Published online Aug 26, 2011. doi: 10.4252/wjsc.v3.i8.70
Revised: January 10, 2011
Accepted: January 20, 2011
Published online: August 26, 2011
AIM: To characterize single-cell-derived mouse clonal mesenchymal stem cells (mcMSCs) established with bone marrow samples from three different mouse strains.
METHODS: We established mcMSC lines using subfractionation culturing method from bone marrow samples obtained from long bones. These lines were characterized by measuring cell growth, cell surface epitopes, differentiation potential, lineage-specific gene expression and T-cell suppression capability. Nonclonal MSCs isolated by the conventional gradient centrifugation method were used as controls.
RESULTS: All mcMSC lines showed typical nonclonal MSC-like spindle shape morphology. Lines differed in optimal growth density requirement. Cell surface epitope profiles of these mcMSC lines were similar to those of nonclonal MSCs. However, some lines exhibited different expression levels in a few epitopes, such as CD44 and CD105. Differentiation assays showed that 90% of the mcMSC lines were capable of differentiating into adipogenic and/or chondrogenic lineages, but only 20% showed osteogenic lineage differentiation. T-cell suppression analysis showed that 75% of the lines exhibited T-cell suppression capability.
CONCLUSION: mcMSC lines have similar cell morphology and cell growth rate but exhibit variations in their cell surface epitopes, differentiation potential, lineage-specific gene expression and T-cell suppression capability.
- Citation: Jeon MS, Yi TG, Lim HJ, Moon SH, Lee MH, Kang JS, Kim CS, Lee DH, Song SU. Characterization of mouse clonal mesenchymal stem cell lines established by subfractionation culturing method. World J Stem Cells 2011; 3(8): 70-82
- URL: https://www.wjgnet.com/1948-0210/full/v3/i8/70.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v3.i8.70
Mesenchymal stem cells (MSCs) were first isolated from the bone marrow by Friedenstein et al[1] in the 1960s and they are now known to be present in a variety of tissues, including adipose tissue, umbilical cord blood and muscle. MSCs can be readily obtained from humans and animals, isolated by means of their ability to adhere to plastic culture plates and can be expanded for many generations in culture.
The culture-expanded MSCs appear to have multilineage differentiation potential, including at least adipogenic, chondrogenic and osteogenic differentiation under appropriate induction conditions[2-4]. Recently, there have been reports regarding additional potential areas for lineage differentiation, such as neurogenic, cardiogenic, myogenic and hepatogenic differentiation[5-10]. Therefore, these cells are now also called multipotent stem/stromal cells since they are capable of differentiating into mesodermal, ectodermal and/or endodermal origin cells. In addition, more experimental data have become available about the immune modulation and hypoimmunogenic property of MSCs[11-16] and there has been a surge of interest in both the basic biology and the potential clinical applications of these cells.
The conventional isolation method of MSCs relies on the fractionation of mononuclear cells by gradient centrifugation and selection of fibroblast-like cells adhering to the culture plate surface by removing nonadherent floating cells. Experimental and clinical data over the last decade have indicated that cultured adherent MSCs are heterogeneous and exhibit variations in terms of their differentiation potentials and clinical outcomes[17,18]. In order to obtain more homogeneous MSC populations, fluorescence-activated cell sorting (FACS) isolation[19] and specific cell-surface antibody selection[20,21] have been applied, improving the purity of the final MSC population. However, although significant improvements have been made in purification techniques, these isolation methods have not yet been sufficient to isolate completely homogeneous populations of MSCs. The limiting dilution method has been the only option to generate single-cell-derived clonal MSC lines. Since a mononuclear cell fraction produced by gradient centrifugation or a culture-expanded population of MSCs was the usual source for the limiting dilution method, there is still a possibility of losing some “real” MSCs in the isolation of mononuclear cells and culture expansion of the cells.
Recently, we developed a new protocol, called the subfractionation culturing method (SCM), to generate single-cell-derived clonal MSC lines from whole bone marrow aspirate without any centrifugation step for mononuclear cells or enzyme treatment process. Using this method, we identified human bone marrow-derived, colony-forming fibroblastic cells as MSCs from relatively small amounts of bone marrow aspirates. This method allowed us to rapidly establish single-cell-derived human clonal MSC (hcMSC) lines from raw bone marrow aspirates and to establish a library of these hcMSC lines[22]. The rationale behind the SCM is that different types of mesenchymal stem or stromal cells can be isolated based on their cell densities and/or adherence to a plastic culture plate produced by a series of transfers of the culture supernatant to subsequent dishes; thus, these low-density stem or stromal cells may have a higher proliferation and differentiation potential.
In the present work, we established single-cell-derived mouse clonal MSC (mcMSC) lines from mouse bone marrow samples from 3 different mouse strains and investigated their cell growth rate, cell surface epitopes, differentiation potentials, lineage-specific gene expression and T-cell suppression capability. The newly established mcMSC lines showed characteristics of progenitors and varied in terms of cell growth rate, cell surface epitopes, differentiation potential, lineage-specific gene expression and T-cell suppression ability, suggesting that variations of progenitor cells exist in mouse bone marrow. These kinds of highly homogeneous, single-cell-derived cMSCs will be very useful for both basic research and clinical applications of such cells.
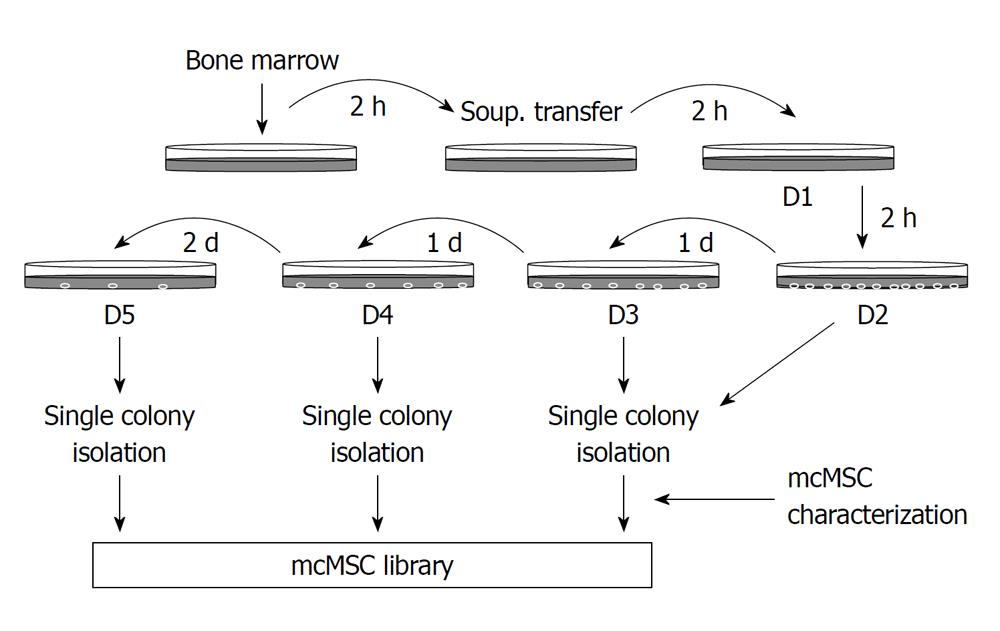
Mouse bone marrow samples were harvested from tibiae and femurs of 5-wk-old BALB/c, C3H and C57/BL6 mice by flushing out the bone marrow cavity with 5-mL culture medium that comprised Dulbecco’s modified Eagle’s Medium (DMEM)-low glucose (GIBCO-BRL, Life Technologies, Gaithersburg, MD), 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Then 15-mL of the complete growth medium was added to the mixture. This mixture was then incubated in a 100-mm culture dish. As shown in Figure 1, after incubation for 2 h at 37°C with 50 mL/L CO2, the supernatant only was transferred to a new, intermediate 100-mm dish. After the second 2-h incubation, the supernatant was transferred from the intermediate dish to a new dish (D1) and incubated for another 2 h. The supernatant was then transferred from D1 to a new dish (D2), incubated for 1 d, then transferred from D2 to a new dish (D3) and incubated for 1 d. This process was repeated 2 more times with 1- and 2-d incubation sequentially (for D4 and D5, respectively). After incubation for 7-14 d, the well-separated single-cell-derived colonies that appeared in the culture dishes were detached and isolated after a 2-3 min treatment with trypsin/ethylene diamine tetraacetic acid (EDTA) in cloning cylinders (GIBCO-BRL), transferred to a six-well plate, and then to larger culture flasks where they continued to expand. Once the cells reached approximately 80%-90% confluence, they were recovered with trypsin/EDTA and passaged at 50-100 cells/cm2. We obtained ten to twenty colonies on D1 to D3 dishes using bone marrow samples out of five mice and then were able to establish a total of seven, four and six mcMSC lines from each C3H, C57/BL6, Balb/c strain respectively. Out of these mcMSC lines, two lines from each strain were selected for this study.
For analysis of proliferation ability, the established mcMSC lines and nonclonal MSCs were seeded at a density of 1 × 103, 5 × 103, or 1 × 104 cells/60-mm dish and cultured in complete growth medium for 3, 7, 10 and 14 d. The culture medium was replaced every 3 d. The cell numbers were counted at each time point with a hemocytometer.
For phenotyping of cell surface antigens, the established mcMSC lines at 5-7 passages were harvested from the 175-cm2 flasks by treatment with trypsin/EDTA and washed twice with phosphate-buffered saline (PBS). The cells were incubated with fluorescein isothiocyanate (FITC)-conjugated antibodies for 30 min at 4°C. The cells were washed twice with PBS consisting of 0.1% bovine serum albumin (BSA). The following antibodies were used for flow cytometry: CD34 (Serotec, Oxford, UK), CD44 (Serotec), CD45 (BD Biosciences Pharmingen, San Diego, CA, USA), CD103 (BD Biosciences Pharmingen), CD105 (R&D Systems, Minneapolis, MN, USA), CD117 (BD Biosciences Pharmingen) and Sca-1 (BD Biosciences Pharmingen). The cells were washed twice with PBS consisting of 0.1% BSA and analyzed using flow cytometry with a 525-nm filter for green FITC fluorescence.
To compare the differentiation potential, the established mcMSC lines and nonclonal MSCs were induced to differentiate into adipogenic, osteogenic or chondrogenic lineage by using the following method.
For chondrogenic differentiation, a pellet culture system was used. Approximately 2 × 105 mcMSCs or nonclonal MSCs were placed in a 15-mL polypropylene tube (Falcon, Bedford, MA, USA) and centrifuged to a pellet. The pellet was cultured at 37°C with 50 mL/L CO2 in 500 μL chondrogenic differentiation medium composed of α-MEM supplemented with 10 ng/mL TGF-β1 (R&D Systems), 10 ng/mL TGF-β3 (R&D Systems), 50 μg/mL ascorbic acid (Sigma Chemical Co., St Louis, MO, USA), 10-7 mol/L dexamethasone (Sigma Chemical Co.), 40 μg/mL proline (Sigma Chemical Co.) and 1% ITS+ Premix (Becton Dickinson, Bedford, MA, USA; 6.25 μg/mL insulin, 6.25 μg/mL transferrin, 6.25 ng/mL selenious acid, 1.25 mg/mL BSA and 5.25 mg/mL linoleic acid). The chondrogenic differentiation medium was changed every 3 d for 3 wk. For microscopy, the pellets were embedded with an OCT compound (Sakura Finetek, Torrance, CA, USA), frozen sectioned into 8-μm sections and stained with toluidine blue or safranin O.
For investigating osteogenic and adipogenic differentiation, a monolayer culture system was used. For osteogenic differentiation, mcMSCs or nonclonal MSCs were plated at 50 cells/cm2 in a 12-well plate and incubated until they reached confluence. The cultures were then incubated in osteogenic differentiation medium composed of α-MEM supplemented with 10% FBS, 50 μg/mL ascorbic acid (Sigma Chemical Co.), 10-8 mol/L dexamethasone (Sigma Chemical Co.), 10 mmol/L β-glycerophosphate (Sigma Chemical Co.), and 1 mmol/L dibutyryl-cyclic AMP (db-cAMP). The db-cAMP was added only for the first 4 d. The medium was changed every 3 d for 3 wk. The cells were fixed with 10% formalin (Sigma Chemical Co.) for 10 min at room temperature and stained with 0.1% alizarin red S (Sigma Chemical Co.).
To evaluate adipogenic differentiation, mcMSCs or nonclonal MSCs were plated at 50 cells/cm2 in a 12-well plate and incubated in α-MEM (20% FBS) until they reached confluence. The cultures were then incubated in adipogenic differentiation medium composed of DMEM-high glucose supplemented with 10% newborn calf serum, 10-7 mol/L dexamethasone (Sigma Chemical Co.), 10 μg/mL insulin (Sigma Chemical Co.), 0.5 μmol/L 1-methyl-3-isobutylxanthine (IBMX; Sigma Chemical Co.) and 50 μg/mL indomethacine (Sigma Chemical Co.). The culture was incubated at 37°C with 50 mL/L CO2 for 4 d. The cells were fixed with 10% formalin (Sigma Chemical Co.) for 10 min and stained with oil-red-O for 10 min at room temperature.
The total RNA was extracted from mcMSC lines grown either in complete growth medium or differentiation medium using easyBLUE® (Intron Biotechnology, Sungnam, Korea) reagent. The complementary DNA was synthesized using an AccuPower Reverse-Transcription kit (Bioneer, Daejeon, Korea) by following the manufacturer’s instructions. Polymerase chain reaction (PCR) was carried out using specific primers designed for each gene as follows: type I collagen α1 chain [ColA1, 587 base pair (bp)], forward primer 5′-ACGTCCTGGTGAAGTTGGTC-3′ and reverse primer 5′-AGCCACGATGACCCTTTATG-3′; type II collagen α1 chain (Col2A1, 418 bp), forward primer 5′-CTACACTCAAGTCACTGAACAACCAGAT-3′ and reverse primer 5′-CTGTTCTTACAGTGGTAGGTGATGTTCT-3′; type X collagen α1 chain (Col10A1, 382 bp), forward primer 5′-GTCCAAGAGGTGAACCTGGA-3′ and reverse primer 5′-GCACCTACTGCTGGGTAAGC-3′; aggrecan (Acan, 564 bp), forward primer 5′-GCGACATCTGGAGTGACTGA-3′ and reverse primer 5′-AGACACAGTGGGGAAACCTG-3′; lipoprotein lipase (LPL, 421 bp), forward primer 5′-GCGTAGCAGGAAGTC TGACC-3′ and reverse primer 5′-CTACAACTCAGGCAGAGCCC-3′; fatty acid-binding protein 4 (FABP4, 319 bp), forward primer 5′-AAAGAAGTGGGAGTGGGCTT-3′ and reverse primer 5′-CTCTTGTGGAAGTCACGCCT-3′; peroxisome proliferator-activated receptor-γ2 (PPAR-γ2, 377 bp), forward primer 5′-CTTATTTATGATAGGTGTGATCTTAACTGC-3′ and reverse primer 5′-GTGATATGTTTGAACTTGATTTTATCTTCT-3′; alkaline phosphatase (ALP, 555 bp), forward primer 5′-AACCCAGACACAAGCATTCC-3′ and reverse primer 5′-ACTGGGCCTGGTAGTTGTTG-3′; Distal-less homeobox 5 (Dlx5, 350 bp), forward primer 5′-
CAGAAGAGTCCCAAGCATCC-3′ and reverse primer 5′-CCACTTCTTTCTCTGGCTGG-3′; Runx2 (469 bp), forward primer 5′-TGCGTATTCCTGTAGATCCG-3′ and reverse primer 5′-TAGTTCTCATCATTCCC GGC-3′; and glyceraldehyde phosphate dehydrogenase (GAPDH; 350 bp), forward primer 5′-AACTCCCTCA AGATTGTCAGCA-3′ and reverse primer 5′-TCCACCACCCTGTTGCTTGTA-3′. PCR was performed for 30-40 cycles with each cycle involving denaturation at 94°C for 30 s, annealing at 58-60°C for 30 s and extension at 72°C for 1 min. The amplified PCR products were run on a 1% agarose gel.
Mouse splenocytes were cultured in RPMI supplemented with 10% FBS, 10 mmol/L HEPES, 2 mmol/L L-glutamine, 1 mmol/L sodium pyruvate, 50 μmol/L β-mercaptoethanol, 100 g/mL penicillin and 100 g/mL streptomycin. Splenocytes were isolated from BALB/c spleen. 2 × 105 cells were stimulated with 1 μg/mL of anti-CD3 and anti-CD28 antibodies (BD Pharmingen, Franklin Lakes, NJ, USA) together with 1 × 104 of indicated mcMSCs for 72 h. Proliferation of T cells was determined by incorporation of [3H]-thymidine (1 μCi/well) for the last 12-16 h of culture. After 48-h stimulation, interferon-γ (IFN-γ) was measured by ELISA according to the protocol of BD Pharmingen.
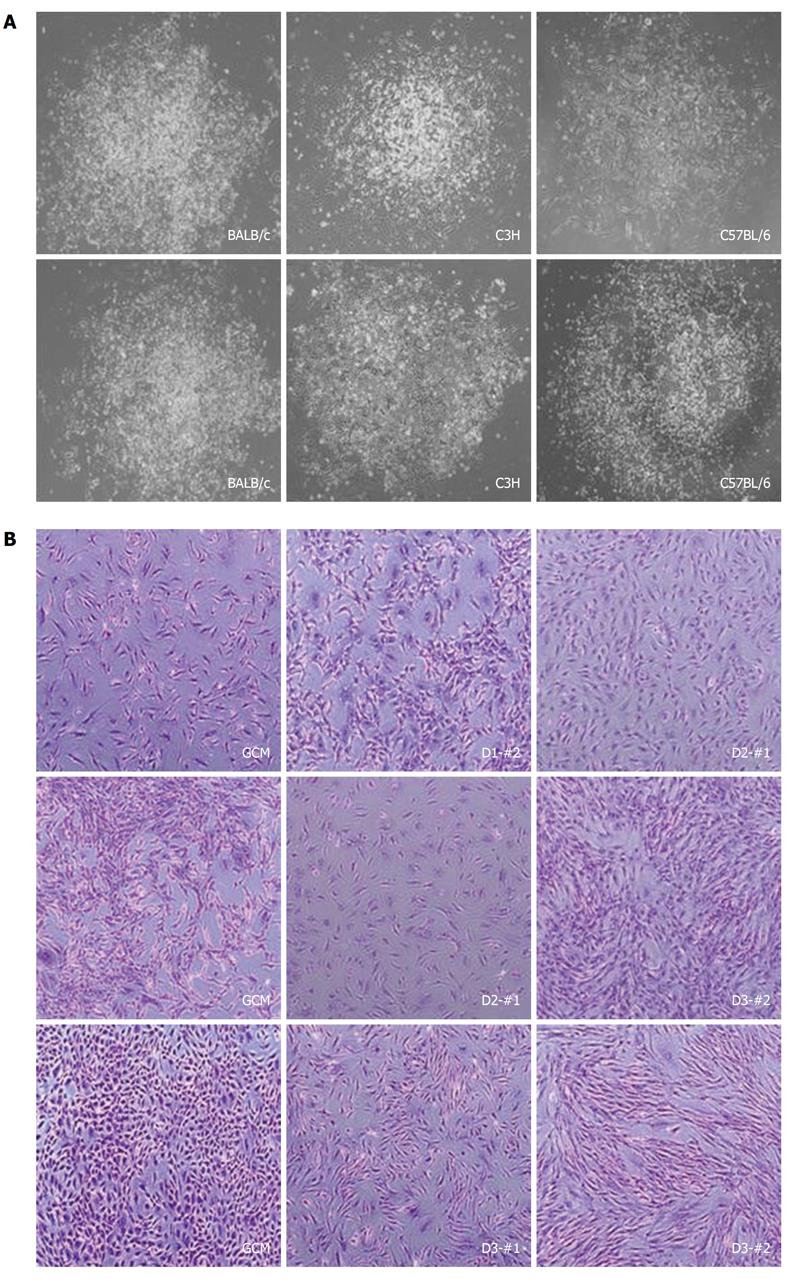
In order to establish mcMSC lines with mouse bone marrow samples from BALB/c, C3H and C57/BL6 strains, as described in the Materials and Methods, approximately 15-mL of each bone marrow mixture with the complete culture medium mixture were plated in a 100-mm culture dish. Subsequently, the supernatant only was transferred to new culture dishes as described in Figure 1. Representative images of two single-cell-derived colonies from each strain and pictures of mcMSC lines and nonclonal MSCs are shown in Figure 2. The nonclonal MSCs isolated by the gradient centrifugation method (GCM) from the strains were used as control cells. These mcMSC lines established by the SCM are spindle-shaped fibroblast-like cells, which are similar to those of nonclonal MSCs.
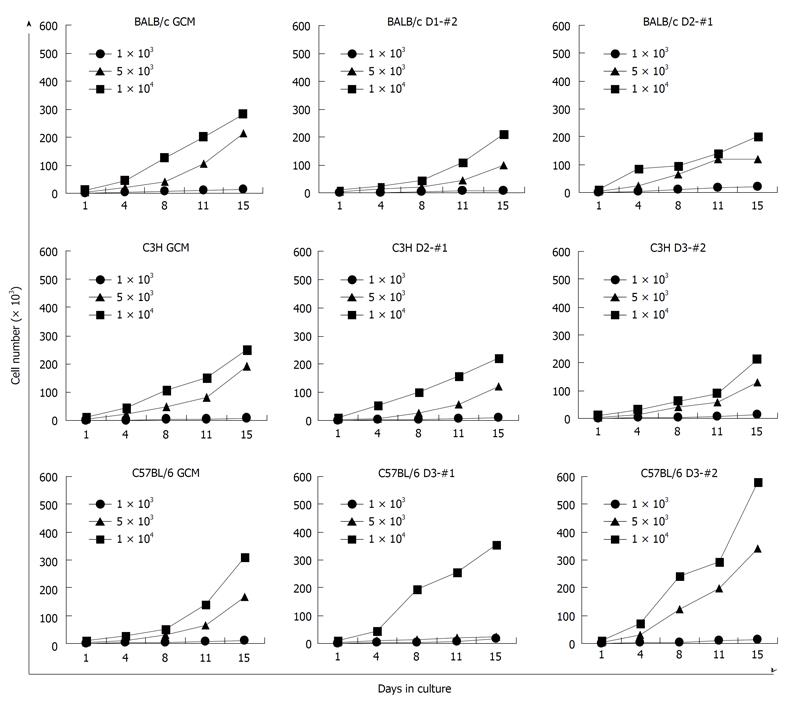
The cell growth rates of the established mcMSC lines and nonclonal MSCs were measured with 3 different starting cell densities, 1 × 103, 5 × 103, and 1 × 104 per 60-mm plate. As shown in Figure 3, all the lines except C57/BL6 D3-#1 showed exponential growth with starting populations of 5 × 103 and 1 × 104 cells. There was no significant difference between the GCM- or SCM-isolated cells in the proliferation rate for the 16 d of culture. No exponential growth was observed with the starting population of 1 × 103 cells, suggesting that a certain initial number of cells are required for attaining proper growth phase of these cells.
The cell surface epitopes of the mcMSC lines and nonclonal MSCs were compared by FACS analysis of a panel of eight cell surface proteins (Table 1). The results showed that all the mcMSC lines and nonclonal MSCs were strongly positive for Sca-1 and negative for CD34, HLA-DR, CD45, CD103 and CD117, as expected. One intriguing finding was that the mcMSC lines showed variations in their expressions of CD44 and CD105, as shown in Table 1. For example, BALB/c D1-#2, C3H D3-#2, C57/BL6 D3-#1 and C57/BL6 D3-#2 showed modest expression of CD44, whereas BALB/s GCM, C3H GCM and C3H D2-#1 were negative. In addition, BALB/c D2-#1 and C57/BL6 D3-#2 were strongly positive and BALB/c GCM, C3H D2-#1 and C3H D3-#2 were modestly positive for the expression of CD105, whereas C3H GCM, C57/BL6 GCM and C57/BL6 D3-#1 were negative. One observation we noticed was that one of the two mcMSC lines, C3H D3-#2, showed both CD44 and CD105 positive exhibited a high level of osteogenic differentiation potential. Other mcMSC lines and nonclonal MSCs showed either CD44 or CD105 positive.
| Sca-1 | CD44 | CD105 | CD34 | HLA-DR | CD45 | CD103 | CD117 | |
| BALB/c GCM | +++ | - | + | - | - | - | - | - |
| BALB/c D1-#2 | +++ | + | - | - | - | - | - | - |
| BALB/c D2-#1 | +++ | - | ++ | - | - | - | - | - |
| C3H GCM | +++ | - | - | - | - | - | - | - |
| C3H D2-#1 | +++ | - | + | - | - | - | - | - |
| C3H D3-#2 | +++ | + | + | - | - | - | - | - |
| C57BL/6 GCM | +++ | + | - | - | - | - | - | - |
| C57BL/6 D3-#1 | +++ | ++ | - | - | - | - | - | - |
| C57BL/6 D3-#2 | +++ | + | ++ | - | - | - | - | - |
These data demonstrate that the mcMSC lines established by the SCM exhibit variations in the expression of certain cell surface epitopes compared to those of nonclonal MSCs. The significance of these variations remains to be studied.
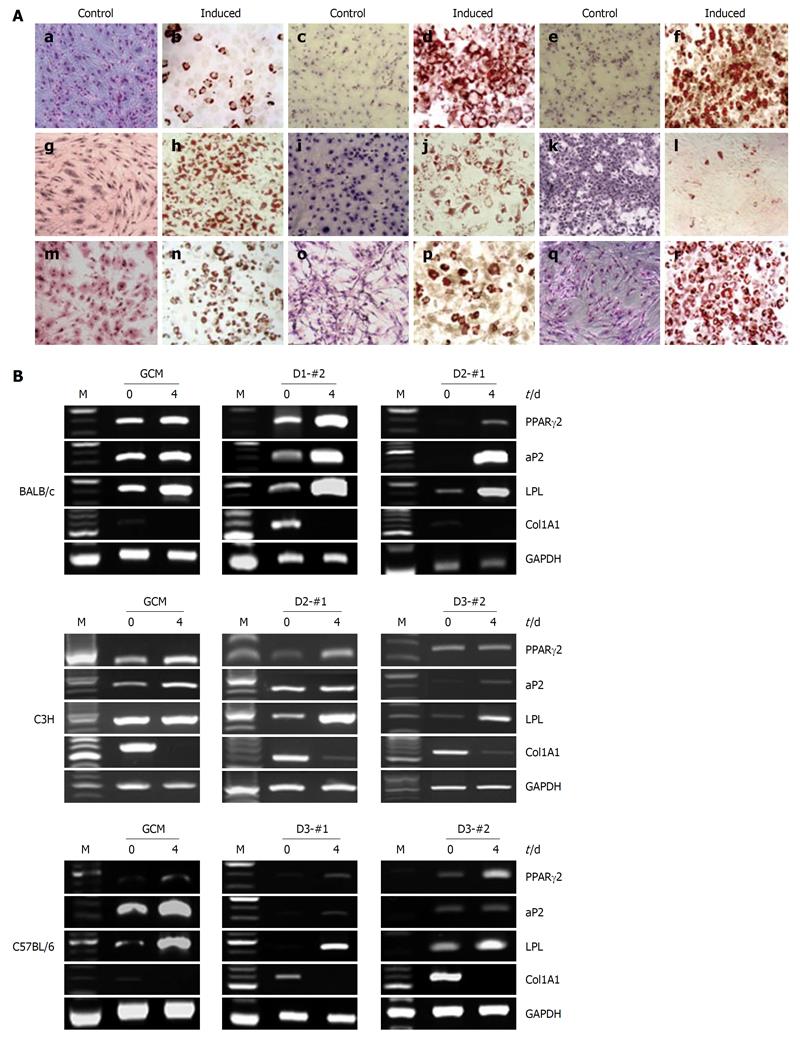
In order to examine the differentiation potential of the established mcMSC lines and nonclonal MSCs, in vitro adipogenic differentiation analysis was performed using a monolayer culture system. Passage 6 mcMSCs from each of the cell lines were induced to differentiate into adipocytes under the conditions described in Materials and Methods. As shown in Figure 4A, every mcMSC line showed a high level of adipogenic differentiation potential except C3H D3-#2. In contrast, the monolayer cells of both nonclonal and mcMSCs grown in the normal control medium showed no oil-red-O staining.
To further confirm the adipogenic differentiation potentials of the established mcMSC lines and nonclonal MSCs, reverse transcriptase-PCR (RT-PCR) assays using total mRNA from the cells were performed before and after induction of differentiation. The lineage-specific expression of adipogenic marker genes, PPAR-γ2, FABP4, LPL and Col1A1 was measured at 0 and 4 d after induction of differentiation. As shown in Figure 4B, the expression of PPAR-γ2, FABP4 and LPL was upregulated in most of the cells, whereas the expression of Col1A1 was downregulated as expected. These results suggest that both mcMSCs and nonclonal MSCs have a high potential for adipogenic differentiation.
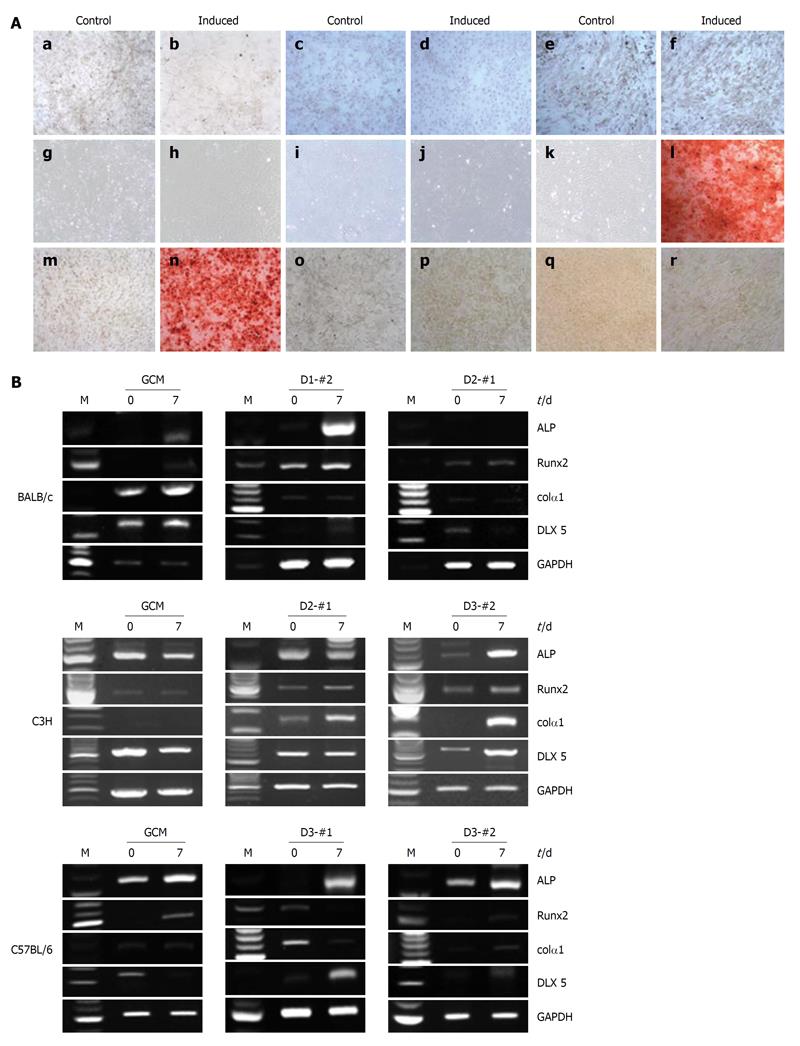
To examine the osteogenic differentiation potential of the established mcMSC lines and nonclonal MSCs, in vitro differentiation analyses were performed using a monolayer culture system for osteogenic differentiation. Passage 6 mcMSCs from each cell line were induced to differentiate into osteoblasts under the conditions described in Materials and Methods. As shown in Figure 5A, among the twelve tested lines, only one nonclonal MSC line, C57/BL6 GCM, and one mcMSC line, C3H D3-#2, exhibited a high level of osteogenic differentiation potential, whereas other nonclonal and mcMSCs showed no differentiation potential. The monolayer cells of both nonclonal and mcMSCs grown in the normal control medium were negative for Alizarin Red S.
To further confirm the osteogenic differentiation capabilities of the mcMSC lines and nonclonal MSCs, RT-PCR assays were performed using total mRNA from cells before and after induction of differentiation. The lineage-specific expression of osteogenic marker genes, ALP, Runx2, Colα1 and Dlx5 were measured at 0 and 7 d after differentiation induction. As seen in Figure 5B, the expression of ALP, Runx2, Colα1 and Dlx5 was upregulated in C3H D3-#2 and C57/BL6 GCM cells.
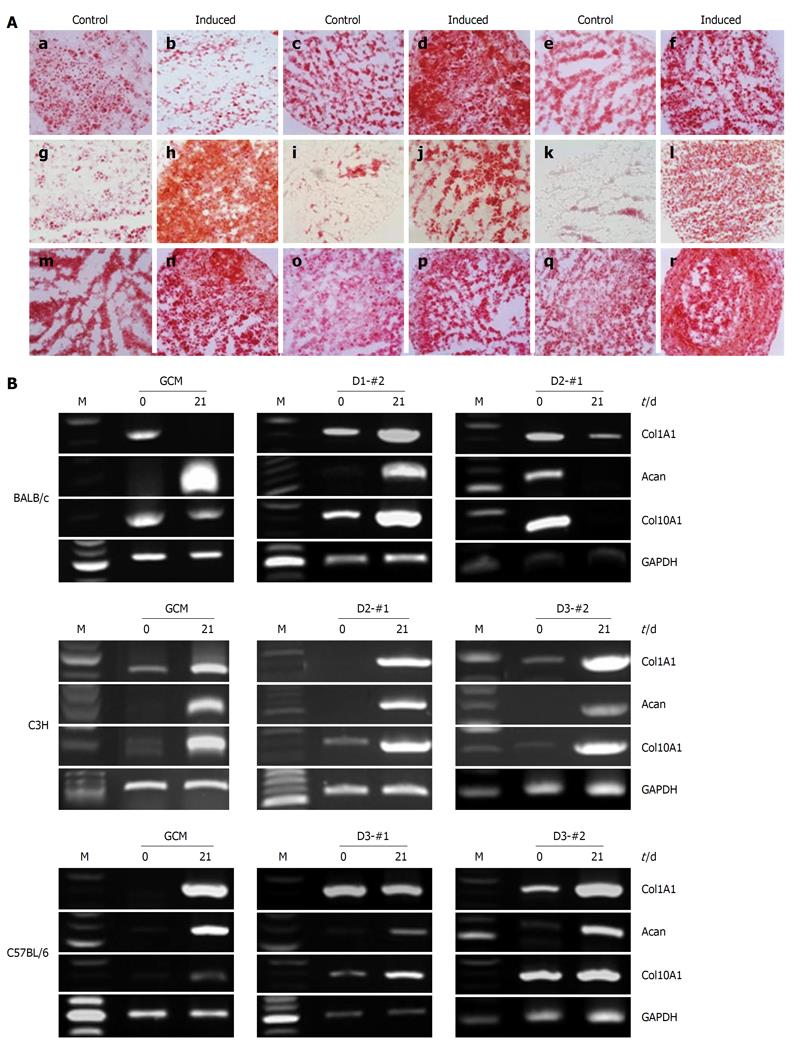
To examine the chondrogenic differentiation potential of the established mcMSC lines and nonclonal MSCs, in vitro chondrogenic differentiation analysis was performed using a pellet culture system. Passage 6 mcMSCs from each cell line were induced to differentiate into chondrocytes under the conditions described in Materials and Methods. As shown in Figure 6A, every mcMSC line showed a high level of chondrogenic differentiation potential except BALB/c GCM and D2-#1. In contrast, the control monolayer cells grown in the normal medium exhibited only slight safranin O staining.
To further confirm the chondrogenic differentiation capabilities of the mcMSC lines and nonclonal MSCs, RT-PCR assays were performed using total mRNA from cells before and after induction of differentiation. The lineage-specific expression of chondrogenic marker genes, Col1A1, Acan and Col10A1 were measured at 0 and 21 d after differentiation induction. As seen in Figure 6B, the expressions of Col1A1, Acan and Col10A1 were observed to be upregulated.
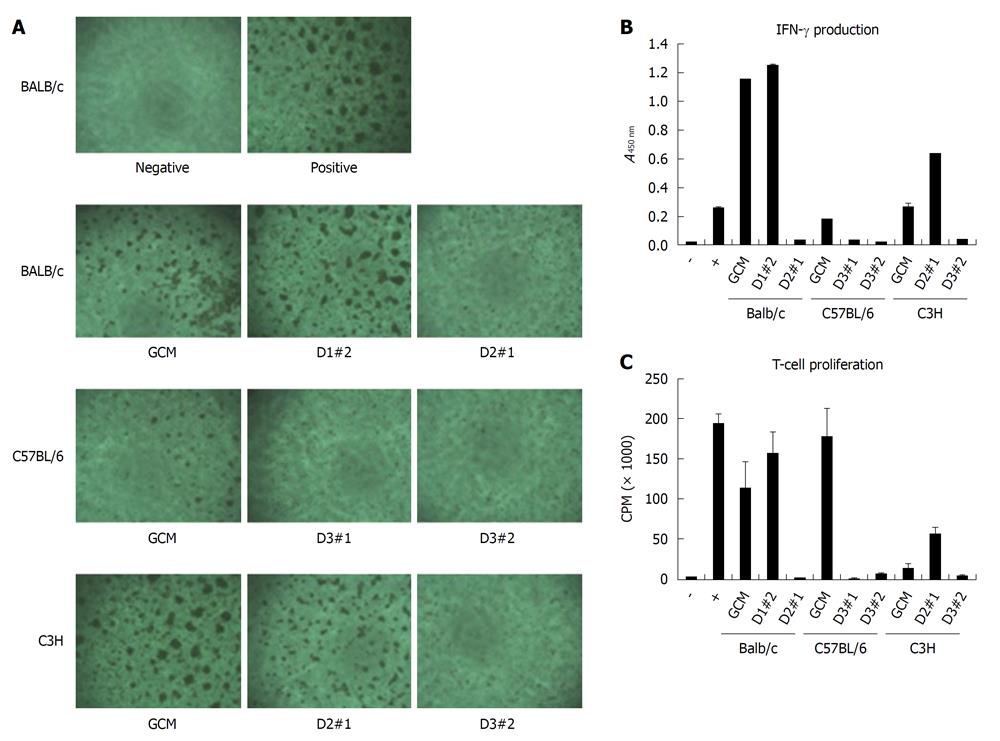
To test the immunosuppressive potential of mcMSC lines and nonclonal MSCs, mixed lymphocyte reaction analysis was performed with the cells. The formation of T-cell receptor (TCR)-activated T-cell clusters, IFN-γ production suppression and T-cell proliferation inhibition were measured with antibody-activated splenocytes in the presence of each mcMSC line or nonclonal MSCs. As shown in Figure 7A, the number of TCR activated T-cell clusters were reduced in BALB/c D2-#1, C57/BL6 GCM, D3-#1, D3-#2 and C3H D3-#2 cells, whereas other MSCs showed less or no reduction. The results of the IFN-γ production suppression and T-cell proliferation inhibition experiments were highly comparable to the T-cell cluster results except for C57/BL6 GCM cells. These data suggest that the T-cell suppression potential may vary from mcMSC line to mcMSC line.
In this paper, single-cell-derived mcMSC lines were established using the SCM and characterized for their proliferation, cell surface epitopes, differentiation potential, lineage-specific gene expression and T-cell suppression capability. The results suggest that mcMSC lines are readily established using the SCM protocol and that some of the newly established mcMSCs have stem/progenitor cell characteristics with variations in their cell surface epitopes, differentiation potential, lineage-specific gene expression and T-cell suppression capability.
In spite of recent high levels of experimental and clinical interest, a standard protocol for producing homogeneous populations of MSCs has not yet been available. The homogeneity of MSCs has become an important issue in terms of understanding the molecular characterization of MSCs and also for applying them in clinical settings. For example, several studies have investigated the gene expression profiles of MSCs to understand the molecular properties of MSCs such as stemness, self-renewal and differentiation potential. Although very informative, most of these studies have limitations due to the heterogeneity of MSCs used in the experiments. In addition, heterogeneous populations of MSCs might possibly generate various efficacies in the treatment of disease. A mixed population of mononuclear cells has been used in most preclinical and clinical studies so far[18]. The benefit of using a mixed population of MSCs vs clonally expanded MSCs from bone marrow for a clinical trial has yet to be adequately investigated. In order to obtain more homogeneous populations of MSCs, antibody selection of specific surface antigen- expressing MSCs[20,21], FACS sorting of specific sizes of MSCs[19] and clonal selection by limiting dilution have been utilized. Recently, we developed the SCM and reported how to establish human bone marrow-derived cMSC lines. This method allows a rapid establishment of single-cell-derived clonal MSC lines with a small amount of bone marrow aspirate and could be a very useful protocol with which to obtain highly homogeneous population of MSCs[22]. The establishment of mcMSC lines from three different mouse strains, as shown in the present study, and our previous success with human bone marrow strongly suggests that the SCM is universally applicable in establishing clonal stem cell lines from relatively small amounts of bone marrow samples regardless of species. We have also succeeded in establishing cMSC lines with rat and rabbit bone marrow aspirates (unpublished data). Therefore, the advantages of the SCM are as follows: Firstly, this method does not require any centrifugation step in the whole procedure. Secondly, it does not require any enzymatic process or filtering procedure. Thirdly, it can generate single-cell-derived homogeneous clonal MSC lines. Fourthly, the protocol works well for bone marrow samples from different species, including human, mouse, rat and rabbit. These advantages suggest that the SCM can generate a homogeneous population of MSCs in a simple, effective and economic way and may allow safer applications in therapeutic settings.
One difference we have observed while establishing and characterizing mouse and human cMSC lines was the number of lines with osteogenic differentiation potential. The frequency of obtaining cMSC lines with osteogenic differentiation potential was approximately five times lower with mouse bone marrow samples than that with human bone marrow. This difference may be just a simple species difference or might be caused by the source of mouse and human bone marrow; the mouse bone marrow was from long bones whereas the human bone marrow was from the iliac crest. Enzymatic treatment of bone marrow, such as with collagenase, might improve the number of mouse lines with osteogenic differentiation potential. Although there was no correlation between the cell surface marker expression and differentiation potential, one observation we noticed was that one of the two mcMSC lines, C3H D3-#2, showed both CD44 and CD105 positive exhibited a high level of osteogenic differentiation potential. Other mcMSC lines and nonclonal MSCs showed either CD44 or CD105 positive.
The rapid establishment of mcMSC lines and variations of the lines in phenotype, differentiation potential, gene expression and T-cell suppression capacity encouraged us to establish a mcMSC library of a variety of different mcMSC lines. This kind of library will be very useful for both the basic study of bone-marrow-derived stem cells and for clinical study of specific target disease treatment. For example, a cMSC line exhibiting great capability for inhibition of T-cell proliferation would be an excellent candidate stem cell for the development of a stem cell product to treat graft-versus-host disease. One potential concern of using clonal MSC lines is their tumorigenicity potential. Thus, we have tested the transformation potential of seven hcMSC lines by in vitro-transformation assay with human HeLa tumor cell line as a positive control and mouse NIH3T3 cell line as a negative control. The results showed that our hcMSC lines produces a smaller number of colonies than do NIH3T3 cells (data not shown), suggesting that the hcMSC lines are probably not tumorigenic.
In conclusion, this study reports a protocol that allows the rapid establishment of mcMSC lines with a small amount of mouse bone marrow. Some of the newly established mcMSC lines exhibit stem/progenitor cell characteristics with some variations in their cell surface epitopes, differentiation potential, lineage-specific gene expression and T cell suppression capability, indicating the presence of different types of stem/progenitor cells in the bone marrow. This protocol will allow easy construction of a mcMSC library containing a variety of different types of bone marrow mcMSC lines. These types of different homogeneous populations of cMSC lines will likely have useful applications in both basic research and clinical applications.
The conventional isolation method of mesenchymal stem cells (MSCs) relies on the fractionation of mononuclear cells by gradient centrifugation and selection of fibroblast-like cells adhering to the culture plate surface by removing nonadherent floating cells. Experimental and clinical data over the last decade have indicated that cultured adherent MSCs are heterogeneous and exhibit variations in terms of their differentiation potentials and clinical outcomes. In order to obtain more homogeneous MSC populations, fluorescence-activated cell sorting (FACS) isolation and specific cell-surface antibody selection have been applied, improving the purity of the final MSC population. However, although significant improvements have been made in purification techniques, these isolation methods have not yet been sufficient to isolate completely homogeneous populations of MSCs.
In order to obtain more homogeneous MSC populations, FACS isolation and specific cell-surface antibody selection have been applied, improving the purity of the final MSC population. However, although significant improvements have been made in purification techniques, these isolation methods have not yet been sufficient to isolate completely homogeneous populations of MSCs.
The advantages of the subfractionation culturing method (SCM) are as follows: Firstly, this method does not require any centrifugation step in the whole procedure. Secondly, it does not require any enzymatic process or filtering procedure. Thirdly, it can generate single-cell-derived homogeneous clonal MSC lines. Fourthly, the protocol works well for bone marrow samples from different species, including human, mouse, rat and rabbit. These advantages suggest that the SCM can generate a homogeneous population of MSCs in a simple, effective and economic way and may allow safer applications in therapeutic settings.
The results in this study suggest that SCM allows the rapid establishment of mouse clonal MSC (mcMSC) lines with a small amount of mouse bone marrow. This protocol will allow easy construction of a mcMSC library containing a variety of different types of bone marrow mcMSC lines. These types of different homogeneous populations of cMSC lines will likely have useful applications in both basic research and clinical applications.
MSC: These stem cells were first isolated from the bone marrow in the 1960s and they are now known to be present in a variety of tissues, including adipose tissue, umbilical cord blood and muscle. They can be differentiated into adipocytes, chondrocytes, osteoblasts, neurons, cardiocytes and hepatocytes. SCM: A newly developed method to isolate clonal MSCs which are derived from a single stem cell so that these cells can be highly homogeneous.
The authors described a simplified protocol to obtain clonal mouse MSCs. The paper is of interest since a lot of data have been collected on the set up of a proper method to isolate MSCs. The paper is well written and the study well conducted.
Peer reviewer: Umberto Galderisi, PhD, Associate Professor, Department of Experimental Medicine, Second University of Naples, Via L. De Crecchio 7, 80138 Napoli, Italy
S- Editor Wang JL L- Editor Roemmele A E- Editor Zheng XM
| 1. | Friedenstein AJ, Petrakova KV, Kurolesova AI, Frolova GP. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation. 1968;6:230-247. [PubMed] |
| 2. | Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143-147. [PubMed] |
| 3. | Phinney DG, Kopen G, Isaacson RL, Prockop DJ. Plastic adherent stromal cells from the bone marrow of commonly used strains of inbred mice: variations in yield, growth, and differentiation. J Cell Biochem. 1999;72:570-585. [PubMed] |
| 4. | Schwarz EJ, Alexander GM, Prockop DJ, Azizi SA. Multipotential marrow stromal cells transduced to produce L-DOPA: engraftment in a rat model of Parkinson disease. Hum Gene Ther. 1999;10:2539-2549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 115] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Woodbury D, Schwarz EJ, Prockop DJ, Black IB. Adult rat and human bone marrow stromal cells differentiate into neurons. J Neurosci Res. 2000;61:364-370. [PubMed] |
| 6. | Wakitani S, Saito T, Caplan AI. Myogenic cells derived from rat bone marrow mesenchymal stem cells exposed to 5-azacytidine. Muscle Nerve. 1995;18:1417-1426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 816] [Cited by in RCA: 752] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 7. | Prockop DJ, Azizi SA, Colter D, Digirolamo C, Kopen G, Phinney DG. Potential use of stem cells from bone marrow to repair the extracellular matrix and the central nervous system. Biochem Soc Trans. 2000;28:341-345. [PubMed] |
| 8. | Toma C, Pittenger MF, Cahill KS, Byrne BJ, Kessler PD. Human mesenchymal stem cells differentiate to a cardiomyocyte phenotype in the adult murine heart. Circulation. 2002;105:93-98. [PubMed] |
| 9. | Liechty KW, MacKenzie TC, Shaaban AF, Radu A, Moseley AM, Deans R, Marshak DR, Flake AW. Human mesenchymal stem cells engraft and demonstrate site-specific differentiation after in utero transplantation in sheep. Nat Med. 2000;6:1282-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 929] [Cited by in RCA: 848] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 10. | Petersen BE, Bowen WC, Patrene KD, Mars WM, Sullivan AK, Murase N, Boggs SS, Greenberger JS, Goff JP. Bone marrow as a potential source of hepatic oval cells. Science. 1999;284:1168-1170. [PubMed] |
| 11. | Di Nicola M, Carlo-Stella C, Magni M, Milanesi M, Longoni PD, Matteucci P, Grisanti S, Gianni AM. Human bone marrow stromal cells suppress T-lymphocyte proliferation induced by cellular or nonspecific mitogenic stimuli. Blood. 2002;99:3838-3843. [PubMed] |
| 12. | Le Blanc K, Tammik C, Rosendahl K, Zetterberg E, Ringdén O. HLA expression and immunologic properties of differentiated and undifferentiated mesenchymal stem cells. Exp Hematol. 2003;31:890-896. [PubMed] |
| 13. | Götherström C, Ringdén O, Westgren M, Tammik C, Le Blanc K. Immunomodulatory effects of human foetal liver-derived mesenchymal stem cells. Bone Marrow Transplant. 2003;32:265-272. [PubMed] |
| 14. | Le Blanc K, Rasmusson I, Götherström C, Seidel C, Sundberg B, Sundin M, Rosendahl K, Tammik C, Ringdén O. Mesenchymal stem cells inhibit the expression of CD25 (interleukin-2 receptor) and CD38 on phytohaemagglutinin-activated lymphocytes. Scand J Immunol. 2004;60:307-315. [PubMed] |
| 15. | Aggarwal S, Pittenger MF. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood. 2005;105:1815-1822. [PubMed] |
| 16. | Krampera M, Pasini A, Pizzolo G, Cosmi L, Romagnani S, Annunziato F. Regenerative and immunomodulatory potential of mesenchymal stem cells. Curr Opin Pharmacol. 2006;6:435-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 122] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 17. | Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8:726-736. [PubMed] |
| 18. | Rosenzweig A. Cardiac cell therapy--mixed results from mixed cells. N Engl J Med. 2006;355:1274-1277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 281] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 19. | Zohar R, Sodek J, McCulloch CA. Characterization of stromal progenitor cells enriched by flow cytometry. Blood. 1997;90:3471-3481. [PubMed] |
| 20. | Joyner CJ, Bennett A, Triffitt JT. Identification and enrichment of human osteoprogenitor cells by using differentiation stage-specific monoclonal antibodies. Bone. 1997;21:1-6. [PubMed] |
| 21. | Waller EK, Olweus J, Lund-Johansen F, Huang S, Nguyen M, Guo GR, Terstappen L. The "common stem cell" hypothesis reevaluated: human fetal bone marrow contains separate populations of hematopoietic and stromal progenitors. Blood. 1995;85:2422-2435. [PubMed] |
| 22. | Song SU, Kim CS, Yoon SP, Kim SK, Lee MH, Kang JS, Choi GS, Moon SH, Choi MS, Cho YK. Variations of clonal marrow stem cell lines established from human bone marrow in surface epitopes, differentiation potential, gene expression, and cytokine secretion. Stem Cells Dev. 2008;17:451-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |