INTRODUCTION
Hepatocellular carcinoma (HCC) is the major histological subtype of liver cancer, accounting for approximately 75%-85% of primary liver cancer cases[1]. Most HCC cases are diagnosed at an advanced stage when the surgical resection opportunity is lost and there are few other therapeutic options[1]. The aggressiveness of HCC is partially due to the presence of cancer stem cells (CSCs), also known as tumor-initiating cells. CSCs are highly tumorigenic and resistant to chemotherapy and radiotherapy[2]. They are a small subset of cancer cells with stem cell properties and capable of self-renewal. CSCs can initiate tumorigenesis and proliferate, leading to the production of a heterogeneous population of cancer cells. They may originate from transformed normal stem cells or from differentiated cells[2]. Like CSCs of other tumor types, liver CSCs (LCSCs) may also be derived from transformed liver progenitor cells or from the reprogramming of hepatocytes and biliary cells. LCSCs are also regulated by their own factors, such as pluripotency transcription factors (pTFs), CSC-related signaling pathways, genetic elements, epigenetic elements, and microenvironmental factors (i.e., the tumor microenvironment). In this review, we will focus on the factors that regulate the stemness properties of LCSCs, as the crosstalk between the tumor microenvironment and LCSCs had been discussed extensively by others[3,4].
ROLE OF PTFS IN THE REGULATION OF LCSCS
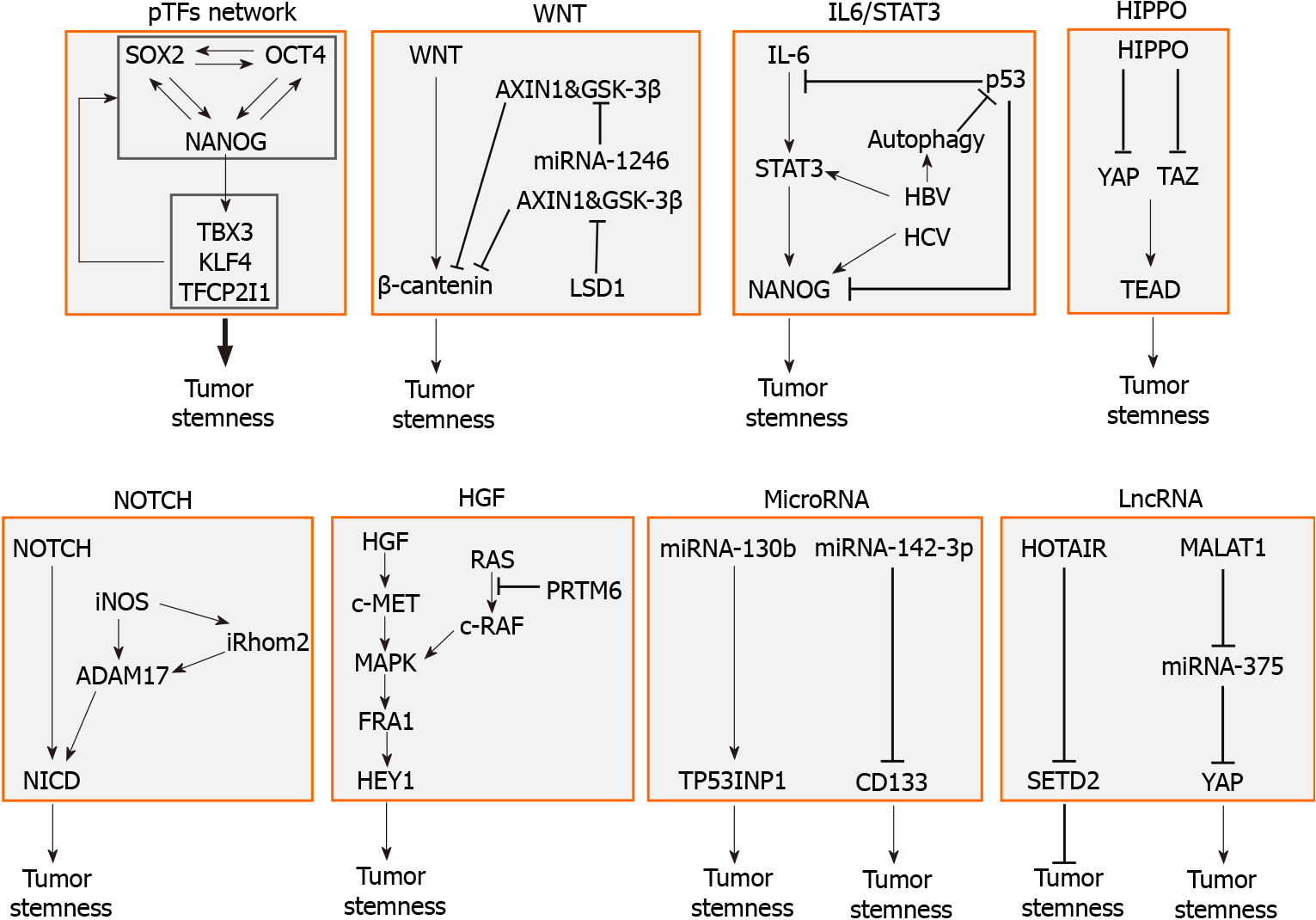
Normal somatic cells can be reprogrammed into induced pluripotent stem cells using four pTFs known as Yamanaka factors, which are OCT4, SOX2, c-Myc, and KLF4[5]. These factors play critical roles in the regulation of cell stemness. The effect of pTFs on CSCs, including LCSCs, has also received a lot of attention in recent years. In addition to Yamanaka factors, pTFs such as NANOG, SALL4, and SOX9 also promote the stemness of LCSCs[6,7]. The expressions of these pTFs are required for LCSCs to maintain their self-renewing ability and pluripotency, as well as to prevent differentiation[6,7]. The induction of one or more pTFs promotes the conversion of HCC cells into tumor cells with CSC properties[7,8]. However, the silencing of only one pTF such as NANOG cannot completely impair the tumorigenic ability of LCSCs[9], indicating that the loss of a limited number of pTFs is insufficient to destroy the properties of LCSCs. This may be explained by the fact that pTFs form a regulatory network, in which they regulate each other to direct the self-renewal of stem cells and to maintain their stemness properties[10,11]. In the pTF network, for example, OCT4 and SOX2, can promote each other’s expression and also form a heterodimeric protein complex to transactivate NANOG expression. NANOG can also induce the expression of OCT4 and SOX2 to create a positive feedback regulation[12]. Through amplifying leukemia inhibitory factor (LIF) signaling, NANOG also indirectly induces the expressions of other pTFs such as TBX3, KLF4, and TFCP2I1, which in turn induce the expression of NANOG, OCT4, and SOX2[12,13]. Therefore, the induction of only one pTF is sufficient to activate the whole network whereas the silencing of one pTF is insufficient to destroy the functions of the entire network[10]. It should be noted that the functions of pTFs mentioned above are uncovered mostly using established HCC cell lines. Whether and how these pTFs affect hepatocarcinogenesis in vivo were rarely reported and thus remain largely unclear. Moreover, although the main function of the established pTF network is to maintain the pluripotency of CSCs, the precise mechanisms involved in the establishment of this network during hepatocarcinogenesis and the tumorigenesis of other tumor types are still unknown. The understanding of the molecular details of these questions will help to identify novel targets for the prevention and treatment of HCC.
LCSC-RELATED SIGNALING PATHWAYS
Several CSC-related signaling pathways are involved in the induction of LCSCs and the maintenance of their properties during hepatocarcinogenesis. Here, we will mainly discuss the effects of Wnt/Catenin, IL-6/STAT3, Hippo, Notch, and MAPK signaling pathways on the induction and maintenance of LCSCs. Among these pathways, Wnt/Catenin and IL-6/STAT3 are two well-known signaling pathways that support the stemness of HCC cells. Somatic mutations of genes in the Wnt pathway are commonly observed in HCC, which will be discussed in detail in the genetic elements section below. STAT3 is often activated in HCC and can transactivate the expression of many genes including NANOG, SOX2, and OCT4, which are pTFs that promote the development of LCSCs[14]. The cytokine interleukin (IL)-6, an upstream regulator of STAT3, is not only produced by immune cells but can also be produced by hepatocytes and hepatoma cells[15,16]. The production of IL-6 by hepatocytes to exert an autocrine effect to activate STAT3 in hepatocytes is noteworthy, as this effect may reprogram hepatocytes into stem-like cells and induce hepatocarcinogenesis in an immune cell-independent manner, even although the development of HCC is often associated with chronic liver inflammation.
Hippo signaling is a tumor-suppressive pathway that inhibits the proliferation of adult stem cells and progenitor cells[17]. The activation of Hippo signaling leads to the phosphorylation of transcriptional coactivators Yes-associated protein 1 (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ), thereby sequestering them in the cytoplasm. This prevents YAP and TAZ from interacting with transcription factors of the TEA domain (TEAD) family in the nucleus to activate gene expression. The inhibition of TAZ expression was found to suppress the proliferation of HCC cells. However, it was also found to induce the compensatory expression of YAP, increase chemoresistance of tumor cells, and upregulate the expression of CD90, an HCC-specific CSC marker. Thus, when targeting the Hippo signaling pathway, it is necessary to simultaneously suppress both YAP and TAZ, as these two transcriptional coactivators are functionally distinct[18].
NOTCH signaling in CSCs promotes cancer progression and requires the cleavage of NOTCH by a disintegrin and metalloprotease (ADAM). The inducible nitric oxide synthase (iNOS) activates the NOTCH signaling by activating ADAM17/TACE, which leads to the production of activated NOTCH intracellular domain (NICD)[19]. iNOS also activates NOTCH signaling by inducing rhomboid protein 2, which promotes ADAM17/TACE-mediated NICD production[19]. Indeed, the expression of iNOS is associated with the expression of CD24 and CD133 stem cell markers in the tumor tissues of HCC patients and poor disease outcomes. Curiously, in a separate study it was found that the activation of the NOTCH pathway reduced HCC growth when the expression of RB, p107 and p130, three members of the retinoblastoma (RB) family, in the liver was abolished. This observation indicates that the role of NOTCH in hepatocarcinogenesis is complicated and may be context-dependent[20].
HCC often develops in association with liver cirrhosis, and activated fibroblasts (i.e., hepatic stellate cells) play a key role in the development of liver cirrhosis. Cancer-associated fibroblasts isolated from HCC secret hepatocyte growth factor (HGF), which upon binding to its receptor c-MET on hepatoma cells can stimulate MAPK signaling to activate the transcription factor FRA1 (FOS-related antigen 1) to induce the expression of HEY1 (transcription of hairy/enhancer-of-split related with YRPW motif 1). HEY1 is also a downstream target of the NOTCH pathway and can increase the population of LCSCs and chemoresistance of hepatoma cells[21].
Protein arginine methyltransferase 6 (PRMT6) methylates and binds to arginine-100 (R100) of c-Raf to disrupt its interaction with Ras to inhibit MAPK signaling. The silencing of PRMT6 activates MAPK signaling and enhances the stemness properties of CD133+ LCSCs[22].
GENETIC ELEMENTS THAT REGULATE LCSCS
The accumulation of oncogenic mutations can serve as the driver for the initiation of hepatocarcinogenesis. p53, the best-known tumor suppressor, plays a crucial role in preventing the malignant transformation of differentiated hepatocytes. It accomplishes this through multiple mechanisms, including surveillance of genome stability, arrest of the cell cycle, induction of apoptosis and suppression of cell stemness[16,23]. The p53 dysfunction by itself is insufficient to initiate hepatocarcinogenesis, and p53 by itself is often also sufficient to prevent hepatocytes from oncogenic transformation and acquisition of CSC properties[24]. When hepatocytes with mutations override p53-mediated tumor suppression, they are converted to HCC progenitor cells (HcPCs) with stemness properties and tumorigenic potentials. HcPCs produce IL-6 to stimulate autocrine IL-6 signaling, cell growth and malignant progression[16,23]. Our recent studies also demonstrated that p53 could suppress the production of CD133+ LCSCs by preventing the OCT4-SOX2 complex from binding to the NANOG promoter and hence inhibiting the expression of NANOG[25].
The mutations of canonical Wnt/β-cantenin signaling-related genes are often observed in HCC. Activating mutations in β-catenin directly activate the Wnt signaling pathway, and inactivating mutations in AXIN 1 and glycogen synthase kinase 3 (GSK-3β), the protein subunits of the functional destruction complex for β-catenin, indirectly activate this pathway[26,27].
The infection of hepatocytes by hepatitis B virus (HBV) or hepatitis C virus (HCV) can also reprogram hepatocytes into stem-like cells to promote hepatocarcinogenesis. The HBV X protein (HBx) reprograms hepatocytes by activating STAT3 signaling and increasing the expression of farnesoid X receptor (FXR) and NANOG[28,29]. In addition, HBV infection induces autophagy to enhance its replication[30]. The selective removal of mitochondria by autophagy, known as mitophagy, can cause degradation of mitochondria-associated p53 to positively regulate CD133+ LCSCs[25]. Thus, HBV-induced autophagy and mitophagy can also contributes to HCC stemness[31]. HCV can also induce autophagy and mitophagy and thus may also induce LCSCs via a similar mechanism[32,33]. The expression of the HCV NS5B polymerase can induce the expression of many stemness-related genes in HCC[34]. The HCV NS5A protein also induces the expression of toll-like receptor 4 (TLR4) in hepatocytes/hepatoblasts. The activation of TLR4 by circulating endotoxin can induce the expression of NANOG, leading to the generation of chemoresistant CD133+ LCSCs[35]. As alcohol increases the circulating endotoxin levels and HCV induces the expression of TLR4, these observations provide an explanation as to why alcohol and HCV synergistically activate LCSCs and induce HCC[36].
EPIGENETIC ELEMENTS THAT REGULATE LCSCS
Epigenetic regulators include DNA methylation, histone methylation and gene silencing by microRNAs. Epigenetic changes can predispose hepatocytes to oncogenic mutations and transformation into LCSCs[37,38]. For example, the treatment of primary human cancer cells and liver cancer cell lines with zebularine, a potent DNA methyltransferase-1 inhibitor, induced global DNA hypomethylation and dramatically increased the frequency of cells with CSC properties, as judged by their self-renewal and superior tumor-initiating abilities[39]. Lysine-specific demethylase 1 (LSD1) suppresses the expression of Prickle 1 and APC, two suppressors of β-catenin, by inhibiting mono- and di-methylation of histone H3K4 at the promoters of these two genes. This leads to the activation of Wnt/β-catenin and enhances the stemness and chemoresistance of Lgr5+ LCSCs[40]. Some microRNAs can also regulate LCSCs. For examples, microRNA-130b suppresses the expression of tumor protein p53-induced nuclear protein 1 (TP53INP1) to promote CD133+ LCSC growth and self-renewal[41], and microRNA-1246 suppresses AXIN2 and GSK-3β to activate Wnt signaling to induce CD133+ LCSCs[42]. In contrast, microRNA-142-3p directly targets the mRNA of CD133, a surface marker of CSCs that plays an important role in the maintenance of CSC properties by regulating neurotensin, IL-8, CXCL1, and MAPK signaling, resulting in the reduction of CD133+ LCSCs[43,44]. LncRNAs also affect the generation of LCSCs. For examples, lncRNA HOTAIR promotes the malignant growth of LCSCs through the downregulation of SETD2, a histone lysine methyltransferase[45], lncRNA MALAT1 promotes LCSC properties by upregulating YAP1 expression via microRNA-375 sponging[46], and lncRNA lncTCF7 promotes self-renewal of LCSCs through activation of Wnt signaling[47].
CONCLUSION
LCSCs are regulated by multiple factors including pTFs, signaling pathways, genetic elements, and epigenetic elements. A summary of these factors is illustrated in Figure 1. These factors affect one another and form an integrated network to regulate the self-renewal and other properties of LCSCs. In-depth understanding of how LCSCs are regulated will be important for the identification of novel therapeutic targets for improving the treatments for HCC.
Figure 1 Molecular factors that regulate the stemness of liver cancer stem cells.
The factors that regulate liver cancer stem cells (LCSCs) include the pluripotency transcription factor (pTF) network, in which pTFs affect the expression of one another to promote the development of LCSCs, the WNT signaling pathway, which is often activated by gene mutations, the IL6/STAT3 pathway, which can also be stimulated by hepatitis B virus or hepatitis C virus, the tumor-suppressive HIPPO pathway, the NOTCH signaling pathway, and hepatocyte growth factor and its downstream MAPK signaling pathway. Specific microRNAs such as miRNA-130b and miRNA-142-3p, and lncRNAs such as HOTAIR and MALAT1 can also promote the stemness of LCSCs. pTF: Pluripotency transcription factor; IL: Interleukin; HBV: Hepatitis B virus; HCV: Hepatitis C virus; YAP: Yes-associated protein; TEAD: TEA domain; iNOS: Inducible nitric oxide synthase; HGF: Hepatocyte growth factor.
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Janvilisri T, Shen ZY S-Editor: Gao CC L-Editor: A P-Editor: Yuan YY