cited by in RCA: 798] [Article Influence: 53.2] [Reference Citation Analysis (0)] |
| 58. | Li F, He Z, Shen J, Huang Q, Li W, Liu X, He Y, Wolf F, Li CY. Apoptotic caspases regulate induction of iPSCs from human fibroblasts. Cell Stem Cell. 2010;7:508-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 59. | Kareta MS, Gorges LL, Hafeez S, Benayoun BA, Marro S, Zmoos AF, Cecchini MJ, Spacek D, Batista LF, O’Brien M. Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell. 2015;16:39-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 153] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 60. | Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, Vinocour M, Levitt M, Pass H, Gazdar AF, Minna JD. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246:491-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 792] [Cited by in RCA: 843] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 61. | Wistuba II, Berry J, Behrens C, Maitra A, Shivapurkar N, Milchgrub S, Mackay B, Minna JD, Gazdar AF. Molecular changes in the bronchial epithelium of patients with small cell lung cancer. Clin Cancer Res. 2000;6:2604-2610. [PubMed] |
| 62. | George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, Leenders F, Lu X, Fernández-Cuesta L, Bosco G. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1125] [Cited by in RCA: 1666] [Article Influence: 166.6] [Reference Citation Analysis (0)] |
| 63. | Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 916] [Cited by in RCA: 1095] [Article Influence: 84.2] [Reference Citation Analysis (0)] |
| 64. | Coe BP, Thu KL, Aviel-Ronen S, Vucic EA, Gazdar AF, Lam S, Tsao MS, Lam WL. Genomic deregulation of the E2F/Rb pathway leads to activation of the oncogene EZH2 in small cell lung cancer. PLoS One. 2013;8:e71670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 65. | Hubaux R, Thu KL, Coe BP, MacAulay C, Lam S, Lam WL. EZH2 promotes E2F-driven SCLC tumorigenesis through modulation of apoptosis and cell-cycle regulation. J Thorac Oncol. 2013;8:1102-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 66. | Niederst MJ, Sequist LV, Poirier JT, Mermel CH, Lockerman EL, Garcia AR, Katayama R, Costa C, Ross KN, Moran T. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun. 2015;6:6377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 376] [Cited by in RCA: 499] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 67. | Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9574] [Cited by in RCA: 9344] [Article Influence: 718.8] [Reference Citation Analysis (0)] |
| 68. | Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, Chew K, Moore D, Rabban J, Chen YY, Kerlikowske K, Tlsty TD. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479-491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 175] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 69. | Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10:R75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 205] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 70. | Bosco EE, Wang Y, Xu H, Zilfou JT, Knudsen KE, Aronow BJ, Lowe SW, Knudsen ES. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J Clin Invest. 2007;117:218-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 153] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 71. | Jiang Z, Jones R, Liu JC, Deng T, Robinson T, Chung PE, Wang S, Herschkowitz JI, Egan SE, Perou CM. RB1 and p53 at the crossroad of EMT and triple-negative breast cancer. Cell Cycle. 2011;10:1563-1570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 72. | Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 1109] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 73. | Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143-1158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 386] [Cited by in RCA: 424] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 74. | Arima Y, Inoue Y, Shibata T, Hayashi H, Nagano O, Saya H, Taya Y. Rb depletion results in deregulation of E-cadherin and induction of cellular phenotypic changes that are characteristic of the epithelial-to-mesenchymal transition. Cancer Res. 2008;68:5104-5112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 122] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 75. | Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2720] [Cited by in RCA: 2974] [Article Influence: 198.3] [Reference Citation Analysis (0)] |
| 76. | Friedlander TW, Roy R, Tomlins SA, Ngo VT, Kobayashi Y, Azameera A, Rubin MA, Pienta KJ, Chinnaiyan A, Ittmann MM. Common structural and epigenetic changes in the genome of castration-resistant prostate cancer. Cancer Res. 2012;72:616-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 77. | Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CE, Witkiewicz AK. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 2010;120:4478-4492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 260] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 78. | Bohrer LR, Chen S, Hallstrom TC, Huang H. Androgens suppress EZH2 expression via retinoblastoma (RB) and p130-dependent pathways: a potential mechanism of androgen-refractory progression of prostate cancer. Endocrinology. 2010;151:5136-5145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 79. | Gordon CA, Gulzar ZG, Brooks JD. NUSAP1 expression is upregulated by loss of RB1 in prostate cancer cells. Prostate. 2015;75:517-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 80. | Guillou L, Aurias A. Soft tissue sarcomas with complex genomic profiles. Virchows Arch. 2010;456:201-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 81. | Choi J, Curtis SJ, Roy DM, Flesken-Nikitin A, Nikitin AY. Local mesenchymal stem/progenitor cells are a preferential target for initiation of adult soft tissue sarcomas associated with p53 and Rb deficiency. Am J Pathol. 2010;177:2645-2658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 82. | Zhu SJ, Pearson BJ. The Retinoblastoma pathway regulates stem cell proliferation in freshwater planarians. Dev Biol. 2013;373:442-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 83. | Desvoyes B, de Mendoza A, Ruiz-Trillo I, Gutierrez C. Novel roles of plant RETINOBLASTOMA-RELATED (RBR) protein in cell proliferation and asymmetric cell division. J Exp Bot. 2014;65:2657-2666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 84. | Conklin JF, Sage J. Keeping an eye on retinoblastoma control of human embryonic stem cells. J Cell Biochem. 2009;108:1023-1030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 85. | Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051-3064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 330] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 86. | Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037-3050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 474] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 87. | Conklin JF, Baker J, Sage J. The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat Commun. 2012;3:1244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 88. | Miura T, Luo Y, Khrebtukova I, Brandenberger R, Zhou D, Thies RS, Vasicek T, Young H, Lebkowski J, Carpenter MK. Monitoring early differentiation events in human embryonic stem cells by massively parallel signature sequencing and expressed sequence tag scan. Stem Cells Dev. 2004;13:694-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 73] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 89. | Viatour P, Somervaille TC, Venkatasubrahmanyam S, Kogan S, McLaughlin ME, Weissman IL, Butte AJ, Passegué E, Sage J. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell. 2008;3:416-428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 135] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 90. | Narita M, Nũnez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703-716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1626] [Cited by in RCA: 1694] [Article Influence: 77.0] [Reference Citation Analysis (0)] |
| 91. | Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1435] [Cited by in RCA: 1567] [Article Influence: 78.4] [Reference Citation Analysis (0)] |
| 92. | Young AP, Schlisio S, Minamishima YA, Zhang Q, Li L, Grisanzio C, Signoretti S, Kaelin WG. VHL loss actuates a HIF-independent senescence programme mediated by Rb and p400. Nat Cell Biol. 2008;10:361-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 188] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 93. | Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459-472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 416] [Cited by in RCA: 410] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 94. | Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136-1139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 793] [Cited by in RCA: 763] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 95. | Manning AL, Benes C, Dyson NJ. Whole chromosome instability resulting from the synergistic effects of pRB and p53 inactivation. Oncogene. 2014;33:2487-2494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 96. | Manning AL, Dyson NJ. pRB, a tumor suppressor with a stabilizing presence. Trends Cell Biol. 2011;21:433-441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 97. | Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 602] [Cited by in RCA: 658] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 98. | Lin W, Cao J, Liu J, Beshiri ML, Fujiwara Y, Francis J, Cherniack AD, Geisen C, Blair LP, Zou MR. Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc Natl Acad Sci USA. 2011;108:13379-13386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 99. | Váraljai R, Islam AB, Beshiri ML, Rehman J, Lopez-Bigas N, Benevolenskaya EV. Increased mitochondrial function downstream from KDM5A histone demethylase rescues differentiation in pRB-deficient cells. Genes Dev. 2015;29:1817-1834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 100. | Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell. 2005;18:623-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 190] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 101. | Pajcini KV, Corbel SY, Sage J, Pomerantz JH, Blau HM. Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell. 2010;7:198-213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 152] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 102. | Jung S, Park RH, Kim S, Jeon YJ, Ham DS, Jung MY, Kim SS, Lee YD, Park CH, Suh-Kim H. Id proteins facilitate self-renewal and proliferation of neural stem cells. Stem Cells Dev. 2010;19:831-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 103. | Takahashi C, Bronson RT, Socolovsky M, Contreras B, Lee KY, Jacks T, Noda M, Kucherlapati R, Ewen ME. Rb and N-ras function together to control differentiation in the mouse. Mol Cell Biol. 2003;23:5256-5268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 104. | Takahashi C, Contreras B, Bronson RT, Loda M, Ewen ME. Genetic interaction between Rb and K-ras in the control of differentiation and tumor suppression. Mol Cell Biol. 2004;24:10406-10415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 105. | Schoeftner S, Scarola M, Comisso E, Schneider C, Benetti R. An Oct4-pRb axis, controlled by MiR-335, integrates stem cell self-renewal and cell cycle control. Stem Cells. 2013;31:717-728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 106. | Sebastian C. Tracking down the origin of cancer: metabolic reprogramming as a driver of stemness and tumorigenesis. Crit Rev Oncog. 2014;19:363-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 107. | Menendez JA, Alarcón T. Metabostemness: a new cancer hallmark. Front Oncol. 2014;4:262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 84] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 108. | Clem BF, O’Neal J, Klarer AC, Telang S, Chesney J. Clinical development of cancer therapeutics that target metabolism. QJM. 2015;Oct 1; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 109. | Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810-2826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 530] [Cited by in RCA: 554] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 110. | Wong S, Weber JD. Deacetylation of the retinoblastoma tumour suppressor protein by SIRT1. Biochem J. 2007;407:451-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 104] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 111. | Dasgupta B, Milbrandt J. AMP-activated protein kinase phosphorylates retinoblastoma protein to control mammalian brain development. Dev Cell. 2009;16:256-270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 113] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 112. | Nicolay BN, Danielian PS, Kottakis F, Lapek JD, Sanidas I, Miles WO, Dehnad M, Tschöp K, Gierut JJ, Manning AL. Proteomic analysis of pRb loss highlights a signature of decreased mitochondrial oxidative phosphorylation. Genes Dev. 2015;29:1875-1889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 113. | Blanchet E, Annicotte JS, Lagarrigue S, Aguilar V, Clapé C, Chavey C, Fritz V, Casas F, Apparailly F, Auwerx J. E2F transcription factor-1 regulates oxidative metabolism. Nat Cell Biol. 2011;13:1146-1152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 219] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 114. | Hansen JB, Jørgensen C, Petersen RK, Hallenborg P, De Matteis R, Bøye HA, Petrovic N, Enerbäck S, Nedergaard J, Cinti S. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc Natl Acad Sci USA. 2004;101:4112-4117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 227] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 115. | Imai Y, Takahashi A, Hanyu A, Hori S, Sato S, Naka K, Hirao A, Ohtani N, Hara E. Crosstalk between the Rb pathway and AKT signaling forms a quiescence-senescence switch. Cell Rep. 2014;7:194-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 116. | Reynolds MR, Lane AN, Robertson B, Kemp S, Liu Y, Hill BG, Dean DC, Clem BF. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene. 2014;33:556-566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 170] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 117. | El-Naggar S, Liu Y, Dean DC. Mutation of the Rb1 pathway leads to overexpression of mTor, constitutive phosphorylation of Akt on serine 473, resistance to anoikis, and a block in c-Raf activation. Mol Cell Biol. 2009;29:5710-5717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 118. | Hsieh MC, Das D, Sambandam N, Zhang MQ, Nahlé Z. Regulation of the PDK4 isozyme by the Rb-E2F1 complex. J Biol Chem. 2008;283:27410-27417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 92] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 119. | Dali-Youcef N, Mataki C, Coste A, Messaddeq N, Giroud S, Blanc S, Koehl C, Champy MF, Chambon P, Fajas L. Adipose tissue-specific inactivation of the retinoblastoma protein protects against diabesity because of increased energy expenditure. Proc Natl Acad Sci USA. 2007;104:10703-10708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 89] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 120. | Scimè A, Grenier G, Huh MS, Gillespie MA, Bevilacqua L, Harper ME, Rudnicki MA. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab. 2005;2:283-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 170] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 121. | Goto TM, Arima Y, Nagano O, Saya H. Lysyl oxidase is induced by cell density-mediated cell cycle suppression via RB-E2F1-HIF-1α axis. Cell Struct Funct. 2013;38:9-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 122. | Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci USA. 2004;101:6570-6575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 549] [Cited by in RCA: 549] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 123. | MacLellan WR, Xiao G, Abdellatif M, Schneider MD. A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol Cell Biol. 2000;20:8903-8915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 102] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 124. | Ciavarra G, Zacksenhaus E. Rescue of myogenic defects in Rb-deficient cells by inhibition of autophagy or by hypoxia-induced glycolytic shift. J Cell Biol. 2010;191:291-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 125. | Liu W, Long Q, Chen K, Li S, Xiang G, Chen S, Liu X, Li Y, Yang L, Dong D. Mitochondrial metabolism transition cooperates with nuclear reprogramming during induced pluripotent stem cell generation. Biochem Biophys Res Commun. 2013;431:767-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 126. | Prigione A, Rohwer N, Hoffmann S, Mlody B, Drews K, Bukowiecki R, Blümlein K, Wanker EE, Ralser M, Cramer T. HIF1α modulates cell fate reprogramming through early glycolytic shift and upregulation of PDK1-3 and PKM2. Stem Cells. 2014;32:364-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 208] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 127. | Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, Hubaud A, Stadler B, Choi M, Bar M. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640-4652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 418] [Cited by in RCA: 416] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 128. | Takubo K, Nagamatsu G, Kobayashi CI, Nakamura-Ishizu A, Kobayashi H, Ikeda E, Goda N, Rahimi Y, Johnson RS, Soga T. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell. 2013;12:49-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 514] [Cited by in RCA: 618] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 129. | Nicolay BN, Gameiro PA, Tschöp K, Korenjak M, Heilmann AM, Asara JM, Stephanopoulos G, Iliopoulos O, Dyson NJ. Loss of RBF1 changes glutamine catabolism. Genes Dev. 2013;27:182-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 130. | Alberghina L, Gaglio D. Redox control of glutamine utilization in cancer. Cell Death Dis. 2014;5:e1561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 109] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 131. | Oburoglu L, Tardito S, Fritz V, de Barros SC, Merida P, Craveiro M, Mamede J, Cretenet G, Mongellaz C, An X. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell. 2014;15:169-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 211] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 132. | Tanaka K, Sasayama T, Irino Y, Takata K, Nagashima H, Satoh N, Kyotani K, Mizowaki T, Imahori T, Ejima Y. Compensatory glutamine metabolism promotes glioblastoma resistance to mTOR inhibitor treatment. J Clin Invest. 2015;125:1591-1602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 187] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 133. | Hernandez-Davies JE, Tran TQ, Reid MA, Rosales KR, Lowman XH, Pan M, Moriceau G, Yang Y, Wu J, Lo RS. Vemurafenib resistance reprograms melanoma cells towards glutamine dependence. J Transl Med. 2015;13:210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 134. | Saiz-Ladera C, Lara MF, Garín M, Ruiz S, Santos M, Lorz C, García-Escudero R, Martínez-Fernández M, Bravo A, Fernández-Capetillo O. p21 suppresses inflammation and tumorigenesis on pRB-deficient stratified epithelia. Oncogene. 2014;33:4599-4612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 135. | Zheng T, Hong X, Wang J, Pei T, Liang Y, Yin D, Song R, Song X, Lu Z, Qi S. Gankyrin promotes tumor growth and metastasis through activation of IL-6/STAT3 signaling in human cholangiocarcinoma. Hepatology. 2014;59:935-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 80] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 136. | Libertini SJ, Chen H, al-Bataina B, Koilvaram T, George M, Gao AC, Mudryj M. The interleukin 6 receptor is a direct transcriptional target of E2F3 in prostate tumor derived cells. Prostate. 2012;72:649-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 137. | Brady JJ, Li M, Suthram S, Jiang H, Wong WH, Blau HM. Early role for IL-6 signalling during generation of induced pluripotent stem cells revealed by heterokaryon RNA-Seq. Nat Cell Biol. 2013;15:1244-1252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 138. | Hasegawa Y, Takahashi N, Forrest AR, Shin JW, Kinoshita Y, Suzuki H, Hayashizaki Y. CC chemokine ligand 2 and leukemia inhibitory factor cooperatively promote pluripotency in mouse induced pluripotent cells. Stem Cells. 2011;29:1196-1205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 139. | Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1097] [Cited by in RCA: 1164] [Article Influence: 72.8] [Reference Citation Analysis (0)] |
| 140. | Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 713] [Cited by in RCA: 710] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 141. | Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci USA. 2011;108:1397-1402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 511] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 142. | He G, Dhar D, Nakagawa H, Font-Burgada J, Ogata H, Jiang Y, Shalapour S, Seki E, Yost SE, Jepsen K. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 2013;155:384-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 358] [Cited by in RCA: 366] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 143. | Taura M, Suico MA, Koyama K, Komatsu K, Miyakita R, Matsumoto C, Kudo E, Kariya R, Goto H, Kitajima S. Rb/E2F1 regulates the innate immune receptor Toll-like receptor 3 in epithelial cells. Mol Cell Biol. 2012;32:1581-1590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 144. | Longworth MS, Walker JA, Anderssen E, Moon NS, Gladden A, Heck MM, Ramaswamy S, Dyson NJ. A shared role for RBF1 and dCAP-D3 in the regulation of transcription with consequences for innate immunity. PLoS Genet. 2012;8:e1002618. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 145. | Hutcheson J, Witkiewicz AK, Knudsen ES. The RB tumor suppressor at the intersection of proliferation and immunity: relevance to disease immune evasion and immunotherapy. Cell Cycle. 2015;14:3812-3819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 146. | Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Pera RR, Yakubov E, Cooke JP. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 278] [Article Influence: 23.2] [Reference Citation Analysis (0)] |