INTRODUCTION
Gliomas can be classified as many different genetically-driven diseases that manifest under the guise of only a few histological variations[1-3]. Our understanding of glioma biology has grown immensely with the advent of cancer genetics and molecular characterization. Large-scale multi-platform characterization of gliomas has revealed strong relationships that tie certain combinations of genetic changes with characteristic epigenetic modifications, transcriptome alterations and clinical presentations to define subtypes[4-7]. Ultimately these findings suggest that the cancer biology in each molecular subclass varies to an extent that remains to be seen. Among the different genetic subclasses of gliomas there is reason to believe that the process of gliomagenesis may also vary. There are many aspects of gliomagenesis to consider: what cell type gives rise to the tumor What genetic changes are compatible with initiating gliomagenesis Are there non-cell autonomous factors that play a role in gliomagenesis, such as microenvironment changes Understanding these tumor-initiating events will allow insight into the spatiotemporal progression of gliomas, the identification of key driver mutations and discovery of early biomarkers.
The cell of origin is the cell type that initiates tumor formation. This differs from the cell of mutation, which is the cell type that acquires oncogenic changes but may not necessarily proliferate until it moves to another point in its respective cellular hierarchy. It is thought that the cell of mutation may either differentiate or de-differentiate to a different cell type, which may then act as the cell of origin via uncontrolled growth[8]. It is unclear if more than one cell of origin or cell of mutation may exist for a single type of tumor. Furthermore, the cells of origin of the different genetic subtypes of glioma are still either a matter of debate or left unexplored. Most of what we know about the potential cells of origin as a function of different combinations of oncogenic mutations in glioma comes from a variety of mouse models. This review will focus on the cell of origin in gliomas by reviewing the different cell types of the neuroglial lineage, exploring cell of origin glioma models and discussing clinical data that suggest differing cells of origin per glioma subtype.
Before proceeding, it is important to recognize the difference between the stem-like cells in a mature tumor and the cell of origin. These stem-like cells are commonly referred to as cancer stem cells (CSCs), brain tumor stem cells (BTSCs), or tumor-initiating cells. For the purposes of this review, the term “tumor-initiating cells” will not be used, as it does not distinguish between the re-initiation of a mature tumor and the initiation of a tumor from its cell of origin. For clarity, we will refer to these cancer stem-like cells as BTSCs or CSCs in this text. In addition, it is also necessary to consider the different context in which we discuss a “stem cell” and “differentiated cell”. When discussing normal human cellular biology, a stem cell is capable of self-renewal and asymmetric differentiation. Progenitors downstream of stem cells may symmetrically differentiate following proliferation. When a fully differentiated stage is reached, the cell typically has limited proliferation potential. Within a tumor, CSCs carry over the same definitions as normal stem cells. It is still a matter of debate as to whether or not the more differentiated cancer cells have limited or unlimited proliferation potential.
There are two prevalent models for the propagation of tumors: the clonal model and cancer stem cell model[9,10]. In the clonal model, single cells within a tumor progressively acquire competitively advantageous genetic changes, accounting for the cellular and genetic heterogeneity observed in tumors. In the cancer stem cell model, there are thought to be CSCs within the tumor that have the ability to self-renew and differentiate. By definition, CSCs can be seeded into another organism and give rise to the tumor it was isolated from, while the non-CSCs either cannot do so, or can do so only with much lower efficiency. In the CSC model, CSCs are thought to give rise to a cellular hierarchy via their differentiation and self-renewal abilities. Both CSCs and non-CSCs acquire genetic mutations, leading to the observed cellular and genetic heterogeneity. BTSCs identified in gliomas are thought to play a key role in the maintenance and virulence of the tumor. How and when the BTSCs arise in the tumor remains a mystery, although at least two possibilities exist. We can hypothesize that differentiated cells in the early tumor eventually de-differentiated to form BTSCs. Conversely, the other possibility is that BTSCs are derivatives of a cell of origin that was once a normal stem cell or progenitor cell. The missing links between cell types in the early tumor and mature tumor are yet to be uncovered. Cell of origin models must be used to explore the developmental arc of a mature tumor that contains a complex cellular hierarchy from a single clone. As was previously mentioned, two major variables are at play in these modeling efforts: the oncogenic mutations and the plethora of cell types found in the brain. In this review we begin with an overview of neurogenesis in the adult brain and follow with a discussion of glioma genetics, glioma cell of origin models and clinical evidence for stem cells as the cells of origin in glioma.
NEUROGENESIS IN THE ADULT BRAIN
Neural stem cells and their progeny have become candidates for the cell of origin of glioma since the discovery of neurogenesis in the adult brain. It is necessary to recognize the variety of cell types in the brain, when they are present and how they arise when discussing the cell of origin of gliomas. Neurogenesis in adults is thought to be responsible for the replacement of neurons and glia for the purposes of cellular replenishment, remodeling and response to injury[11]. We know that adult gliomas arise from the neuroglial lineage during post-natal life due primarily to strong evidence from the histological characteristics of glioma, their molecular signature and mouse glioma models that target the neuroglial lineage. Accordingly, this introduction is mostly limited to adult neurogenesis (vs embryonic or pre-natal neurogenesis) and excludes extensive discussion of other central nervous system (CNS) and non-CNS cell types found in the brain (such as the meninges, endothelium, ependyma and microglia).
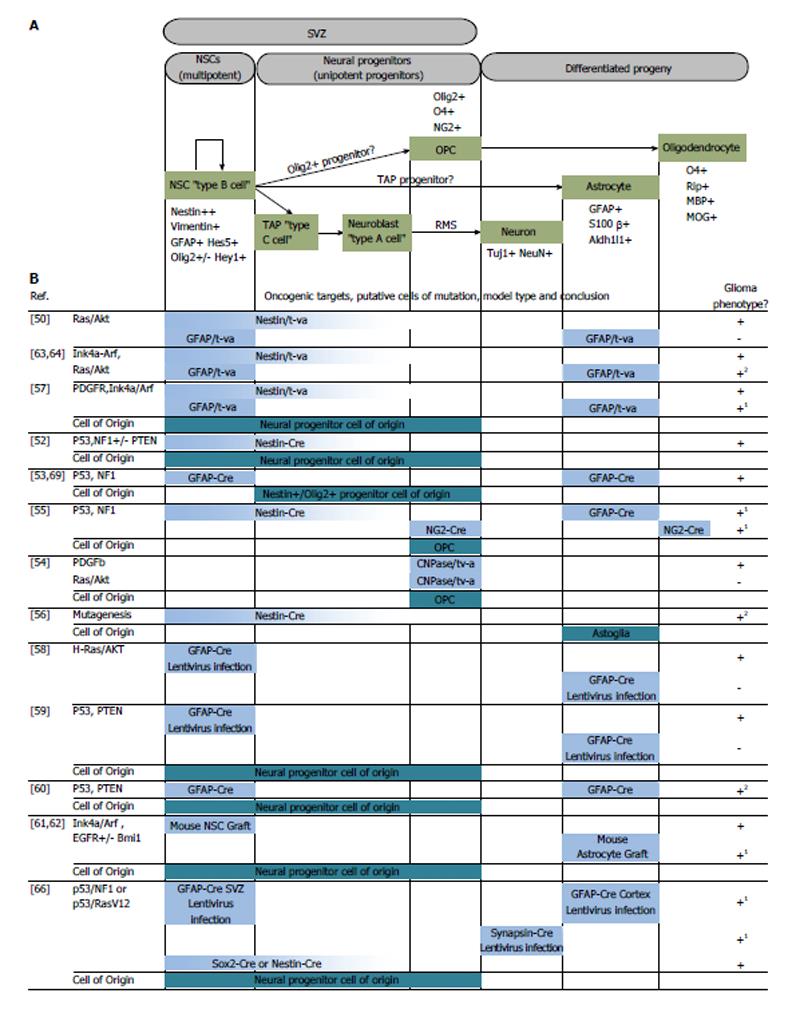
There are two identified neurogenic niches in the adult mammalian brain: the subventricular zone (SVZ) and the subgranular zone (see review by Alvarez-Buylla[11]). Ciliated ependymal cells that encase the cerebrospinal fluid line the lateral ventricles and this monolayer of cells is contained within the ventricular zone[12,13]. On the lateral surfaces of the ventricles, the ependymal cells are laterally lined by neural stem cells (NSCs), or type B NSCs, in a second layer of cellular stratification within the SVZ[13-15]. These type B NSCs arise from neuroepithelium-derived radial glia that are responsible for the stratified organization of the cortex[16-18]. During the transition to post-natal life, radial glia differentiate into type B NSCs that extend a small process to make contact with the cerebrospinal fluid in the ventricular zone. Their cell bodies are mostly confined in the SVZ, with an apical process that extends laterally to contact blood vessels. The type B NSCs in the SVZ are capable of asymmetric division leading to the production of glia or neurons (Figure 1). To produce neurons, the type B cells give rise to transit amplifying cells, or type C cells, which proliferate and progress to type A cells, or neuroblasts. These neuronal precursors are known to migrate through the rostral migratory stream (RMS) in the frontal cortex to replenish interneurons in the olfactory bulb, becoming granule or periglomerular neurons[19-22]. Depending on the regulatory signals in the SVZ niche, type B cells may also generate cortical astrocytes or oligodendrocyte precursors cells (OPCs), which mature to oligodendrocytes[11,23,24].
Figure 1 Targeted oncogenesis in the neuroglial cellular hierarchy.
(A) Neurogenesis in the adult mammalian brain; (B) Summary of murine models that targeted selected cell types for the purposes of discovering the glioma cell of origin. Horizontal bars in (B) correlate to the representative cell types in (A). 1Experimental results by authors suggest differentiated progeny acted as the cell of mutation and de-differentiated before forming a tumor; 2Lack of lineage trace, distinguishing cell of mutation from cell of origin is inconclusive despite tumor forming capacity of differentiated progeny. Partially adapted from Ref. [8,48]. SVZ: Subventricular zone; NSCs: Neural stem cells; TAP: Transit amplifying progenitors.
In the hippocampal formation, radial astrocytes (type 1 cells) serve as stem cells[25]. Type 1 NSCs differentiate into intermediate progenitor cells (type 2 cells), which form immature granule cells (type 3 cells). Subsequently, type 3 cells will mature into the granule neurons found in the hippocampus[26].
Because most of what we know about post-natal neurogenesis and its cellular hierarchy in the brain comes from the study of rodents, there has been intense speculation as to whether human brains harbor active NSCs that generate progenitors and what their subsequent roles are during adult life. The implication of active neurogenesis in adult humans suggests that a decline or defect in the process may play a role in neurodegenerative disorders or glioma formation, respectively. The quest for uncovering neurogenesis in higher organisms consisted mostly of labeling studies in post-mortem brains of monkeys and human patients. Through these studies we have gained substantial evidence for the presence of post-natal human neurogenesis, although their roles in maintaining the human brain’s function remain matters of ongoing study.
Mounting evidence for two neurogenic regions in the rodent brain led to the search for their human homologues. Explant culture and labeling experiments of human brain surgical specimens generated new neurons and glia[27,28]. This was the first direct observation and in vitro generation of human neuronal cell types. Shortly thereafter, many others demonstrated that multipotent or neurosphere-forming cells could be isolated and cultured from the human SVZ and subgranular zone (SGZ). Such cultures were extremely heterogeneous, but they were shown to be capable of directed differentiation in vitro to both glia and neurons, indicating that they contained either undifferentiated precursors or NSCs[29-34]. In a rare form of scientific inquiry, human cancer patients were injected with Bromodeoxyuridine (BrdU), a mitotic marker, as a part of a diagnostic procedure. Post-mortem examination of their hippocampi revealed BrdU-labeled neural and glial cell types, and a small population of BrdU-positive cells that did not co-stain for differentiation markers. These unidentifiable cell types were presumed to be the undifferentiated stem cells or progenitors[35]. Interestingly, BrdU-positive cells in the SVZ were also noted in all five patients examined, who were between the ages of 58 and 72 years old at time of death, indicating that neurogenesis may continue late into adult life.
The evidence supporting neurogenic activity in the human brain raises other important questions: where do stem cells reside How does the cellular hierarchy operate in the primate brain The first identification of neurogenesis in monkeys was made in the hippocampus structure. Kornack et al[36] and Gould et al[37] observed that the rate of formation of new granule neurons in the SGZ could be modulated by stress and that the primate brain was also capable of generating astrocytes and oligodendrocytes, a process that continued even as the monkeys increased in age. Neuroblast (type A cell) formation was also observed in the adult forebrain of monkeys, lending further evidence for adult SVZ neurogenesis in primates[38]. These neuroblasts were also found to travel along the RMS[39], as observed in their mammalian rodent counterparts[19,40]. The first evidence for the existence of human neuroblasts (type A cells) in the olfactory bulb came from examination of post-mortem brains, which showed immuno-positivity for neuroblast markers[21]. Following this study, three separate groups provided evidence, once again through immunostaining and ultrastructural studies of post-mortem human tissue, for neuroblast chain migration through the RMS[22,41-43]. In addition, Alvarez-Buylla and colleagues claim to have identified the Medial Migratory Stream, an additional migratory pathway for neuroblasts that extends medially to the pre-frontal cortex[42]. They indicated, however, that chain migration through this region ends after approximately 18 mo of age. The direct identification of multipotent NSCs (type B cells) in the adult human SVZ has provided us with evidence that humans do harbor NSCs and that they are capable of producing both glia and neurons in a fashion similar to other mammals[12,44]. Given the hypothesis that tumorigenesis is more likely to occur in mitotically active cells rather than in quiescent cell types, it will be interesting to explore if tumor incidence, and type, vary with neuronal developmental stages in a child or adolescent, or with stress, injury and increased age.
GLIOMA MODELS AND THE GLIOMA CELL OF ORIGIN
The discovery of human NSCs and their progeny has led to the question of whether or not they act as the cell of origin in glioma. A number of mouse models have been developed to explore this topic. Mouse models recapitulate a small number of genetic mutations found in human glioma by functionally expressing an oncogene or inactivating a tumor suppressor. Genetically engineered mice or targeted lentiviral transduction systems are used for the purposes of modeling gliomagenesis. The genetic targets in these models, although found to be mutated in human gliomas, are not necessarily driver mutations in glioma development, but we are limited in our ability to identify driver mutations from human gliomas. This is also evident by the fact that some mutations, in the context of mouse models, do not produce tumors or fail to produce appropriate phenotypes when mutated alone[45]. There are very limited mechanisms by which we can infer or identify driver mutations from human cancers. One way is to see which types of mutations occur at highest frequencies within a subclass of tumor; another method is to determine what percentage of the cell population carries the mutation. When a mutation is found in nearly every tumor cell, it implies that a disproportionate growth advantage is conferred or that a particular mutation occurred very early in tumor development. Regardless of what we can infer from clinical data, we do have a good understanding of the most common genetic lesions found in gliomas and modeling efforts have focused on dissecting the role of these common culprits.
There are a number of ways to model gliomagenesis. Some model systems aim to create a “mature” glioma, while others aim to identify how limited and defined oncogenic mutations drive initial glioma formation, or gliomagenesis (see review[45]). The most genetically faithful models of glioma are xenografts of human brain tumors in the mouse brain. Xenografts of primary tumors have been used successfully to study glioma biology and genetics because they are very close representations of the mature tumor that is removed during surgery[46]. The drawbacks of such systems are the selection process during cell line derivation, the need to culture these cells ex vivo (which over time leads to epigenetic and genetic alteration), and the need to grow tumors in immune deficient mice. Although human glioma xenografts replicate human pathology, they do not represent the earliest stages of glioma formation. For example, glioma xenografts do not recapitulate the transformation of a normal endogenous neuroglial cell type to neoplastic stages and beyond. Furthermore, glioma xenografts cannot be used to identify the cell of origin or cell of mutation. Explants of glioma have also been used to study glioma biology, although such systems are technically challenging and are limited to the tissue obtained after surgery[47]. Since human tumors cannot be used to understand the beginning stages of gliomagenesis, approaches that involve selective mutation of tumor suppressor genes or induction of oncogenes in model organisms are used to dissect oncogenic transformation.
The most commonly used model system to study gliomagenesis has been genetically engineered mice that form tumors either spontaneously or after induction. One of the main advantages in using mice is the scale and reproducibility in which genetic alterations can be studied, which has proved to be a powerful tool in understanding cancer genetics. The disadvantage of using mice is their species difference from humans, which obviously translates to differing genetics, physiology and anatomy, as well as the failure of some of these models to capture the molecular diversity and heterogeneity of human tumors.
Virally mediated oncogenic transduction is also used to target specific areas and cell types in the mouse brain for gliomagenesis. Such an approach allows the localization of genetic alterations to specific areas within the brain and selective targeting of cell types within that region depending on the type of model used. The drawback of this system is the need for invasive injection of viruses or virus producing cells. Nevertheless, functional mutations in these model systems have provided the platform to study the cell of origin in cancers (see reviews[8,48,49]).
The genetic targets used for these studies are primarily those that are mutated in Glioblastoma Multiforme (GBM), or World Health Organization (WHO) grade IV gliomas. Gliomas are graded based on histological characteristics on a WHO grading scale of I-IV[3]. In GBM, the most common and deadly of the glioma subtypes, a number of high frequency alterations have been found most commonly in the tumor suppressors p53, PTEN, CDK2A/p16INK4A/p14ARF, CDK4, RB and in proto-oncogenes EGFR, PDGFR, PIK3CA, PIKR1, Kras and IDH1[4-7]. The models discussed here have dually aimed to recreate functional recapitulations of genetic alterations to these genes and to understand in what cell type they initiate gliomagenesis.
One of the landmark papers in modeling the cell of origin in glioma came from Holland et al[50]. This unique mouse model employed a genetically engineered strain that expressed a receptor for a retrovirus that harbored either a mutant form of Kras or Akt. Retroviruses were produced by xenografts of chicken cell lines harboring Replication-Competent ALV Splice-acceptor (RCAS) viral vectors[51]. The receptor for these retroviruses is expressed under the control of tissue-specific promoters, such as glial fibrillary acidic protein (GFAP) (expressed primarily in glia, but also NSCs) or nestin (expressed in NSCs and early progenitors). The novelty of this approach lied in targeting of two different cell populations in the neural lineage that were either neural progenitors (using the nestin promoter), or differentiated astrocytes (using the GFAP promoter). When Kras and Akt were targeted to nestin-expressing cells, high-grade glioma formation was observed. Conversely, targeting GFAP-expressing cell types did not yield tumors. This was the first example of a glioma model that differentiated between the oncogenic potential of two different populations of cells along the same neuroglial axis. One weakness of this model was that, by virtue of the nestin promoter being active in both NSCs and lineage-restricted progenitor cells, the exact cell of origin could not be pinpointed still.
Many mouse models followed in dissecting the relationships between genetic lesions, cell types targeted and tumor phenotype produced. Tumor suppressor models produced by Alcantara Llaguno et al[52] aimed to recreate some of the most common genetic lesions in GBM using combinations of p53, phosphatase and tensin homolog (PTEN) and neurofibromin 1 (NF1) knockout in mice. With their models, they concluded that nestin-positive NSCs or their progenitor cells found in the SVZ harbored the ability to initiate high-grade glioma[52]. Using a mutated p53 model that allowed the tracking of p53 mutant cells, Wang et al[53] observed that type B, C and A cells were capable of accumulating mutant p53. However, it was a nestin/olig2-positive population that resembled type C cells, which was thought to initiate the high-grade glioma. Interestingly, they noted that some SVZ type A neuroblasts that harbored the p53 mutation traveled to the olfactory bulb, but no glioma formation was observed[53]. Additionally, two separate groups generated p53 and NF1 knockout mouse models of glioma and also claimed that it was the OPCs that served as the cell of origin in the production of high-grade tumors[54,55]. Koso et al[56] used a transposon-mediated mutagenesis approach in isolated mouse NSCs. Their model revealed dozens of mutations in combination that could sensitize NSCs to immortalization and tumor formation. Interestingly their mutagenized NSCs were most sensitive to oncogenic transformation after differentiation to the astrogial lineage. Other models, such as PDGFR activation via RCAS-tVA[57], lentiviral delivery of Kras/Akt oncogenes[58], and PTEN/p53 inactivation[59,60] suggested multipotent progenitors found in the SVZ as potential cells of origin for glioblastoma (Figure 1).
Cell types found outside of the neurogenic niches were also found to harbor tumor-initiating potential in mouse models. The demarcation between cell of mutation and cell of origin is less commonly explored due to lack of lineage tracing in many of these models however. In cases where lineage tracing has been used, differentiated progeny were found to de-differentiate to a stem cell state preceding tumor growth. These mouse models include Ink4a-ARF knockout[61], Bmi knockout[62], combined Ink4a-ARF knockout and Kras activation[63,64] and aberrant platelet-derived growth factor signaling[57,65], all of which initiated tumors in areas and cell types both outside and inside the neurogenic regions.
Interestingly, there is also evidence that neurons can act as the cell of mutation in a mouse GBM model when they acquire p53/NF1 mutations after undergoing de-differentiation[66]. The implication for this is that non-neurogenic regions of the brain containing quiescent neurons may be capable of gliomagenesis as well. As mouse modeling continues, emphasis will likely be placed on lineage tracing of defined cell types to understand the plasticity of the cell of mutation and the relationship, if any, between genetic lesions and cell of origin. In addition, the field is faced with the challenge of correlating the mouse glioma cells of origin to the likely cell of origin in human glioma. By drawing parallels between the cell of origin and the restricted number of genetic events that must occur in early tumorigenesis we may one day be able to discover early tumor biomarkers, target tumors when they are exponentially more sensitive to therapy and develop therapies that target the unique stem cell biology of tumor formation and propagation.
CLINICAL EVIDENCE FOR STEM CELLS AS THE CELL OF ORIGIN
The presence of CSCs in human glioma specimens and other tumors from the clinic raises the question of why they are present in the tumors to begin with. The interesting aspect of the presence of CSCs in glioma, as it relates to the cell of origin, is that CSCs are thought to be multipotent and capable of self-renewal, as well as express markers of NSCs. CSCs were identified in human GBM and have been shown to re-initiate mature tumors when seeded into the mouse brain at a much higher efficiency then their non-stem counterparts in the tumor. Did CSCs come from a stem cell of origin Did CSCs de-differentiate from differentiated cell types in the tumor At what point during gliomagenesis do CSCs appear These questions remain entirely unanswered as the continuum between the origination of a tumor and its mature form has not been explored. Here, we will present epidemiological and radiographic evidence that cell types with stem cell properties in the tumor may have originated from NSCs or lineage-restricted progenitors.
A number of radiographic studies have shown that there is a tendency for glioma formation to occur near the periventricular area of the brain[67-69], although not all gliomas are necessarily in contact with the ventricles. GBM (WHO grade IV) appears to occur mostly supratentorialy, with tumor epicenters around the ventricles and frontal lobe propensity before the age of 65 and temporal lobe propensity after the age of 65[67,70-72]. Once again, this discrepancy can be accounted by two possibilities: gliomas may initiate from many different cell types found in all areas of the cortex, or gliomagenesis may be preceded by migration of the cell of mutation/cell of origin. Correlating radiographic evidence to molecular subtypes of GBM has yielded interesting patterns in anatomical distribution, but most of these imaging studies are conducted after the tumor has had months or even years to grow. In such cases, the large size of tumors precludes the exact localization of its epicenter.
WHO grade II and III gliomas have a very different anatomical distribution then their grade IV counterparts and a much more “compact” set of associated genetic lesions. The exact reason for this is unknown, but one logical possibility is that these lower grade gliomas have different cells of origin. Up to 80% of the low-grade gliomas are mutated in Isocitrate Dehydrogenase (IDH) with an accompanying p53 mutation or 1p19q deletion[73]. IDHs normally convert isocitrate to α-ketoglutarate and produce a nicotinamide adenine dinucleotide phosphate (or NADH) molecule, an essential metabolic process that occurs in the mitochondrial Krebs (tricarboxylic acid cycle) cycle, cytosol and peroxisomes[74,75]. The mutated form found in glioma and leukemia is a gain-of-function mutation that causes the conversion of α-ketoglutarate to 2-Hydroxyglutarate, a so called “oncometabolite” due to its ability to cause epigenetic reprogramming that is thought to drive tumorigenesis[76-79]. The low-grade IDH mutated gliomas are often found supratentorially in the frontal lobe in young adults[67,80]. Interestingly, this area overlaps with the SVZ in the frontal lobe that generates neuroblasts destined for the olfactory bulb and possibly the medial pre-frontal cortex. However the significance of this finding remains to be understood as the only known mouse models of IDH have failed to produce appropriate human tumor phenotypes in both brain and myeloid neoplasm contexts[81-83].
CONCLUSION
How glial tumors form and develop into their lethal variety remains a standing question in glioma biology. Our understanding of the molecular events, niche changes and cell types involved has brought us closer to this goal. Since the discovery of neurogenesis in the adult brain and the growing body of work on cancer genetics, we have both cellular and genetic candidates to pursue to this end. Many have employed murine models that target the neuroglial cell population with genetic changes akin to those found in human glioma in search of the cell of origin and to understand key initiating genetic changes. These advances have produced mixed results, with the majority of models pointing to the neuronal progenitors or NSCs as the most likely cell of origin. However, other models have found that differentiated cell types may be capable of tumor initiation as well. The models described varied significantly in the type of genetic mutation made, cell population targeted and lineage tracing technique, if any. Furthermore, it is difficult to infer which of the many genetic mutations identified in glioma act as the initiating event. It is quite possible that more than one cell of origin may exist for the various subtypes of glioma and that more than one genetic change is capable of sending a cell down the path to carcinogenesis. As we dissect the roles of these oncogenes and tumor suppressor genes in glioma initiation, we will gain a broader understanding of gliomagenesis as a process rather than a random event. Through these ongoing efforts it is possible that we will identify very early biomarkers and develop an understanding of what type of restricted changes a young tumor must make to progress to a more malignant state, presumably at a stage where these pre-malignant cells are most susceptible to therapies.
P- Reviewers: Ho I, Pajonk F S- Editor: Wen LL L- Editor: A E- Editor: Liu SQ