Published online Jan 26, 2022. doi: 10.4252/wjsc.v14.i1.92
Peer-review started: February 26, 2021
First decision: June 16, 2021
Revised: July 7, 2021
Accepted: January 5, 2022
Article in press: January 5, 2022
Published online: January 26, 2022
Processing time: 327 Days and 18.9 Hours
Bone is a complex tissue that undergoes constant remodeling to maintain homeostasis, which requires coordinated multilineage differentiation and proper proliferation of mesenchymal stromal cells (MSCs). Mounting evidence indicates that a disturbance of bone homeostasis can trigger degenerative bone diseases, including osteoporosis and osteoarthritis. In addition to conventional genetic modifications, epigenetic modifications (i.e., DNA methylation, histone modifications, and the expression of noncoding RNAs) are considered to be contributing factors that affect bone homeostasis. Long noncoding RNAs (lncRNAs) were previously regarded as ‘transcriptional noise’ with no biological functions. However, substantial evidence suggests that lncRNAs have roles in the epigenetic regulation of biological processes in MSCs and related diseases. In this review, we summarized the interactions between lncRNAs and epigenetic modifiers associated with osteo-/adipogenic differentiation of MSCs and the pathogenesis of degenerative bone diseases and highlighted promising lncRNA-based diagnostic and therapeutic targets for bone diseases.
Core Tip: In this review, we summarized the roles of long noncoding RNAs (lncRNAs) played in mesenchymal stromal cells (MSCs) differentiation and common degenerative bone diseases through reciprocal interactions between lncRNAs and epigenetic modifiers, focusing on the most common epigenetic mechanisms: DNA methylation and histone modifications. It is our hope that this review may provide an updated summary that sheds light on the lncRNA-based precise regulation of the MSC differentiation process and highlights possible therapeutic targets of degenerative bone diseases.
- Citation: Xia K, Yu LY, Huang XQ, Zhao ZH, Liu J. Epigenetic regulation by long noncoding RNAs in osteo-/adipogenic differentiation of mesenchymal stromal cells and degenerative bone diseases. World J Stem Cells 2022; 14(1): 92-103
- URL: https://www.wjgnet.com/1948-0210/full/v14/i1/92.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v14.i1.92
The skeletal system contains bones, joints, and ligaments that function together as a locomotive organ and provide structural support. Originating from mesenchymal progenitors during embryogenesis, the skeletal system undergoes modeling and remodeling throughout life[1]. Mesenchymal stromal cells (MSCs) refer to a heterogeneous unfractionated population of cells, which include fibroblasts, myofibroblasts, and progenitor cells[2,3]. MSCs are able to differentiate into chondrocytes or osteoblasts to comply with bone formation and regeneration needs[4]. It is worth mentioning that adipocytes, as well as osteoblasts, derive from the same population of MSCs. A shift in the osteoadipogenic differentiation balance may lead to bone diseases, such as osteoporosis, which typically manifests as a shift toward adipogenesis[5,6]. Likewise, osteoarthritis is usually characterized by impairment of cartilage regeneration due to the attenuated chondrogenic capacity of MSCs[7,8]. Therefore, the differentiation of MSCs, which proceeds under the control of various transcription factors, influences the pathogenesis of common bone diseases[9-11].
In addition to conventional genetic and environmental factors, epigenetic modifications can influence the bone phenotype and the development of skeletal diseases[12,13]. Epigenetic mechanisms alter gene expression patterns without changing the DNA sequence by three major mechanisms, including DNA methylation, histone modifications, and altered expression of noncoding RNAs[14]. With the rapid development of next-generation sequencing (NGS) and advanced bioinformatic tools, the crucial roles of epigenetic mechanisms in the differentiation of MSCs and the pathogenesis of bone diseases have begun to be elucidated[15-17].
Long noncoding RNAs (lncRNAs) are defined as a set of noncoding RNAs longer than 200bp that have no protein-coding ability. Evidence is rapidly accumulating on the functions of lncRNAs in epigenetic regulation in the differentiation of MSCs and the occurrence of many diseases[18-21]. In this review, we revisit the epigenetic regulatory mechanisms of lncRNAs involved in DNA methylation and histone modifications and summarize the biological functions of lncRNAs in regulation crucial differentiation- and bone disease-related genes by interacting with key epigenetic modifiers. It is our hope that this review may provide an updated summary that sheds light on the lncRNA-based precise regulation of the MSC differentiation process and highlights possible therapeutic targets of degenerative bone diseases.
DNA methylation functions as a regulator of osteogenesis and adipogenesis of MSCs and is involved in common bone diseases[22-24]. In humans, the majority of DNA methylation occurs at cytosines in cytosine-phospho-guanosine (CpG) dinucleotides[25,26]. Approximately 75% of all gene promoters are within CpG-rich regions, known as CpG islands, that are mostly unmethylated[27]. It is generally accepted that the methylation of these CpG islands is associated with the repression of gene expression[28]. Nevertheless, it is worth mentioning that DNA methylation is also associated with upregulated gene expression under certain circumstances[29].
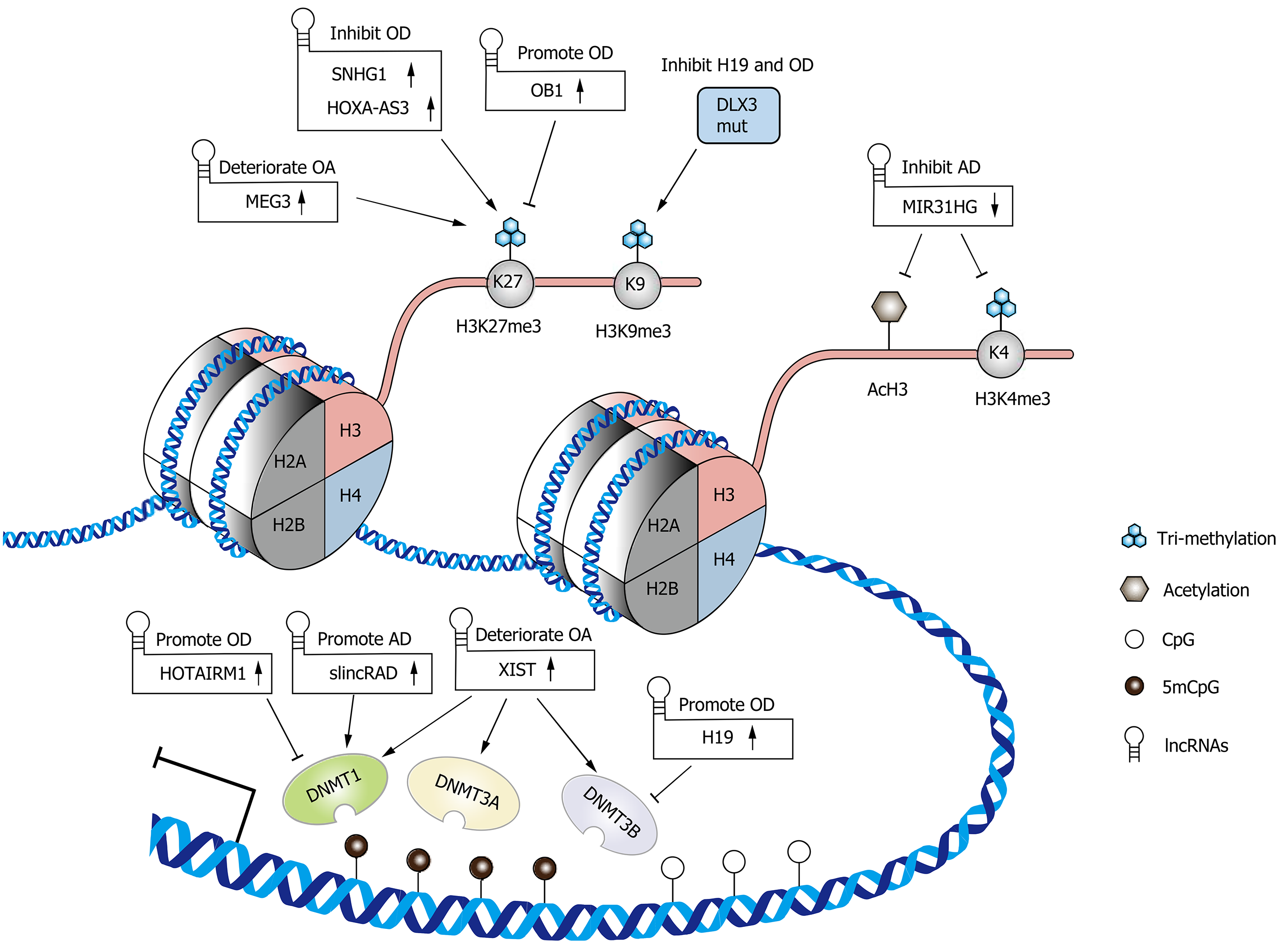
As writer enzymes, DNA methyltransferases (DNMTs) catalyze DNA methylation by transferring a methyl group onto the C5 position of a cytosine at CpG dinucleotide sites to form 5mCpG[30]. A member of the DNMT family, DNMT1, which is also called the maintenance DNMT, maintains the original methylation pattern during DNA replication, while DNMT3a and DNMT3b are involved in de novo methylation[30,31]. The interaction of lncRNAs with DNMTs is varied and reciprocal. For example, lncRNAs can recruit DNMTs to the promoters of target genes and regulate their expression patterns. In turn, the changes in the methylation level of specific lncRNA gene promoters can alter the expression of lncRNAs, including downstream lncRNA-regulated genes[32,33]. In MSCs, lncRNAs, as regulators of DNA methylation, have received increasing attention due to their great importance in the regulation of differentiation and bone-related diseases (Figure 1).
H19, a well-known lncRNA, plays a crucial role in embryo development, cell differentiation, and the occurrence and development of bone diseases[34-37]. In human dental pulp stromal cells (hDPSCs), H19 positively regulates odontogenic differentiation via hypomethylation of distal-less homeobox 3 (DLX3), a key factor in odontogenic differentiation[32]. H19 decreases SAHH and DNMT3B activity, consequently promoting the expression of DLX3[32]. In turn, a mutation of DLX3 identified in dentin hypoplasia patients could increase DNMT3B activity, and the subsequently repressed H19/miR-675 axis impairs the odontoblastic differentiation of hDPSCs[38]. Similarly, in valve interstitial cells (VICs), which have a mesenchymal origin[39], the knockdown of H19 attenuated their osteogenic differentiation capacity by increasing the transcription of NOTCH1 and decreasing the levels of RUNX2 and BMP2[40]. In mineralized aortic valve tissue, H19 was upregulated as a result of hypomethylation of CpG in its promoter region[40]. These results suggest the possibility that H19 forms a positive feedback loop with DNMTs and promotes the osteogenic differentiation of MSCs.
Another study found an inverse association between the methylation level of perinatal CDKN2A, which encodes the lncRNA antisense noncoding RNA in the INK4 Locus (ANRIL), and bone mass at ages 4 and 6 years[41]. Considering that transitional hypomethylation of CDKN2A has been identified in human bone marrow stromal cells (hBMSCs) during osteogenic differentiation[42], the authors further verified that the methylation of CDKN2A decreased the binding of transcription factors SMAD3/4 and consequently downregulated the expression of ANRIL[41]. In terms of the functional mechanism of ANRIL, it has been demonstrated that the knockdown of ANRIL decreased the number of live cells and induced cell apoptosis of SaOS-2 cells[41].
Given the crucial roles of HOX genes in development and differentiation, it is reasonable to believe that the lncRNAs encoded by the HOX gene cluster could also exert their function as critical biological regulators (i.e., HOTAIR in the HOXC cluster and HOTAIRM1 in the HOXA cluster)[43-45]. In human dental follicle stromal cells (hDFSCs), lncRNA HOTAIRM1 promoted osteogenesis by inhibiting the enrichment of DNMT1 in the HOXA2 promoter region and subsequently maintaining two CpG islands in a hypomethylated state, which guaranteed the transcriptional activation of HOXA2[17].
lncRNA HOTAIR, encoded by the HOXC gene cluster as mentioned above, could also inhibit the adipogenic differentiation of hBMSCs[46]. In this process, HOTAIR probably directly interacts with DNMTs or is involved in gene regulation by triple helix formation[46].
Peroxisome proliferator-activated receptor-gamma (PPAR-γ) and CCAAT enhancer binding protein-alpha (C/EBP-α) are key transcription factors involved in adipogenesis. They synergistically promote the transcriptional activation of genes that induce the adipocyte phenotype and maintain their expression throughout the entire differentiation process and the entire life of the adipocytes[47,48]. In mouse ST-2 cells (bone marrow stromal cells), 3T3-L1 cells (committed preadipocytes derived from MSCs), and C3H10T1/2 cells (embryonic stem cells) as well as in bone marrow stromal cells, lncRNA Plnc1 promotes adipogenesis by increasing Ppar-γ2 transcription through reducing the DNA methylation level on its promoter[49].
Upregulation of lncRNA slincRAD is also observed in the early stages of adipocyte differentiation in 3T3-L1 cells[50]. LncRNA slincRAD guides Dnmt1 to translocate to the perinuclear region in S phase and direct Dnmt1 to the promoter of cell cycle-related genes, including p21 (Cdkn1a)[50]. As p21 is a cyclin-dependent kinase inhibitor that plays an important role in the differentiation of 3T3-L1 cells, this effect facilitates the progression of differentiation[50,51].
The building block of chromatin is the nucleosome, which consists of a complex of DNA and four types of core histone subunits (H2A, H2B, H3, and H4)[52]. Histone proteins are subject to a variety of modifications, with most studies focusing on methylation and acetylation. Lysine (K) residues in histone H3 are commonly modified by methylation, which is orchestrated by histone methyltransferases (HMTS) and histone demethylases (HDMs)[53,54]. Previous studies have revealed that trimethylation of H3K4 (H3K4me3) promotes transcription, whereas H3K9me3 and H3K27me3 restrict gene expression[53]. Likewise, acetylation and deacetylation of lysine residues in histones are regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. It is believed that the addition of an acetyl group to lysine residues alters the structure and folding of the nucleosome and consequently loosens the chromatin to enable transcription[55]. During cellular biological and pathologic processes, including cell differentiation, bone regeneration and disease, histone modifications are dynamically changed[53,56]. This process is at least in part mediated by lncRNAs that recruit histone-modifying enzymes to targeted gene promoters and alter histone modification enrichment (Figure 1).
As mentioned earlier, a mutation of DLX3 identified in dentin hypoplasia patients could increase DNMT3B activity[38]. This study also reported that this mutation was capable of repressing H19 expression by increasing the enrichment of H3K9me3 in the promoter region of the H19 gene and retarding the odontoblastic differentiation of hDPSCs[38].
Similar to RUNX2, Osterix (OSX) is considered a master transcription factor that regulates the osteogenic differentiation of MSCs and it is required for the maturation of functional osteoblasts[57]. lnc-OB1 promotes osteogenic differentiation of MSCs, probably by upregulating OSX via the inhibition of H3K27me3 in the OSX promoter region[58]. In human osteoblast cells, this regulation might be mediated by an interaction between lnc-OB1 and SUZ12, which is an integral component of polycomb repressive complex 2 (PRC2), responsible for H3K27me3[58,59].
Another core part of PRC2, EZH2[59], was also found to interact with lncRNAs and regulate osteogenic differentiation. It has been shown that lncRNA SNHG1 inhibits the osteogenic differentiation of human periodontal ligament stromal cells by repressing the expression of KLF2, a positive regulator of osteoblast differentiation[60], through EZH2-mediated H3K27me3 of its promoter[61]. Likewise, lncRNA HOXA-AS3 inhibits hBMSC osteogenesis, possibly via EZH2-dependent H3K27me3, and represses RUNX2 expression[62].
As a critical transcription factor for adipogenesis, C/EBP-α was found to be upregulated via the recruitment of the MLL3/4 complex to its promoter, which is guided by the binding of PA1 (a component of the MLL3/4 complex) to lncRNA ADINR during adipogenic differentiation of human adipose-derived stromal cells (hASCs)[63]. It is believed that MLL3/4 complexes are involved in the maintenance of H3K4me3 and the removal of H3K27me3, thereby regulating downstream gene expression[64,65].
Adipocyte fatty acid-binding protein (A-FABP, also known as FABP4 or aP2), a downstream target gene of PPAR-γ and C/EBP-α, is considered a marker of adipogenic differentiation[66,67]. The knockdown of lncRNA MIR31HG suppressed FABP4 expression by reducing the enrichment of acetylated histone 3 (AcH3) and H3K4me3 in the FABP4 promoter, leading to the inhibition of adipogenic differentiation of hASCs[16].
H19 and miR-675 (derived from H19) inhibited the adipogenic differentiation of hBMSCs through the miRNA-mediated repression of HDAC4, 5 and 6. In turn, the inhibition of HDACs decreased CCCTC-binding factor (CTCF) occupancy on the imprinting control region (ICR) of H19 and reduced H19 expression[68]. This evidence, combined with that mentioned in an earlier section that H19 is considered a positive regulator of osteogenic differentiation, suggests that DNA methylation and histone modifications might be linked together by H19 and shift the osteoadipogenic differentiation balance toward osteogenesis.
More recently, epigenetic regulation of bone homeostasis has been considered as an important factor in the pathogenesis of degenerative bone diseases, such as osteoporosis, arthritis, post menopausal osteoporosis, etc.[69,70]. As mentioned above, lncRNAs have attracted considerable attention in the epigenetic regulation of bone homeostasis. The potential link between degenerative bone diseases and lncRNAs at the epigenetic level is also an intriguing area for exploration.
Osteoarthritis (OA) is a common degenerative joint disease that is associated with the impairment of cartilage regeneration, chondrocyte apoptosis, and the degradation of the cartilage extracellular matrix (ECM)[71,72]. In this sophisticated balance between biosynthesis and degradation, lncRNAs play a role in the survival of chondrocytes and the regulation of arthritis-associated factors[73].
It has been reported that the overexpression of lncRNA CTBP1-AS2 downregulates miR-130a by increasing the methylation level of the miR-130a gene, which finally leads to a decreased proliferation rate of chondrocytes in OA patients[74].
As a natural inhibitor of matrix metalloproteinases (MMPs), TIMP-3 deficiency can lead to mild cartilage degeneration in patients with OA[75]. lncRNA XIST is capable of downregulating the expression of TIMP-3 through the recruitment of DNMT1, DNMT3A, and DNMT3B, which increased the methylation ratio of the CpG island in the TIMP-3 promoter region, and consequently increased collagen degradation in OA chondrocytes[76].
Increasing evidence suggests that small nucleolar RNA host gene (SNHG) family members are involved in the pathogenesis of OA[77-79]. The overexpression of lncRNA SNHG15 alleviated ECM degradation and promoted chondrocyte formation via competing endogenous RNA (ceRNA) SNHG15/miR-7/KLF4 axis[33]. In human OA cartilage tissues, however, the promoter region of lncRNA SNHG15 had a higher level of methylation than in normal cartilage tissues, and this might be a promising therapeutic target for OA[33]. Another SNHG family member, lncRNA SNHG9, was found to be downregulated in chondrocytes from OA patients[80]. Functional studies indicated that the overexpression of SNHG9 led to a decreased apoptotic rate through increased methylation of the miR-34a gene that suppressed the expression of miR-34a[80].
Osteoporosis is characterized by a loss of bone mass and microarchitectural deterioration of the skeletal structure[81]. The imbalance of bone homeostasis between osteoblastic bone formation and osteoclastic bone resorption plays a fundamental role in the pathogenesis of osteoporosis[82]. Emerging evidence suggests that epigenetic modifications are deeply involved in bone metabolism, which contributes to the development of osteoporosis.
The ERK-MAPK signaling pathway is a well-established pathway with critical roles in immune responses and embryonic development, including the regulation of bone mass via controlling osteoblast differentiation[83]. A previous study suggested that lncRNA H19 promoted tension-induced osteogenesis of hBMSCs through the FAK-ERK1/2-RUNX2 signaling pathway[84]. Likewise, an alteration in H19 methylation may also be involved in the disruption of bone formation in disuse osteoporosis. It has been shown that DNMT1-induced hypermethylation of the H19 promoter results in H19 downregulation and ERK-MAPK signaling inhibition, which leads to osteogenesis impairment both in vivo and in vitro (rat osteoblast/osteocyte-like UMR-106 cells)[85].
An abnormality of cartilage regeneration can be related to attenuated chondrogenic differentiation of MSCs in OA patients[8]. Similar to other MSCs derived from other tissues, synovium-derived mesenchymal stromal cells (SMSCs) are multipotent but have the greatest chondrogenesis potential, representing a promising stem cell source for cartilage repair in OA patients[86]. lncRNA MEG3 was reported to have the ability to inhibit the chondrogenic differentiation of SMSCs and the expression of cartilage-associated genes (aggrecan and Col2A1) by inhibiting TRIB2 expression through EZH2-mediated H3K27me3[87].
LncRNAs are extensively involved in various types of epigenetic modifications, including DNA methylation, histone modifications, and noncoding RNA interactions, during MSC differentiation and the occurrence and progression of degenerative bone diseases. Concerning the large body of available literature and comprehensive reviews on the RNA-RNA interactions of lncRNAs (i.e., ceRNA mechanisms)[88,89], this topic of epigenetics is not discussed in this review, but it is worth mentioning that in some cases, ceRNA mechanisms act as mediators between lncRNAs and epigenetic modifiers. Another potential involvement of lncRNAs in epigenetics is the interaction with the key enzyme of methyl metabolism. It is known that DNMT and HMT utilize S-adenosylmethionine (SAM) as a major methyl-group donor in mammals, which is consumed and regenerated in one-carbon metabolism[90,91]. Several studies have shown that lncRNAs play a role in SAM-dependent methylation through regulating enzymes related to the metabolism[92,93]. However, similar studies on differentiation and bone diseases are lacking. Further studies are needed to assess the potential importance of lncRNAs on the methyl metabolism.
Although it seems that DNA methylation and histone modification are two different types of epigenetic modification, these two systems can be dependent on and influence one another during organism development[94]. However, the underlying molecular mechanisms are complicated and remain vague. Intriguingly, lncRNAs are capable of regulating gene expression either in a cis- or trans- manner by guiding or serving as scaffolds for transcription factors or epigenetic modifiers to specific gene loci[95]. This raises the possibility that lncRNAs could be coordinator of these processes. In this review, we summarized the roles of lncRNAs played in MSC differentiation and common degenerative bone diseases through reciprocal interactions between lncRNAs and epigenetic modifiers. A complete list of the epigenetic regulatory mechanisms of lncRNAs discussed in this review is available in Tables 1-3.
| LncRNAs | Samples | Expression | Epigenetic regulatory mechanisms | Target genes | Effects | Ref. |
| H19 | hDPSCs | Up | Decreasing DNMT3B activity | DLX3 | Promote odontogenic differentiation | Zeng et al[32] |
| H19 | hDPSCs | Down | H19 was inhibited by the recruitment of DNMT3B and the enrichment of H3K9me3 in its promoter | miR-675 (derived from H19) | Inhibit odontogenic differentiation | Zeng et al[38] |
| H19 | VICs | Up | H19 was upregulated by hypomethylation of its promoter | NR | Promote osteogenic differentiation | Hadji et al[40] |
| ANRIL | Umbilical cord | Down | ANRIL was inhibited by methylation of its promoter | NR | Decrease bone mass | Curtis et al[41] |
| HOTAIRM1 | hDFSCs | Up | Inhibiting the recruitment of DNMT1 | HOXA2 | Promote osteogenic differentiation | Chen et al[17] |
| HOXA-AS3 | hBMSCs | Up | Facilitating EZH2-mediated H3K27me3 | RUNX2 | Inhibit osteogenic differentiation | Zhu et al[62] |
| SNHG1 | hPDLSCs | Up | Facilitating EZH2-mediated H3K27me3 | KLF2 | Inhibit osteogenic differentiation | Li et al[61] |
| OB1 | human osteoblasts | Up | Inhibiting H3K27me3 by interacting with SUZ12 (a core part of PRC2) | Osterix | Promote osteogenic differentiation | Sun et al[58] |
| LncRNAs | Samples | Expression | Epigenetic regulatory mechanisms | Target genes | Effects | Ref. |
| HOTAIR | hBMSCs | Up | Interacting with DNMTs | NR | Inhibit adipogenic differentiation | Kalwa et al[46] |
| Plnc1 | BMSCs | Up | Reducing the DNA methylation level | Ppar-γ2 | Promote adipogenic differentiation | Zhu et al[49] |
| slincRAD | 3T3-L1 | Up | Facilitating the recruitment of Dnmt1 | Cdkn1a | Promote adipogenic differentiation | Yi et al[50] |
| ADINR | hASCs | Up | Facilitating the recruitment of MLL3/4 complex (involved in the maintenance of H3K4me3 and the removal of H3K27me3) by binding PA1 | C/EBP-α | Promote adipogenic differentiation | Xiao et al[63] |
| MIR31HG | hASCs | Down | Reducing the enrichment of AcH3 and H3K4me3 | FABP4 | Inhibit adipogenic differentiation | Huang et al[16] |
| H19 | hBMSCs | Up | facilitating miR-675-mediated repression of HDACs | NR | Inhibit adipogenic differentiation | Huang et al[68] |
| LncRNAs | Samples | Expression | Epigenetic regulatory mechanisms | Target genes | Effects | Ref. |
| CTBP1-AS2 | OA chondrocytes | Up | Increasing the methylation level of target gene | miR-130a | Decease proliferation rate of OA chondrocytes | Zhang et al[74] |
| XIST | OA chondrocytes | Up | Facilitating the recruitment of DNMT1, DNMT3A, and DNMT3B | TIMP-3 | Raise collagen degradation in OA chondrocytes | Chen et al[76] |
| SNHG15 | OA cartilage tissues | Down | SNHG15 was inhibited by methylation of its promoter | miR-7/KLF4 | Affect ECM homeostasis | Chen et al[33] |
| SNHG9 | OA chondrocytes | Down | Altering the methylation level of target gene | miR-34a | Affect apoptotic rate of chondrocytes | Zhang et al[80] |
| H19 | UMR-106 and bone tissues from osteoporosis rat model | Down | H19 was inhibited by DNMT1-induced hypermethylation of its promoter | ERK-MAPK signaling-related genes | Impair osteogenic differentiation | Li et al[85] |
| MEG3 | SMSCs | Up | Facilitating EZH2-mediated H3K27me3 | TRIB2 | Inhibit chondrogenic differentiation | You et al[87] |
Taken in combination with previous studies[96-98], the present evidence indicates that lncRNAs could be diagnostic and prognostic biomarkers in degenerative bone diseases. Moreover, as lncRNAs can be manipulated pharmacologically to modulate epigenetic modifications[99], this also opens new avenues for future therapeutic interventions. However, multiple challenges need to be overcome before clinical applications can be achieved. Given that lncRNAs have complex secondary structures, one of the challenges that lies ahead is the off-target possibilities, as a single lncRNA is capable of binding to multiple epigenetic modifiers and targeting several genes. Therefore, more reliable bioinformatic tools in terms of in silico algorithms for comprehensive lncRNA interaction prediction and sequencing technologies are required. Despite these impediments, lncRNA-based epigenetic interventions have shown potential in the regulation of MSC differentiation and therapeutic strategies for bone diseases.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Oliva J S-Editor: Gong ZM L-Editor: A P-Editor: Gong ZM
| 1. | Clarke B. Normal bone anatomy and physiology. Clin J Am Soc Nephrol. 2008;3 Suppl 3:S131-S139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1467] [Cited by in RCA: 1185] [Article Influence: 69.7] [Reference Citation Analysis (0)] |
| 2. | Viswanathan S, Shi Y, Galipeau J, Krampera M, Leblanc K, Martin I, Nolta J, Phinney DG, Sensebe L. Mesenchymal stem versus stromal cells: International Society for Cell & Gene Therapy (ISCT®) Mesenchymal Stromal Cell committee position statement on nomenclature. Cytotherapy. 2019;21:1019-1024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 520] [Article Influence: 86.7] [Reference Citation Analysis (0)] |
| 3. | Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop Dj, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11055] [Cited by in RCA: 12678] [Article Influence: 704.3] [Reference Citation Analysis (2)] |
| 4. | Su P, Tian Y, Yang C, Ma X, Wang X, Pei J, Qian A. Mesenchymal Stem Cell Migration during Bone Formation and Bone Diseases Therapy. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 155] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 5. | Qi M, Zhang L, Ma Y, Shuai Y, Li L, Luo K, Liu W, Jin Y. Autophagy Maintains the Function of Bone Marrow Mesenchymal Stem Cells to Prevent Estrogen Deficiency-Induced Osteoporosis. Theranostics. 2017;7:4498-4516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 139] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 6. | Nuttall ME, Gimble JM. Controlling the balance between osteoblastogenesis and adipogenesis and the consequent therapeutic implications. Curr Opin Pharmacol. 2004;4:290-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 276] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 7. | Murphy JM, Dixon K, Beck S, Fabian D, Feldman A, Barry F. Reduced chondrogenic and adipogenic activity of mesenchymal stem cells from patients with advanced osteoarthritis. Arthritis Rheum. 2002;46:704-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 369] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 8. | Rocha B, Cillero-Pastor B, Eijkel G, Calamia V, Fernandez-Puente P, Paine MRL, Ruiz-Romero C, Heeren RMA, Blanco FJ. Integrative Metabolic Pathway Analysis Reveals Novel Therapeutic Targets in Osteoarthritis. Mol Cell Proteomics. 2020;19:574-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Jiang Y, Tuan RS. Origin and function of cartilage stem/progenitor cells in osteoarthritis. Nat Rev Rheumatol. 2015;11:206-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 288] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 10. | Hao J, Zhang Y, Jing D, Shen Y, Tang G, Huang S, Zhao Z. Mechanobiology of mesenchymal stem cells: Perspective into mechanical induction of MSC fate. Acta Biomater. 2015;20:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 11. | Crane JL, Cao X. Bone marrow mesenchymal stem cells and TGF-β signaling in bone remodeling. J Clin Invest. 2014;124:466-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 337] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 12. | Rice SJ, Beier F, Young DA, Loughlin J. Interplay between genetics and epigenetics in osteoarthritis. Nat Rev Rheumatol. 2020;16:268-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 13. | Grandi FC, Bhutani N. Epigenetic Therapies for Osteoarthritis. Trends Pharmacol Sci. 2020;41:557-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 14. | Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1401] [Cited by in RCA: 1786] [Article Influence: 198.4] [Reference Citation Analysis (0)] |
| 15. | Xin TY, Yu TT, Yang RL. DNA methylation and demethylation link the properties of mesenchymal stem cells: Regeneration and immunomodulation. World J Stem Cells. 2020;12:351-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 16. | Huang Y, Jin C, Zheng Y, Li X, Zhang S, Zhang Y, Jia L, Li W. Knockdown of lncRNA MIR31HG inhibits adipocyte differentiation of human adipose-derived stem cells via histone modification of FABP4. Sci Rep. 2017;7:8080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 17. | Chen Z, Zheng J, Hong H, Chen D, Deng L, Zhang X, Ling J, Wu L. lncRNA HOTAIRM1 promotes osteogenesis of hDFSCs by epigenetically regulating HOXA2 via DNMT1 in vitro. J Cell Physiol. 2020;235:8507-8519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 18. | Chen J, Wang Y, Wang C, Hu JF, Li W. LncRNA Functions as a New Emerging Epigenetic Factor in Determining the Fate of Stem Cells. Front Genet. 2020;11:277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 19. | Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 626] [Cited by in RCA: 810] [Article Influence: 73.6] [Reference Citation Analysis (0)] |
| 20. | Yoshioka H, Yoshiko Y. The Roles of Long Non-Protein-Coding RNAs in Osteo-Adipogenic Lineage Commitment. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 21. | Xia K, Cen X, Yu L, Huang X, Sun W, Zhao Z, Liu J. Long noncoding RNA expression profiles during the NEL-like 1 protein-induced osteogenic differentiation. J Cell Physiol. 2020;235:6010-6022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Yu L, Xia K, Cen X, Huang X, Sun W, Zhao Z, Liu J. DNA methylation of noncoding RNAs: new insights into osteogenesis and common bone diseases. Stem Cell Res Ther. 2020;11:109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 23. | Broholm C, Olsson AH, Perfilyev A, Gillberg L, Hansen NS, Ali A, Mortensen B, Ling C, Vaag A. Human adipogenesis is associated with genome-wide DNA methylation and gene-expression changes. Epigenomics. 2016;8:1601-1617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Shen WC, Lai YC, Li LH, Liao K, Lai HC, Kao SY, Wang J, Chuong CM, Hung SC. Methylation and PTEN activation in dental pulp mesenchymal stem cells promotes osteogenesis and reduces oncogenesis. Nat Commun. 2019;10:2226. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 111] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 25. | van Meurs JB, Boer CG, Lopez-Delgado L, Riancho JA. Role of Epigenomics in Bone and Cartilage Disease. J Bone Miner Res. 2019;34:215-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 26. | Ahuja N, Sharma AR, Baylin SB. Epigenetic Therapeutics: A New Weapon in the War Against Cancer. Annu Rev Med. 2016;67:73-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 252] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 27. | Bestor TH, Edwards JR, Boulard M. Notes on the role of dynamic DNA methylation in mammalian development. Proc Natl Acad Sci USA. 2015;112:6796-6799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 162] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 28. | Edwards JR, Yarychkivska O, Boulard M, Bestor TH. DNA methylation and DNA methyltransferases. Epigenetics Chromatin. 2017;10:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 321] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 29. | Rauluseviciute I, Drabløs F, Rye MB. DNA hypermethylation associated with upregulated gene expression in prostate cancer demonstrates the diversity of epigenetic regulation. BMC Med Genomics. 2020;13:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 30. | Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2332] [Cited by in RCA: 2976] [Article Influence: 248.0] [Reference Citation Analysis (0)] |
| 31. | Jiang W, Agrawal DK, Boosani CS. Non-coding RNAs as Epigenetic Gene Regulators in Cardiovascular Diseases. Adv Exp Med Biol. 2020;1229:133-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 32. | Zeng L, Sun S, Han D, Liu Y, Liu H, Feng H, Wang Y. Long non-coding RNA H19/SAHH axis epigenetically regulates odontogenic differentiation of human dental pulp stem cells. Cell Signal. 2018;52:65-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 33. | Chen Y, Guo H, Li L, Bao D, Gao F, Li Q, Huang Q, Duan X, Xiang Z. Long Non-Coding RNA (lncRNA) Small Nucleolar RNA Host Gene 15 (SNHG15) Alleviates Osteoarthritis Progression by Regulation of Extracellular Matrix Homeostasis. Med Sci Monit. 2020;26:e923868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 34. | Zhou J, Xu J, Zhang L, Liu S, Ma Y, Wen X, Hao J, Li Z, Ni Y, Li X, Zhou F, Li Q, Wang F, Wang X, Si Y, Zhang P, Liu C, Bartolomei M, Tang F, Liu B, Yu J, Lan Y. Combined Single-Cell Profiling of lncRNAs and Functional Screening Reveals that H19 Is Pivotal for Embryonic Hematopoietic Stem Cell Development. Cell Stem Cell. 2019;24:285-298.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 35. | Li Z, Yan M, Yu Y, Wang Y, Lei G, Pan Y, Li N, Gobin R, Yu J. LncRNA H19 promotes the committed differentiation of stem cells from apical papilla via miR-141/SPAG9 pathway. Cell Death Dis. 2019;10:130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 36. | Chan LH, Wang W, Yeung W, Deng Y, Yuan P, Mak KK. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene. 2014;33:4857-4866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 126] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 37. | Li Z, Hong Z, Zheng Y, Dong Y, He W, Yuan Y, Guo J. An emerging potential therapeutic target for osteoporosis: LncRNA H19/miR-29a-3p axis. Eur J Histochem. 2020;64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 38. | Zeng L, Sun S, Dong L, Liu Y, Liu H, Han D, Ma Z, Wang Y, Feng H. DLX3 epigenetically regulates odontoblastic differentiation of hDPCs through H19/miR-675 axis. Arch Oral Biol. 2019;102:155-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 39. | Liu AC, Joag VR, Gotlieb AI. The emerging role of valve interstitial cell phenotypes in regulating heart valve pathobiology. Am J Pathol. 2007;171:1407-1418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 433] [Cited by in RCA: 474] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 40. | Hadji F, Boulanger MC, Guay SP, Gaudreault N, Amellah S, Mkannez G, Bouchareb R, Marchand JT, Nsaibia MJ, Guauque-Olarte S, Pibarot P, Bouchard L, Bossé Y, Mathieu P. Altered DNA Methylation of Long Noncoding RNA H19 in Calcific Aortic Valve Disease Promotes Mineralization by Silencing NOTCH1. Circulation. 2016;134:1848-1862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 188] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 41. | Curtis EM, Murray R, Titcombe P, Cook E, Clarke-Harris R, Costello P, Garratt E, Holbrook JD, Barton S, Inskip H, Godfrey KM, Bell CG, Cooper C, Lillycrop KA, Harvey NC. Perinatal DNA Methylation at CDKN2A Is Associated With Offspring Bone Mass: Findings From the Southampton Women's Survey. J Bone Miner Res. 2017;32:2030-2040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 42. | Kang MI, Kim HS, Jung YC, Kim YH, Hong SJ, Kim MK, Baek KH, Kim CC, Rhyu MG. Transitional CpG methylation between promoters and retroelements of tissue-specific genes during human mesenchymal cell differentiation. J Cell Biochem. 2007;102:224-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 43. | Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526-2534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 300] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 44. | Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311-1323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3533] [Cited by in RCA: 3389] [Article Influence: 188.3] [Reference Citation Analysis (0)] |
| 45. | Mallo M. Reassessing the Role of Hox Genes during Vertebrate Development and Evolution. Trends Genet. 2018;34:209-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 46. | Kalwa M, Hänzelmann S, Otto S, Kuo CC, Franzen J, Joussen S, Fernandez-Rebollo E, Rath B, Koch C, Hofmann A, Lee SH, Teschendorff AE, Denecke B, Lin Q, Widschwendter M, Weinhold E, Costa IG, Wagner W. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016;44:10631-10643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 47. | Chen Q, Shou P, Zheng C, Jiang M, Cao G, Yang Q, Cao J, Xie N, Velletri T, Zhang X, Xu C, Zhang L, Yang H, Hou J, Wang Y, Shi Y. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23:1128-1139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 606] [Cited by in RCA: 911] [Article Influence: 101.2] [Reference Citation Analysis (0)] |
| 48. | Lin FT, Lane MD. CCAAT/enhancer binding protein alpha is sufficient to initiate the 3T3-L1 adipocyte differentiation program. Proc Natl Acad Sci USA. 1994;91:8757-8761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 356] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 49. | Zhu E, Zhang J, Li Y, Yuan H, Zhou J, Wang B. Long noncoding RNA Plnc1 controls adipocyte differentiation by regulating peroxisome proliferator-activated receptor γ. FASEB J. 2019;33:2396-2408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 50. | Yi F, Zhang P, Wang Y, Xu Y, Zhang Z, Ma W, Xu B, Xia Q, Du Q. Long non-coding RNA slincRAD functions in methylation regulation during the early stage of mouse adipogenesis. RNA Biol. 2019;16:1401-1413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 51. | Tsai MS, Su YH, Ho MC, Liang JT, Chen TP, Lai HS, Lee PH. Clinicopathological features and prognosis in resectable synchronous and metachronous colorectal liver metastasis. Ann Surg Oncol. 2007;14:786-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 137] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 52. | Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6680] [Cited by in RCA: 6756] [Article Influence: 241.3] [Reference Citation Analysis (0)] |
| 53. | Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1676] [Cited by in RCA: 1641] [Article Influence: 126.2] [Reference Citation Analysis (0)] |
| 54. | Turner BM. Cellular memory and the histone code. Cell. 2002;111:285-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 848] [Cited by in RCA: 810] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 55. | Javaid N, Choi S. Acetylation- and Methylation-Related Epigenetic Proteins in the Context of Their Targets. Genes (Basel). 2017;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 56. | Yi SJ, Lee H, Lee J, Lee K, Kim J, Kim Y, Park JI, Kim K. Bone Remodeling: Histone Modifications as Fate Determinants of Bone Cell Differentiation. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 57. | Liu Q, Li M, Wang S, Xiao Z, Xiong Y, Wang G. Recent Advances of Osterix Transcription Factor in Osteoblast Differentiation and Bone Formation. Front Cell Dev Biol. 2020;8:601224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 145] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 58. | Sun Y, Cai M, Zhong J, Yang L, Xiao J, Jin F, Xue H, Liu X, Liu H, Zhang Y, Jiang D, Hong A, Ji X, Wang Z, Zhang G, Wang X. The long noncoding RNA lnc-ob1 facilitates bone formation by upregulating Osterix in osteoblasts. Nat Metab. 2019;1:485-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 59. | Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343-349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2760] [Cited by in RCA: 2522] [Article Influence: 180.1] [Reference Citation Analysis (0)] |
| 60. | Hou Z, Wang Z, Tao Y, Bai J, Yu B, Shen J, Sun H, Xiao L, Xu Y, Zhou J, Geng D. KLF2 regulates osteoblast differentiation by targeting of Runx2. Lab Invest. 2019;99:271-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 61. | Li Z, Guo X, Wu S. Epigenetic silencing of KLF2 by long non-coding RNA SNHG1 inhibits periodontal ligament stem cell osteogenesis differentiation. Stem Cell Res Ther. 2020;11:435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 62. | Zhu XX, Yan YW, Chen D, Ai CZ, Lu X, Xu SS, Jiang S, Zhong GS, Chen DB, Jiang YZ. Long non-coding RNA HoxA-AS3 interacts with EZH2 to regulate lineage commitment of mesenchymal stem cells. Oncotarget. 2016;7:63561-63570. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 63. | Xiao T, Liu L, Li H, Sun Y, Luo H, Li T, Wang S, Dalton S, Zhao RC, Chen R. Long Noncoding RNA ADINR Regulates Adipogenesis by Transcriptionally Activating C/EBPα. Stem Cell Reports. 2015;5:856-865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 64. | Mohan M, Herz HM, Shilatifard A. SnapShot: Histone lysine methylase complexes. Cell. 2012;149:498-498.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 65. | Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 981] [Cited by in RCA: 1058] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 66. | Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ Jr, Liu XS, Lazar MA. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941-2952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 695] [Cited by in RCA: 668] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 67. | Tchoukalova YD, Sarr MG, Jensen MD. Measuring committed preadipocytes in human adipose tissue from severely obese patients by using adipocyte fatty acid binding protein. Am J Physiol Regul Integr Comp Physiol. 2004;287:R1132-R1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 64] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 68. | Huang Y, Zheng Y, Jin C, Li X, Jia L, Li W. Long Non-coding RNA H19 Inhibits Adipocyte Differentiation of Bone Marrow Mesenchymal Stem Cells through Epigenetic Modulation of Histone Deacetylases. Sci Rep. 2016;6:28897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 105] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 69. | Huang T, Peng X, Li Z, Zhou Q, Huang S, Wang Y, Li J, Song Y. Epigenetics and bone diseases. Genet Res (Camb). 2018;100:e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 70. | Yang S, Duan X. Epigenetics, Bone Remodeling and Osteoporosis. Curr Stem Cell Res Ther. 2016;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 71. | Zhao X, Petursson F, Viollet B, Lotz M, Terkeltaub R, Liu-Bryan R. Peroxisome proliferator-activated receptor γ coactivator 1α and FoxO3A mediate chondroprotection by AMP-activated protein kinase. Arthritis Rheumatol. 2014;66:3073-3082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 72. | Maneiro E, Martín MA, de Andres MC, López-Armada MJ, Fernández-Sueiro JL, del Hoyo P, Galdo F, Arenas J, Blanco FJ. Mitochondrial respiratory activity is altered in osteoarthritic human articular chondrocytes. Arthritis Rheum. 2003;48:700-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 191] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 73. | Cen X, Huang XQ, Sun WT, Liu Q, Liu J. Long noncoding RNAs: a new regulatory code in osteoarthritis. Am J Transl Res. 2017;9:4747-4755. [PubMed] |
| 74. | Zhang H, Li J, Shao W, Shen N. LncRNA CTBP1-AS2 is upregulated in osteoarthritis and increases the methylation of miR-130a gene to inhibit chondrocyte proliferation. Clin Rheumatol. 2020;39:3473-3478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 75. | Sahebjam S, Khokha R, Mort JS. Increased collagen and aggrecan degradation with age in the joints of Timp3(-/-) mice. Arthritis Rheum. 2007;56:905-909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 114] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 76. | Chen H, Yang S, Shao R. Long non-coding XIST raises methylation of TIMP-3 promoter to regulate collagen degradation in osteoarthritic chondrocytes after tibial plateau fracture. Arthritis Res Ther. 2019;21:271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 77. | Xiao K, Yang Y, Bian Y, Feng B, Li Z, Wu Z, Qiu G, Weng X. Identification of differentially expressed long noncoding RNAs in human knee osteoarthritis. J Cell Biochem. 2019;120:4620-4633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 78. | Lei J, Fu Y, Zhuang Y, Zhang K, Lu D. LncRNA SNHG1 alleviates IL-1β-induced osteoarthritis by inhibiting miR-16-5p-mediated p38 MAPK and NF-κB signaling pathways. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 79. | Shen H, Wang Y, Shi W, Sun G, Hong L, Zhang Y. LncRNA SNHG5/miR-26a/SOX2 signal axis enhances proliferation of chondrocyte in osteoarthritis. Acta Biochim Biophys Sin (Shanghai). 2018;50:191-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 80. | Zhang H, Li J, Shao W, Shen N. LncRNA SNHG9 is downregulated in osteoarthritis and inhibits chondrocyte apoptosis by downregulating miR-34a through methylation. BMC Musculoskelet Disord. 2020;21:511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 81. | Raisz LG. Pathogenesis of osteoporosis: concepts, conflicts, and prospects. J Clin Invest. 2005;115:3318-3325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1185] [Cited by in RCA: 1143] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 82. | Yang TL, Shen H, Liu A, Dong SS, Zhang L, Deng FY, Zhao Q, Deng HW. A road map for understanding molecular and genetic determinants of osteoporosis. Nat Rev Endocrinol. 2020;16:91-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 232] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 83. | Greenblatt MB, Shim JH, Glimcher LH. Mitogen-activated protein kinase pathways in osteoblasts. Annu Rev Cell Dev Biol. 2013;29:63-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 197] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 84. | Wu J, Zhao J, Sun L, Pan Y, Wang H, Zhang WB. Long non-coding RNA H19 mediates mechanical tension-induced osteogenesis of bone marrow mesenchymal stem cells via FAK by sponging miR-138. Bone. 2018;108:62-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 98] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 85. | Li B, Zhao J, Ma JX, Li GM, Zhang Y, Xing GS, Liu J, Ma XL. Overexpression of DNMT1 leads to hypermethylation of H19 promoter and inhibition of Erk signaling pathway in disuse osteoporosis. Bone. 2018;111:82-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 86. | de Sousa EB, Casado PL, Moura Neto V, Duarte ME, Aguiar DP. Synovial fluid and synovial membrane mesenchymal stem cells: latest discoveries and therapeutic perspectives. Stem Cell Res Ther. 2014;5:112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 87. | You D, Yang C, Huang J, Gong H, Yan M, Ni J. Long non-coding RNA MEG3 inhibits chondrogenic differentiation of synovium-derived mesenchymal stem cells by epigenetically inhibiting TRIB2 via methyltransferase EZH2. Cell Signal. 2019;63:109379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 88. | Ratneswaran A, Kapoor M. Osteoarthritis year in review: genetics, genomics, epigenetics. Osteoarthritis Cartilage. 2021;29:151-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 89. | Ju C, Liu R, Zhang YW, Zhang Y, Zhou R, Sun J, Lv XB, Zhang Z. Mesenchymal stem cell-associated lncRNA in osteogenic differentiation. Biomed Pharmacother. 2019;115:108912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 90. | Johnson C, Warmoes MO, Shen X, Locasale JW. Epigenetics and cancer metabolism. Cancer Lett. 2015;356:309-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 91. | Mentch SJ, Locasale JW. One-carbon metabolism and epigenetics: understanding the specificity. Ann N Y Acad Sci. 2016;1363:91-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 285] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 92. | Guo T, Gong C, Wu P, Battaglia-Hsu SF, Feng J, Liu P, Wang H, Guo D, Yao Y, Chen B, Xiao Y, Liu Z, Li Z. LINC00662 promotes hepatocellular carcinoma progression via altering genomic methylation profiles. Cell Death Differ. 2020;27:2191-2205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 93. | Zhou J, Yang L, Zhong T, Mueller M, Men Y, Zhang N, Xie J, Giang K, Chung H, Sun X, Lu L, Carmichael GG, Taylor HS, Huang Y. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun. 2015;6:10221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 191] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 94. | Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1601] [Cited by in RCA: 1640] [Article Influence: 102.5] [Reference Citation Analysis (0)] |
| 95. | Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3236] [Cited by in RCA: 3193] [Article Influence: 245.6] [Reference Citation Analysis (0)] |
| 96. | Chen Y, Lin Y, Bai Y, Cheng D, Bi Z. A Long Noncoding RNA (lncRNA)-Associated Competing Endogenous RNA (ceRNA) Network Identifies Eight lncRNA Biomarkers in Patients with Osteoarthritis of the Knee. Med Sci Monit. 2019;25:2058-2065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 97. | Zhao Y, Xu J. Synovial fluid-derived exosomal lncRNA PCGEM1 as biomarker for the different stages of osteoarthritis. Int Orthop. 2018;42:2865-2872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 94] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 98. | Silva AM, Moura SR, Teixeira JH, Barbosa MA, Santos SG, Almeida MI. Long noncoding RNAs: a missing link in osteoporosis. Bone Res. 2019;7:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 99. | Prabhakar B, Zhong XB, Rasmussen TP. Exploiting Long Noncoding RNAs as Pharmacological Targets to Modulate Epigenetic Diseases. Yale J Biol Med. 2017;90:73-86. [PubMed] |