Published online Jul 26, 2021. doi: 10.4252/wjsc.v13.i7.795
Peer-review started: February 18, 2021
First decision: March 16, 2021
Revised: March 25, 2021
Accepted: May 7, 2021
Article in press: May 7, 2021
Published online: July 26, 2021
Processing time: 155 Days and 0 Hours
Hepatocellular carcinoma (HCC) is a heterogeneous malignancy related to diverse etiological factors. Different oncogenic mechanisms and genetic variations lead to multiple HCC molecular classifications. Recently, an immune-based strategy using immune checkpoint inhibitors (ICIs) was presented in HCC therapy, especially with ICIs against the programmed death-1 (PD-1) and its ligand PD-L1. However, despite the success of anti-PD-1/PD-L1 in other cancers, a substantial proportion of HCC patients fail to respond. In this review, we gather current information on biomarkers of anti-PD-1/PD-L1 treatment and the contribution of HCC heterogeneity and hepatic cancer stem cells (CSCs). Genetic variations of PD-1 and PD-L1 are associated with chronic liver disease and progression to cancer. PD-L1 expression in tumoral tissues is differentially expressed in CSCs, particularly in those with a close association with the tumor microenvironment. This information will be beneficial for the selection of patients and the management of the ICIs against PD-1/PD-L1.
Core Tip: Immune checkpoint inhibitors (ICIs), in particular the ICIs against the programmed death-1/programmed death ligand 1 (PD-L1/PD-L1) axis, have recently been presented for the treatment of hepatocellular carcinoma (HCC). However, despite the success of anti-PD-1/PD-L1 in other cancers, a substantial proportion of HCC patients fail to respond. Here, we gather current information on biomarkers of anti-PD-1/PD-L1 treatment and the contribution of HCC heterogeneity and hepatic cancer stem cells.
- Citation: Sukowati CHC, El-Khobar KE, Tiribelli C. Immunotherapy against programmed death-1/programmed death ligand 1 in hepatocellular carcinoma: Importance of molecular variations, cellular heterogeneity, and cancer stem cells. World J Stem Cells 2021; 13(7): 795-824
- URL: https://www.wjgnet.com/1948-0210/full/v13/i7/795.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i7.795
International epidemiology data Globocan 2018 predicted primary liver cancer to be the sixth most commonly diagnosed cancer and the fourth leading cause of cancer-related mortality worldwide. In the male population, its incidence and mortality were 2 to 3 times higher compared to females, ranking it as fifth in terms of global cases and second in terms of deaths[1]. Hepatocellular carcinoma (HCC) accounts for about 90% of liver cancer cases, with cirrhosis as the strongest underlying condition[2,3].
HCC is caused by various etiological factors. Major risk factors for HCC are liver cirrhosis due to chronic hepatitis B virus (HBV) and/or hepatitis C virus (HCV) infection, which comprised around 80% of HCC cases globally[4], alcoholic liver disease (ALD), non-alcoholic fatty liver disease (NAFLD) leading to non-alcoholic steatohepatitis (NASH), and exposure to aflatoxin B1. It is noteworthy that even though chronic HBV and HCV infection is the current major driver of HCC cases, the rise of liver disease due to metabolic syndrome (NAFLD/NASH) may lead to a high number of HCC cases in the future[5,6].
The international consensus for HCC management[7] recommends surgical intervention as the main curative treatment for HCC, resulting in the best outcomes in well-selected candidates (five-year survival of 60%-80%)[8]. Image-guided radiofrequency ablation is the treatment of choice for HCC patients with early-stage HCC when liver transplantation or hepatectomy are precluded. For patients in an intermediate stage, palliative treatment using trans-arterial chemoembolization (TACE) is recommended[7,9]. For advanced HCC, oral systemic treatment with the tyrosine-kinase inhibitor sorafenib may extend the patient’s overall survival (OS) for around 3 mo[10]. Despite the fast emergence of targeted therapy development, HCC remains largely incurable due to low response rate and therapeutic resistance[11].
Immunotherapy represents an effective and promising option against various types of cancer. Recently, a new immune-based strategy using immune checkpoint inhibitors (ICIs) for HCC therapy was shown to be highly promising compared to chemotherapy and systemic therapy. Immune checkpoints are pathways that inhibit the immune response to maintain self-tolerance and regulate the duration and amplitude of immune responses[12]. The liver tissue is immune tolerant due to its physiological function, and liver sinusoidal endothelial cells are exposed to a significant amount of bacterial antigens from the portal circulation[13].
ICIs against the programmed death-1 (PD-1, CD279) and its ligand, is an important focus in cancer immunology and oncology with FDA approval for various types of cancer. Immunotherapies targeting PD-1/programmed death-ligand 1 (PD-L1) signaling have now become the first-line treatment for some cancers due to their promotion of anti-tumor immune responses in patients with advanced cancers[14].
PD-1 is a cell surface receptor belonging to the extended CD28/CTLA-4 (cytotoxic T-lymphocyte-associated protein 4) family. It is an approximately 55-kDa type I transmembrane glycoprotein that exists as a monomeric receptor. PD-1 is mainly expressed on T cells, B cells, monocytes, dendritic cells (DCs), and natural killer (NK) cells[15,16]. In a phase I/II CheckMate 040 trial in 2017, nivolumab, a checkpoint inhibitor against anti-PD-1 showed promising results in advanced HCC patients[17]. It was well tolerated and patients had a long-lasting response. Nivolumab is a human immunoglobulin G4 (IgG4) monoclonal antibody that binds to the PD-1 receptor and disrupts the interaction with PD-L1 and PD-L2, its ligands in tumor cells. This interaction releases PD-1 pathway-mediated inhibition of the immune response, including the anti-tumor immune response[18]. Other ICIs, pembrolizumab (anti PD-1)[19] and tremelimumab (anti CTLA)[20,21] were under phase 2 trial both as single or as combination therapy. It was shown that anti-PD-1 therapy in HCC patients intolerant to sorafenib resulted in an excellent complete response[22].
A more recent approach is to target the ligand PD-L1. PD-1 has two ligands from the B7 transmembrane proteins family, the PD-L1 (B7-H1) and PD-L2 (CD273, B7-DC)[16]. PD-L2 affinity to PD-1 is three-fold higher than PD-L1; however, PD-L2 is only expressed in antigen-presenting cells[15]. PD-L1 is a 40-kDa type I transmembrane protein, which is expressed in immune cells (ICs) such as T cells, B cells, NK cells, DCs, macrophages, and myeloid-derived suppressor cells. It is also expressed in non-IC types including epithelial, endothelial, and tumor cells[14,23].
The safety and activity of PD-L1 inhibition using the engineered humanized antibody atezolizumab was first reported in lung cancer[24,25]. It is a high-affinity human monoclonal IgG1 antibody that specifically binds to PD-L1 and prevents its interaction with PD-1 and B7.1[26]. In HCC, a phase 1b GO30140 study of atezolizumab plus bevacizumab (a monoclonal antibody against VEGF) in untreated patients with unresectable HCC showed an acceptable low side-effect profile and promising antitumor activity with a median progression-free survival (PFS) of 7 mo[27]. Recently, the IMbrave150 study, a global, multicenter, open-label, phase 3 randomized trial, demonstrated the safety and efficacy of atezolizumab plus bevacizumab as compared with sorafenib. In 501 unresectable HCC patients at 111 sites in 17 countries, PFS was significantly longer with atezolizumab–bevacizumab than with sorafenib with a median of 6.8 vs 4.3 mo. The OS was also significantly longer in this group with the estimated rates of survival at 12 mo was 67% vs 55%[28].
However, despite the success of ICIs against the PD-1/PD-L1, a substantial proportion of patients fail to respond. In 29 unresectable HCC patients, the objective response rate after nivolumab monotherapy was around 21% with an OS of 26 wk. Interestingly, differential responses to nivolumab among multiple tumor nodules in a single HCC patient were found in 18% of total cases, where small metastatic tumors but not large tumors regressed[29]. The efficacy of immunotherapy might depend on different factors, such as patients’ heterogeneity on genomic features, oncogenic pathways, cancer microenvironment, systemic immunity status, microbiome, and metastases, as reviewed in[30].
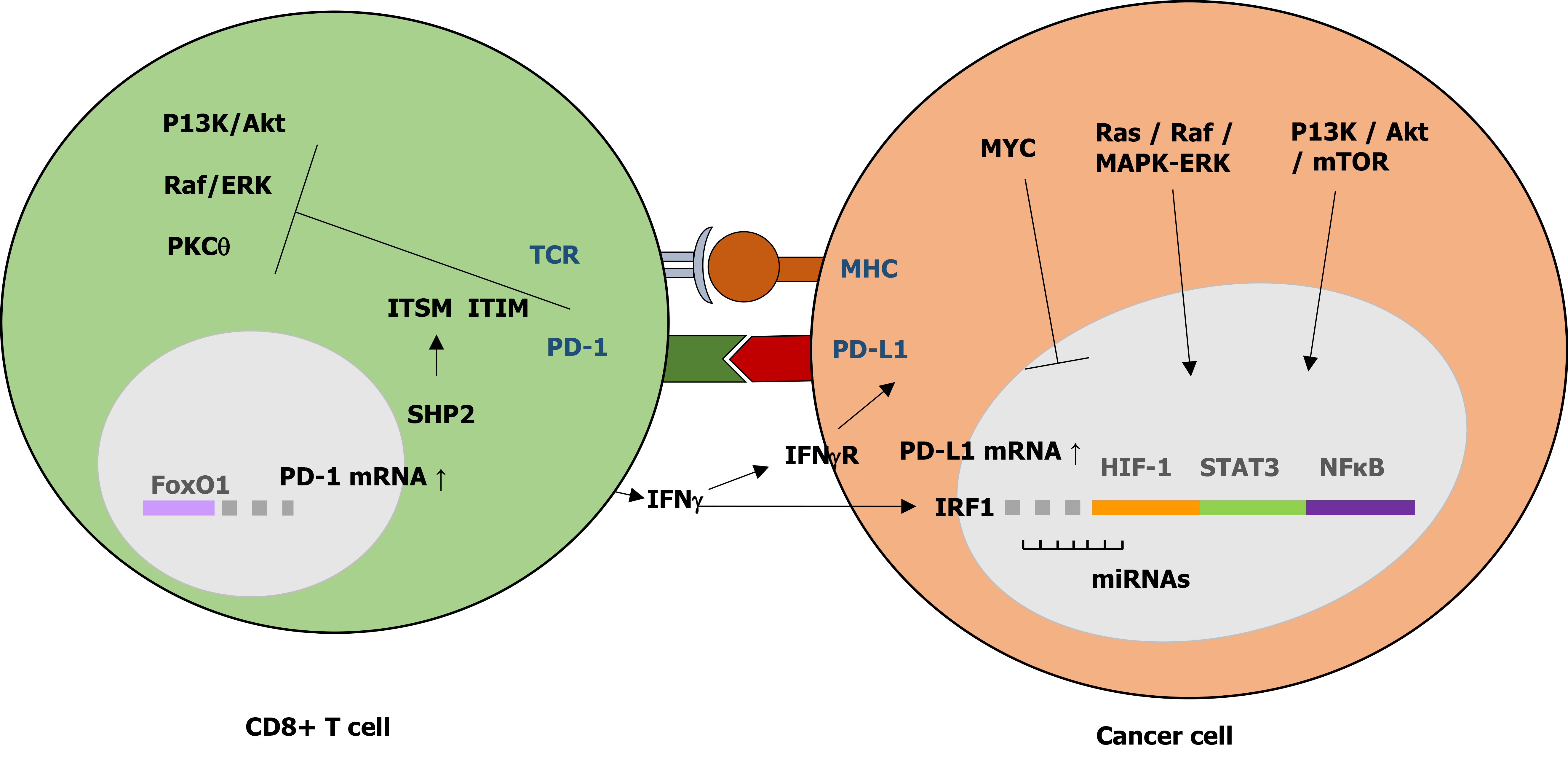
PD-1 transcription may be induced by various transcription factors including the nuclear factor of activated T cells, NOTCH, Forkhead box protein (FOX) O1, and interferon regulatory factor 9 (IRF9)[31]. During antigen stimulation, binding of PD-1 and its ligand leads to phosphorylation of the two PD-1 tyrosine residues, the immunoreceptor tyrosine-based inhibitory motif (ITIM) and immunoreceptor tyrosine based switch motif (ITSM), in the PD-1 cytoplasmic region, followed by recruitment of SHP1 and SHP2 phosphatases to ITIM and ITSM, respectively[32,33]. These interactions resulted in decreased phosphorylation of various signaling molecules including ZAP70/CD3 and downstream PKC signaling, vav, AKT, and ERK in T cells, and Ig and ERK in B cells[34,35].
PD-L1 is activated by pro-inflammatory cytokines such as IFN-γ and IL-4, through its IFN regulatory factor 1 (IRF1) response element in the PD-L1 promoter region[36,37]. PD-1 interaction with either ligand has been shown to inhibit T cell proliferation through cell cycle arrest at G0/G1, promote apoptosis, and stimulate immunosuppressive IL-10 secretion but impair IL-2 secretion[35,38,39]. PD-L1 also regulates macrophage proliferation and activation. PD-L1 negatively regulated macrophage proliferation, survival, and activation by inhibition of the mTOR pathway, resulting in an immunosuppressive phenotype. However, treatment with anti-PD-L1 antibody, but not anti-PD-1 antibody, reversed this phenotype and triggered macrophage-mediated anti-tumor activity instead[40].
The PD-1/PD-L1 axis plays an important role in successful protective cellular immune response and prevention of immune overstimulation and autoimmune disorders through maintenance of T cell homeostasis and control of self-tolerance[14,36,38] (adapted in Figure 1). PD-L1 binding to PD-1 leads to the inhibition of T cell functional activation by: (1) Directly targeting the T cell receptor (TCR) signaling through phosphatases recruitment and inhibition of ZAP70 and PI3K downstream pathways; (2) Indirectly inhibiting TCR signaling and T cell proliferation by regulating CK2 expression and activation; (3) Regulating TCR surface expression by promoting E3 ubiquitin ligases expression, leading to TCRs removal from the T cell surface and loss of T cell response upon antigen stimulation; and (4) Altering T cell metabolism by inhibition of ERK and PI3K/Akt activities, resulting in a long-lived memory T cells phenotype[32,34]. This PD-1 TCR-dependent inhibition of T cell activation occurs with or without CD28 or ICOS (inducible T cell co-stimulator) co-stimulation, although CD28 co-stimulation reduced PD-1 efficiency in inhibiting TCR-dependent T cell activation[34].
The PD-1/PD-L1 dependent T cell activation is often found to be dysregulated or overexpressed in tumor cells and viral-infected cells. Overexpression of PD-1/PD-L1 in tumor and viral-infected cells promotes self-tolerance by inducing PD-1 dependent inhibitory signals, just like in normal cells, thereby escaping T cell-mediated immune response and promoting tumor progression and survival[15].
High PD-L1 expression in cancers is associated with a poor prognosis. Cancer cell’s PD-L1 binding to PD-1 receptor on tumor-infiltrating T cells (TILs) has been shown to induce SHP2 activation resulting in suppression of the TCR pathway and inhibition of T cell activity[41]. During viral infection, PD-1 is expressed transiently by CD8+ T cells, which will gradually decline at the end of acute infection due to lack of specific TCR stimulation[15]. However, in chronic viral infection, exhausted CD8+ T cells maintained PD-1 expression due to continuous TCR-antigen ligation and lack of PD-1 promoter re-methylation[15,42]. The lack of DNA re-methylation left the PD-1 locus ready for rapid PD-1 expression, providing a premature termination of CD8+ T cell antiviral functions[42].
PD-1/PD-L1 overexpression in many cancers causes functionally exhausted T cells, thus blocking the PD-1/PD-L1 pathway may restore anti-tumor immunity by enhancing T cell killing activities and improve cancer prognosis[41]. In addition, PD-1 has been shown to bind and phosphorylate two mTOR downstream effectors, eukaryotic initiation factor 4E (eIF4E) and ribosomal protein S6 (rpS6), to promote tumor growth in HCCs[43].
PD-L1 expression in cancer is complex. It is regulated differently at transcriptional and post-transcriptional levels. PD-L1 can be activated by aberrant oncogenic signaling pathways including Ras/Raf/MEK/MAPK-ERK and PI3K/Akt/mTOR, and various transcription factors such as STAT-3, STAT-1, c-Jun, HIFs, and NF-κβ . PD-L1 was also regulated post-transcriptionally by various microRNAs (miRNAs), which bind to PD-L1 mRNA to either repress or enhance translation[16]. The binding of miR-4717 and miR-570 to PD-L1 3’UTR resulted in downregulation of PD-L1 mRNAs[44,45]. Several recent studies have also reported additional regulatory mechanisms of PD-L1 expression in cancer, including epigenetic regulation through methylation and histone acetylation, post-translational modification on PD-L1 protein including phosphorylation, N-glycosylation, poly-ubiquitination, and palmitoylation, and various genetic alterations[41]. PD-L1 expression in HCCs was epigenetically regulated by EZH2-induced upregulation of H3K27me3 levels on the PD-L1 promoter and IRF1 promoter, an essential transcription factor PD-L1, without affecting the IFNγ-STAT1 pathway[46]. In addition, PKM2-induced phosphorylation of histone H3 was important for EGF-mediated PD-L1 transcription in HCC[47].
Oncogenic Ras signaling upregulated PD-L1 expression in tumor cells through modulation of the AU-rich element-binding protein tristetraprolin (TTP), located in the 3’UTR of PD-L1 mRNA, resulting in stable PD-L1 mRNA expression. Furthermore, MEK signaling, Ras downstream effectors, phosphorylated and inhibited TTP activity through kinase MK2 activation[48]. Downregulation of PD-L1 expression in liver cancer cells was the result of Tyr56 phosphorylation and activation of glycogen synthase kinase 3β (GS3Kβ), part of the Wnt/ β-catenin pathway, by MET (hepatocyte growth factor) receptor tyrosine kinase. Treatment with MET inhibitors in these cells decreased the antitumor activity of T cells in HCCs[49], possibly through PD-L1 effects on CD8+ T cell activity.
The oncogene MYC has been shown to affect PD-L1 expression in cancer cells. In HCC, MYC activation was shown to downregulate PD-L1 expression through the reduced level of STAT-1, a crucial part of the IFN-γ pathway[50]. This observation was in line with a previous study which showed a positive correlation of JAK-2 expression, a STAT-1 upstream effector, with increased PD-L1 expression in nodular sclerosing Hodgkin lymphoma and large B-cell lymphoma[51]. p53 tumor suppressor gene is found frequently mutated in many types of cancer. Increased p53 expression was related to increased PD-L1 in tumor cells of HCC and oral squamous cell carcinoma[52,53]. p53 expression in HCCs was also positively correlated with an increased level of APE-1[52], a multifunctional enzyme involved in the base excision repair pathway. Additionally, p53 level was associated with IFN-γ-induced PD-L1 expression in melanoma through maintenance of JAK-2 expression in tumor cells[54].
IFN-γ induces PD-L1 mRNA and protein expression through upregulation of transcription factor IRF-1, and its binding to the interferon stimulated response element in the PD-L1 gene promoter. This IRF-1 upregulation of PD-L1 expression is antagonized by IRF-2 competitive binding to the PD-L1 promoter[55]. Increased IRF-1 mRNA expression can be observed in patients with well-differentiated or early stages of HCC tumors[55]. IFN-γ upregulation of PD-L1 was associated with epithelial to mesenchymal transition (EMT) in pancreatic ductal adenocarcinoma (PDA), characterized by increased vimentin expression and infiltration of CD8+ T cells and Foxp3+ cells. This EMT promotion of IFN-γ in PDA was inhibited by the treatment of STAT-1 siRNA[56]. In HCC, PD-L1 upregulation also promoted EMT, characterized by increased N-cadherin but reduced E-cadherin levels, through the activation of SREBP-1 of the PI3K/Akt pathway[57,58].
ICs that infiltrate some types of HCC also secrete IFN-γ. CD8+ CTLs secretion of IFN-γ is impaired following CD8+ CTLs upregulation of PD-L1 in HCC tumor cell lines by HLA class-I specificity, which indicates a negative feedback regulatory mechanism of IFN-γ and PD-L1 expression in CD8+ CTLs[59]. Some HCC cells expressed myocyte enhancer factor 2D (MEF2D), which was associated with high PD-L1 expression and shorter survival time. MEF2D expression in HCCs also negatively correlated with lower numbers of CD4+ and CD8+ T cells, attenuating its antitumor activity. IFN-γ treatment of HCC cells resulted in MEF2D acetylation by p300 activation, followed by MEF2D binding to the PD-L1 gene promoter and upregulation of PD-L1. Without IFN-γ, MEF2D acetylation was inhibited by SIRT-7 by forming a complex with MEF2D[60]. Independently, SIRT-7 seemed to promote HCC cell proliferation, as SIRT-7 knockout mice had reduced cell proliferation and tumor growth[60].
Hypoxia commonly occurs in the tumor microenvironment. ICs secrete tumor-promoting inflammatory cytokines under hypoxic conditions, which further activate STAT-3 and NF-κβ transcription factors resulting in tumor proliferation, survival, and invasion[61]. NF-κβ and HIF-1a (hypoxia-inducible factor 1a) were shown to upregulate PD-L1 expression in non-small cell lung carcinoma (NSCLC), through epidermal growth factor receptor activation and phosphorylation of Akt and ERK[62]. Increased HIF-1a level in HCCs was also associated with high PD-L1 expression and increased risk of cancer recurrence and metastasis[63]. HIF-1a level in HCC was also correlated with increased CXCL-12 mRNAs, a ligand for CXCR-4 chemokine receptor, which was known for its positive effect on migration, proliferation, and survival of cancer cells[64]. Accordingly, loss of nuclear CXCL-12 expression was correlated with better OS[64]. HIF-1a also induced TREM-1 (triggering receptor expressed on myeloid cells-1) in tumor-associated macrophages (TAMs), which were found abundantly in advanced stages of HCCs, resulting in impaired cytotoxic functions and induced apoptosis of CD8+ T cells[65]. Blocking of TREM-1+ TAMs led to reversal of its immunosuppressive effect and PD-L1-induced resistance in liver cancer cells[65].
Aberrant PD-1/PD-L1 binding which leads to activation of the self-tolerance pathway has been observed in both ICs and tumor cells. Increased PD-1 and PD-L1 expression was observed in pathological liver specimens[66]. PD-1 was mostly noted in tumor-infiltrating CD8+ T cells. HCCs with a PD-1-high cell population were aggressive and had higher levels of predictive biomarkers of response to anti-PD1 therapy[67,68]. The PD-1 expression was also elevated in monocytes of HCC patients[69].
High CD8+ T cells seemed to result in a favorable outcome for HCC[70]. Low CD8+ T cell infiltration in HCCs has been associated with EMT through increased vimentin expression resulting in poor patient prognosis[56]. CD8+ T cells in HCCs seemed to express PD-1 at different levels. PD-1-high CD8+ T cells which expressed LAG-3 (lymphocyte-activation gene 3), a T cell expression marker, and/or TIM-3 (T-cell immunoglobulin mucin-3), produced low levels of IFN-γ and TNF in response to anti-CD3. Treatment of this subset of cells with antibodies against PD-1 and LAG-3 or TIM-3 restored both cell proliferation and cytokine production[67] making them more susceptible to immune checkpoint blockade therapy.
A higher ratio of LAG-3+ to CD8+ cells was found on HCC tissues compared to adjacent normal tissues in a cohort of 143 Chinese patients. A high level of LAG-3 (T cell expression marker expressed on both CD4+ and CD8+ cells) was also associated with a high level of FGL-1 (fibrinogen-like protein 1), a major ligand for LAG-3, but not PD-L1 level[70]. A previous study in Caucasian HCC patients reported that tumor cells have increased PD-L1+ and LAG-3+ cells but reduced CD8+ T cells expression[71]. Taken together, it seems that PD-1, PD-L1, and LAG-3 expression was regulated differently in different HCC subsets, in association with the regulation of CD8+ T cell tolerance. However, it can be inferred from all studies that high LAG-3 expression in HCCs, which was associated with poor disease outcome[67,70,71], may represent another layer of tumor evasion mechanism based on its effect on T cell activation.
Tumor-infiltrating ICs (TIICs) play crucial roles in the reactivation of effective antitumor responses[72]. TIICs in HCCs have also been shown to express high LAG-3 positivity[71]. Using advanced cytometry by time-of-flight (CyTOF) analysis, detailed TILs profiling in a spontaneous HCC model that was resistant to anti-PD-1 treatment revealed that effector memory CD8+ T cells (CD44+CD62L-KLRGint) had a high level of T cell exhaustion markers, TIGIT (T-cell immunoglobulin and ITIM domain), LAG-3, and CD39. Furthermore, this enhanced TIGIT expression on CD8+ and CD4+ T cells was tumor specific[73]. In addition, they also found a high level of PVRL1 (poliovirus receptor-related 1) mRNA and protein in HCC tissues. This PVRL1 upregulation stabilized cell surface PVR (poliovirus receptor) which interacted with TIGIT, an inhibitory molecule on CD8+ effector memory T cells[73]. Inhibitors of the PVRL1/PVR/TIGIT signaling axis may be beneficial for the development of HCC treatment, in combination with anti-PD-1, due to its induction of the anti-tumor immune response[72,73].
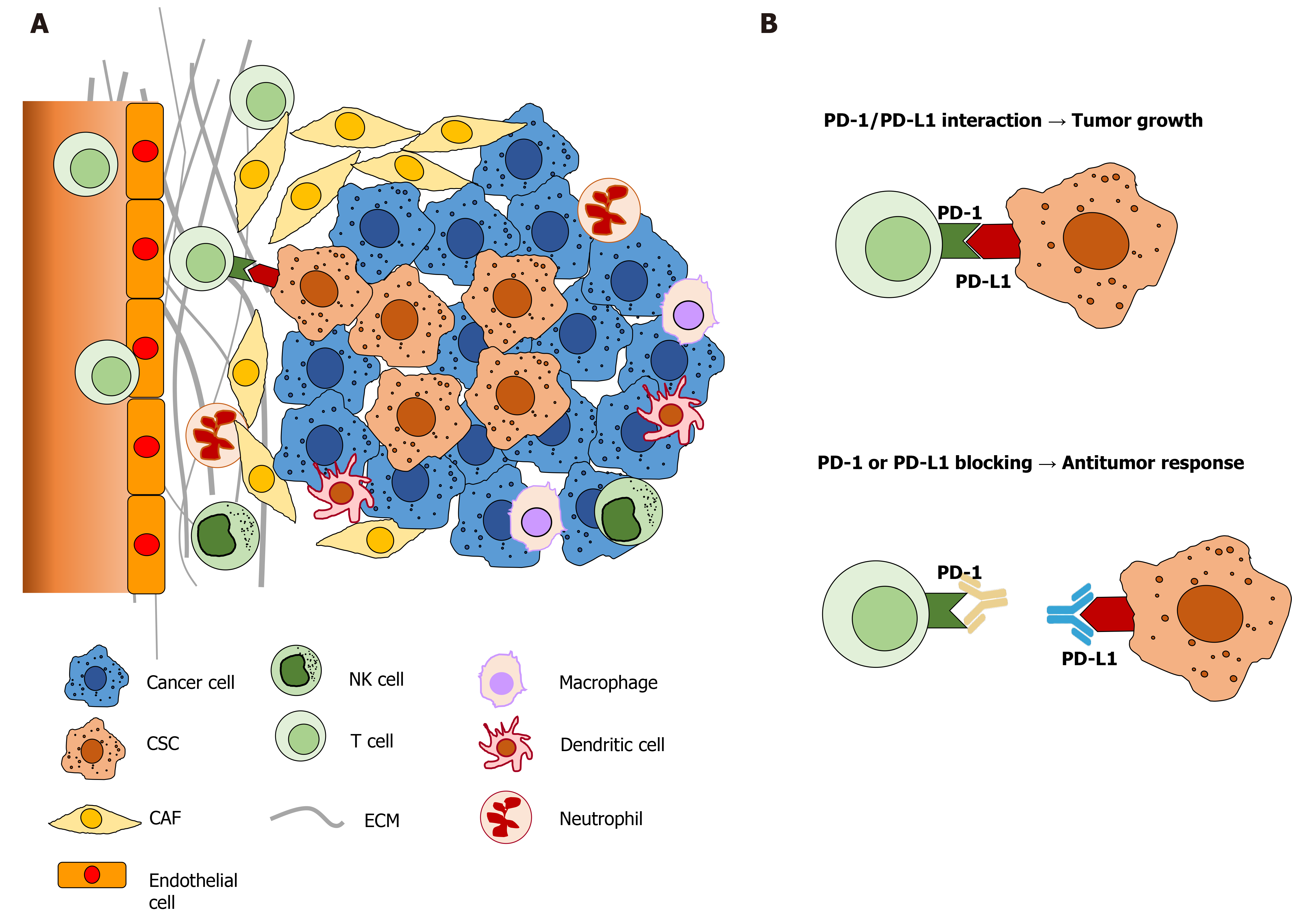
Many tumors overexpress PD-L1 to escape immune surveillance by deregulating the survival and proliferation pathways[74]. Elevated PD-L1 expression has been reported in various cancers and was strongly correlated with advanced disease state and unfavorable prognosis[14]. Blockade of the PD-1/PD-L1 signaling axis by anti-PD-1 and/or anti-PD-L1 antibodies resulted in reactivation of the exhausted ICs in the tumor microenvironment and elimination of cancer cells[41]. In HCC, PD-L1 was stained positive in tumor cells and TILs, but rarely in normal hepatocytes[26]. PD-L1 expression in tumor cells may be constitutive by the regulation of oncogenic events in tumor cells, and inductive, by the stimulation of immune response in the tumor microenvironment[75]. PD-L1 staining showed that the main PD-L1 expression on HCC cells was in the cell membrane with variable staining in the cytoplasm[76-79].
PD-L1 expression was found to be higher in HCC tissue compared to the corresponding non-tumor liver[64,80]. High PD-L1 expression was associated with tumor size and histological grade. Furthermore, Kaplan Meier analysis showed that hepatic membrane-bound PD-L1 expression represented a predictive biomarker for HCC aggressiveness and patient survival[64,76,78], as summarized in a recent meta-analysis of 23 studies with around 3,500 patients[81].
Higher expression of PD-1/PD-L1 was associated with shorter OS and tumor-free survival. Furthermore, circulating PD-1/PD-L1 expression was also closely correlated with intratumoral PD-L1 expression[82]. Overexpression of both PD-L1 and PD-L2 was also observed in resected HCC tissues, which were also associated with poor survival[78]. In addition, increased PD-1 expression in HCC is associated with the promotion of tumor growth[43]. There are several anti-PD-1 and anti-PD-L1 drugs on trial as potential ICIs for HCC patients including nivolumab and atezolizumab[57], but not all HCC tumors respond well to anti-PD-1 inhibitors[73] which suggests complex PD-1/PD-L1 regulation in different HCC subsets.
In a Caucasian HCC cohort (n = 217), PD-L1 was expressed in 17% of tumors, with a positivity rate ranging from 1% to 30%. This PD-L1 expression was associated with HCC progenitor subtype with all the common markers for tumor aggressiveness including high alpha-fetoprotein (AFP) levels, satellite nodule, macrovascular and microvascular invasion, poor differentiation, and CK-19 expression[83]. Similarly, in a Chinese HCC cohort (n = 411), only 19% of tumor cells expressed PD-L1 positivity, which was correlated with high CD8+ T cell densities. The high level of CD8+ T cells was associated with better OS and recurrence-free survival[59]. In a different Chinese HCC cohort (n = 304), only increased PD-L2 expression was observed in 19% of tumor cells. A high proportion of PD-L1 and PD-L2 was also observed in TIICs in HCC stroma, which was correlated with higher CD8+ T cells[84].
PD-L1 is expressed by stromal cells in the HCC microenvironment, especially in tissue adjacent to carcinoma portal exchange and endothelial cells[52]. PD-L1-expressing inflammatory cells were identified in 76% of tumors (n = 217, Caucasian cohort), and was associated with high AFP levels, macrovascular invasion, poor differentiation, high PD-1 expression, and lymphoepithelioma-like histological subtype of HCC[83]. Positive PD-L1 expression was also identified on sinusoidal lining cells (mostly Kupffer cells), endothelial cells, and ICs in adjacent non-HCC parenchyma and non-cirrhotic liver (n = 68); with most PD-L1 positive cells identified as ICs[85]. CD8+CD68+Foxp3+ ICs were associated with HCC especially in the invasive margin, while CD8+ cells were correlated with PD-L1 positive cells[85]. Similarly, high expression of PD-1 and PD-L1 in both tumor interior and invasive margin were correlated with high densities of CD3+ and CD8+ T cells; which was associated with a low rate of occurrence and prolonged RFS (relapse-free survival)[86].
In a Chinese HCC cohort (n = 90), high PD-L1 positive expression (31%) was found in HCC peritumoral tissues, which was related to more vascular invasion, lower albumin level, and worse OS[63]. Another study with a bigger HCC cohort (n = 304) also found high PD-L1 expression in immune stroma (which was identified as mostly macrophages). This high PD-L1 expression was correlated with CD8+ T cells infiltration, but interestingly resulted in poorer OS and DFS (disease-free survival) outcome[84]. The difference between CD8+ T cells infiltration and disease outcome in Caucasian vs Chinese cohorts may be attributed to TGF-β expression. Increased TGF-β and TGF-β+ Tregs were identified in the peripheral blood of HCC patients (n = 100), and patients with high TGF-β and TGF-β+ Tregs had a lower OS rate. TGF-β may promote IL-6 production thereby promoting tumor growth and proliferation[87]. In addition, high TGF-β level also reduced IFN-γ secretion by CD8+ T cells[88]. Lowering TGF-β level in tumor cells with high CD8+ T cells seemed to improve the outcome of the disease[59].
High PD-L1 positivity rate in various cancers was identified in 36% of TIICs based on a meta-analysis study[89], and this high PD-L1 positivity showed a good correlation with a lower risk of death and better cancer survival. Increased PD-L1 level in TIICs was also found in post-sorafenib HCC tissues compared to pre-sorafenib tissues, and these TIICs were mostly identified as CD68+ macrophages[90]. Macrophage activation in HCC has been attributed to hypoxia in the tumor microenvironment. Moreover, treatment with sorafenib has been shown to induce hypoxia in the tumor microenvironment, further inducing activation of macrophages and other ICs[64]. In addition, M1 macrophages (CD68+HLA-DR+) in HCCs can induce PD-L1 expression through IL-1β signaling[91].
TAMs were regulated by PD-L1 and osteopontin (OPN) in HCCs, and high OPN level was associated with TAMs infiltration in HCC tumor cells. High OPN expression in HCCs provides alternative activation of macrophages and facilitates the chemotactic migration of macrophages. In addition, OPN also upregulated PD-L1 expression in HCC via CSF-1 (colony stimulating factor 1)-CSF-1R (colony stimulating factor receptor 1) pathway activation in macrophages[92]. TAMs trafficking in HCCs was blocked by inhibiting CSF-1/CSF-1R leading to enhancement of ICI efficacy in HCC treatment[92].
PD-L1 expression in HBV-related HCCs may affect the activation of follicular helper T (Tfh) cells leading to impaired B cell antibody responses. Increased PD-L1 in this cohort is accompanied by reduced expression of ICOS, secretion of IL-10 and IL-2, and Tfh proliferation. The PD-L1 effect on Tfh cells increased gradually through different HCC stages, with Tfh cells from stage III patients showing a lower effectiveness in inducing naïve B cells differentiation into plasmablasts. PD-1 blockade only partially rescued Tfh functions in HCC stage I and II, but not in stage III HCC. On the other hand, treatment with recombinant PD-L1 strongly suppressed Tfh functions in all HCC stages[93]. This study clearly defined the PD-1/PD-L1 upregulation effect on Tfh cells exhaustion in HCCs.
A novel subset of protumorigenic PD-1+ B cells was identified in HCCs. These B cells expressed CD5hiCD24-/+CD27hi/+CD38dim, different to the usual CD24hiCD38hi phenotype of the peripheral regulatory B cells[94]. Upregulation of TLR4-mediated BCL-6 induced this B cell subset, while STAT-6 phosphorylation by IL-4 abolished them. This B cell subset interacted with PD-1, suppressed tumor-specific T-cell immunity, and promoted cancer growth via the IL-10 pathway[94].
NK cells which play an important role in tumor immunosurveillance, were found in low abundance in HCC tissues compared to the adjacent normal liver tissues. However, the abundance of NK cells in HCCs was associated with various immune checkpoint proteins including PD-1, PD-L1, KLRD-1, CTLA-4, and CD86. Higher abundance of NK cells resulted in a better response to sorafenib and OS of HCC patients[95].
PD-L1 expression in HCC can also be induced by monocytes. Monocytes may greatly enhance the glycolysis process at the HCC peritumoral region, inducing PD-L1 expression in these cells but attenuating cytotoxic T lymphocyte responses in HCC tumor tissues[96]. This increased glycolysis rate was enabled by the upregulation of PFKFB-3, a glycolytic enzyme, in tumor-associated monocytes by tumor-derived soluble factors such as hyaluronan fragments. Increased PFKFB-3+CD68+ cell infiltration in peritumoral HCC tissues was negatively correlated with OS[96]. In addition, PFKFB-3 also induced direct PD-L1 expression by activating the NF-κβ pathway[96]. The increase in PD-L1 and PD-L2 on monocytes (CD14+) in HCC patients was also associated with a poor prognosis[97].
Apart from its membrane-bound forms, PD-1 and PD-L1 can exist in soluble forms, sPD-1 and sPD-L1, respectively[98]. sPD-1 resulted from alternative splicing of the transmembrane domain exon 3 from the PD-1 gene. The sPD-1 level was found to be increased following PBMCs activation with anti-CD3+ and anti-CD28 monoclonal antibodies, in parallel with an increased level of full-length PD-1. This observation suggests an interplay between sPD-1 and PD-1 in the maintenance of peripheral self-tolerance and prevention of autoimmunity[99]. sPD-L1 possibly resulted from cleavage of membrane PD-L1 (mPD-L1) by matrix metalloproteinases; however, it still retained the IgV ligand-binding domain required for PD-1 interaction and subsequent inhibition of T cell activation[100,101]. sPD-L1 is mainly produced by myeloid-derived cells such as monocytes, macrophages, and DCs, but has also been found in several human cancer cell lines[100]. Increased sPD-L1 levels in blood were associated with metastasis and poor prognosis in breast cancer, diffuse large B cell lymphoma, and clear cell renal cell carcinoma (ccRCC)[100,101]. High sPD-L1 level was also associated with increased mortality risks in ccRCC and HCC[101,102].
The impact of sPD-1 levels on long-term dynamics of HBV load and HCC risk in 2903 Chinese HBV patients showed that sPD-1 levels were associated with higher viral load for more than four consecutive years and increased risk of HCC, especially in male patients. The high levels of sPD-1 and HBV load was also associated with a 6-fold increase in HCC risk[103], showing an association between high sPD-1 level and HCC development. Similarly, sPD-L1 levels were positively correlated with stages of liver cirrhosis and HCC in a cohort of 215 Caucasian HCC patients. Patients with a high sPD-L1 level had an increased risk of mortality, while those with low sPD-L1 had a better prognosis[102].
It is interesting to note that a recent study on soluble PD-1 and PD-L1 in HCC patients, concluded that sPD-1 and sPD-L1 were independent prognostic biomarkers with opposite effects in HCC, with sPD-1 level a favorable prognostic factor for HCC patients[98]. They found detectable sPD-1 in all HCC patients’ sera (n = 120), while sPD-L1 level was only detectable in two-thirds of the patients, although sPD-L1 level seems to be positively correlated with sPD-1 level[98]. Furthermore, there was no association found between sPD-L1/sPD-1 level and intratumoral expression of PD-L1 level or numbers of CD4+ and CD8+ TILs[98]. Several studies have reported that sPD-1 may suppress the PD-1/PD-L1 pathway leading to restored T cell function and enhanced antitumor immunity[104,105], which may explain the results by Chang et al[98], which showed a favorable sPD-1 level with HCC progression. More studies are needed to elucidate the role of sPD-1 and sPD-L1 in HCC development.
Since the benefit of ICIs targeting the PD-1/PD-L1 is restricted to a subset of patients, predictive biomarkers are essential for patient selection[106]. Several putative markers have been proposed with predictive potential, but the strongest proven marker to date is the expression of PD-L1 assessed by immunohistochemistry (IHC)[106].
Further study on tumor samples from dose-escalation and dose-expansion phases of the anti-PD-1 trial, the CheckMate 040[107], showed that tumoral expression of PD-1 and PD-L1 was associated with improved OS. The percentage of PD-1+ cells was higher in responders or partial responders compared to patients with stable or progressive disease, where tumor PD-L1 expression ≥ 1% was associated with improved OS. Gene analysis by RNA-seq also showed that the inflammatory signature consisting of CD274 (PD-L1), CD8A, LAG-3, and STAT-1 was associated with improved patient survival and response to anti-PD-1[108]. In a study of cytokine-induced killer (CIK) cell immunotherapy, patients with higher PD-L1 expression were those who exhibited long-term survival benefit post CIK[76].
Higher response to anti-PD-1 was observed among patients with a high intratumoral CD38+ cell proportion in the tumor microenvironment[109]. Previously it was demonstrated that TILs expressing activation marker CD38 in the tumor was correlated with patient survival, indicating that enhanced local immune activation contributes to a better prognosis for patients with HCC[110].
In line with the above studies, anti-PD-1 treatment failure was associated with the upregulation of alternative immune checkpoints that limit the antitumoral immune response[111]. Adaptive resistance to anti-PD-1 treatment was shown to correlate with the upregulation of indoleamine 2,3-dioxygenase (IDO) and alternative checkpoints for TIM-3[112,113]. In their study, Koyama et al[113] sorted T cells and tumor cells by mRNA sequencing and flow cytometry of anti-PD-1-resistant cells vs untreated tumors. TIM-3, LAG-3, and CTLA-4 were expressed at higher levels in PD-1-resistant cells, but only TIM-3 showed a significant increase. In a mouse model, resistance to PD-1 blockade was overcome by the addition of TIM-3 antibody[113]. Higher serum levels of TIM-3 have been correlated with advanced HCC stage, poor prognosis, and patient’s response to TACE[114,115]. Univariate logistic regression showed that higher serum TIM-3 values were associated with a higher probability of serum PD-L1 detection[115], which might be related to the simultaneous activation of both immune checkpoints in advanced HCCs.
Systematic interrogation of TILs is key to the development of immunotherapy and the prediction of their clinical responses in cancers[116]. In the study by Zheng et al[116], single-cell RNA-sequencing analyses of > 5,000 single T cells isolated from HCC patients showed that specific subsets such as exhausted CD8+ T cells and Tregs, with high expression of PDCD1 were preferentially enriched and potentially clonally expanded. Layilin (LAYN) was upregulated on activated CD8+ T cells and Tregs. In vitro, LAYN overexpression in primary CD8+ T cells resulted in inhibition of IFN-γ production, suggesting a regulatory function of LAYN[116].
Genomic mutations including single nucleotide polymorphisms (SNPs) have been associated with HCC risk, including predisposition to risk factors, the severity of liver disease, malignant transformation, and tumor progression[117]. Tumor mutational burden (TMB), defined as the total number of somatic mutations per megabase or the nonsynonymous mutations in tumor tissues, including replacement and insertion-deletion mutations[118], has been associated with the success of ICIs therapy. In a meta-analysis report of 2,661 patients from 8 trials (mostly of lung cancer), patients with high TMB showed significant benefits from PD-1/PD-L1 inhibition compared to patients with low TMB[118]. The significance of TMB as a biomarker in anti-PD-1/PD-L1 treatment was also reported in several independent studies[119-121].
However, this information is still lacking for HCC. The evaluation of the frequency of genomic biomarkers including the TMB in 755 patients of advanced HCC showed no significant genomic or TMB differences between responsive patients and those with progressive or stable disease. Furthermore, PD-L1 positivity was not associated with high TMB, where several patients with high positive PD-L1 were also TMB low (2-5 mutations/Mb)[122]. These data were confirmed by more recent studies showing that TMB could not predict OS and patient’s responsiveness to anti-PD-1[123-125]. Besides, HCC had low levels of microsatellite instability, a phenotype due to accumulated mutations resulting from a defect in mismatch repair[126].
PD-1 is encoded by the PDCD1 gene located on chromosome 2 which contains five exons, while PD-L1 is encoded by the CD274 gene located on chromosome 9 which contains seven exons[36]. Various genetic aberrations that can affect PD-1/PD-L1 gene expression have been identified, including SNPs, copy number alterations (CNAs), amplifications, deletions, mutations, and spliced variants (Table 1).
| Variants | Relevance | Ref. | |
| PD-1 | rs7421861 | Increases cancer risk, especially in NAFLD-HCC | [127,140] |
| rs2227981 | Reduces cancer risk | [127] | |
| rs42386439 | Low HBV viral load | [129] | |
| rs36084323 | Worse disease progression in chronic HBV, low TNF-α and IFN-γ levels | [15,131,132] | |
| rs10204525 | Disease progression in chronic HBV, high TNF-α and IFN-γ levels, longer overall survival | [131,133-135] | |
| rs11568821 | Reduces cancer risk | [23,127,136] | |
| PD-L1 | rs4143815 | Increases cancer risk | [141,143] |
| rs17718883 | Reduces cancer risk | [142] | |
| rs10815225 | Increases cancer risk | [143] | |
| rs2297136 | Increases cancer risk | [142] | |
| rs2890658 | Not associated with cancer risk | [142] | |
| PD-1 and PD-L1 | PD-1 rs11568821 and PD-L1 rs4143815 | Liver transplantation setting | [148] |
Several PD-1 gene polymorphisms have been identified, including six of the most studied PD-1 SNPs concerning various cancers. One is located in-frame rs2227982 (C>T), resulting in amino acid mutation from alanine to valine (A215V), two located upstream in the promoter region rs2227981 (C>T) and rs36084323 (G>A), two in the intron region rs7421861 (T>C) and rs11568821 (G>A), and one in the 3’UTR region [rs10204525 (G>A)][15]. A meta-analysis of these SNPs on various human cancers showed that rs7421861 polymorphism was associated with increased risk of developing cancer, while rs2227981 and rs11568821 polymorphisms were associated with overall reduced cancer risk[127]. The cancer-protective effect of PD-1 genotype can be seen in both the Asian and Caucasian populations[128].
Earlier studies have indicated the association of several PD-1 gene polymorphisms with chronic HBV liver disease progression in Chinese patients. PD-1 rs42386439 T allele, located in intron 4, which acts as a negative cis-element for gene transcription, was significantly associated with lower HBV viral load[129]. PD-1 rs36084323 SNP was associated with liver disease progression to cirrhosis and HCC in chronic HBV patients[130]. In addition, rs36084323 AA genotype in chronic HBV patients was associated with overall lower TNF-α and IFN-γ levels[131]. Higher PD-1 rs36084323 AA genotype but lower rs2227981 TT genotype frequencies were also found in chronic HBV patients compared to the spontaneously recovered control group[132]. PD-1 rs36084323 polymorphism, located in the PD-1 promoter region, may interrupt PD-1 gene activation and transcription, thus affecting T cell activation and function and altered cytokine secretion, possibly resulting in a worse prognosis of liver diseases including cancer development[15,131].
PD-1 rs10204525 polymorphism, located in the 3’UTR, was also associated with progression of HBV-related liver disease[133]. PD-1 rs10204525 GG genotype HBV patients had a higher level of TNF-α, possibly conferring a strong inhibitory effect on PD-1 function and subsequent T-cell activation[131]. A functional study on lympho
PD-1 rs11568821 variants, an intronic SNPs, affect PD-1 mRNA level by changing its binding affinity to RUNX (a PD-1 transcriptional factor), resulting in impaired PD-1 inhibitory effect and subsequent positive regulation of cytotoxic T lymphocyte activity[15,23]. As such, PD-1 rs11568821 GG genotype has been associated with decreased cancer risk[23,136]. However, a 2012 Turkish study found no significant differences in PD-1 rs11568821 genotype between HCC cases and control subjects[137]. A study performed in 2015 also found no significant differences in the genotype distributions of PD-1 rs11568821 and rs41386439 in chronic HBV patients compared to the spontaneously recovered control group[138], indicating the possibility of lack of association between PD-1 gene polymorphisms and HBV infection susceptibility and HBV-related HCC progression in Turkish patients. These findings were confirmed in a more recent study investigating three PD-1 gene polymorphisms and HCC progression; again no significant distribution of rs36084323, rs2227981, and rs10204525 genotypes were observed in HCC cases in Turkish patients[139].
PD-1 rs7421861 SNP, located in intron 1, may disrupt the putative alternative splice site and promote full-length transcript expression instead[15,140]. An earlier study showed no association between rs7421861 variants and cancer risks[136]. However, a more recent study in NAFLD-HCC European cohorts revealed that PD-1 rs7421861 allele A was significantly associated with HCC, independent of age, sex, cirrhosis, and diabetes. Furthermore, allele A of rs10204525 in this cohort was also associated with increased risk of NAFLD-HCC, especially in female patients[140]. These findings revealed the association between PD-1 gene polymorphisms not only in HBV-related HCC but also in NAFLD-HCC.
Similar to PD-1 polymorphisms, several PD-L1 polymorphisms, rs4143815 (C>G), rs2890658 (A>C), rs2297136 (C>T), rs17718883 (C>G), and rs10815225 (G>C), have been studied in association with the development of various cancers. A meta-analysis showed that PD-L1 rs4143815 polymorphism was associated with protection against various cancers[127]. However, different studies have shown that carriers of PD-L1 rs4143815 GG genotype have a higher risk of developing gastric adenocarcinoma[44] and HCC[141].
A 2018 study examining several PD-L1 variants in 225 Chinese HCC patients confirmed that PD-L1 rs4143815 GG and rs2297136 TT genotypes were associated with increased HCC risks. On the other hand, PD-L1 rs17718883 CG+GG genotypes reduced the risk of HCC occurrence, while rs2890658 SNPs were not associated with HCC risks[142]. PD-L1 rs4143815 polymorphisms, located in the 3’UTR, resulted in elevated PD-L1 protein expression via disruption of miR-570 binding to PD-L1 mRNA[44]. This polymorphism was found in disequilibrium with PD-L1 rs10815225 polymorphism in the PD-L1 promoter region, which serves as a binding site for Sp1 transcription factor (SP1). The G allele of rs10815225 bound more effectively to SP1 resulting in an increased level of PD-L1 mRNA level. PD-L1 rs10815225 GG genotype was associated with increased risk of gastric cancer, and the haplotype of rs10815225 and rs4143815 polymorphisms were found to greatly increase gastric cancer risk[143].
Aside from the association of singular PD-1 and/or PD-L1 polymorphisms and cancer progression, the interaction between multiple SNPs of PD-1/PD-L1 with various genes has also been shown to be associated with HBV infection and related cancer development. A combination of PD-1 rs10204525 GG and rs2227982 CC genotypes in Chinese patients has been shown to result in better protection from HBV infection and lower HBV viral load in asymptomatic carriers[144]. On the other hand, the interactions between PD-1 rs41386349 and rs6710479 with TIM-2 rs246871 variant were shown to affect susceptibility to chronic HBV infection and may influence later hepatocarcinogenesis[145]. Furthermore, PD-1 rs10204525 AA genotype and TIM-3 rs10053538 GT or TT genotypes were more frequently found in HBV-associated HCC patients[134].
Similarly, PD-1 rs11568821 synergy with CTLA-4 49AG:CT60 A:A haplotype has been associated with an increased risk of primary biliary cancer (PBC)[146]. Interactions between PD-L1 rs10815225 and PD-1 rs7421861 polymorphisms were also associated with the development and outcome of ccRCC[147]. No such observation has been reported so far for HCC, although the interaction between PD-L1 rs4143815 and PD-1 rs11568821 variants was found to be important in the liver transplantation setting. PD-L1 rs4143815 was associated with different PD-L1 expression on donor hepatic DCs upon IFN-γ stimulation, and PD-1 rs11568821 A allele recipients receiving donors from PD-L1 rs4143815 GG genotypes had a higher risk for late acute rejection after liver transplantation[148].
Aberrant PD-L1 expression may be caused by PD-L1 genetic alterations affecting the PD-L1 locus. PD-L1 CNAs affecting either the focal regions, chromosome 9p24.1, or the whole chromosome 9 have been identified in 22 major cancer types, resulting in changes in PD-L1 mRNA expression[74]. These PD-L1 variants significantly affected PD-L1 expression, and a higher PD-L1 expression was observed in cancer patients with altered PD-L1 variants, with PD-L1 gene fusion and amplification showing the highest increase in PD-L1 expression[149]. PD-L1 copy number gains and deletions were associated with higher mutational loads, while PD-L1 amplifications and deletions of core regions were associated with a more dismal cancer prognosis[150]. PD-L1 deletions were more frequent in solid tumors, especially in melanoma and NSCLC where more than half of the tumors had PD-L1 deletions[14,150].
In liver cancer, PD-L1 copy number gains were associated with increased JAK-2 mRNA expression[150]. It is interesting to note that JAK-2 and PD-L1 encoding genes are both in chromosome 9p, with both having high alternation rates. JAK-2 amplification and mutation which increased JAK-2 and its downstream STAT effectors expression has also been shown to upregulate PD-L1 expression[41,51]. A 2018 study on Chinese HCC patients revealed a significant proportion of chromosome 9p24.1 polysomy (16%-31%) and amplification (7%-15%). Furthermore, these PD-L1 genetic alterations were significantly associated with upregulation of both PD-L1 and PD-L2 expression, high infiltration of PD-1+ ICs, and overall poor cancer survival[151].
A high expression level of PD-L1 truncated form was first identified in a head and neck squamous cell carcinoma (HNSCC), as a result of the human papillomavirus integration into the PD-L1 locus upstream of the transmembrane domain-encoding region[152]. A follow-up study on 33 cancer types and human cancer cell lines identified additional PD-L1 truncated forms in 20 cancers and human cancer cell lines, characterized by exon 4 enrichment. This truncated PD-L1 was preferentially secreted but still maintained its binding ability to PD-1 and served as a negative regulator for T cell activation by inhibiting IL-2 and IFN-γ secretion[153].
A different secreted splice variant of PD-L1 (secPD-L1) has been identified in various tumor types and malignant cell lines. This secPD-L1 contains the first exons of PD-L1 but lacks exon 5 and cannot splice into the transmembrane domain. However, due to its 18 amino acids tail containing tyrosine, this variant can homodimerize and inhibit T-cell proliferation and IFN-γ production, retaining its immunosuppressive effect[154]. PD-L1 secreted (secPD-L1) splice variants lacking the transmembrane domain were identified in NSCLC patients. These secPD-L1 variants mediated resistance to PD-L1 blockade therapy by acting as a decoy to PD-L1 antibody, which was found to be constrained by treatment with anti-PD-1 antibody[155].
HCC is a vast heterogeneous malignancy, either within an individual (intratumoral heterogeneity) or among subjects (intertumoral heterogeneity). Besides its various underlying etiologies, long-term development affects its different profiles and distinct progression of the disease. In the ‘-omics’ era with abundant studies of global gene screening and genetic array, HCC classifications have shifted from histology[156] to molecular-typing-based. HCC (sub)types can arise from dysregulations of various oncogenic pathways and/or different cells of origin[157]. Starting in the 2000s, several studies demonstrated that HCCs could be categorized into specific subclasses based on their distinct molecular signatures (Table 2).
| Ref. | Subclass | Marker | Main methods |
| Yamashita et al[158] | Type A-D | EpCAM, AFP | cDNA microarray, IHC |
| Lee et al[157,159] | Cluster A and B, and 3 distinct phenotypes | Sets of survival genes, AFP | Integrated cDNA microarray in human specimens and animal models |
| Boyault et al[160] | Groups G1-G6 | Tumor heterozygosity, gene mutations, methylation, and HBV DNA | cDNA microarray, IHC, qRT-PCR |
| Hoshida et al[161] | Subclasses S1-S3 | Cellular differentiation, serum AFP, signaling pathways | Meta-analysis of gene expression profiles datasets, IHC |
| Sia et al[163] | Immune classes active and adaptive | Immune response genes | cDNA microarray, IHC, TCGA analysis |
| Zhang et al[164] | Immunophenotypic subtypes 1-3 | Immune response genes and antitumoral immunity | Whole-exome and RNA sequencing, mass spectrometry-based proteomics and metabolomics, CyTOF, single-cell analysis |
HCC classification based on progenitor cells was identified using the CSC marker EpCAM via cDNA microarray analysis and IHC analysis. EpCAM+ HCC displayed the features of hepatic progenitor cell markers (e.g., CK-19, c-Kit, EpCAM, and activated Wnt/β-catenin), whereas EpCAM- HCC displayed genes with features of mature hepatocytes. The two groups were then classified into four subgroups with a high or low level of AFP. Kaplan-Meier analysis showed that EpCAM+AFP+ (type B) and EpCAM−AFP+ (type C) HCCs were correlated with poor prognosis, whereas EpCAM−AFP− (type D) HCCs was correlated with an intermediate prognosis. Interestingly, EpCAM+AFP− (type A) HCCs was correlated with a good prognosis[158].
In 2004, using cDNA microarray, the Thorgeirsson group classified HCCs into clusters A and B. Cluster A, in combination with low serum AFP, was associated with low survival[159]. Using integrated gene expression data between human specimens and animal models, the group further classified HCC into three distinct biological phenotypes with significant differences in clinical outcome. The hepatoblast (HB) subtype belonged to cluster A, while the hepatocyte (HC) subtype belonged to both cluster A and B. Differential expression of around 1500 genes showed exclusive differences between HB and HC subtypes, especially in the pathway of the JUN-FOS heterodimers–AP-1 complex. Individuals with HCC who shared a gene expression pattern with HB subtype had a poor prognosis[157].
Also using global transcriptome analysis, Boyault et al[160] classified HCCs into subgroups G1 to G6, each associated with specific clinical and genetic characteristics, especially tumor heterozygosity, gene mutations, promoter DNA methylation, and HBV DNA copy number. The subgroup G1 and G2 were related to activation of the AKT pathway, while G5 and G6 to β-catenin mutations leading to Wnt pathway activation. In brief, the characterization of G1 was related to HBV low copy number and fetal liver gene expression, G2 was associated with HBV high copy number, PIK3CA, and TP53 mutation, and G3 was associated with TP53 mutation and overexpression of cell-cycle genes. Subgroup G4 heterogeneously comprised TCF-1 mutated hepatocellular adenomas and carcinomas. Subgroup G5 was associated with stress and immune response, and G6 with amino acid metabolism and satellite nodules[160].
In 2009, Hoshida et al[161] performed a meta-analysis of gene expression profiles from 8 independent datasets, validated by immunohistostaining of clinical specimens. Here, they classified HCC into 3 robust subclasses S1 to S3 which were correlated with clinical parameters. Subclass S1 was characterized by stromal cells with TGF-β activation, S2 by stem-angiogenic cells with MYC and AKT activation, and S3 by mature hepatocyte differentiation. The subclass S2 with EpCAM positive cells, categorized as stem cells-like and hepatoblast-like HCC, was also highly aggressive[161], as noted in previous studies[157,159]. A more recent study from the group showed that HCC S1 and S2 were found in various established cell lines, thus indicating the appropriate use of in vitro models to evaluate the effectiveness of subtype-specific drug response. For example, (+)-JQ1, an anti-MYC compound, was highly sensitive in S2 cell lines HepG2 and Huh7[162].
From an immunological point of view, several studies have categorized HCC based on the tumor microenvironment via the infiltration of ICs. By using gene expression profiles from the tumor, stromal, and ICs, followed by immunohistochemical analysis, Sia et al[163] identified two robust HCC immune classes. HCC data in The Cancer Genome Atlas (TCGA) were analyzed to correlate the ICs gene expression profiles with chromosomal aberrations and mutations. The active immune response subtype (approximately 65%) was characterized by overexpression of adaptive immune response genes, while the exhausted immune response subtype (approximately 35%) was characterized by the presence of immunosuppressive signals (e.g., TGF-β, M2 macrophages)[163]. This finding indicated the susceptibility of HCC upon immune modulation therapy against T cells.
A recent paper by Zhang et al[164] further confirmed the heterogeneity of the HCC microenvironment in a complex and integrated multiomics analysis. The immune status of the HCC microenvironment was relatively less heterogeneous, thus rendering the significance of HCC immunophenotypic classification[164]. Using whole-exome sequencing, RNA sequencing, mass spectrometry-based proteomics and metabo
The population of cancer stem cells (CSCs), also known as tumor-initiating cells (TICs) or side population (SP), has been recognized as one if not the most important cells in cancer. They are responsible for the initiation and the maintenance of various types of cancer, while also contributing to tumor resistance during treatment.
In HCC, various protein markers, including the CD133/Prom-1, CD90/Thy-1, EpCAM, CD24, CD13/ANPEP, ABCG2/BCRP, aldehyde dehydrogenase/ALDH, CD44, and many more, have been proposed to define and to isolate the CSCs from the tumor ‘bulk’ populations[165]. Furthermore, the combinations of using two or more markers added to the variations of CSC populations. In fact, until now there is no consensus on the use of CSC markers for HCC. It is important to note that each CSC population had its distinct characteristics[166].
The origin of hepatic CSCs is various, thus further increasing the vast cellular variations within the tumor. In the beginning, it was thought that CSCs were derived exclusively from oncogenic transformation in normal stem/progenitor cells[167]. Due to the complexity and physiology of the liver, however, the origin of hepatic CSCs can be traced into multiple lineages of liver maturation. In 2013, the Thorgeirsson group provided strong direct evidence on various sources of hepatic CSCs. Upon controlled oncogenic transformation with H-ras and SV40LT, they noted that adult hepatocytes, hepatoblasts, and hepatic progenitor cells (HPC) could be oncogenically reprogra
Besides its main capacity to induce tumor, the main feature of the CSC population is their resistance to various treatments. The CSC populations (e.g., CD133, CD13, EpCAM) are highly resistant to chemotherapy and radiotherapy[171-175], and sorafenib[176-181]. On the other hand, the preferential expression of CSCs and their response to immune therapy is still unclear.
Immunotherapy is a rather new field in cancer study. It is still unclear whether the success of ICIs has any association with HCC cellular hierarchy. CSCs have been demonstrated to have a preferential role in therapy resistance, including in chemotherapy, radiotherapy, and molecular therapy, thus it is reasonable to investigate the potential relevance of CSCs against immunotherapy. CSC tumoral heterogeneity might have a close association with the intrinsic PD-L1 properties in cancer cells.
Until now, contrasting studies showed the association of PD-L1 to tumorigenesis and CSCs (Table 3). In breast and colorectal cancer, PD-L1 was positively correlated with CSC populations, regardless of their phenotypic markers. Using flow cytometry analysis, authors have shown that PD-L1 expression was higher in CSCs of both cancers compared to non-stem like cancer cells. High PD-L1 expression was noted in CSC subpopulations expressing CD44+, ALDH+, CD44hiCD24lo, EpCAM+CD90hi, and EpCAM+CD44hiCD24lo[182-184]. For example, in CSC EpCAM+CD44hiCD24lo, PD-L1 was overexpressed up to 3-fold compared to more differentiated-like cancer cells. Functional in vitro and in vivo assays also showed higher stemness of PD-L1hi as compared to PD-L1lo cells. Among the different pathways examined, PD-L1 expression on CSCs was partly dependent on Notch and/or PI3K/AKT pathway activation[184].
| Malignancy | Stemness/CSC markers | Method | Relevance | Ref. |
| HCC | CD133+ interaction with lymphatic endothelial cells | Co-culture | Upregulates PD-L1 | [196] |
| HCC | CK-19, SALL-4 | IHC | Positively associated with PD-L1 | [194] |
| HCC | EpCAM | IHC | Negatively associated with PD-L1 | [194] |
| Breast cancer | CD44hiCD24lo | Flow cytometry | High PD-L1 expression | [182] |
| Breast cancer | EpCAM+CD44hi CD24lo | Flow cytometry | High PD-L1 expression | [184] |
| Breast cancer | EpCAM+CD90hi | Flow cytometry | High PD-L1 expression | [184] |
| Breast cancer | EpCAM+CD44hiCD24lo | Flow cytometry, IF | High PD-L1 expression; nuclear PD-L1 | [185] |
| Breast cancer | Stemness score | mRNA from TCGA | Significant correlation to PD-L1 | [185] |
| Breast cancer | ALDH+, CD44hi | Flow cytometry | High PD-L1 expression | [183] |
| Colorectal cancer | CD44hiCD133hi | Flow cytometry | High PD-L1 expression | [182] |
| Colorectal cancer | CD133+CD44+ | Flow cytometry | High PD-L1 expression | [189] |
| Ovarian cancer | ALDH | qRT-PCR, IF | High PD-L1 expression | [188] |
| HNSCC | CD44+ | Flow cytometry, qRT-PCR, RNA in situ hybridization | High PD-L1 expression | [191] |
| Lung adenocarcinoma | CD44 | mRNA in TIMER datasets | Positively associated with PD-L1 | [186] |
| NSCLC | ALDH | mRNA from TCGA, IHC | Negatively associated with PD-L1 | [192] |
| Cholangiocarcinoma | ALDH | Sorting of PD-L1 cells | High ALDH+ in PD-L1lo | [193] |
| Pancreatic cancer | CD44+CD133+ | IF | Positively associated with PD-L1 | [190] |
Analysis of stemness score from TCGA data of 530 breast cancer patients on PD-L1 expression and CSCs, also indicated a strong association between PD-L1 expression and stem-like cells[185]. EpCAM+CD44hiCD24lo CSCs had a higher level of PD-L1 compared to their differentiated counterparts (EpCAMlo/negCD44loCD24hi). Immunofluorescence results also confirmed the higher level of PD-L1 expression in CSCs compared to the more-differentiated breast cancer cells. Interestingly, in addition to membranous PD-L1 there was also a PD-L1 nuclear fraction in CSCs. PD-L1 knockdown on the expression of stem-related molecules suggests a direct role for this molecule in CSC maintenance[185]. A positive correlation between mRNA expression of CD44 and PD-L1 was observed in lung adenocarcinoma using Tumor Immune Estimation Resource (TIMER) datasets, which was validated using IHC[186].
In gastric cancer, a CSC cell line, the NCC-S1M, overexpressed PD-L1 compared to normal gastric tissue. Furthermore, anti-PD-1 treatment suppressed in vivo growth of CSC-like cell allografts in syngeneic mice. PD-L1 was controlled by transcriptional factor Smad-4[187]. In ovarian cancer, in both in vitro model and in vivo mouse model, higher expression of PD-L1 was observed in CSCs (ALDH+) than in non-CSC cells[188]. In colorectal and pancreatic cancer, CD44hiCD133hi CSCs also expressed high PD-L1[182,189,190]. Overexpression of PD-L1 promoted colorectal CSCs self-renewal in vitro and in vivo, and increased its chemoresistance[189]. In HNSCC, PD-L1 was also highly expressed in CD44+ cells[191].
However, in NSCLC and cholangiocarcinoma, the expression of PD-L1 was inversely associated with ALDH-expressing cells[192,193]. mRNA analysis of TCGA data showed that PD-L1 expression was negatively correlated with ALDH-1 expression in adenocarcinoma, also observed by IHC[192]. In cholangiocarcinoma, PD-L1lo cells isolated from cell lines were highly tumorigenic compared to PD-L1hi cells. These cells had high ALDH activity, reduced reactive oxygen species production, and were in a dormant state of the cell cycle. Furthermore, in clinical specimens, the low expression of PD-L1 was well-correlated with poor prognosis of patients[193].
With regard to HCC, to date, information on the direct association between PD-L1 and hepatic CSCs is still very limited. Nishida et al[194] analyzed 154 HCCs and their noncancerous liver tissue counterparts for the expression of PD-L1 and stemness. They showed that PD-L1 was frequently expressed in stem cell features of HCC. The expression of PD-L1 was associated with aggressive high-grade tumors. Using IHC, the presence of PD-L1 was positively associated with cytokeratin 19 (CK-19) and Sal-like protein 4 (SALL-4), but not with EpCAM[194]. Previously, it was shown that SALL-4 regulated the stemness of EpCAM-positive HCC. The activation of SALL-4 enhanced CSC spheroid formation and invasion capacities and upregulated the expression of CK-19, EpCAM, and CD44 in cell lines[195].
Another study showed that HCC CSC CD133+ preferentially interacts with lymphatic endothelial cells. Lymphatic endothelial cells create a CSC-microenvironment through direct contact with CSCs. Co-culture of CD133+ cells with lymphatic endothelial cells stimulated IL-17A expression that further promoted the immune escape of CD133+ cells through the upregulation of PD-L1. These data showed that the tumor niche promoted the self-renewal and immune escape of CSCs via PD-L1[196].
The presence of PD-L1 in the circulating tumor cells (CTCs) was demonstrated to be a prognostic and predictive biomarker for HCC patients. CTCs expressed various phenotypic profiles such as EMT and stem cell markers. Phenotype profiling of HCC CTCs in patients was performed by CTCs isolation and enrichment with an HCC-specific antibody cocktail including CSC marker EpCAM, and stained with antibodies against pan-cytokeratin (CK), CD45, and PD-L1, together with DAPI. Survival analysis showed that patients with PD-L1+ CTCs (DAPI+CK+PD-L1+CD45-) had significantly worse OS compared to patients without PD-L1+ CTCs (DAPI+CK+PD-L1-CD45-)[197].
How the PD-L1 pathway is involved in the tumorigenicity of hepatic CSCs is still unclear. PD-L1 is known to be transcriptionally upregulated upon EMT in the cancer microenvironment. By overexpressing and knocking-down the PD-L1 in sorafenib-resistant cells, PD-L1 expression promoted EMT and cellular migratory and invasive abilities via the PI3K/Akt pathway[58].
The tumor microenvironment (Figure 2) is crucial for the self-renewal and maintenance of hepatic stem cells, which may lead to the development of HCC[196]. Immunologic mechanisms such as chronic inflammation due to chronic viral hepatitis or metabolic diseases play a crucial role in the initiation, development, and progression of HCC. Thus, it is important to understand the underlying mechanisms shaping the unique HCC tumor microenvironment[198].
Many of the above studies showed that PD-L1 was preferably expressed in CSCs with the phenotypic marker CD44[182-185,191]. CD44 was previously shown to function as an independent marker of hepatic CSCs[199-203]. CD44 expression was also associated with the EMT phenotype in HCC cell lines, and knocking down CD44 resulted in the switch back to the mesenchymal-epithelial-transition (MET)[199]. CD44 is a multidomain, transmembrane platform, a major adhesion molecule of the extracellular matrix. It is a signaling molecule that connects the microenvironment with growth factor and cytokine signals and regulates a variety of gene expression levels related to cell-matrix adhesion, cell migration, proliferation, differentiation, and survival[204,205]. Its ligation to growth factors was demonstrated to be able to induce partial or full EMT[206]. Our previous study demonstrated that the inhibition of hyaluronic acid in the HCC microenvironment resulted in the decreased expression of CD44 in a transgenic mouse model and HCC cell lines[207].
CSC CD44+ cells were less immunogenic than CD44- cells when cultured with autologous CD8+ TILs. IFN-γ treatment preferentially induced even further PD-L1 expression on CD44+ cells and was associated with enhanced IFN-γ receptor expression and phosphorylation of STAT-1. Long-lived CD44+ TICs can selectively evade host immune responses[191]. A subsequent study showed that EMT preferably enriched the PD-L1 in CSCs compared to the general cancer population through the EMT/β-catenin/STT-3/PD-L1 signaling axis. EMT transcriptionally induced N-glycosyltransferase of STT-3 through β-catenin, and subsequent STT-3-dependent PD-L1 N-glycosylation stabilizes and upregulates PD-L1[208].
Several studies also hinted at the central role of P13K/AKT and mTOR molecular pathways in the biology of PD-L1-expressing CSCs[184,185,209]. The inhibition of STAT-1 and STAT-3, AKT downstream transcription factors, downregulated PD-L1 expression[210]. In HCC, genetic alterations involved in the PI3K/AKT pathway were significantly associated with PD-L1 positivity whereas mutations in the β-catenin pathway were inversely correlated with PD-L1 in HCC. Comparisons in the TCGA cohort showed that mutations in the PI3K/AKT pathway could positively affect the expression of PD-L1, while mutations in the β-catenin pathway were related to the absence of PD-L1 expression[194].
HCC is an immunologic cancer; therefore, immunotherapy is one of the potential treatment methods. On the other hand, HCC is vastly heterogeneous which might hamper the efficacy of therapy, especially in patients who cannot receive surgical interventions. Genetic variations in PD-1 and PD-L1 genes have been associated with the progression of liver disease. Even though its relevance in anti-PD-1/PD-L1 therapy is still lacking, these variations could be useful in determining patient acceptance of HCC treatment, also for sorafenib.
Targeting immune checkpoint ligands present in tumor cells and the microenvironment (e.g., PD-L1) can be an interesting approach. Anti-PD-L1 inhibits both the cell’s constitutive expression and inductive stimulation caused by binding of the ligand to the immunomodulatory molecule (e.g., PD-1/PD-L1). It can be given in combination with other molecular targeted therapies, which increase the ‘targeting’ of the therapy, as had been demonstrated in the co-treatment approach of atezolizumab (anti-PD-L1) plus bevacizumab (targeting VEGF)[28].
High expression of PD-L1 protein in tumoral cells and/or tumor microenvironment is indicated as a strong biomarker for the success of the ICIs against PD-1/PD-L1. Several important findings, however, need to be elucidated to measure the real efficacy of the treatment. Technical variations among laboratories may influence the results. For example, the use of the correct antibody for PD-L1 detection. A recent study showed that PD-L1 was expressed in inflammatory cells within the HCC tissue and cirrhotic parenchyma, but not in neoplastic cells[211]. Upon comparison of several anti-PD-L1 clones, the authors did not find PD-L1 immunoreactivity in both neoplastic and normal hepatocytes[211]. Different clones of anti-PD-L1 give different results thus affecting its efficiency and detection results[211-214].
Another important matter that must be considered is the use of the proper marker to characterize the hepatic CSCs phenotypes. Susceptibility and resistance to treatments have been widely attributed to cellular heterogeneity and hepatic CSCs in HCC. PD-L1 expression was positively correlated with EpCAM, CD44, and CD133 in breast and colon cancer, but it was negatively correlated with ALDH in cholangiocarcinoma. These four markers are considered CSCs markers in HCC. In vitro, it is beneficial to perform isolation of different (sub)populations from a single HCC cell line, even by single-cell sequencing, to define the preference of the expression of PD-L1. Data from one cell line should be compared with others to comprise HCC multiple classifications. The use of animal and various in vitro models is also crucial to study the intrinsic PD-L1 in its correlation with hepatic CSCs and cellular heterogeneity. A recently described orthotopic HCC mouse inoculated with PD-L1-expressing liver cells could give more information[215].
In summary, the study on PD-1/PD-L1 immunotherapy could be an emerging and promising approach for HCC therapy. However, the understanding of cancer heterogeneity must be clarified for better selection of patients who are eligible to receive treatment. Comprehensive translational scientific information from cell and animal models, and clinical samples will help the progress of the development and application of immunotherapy in the future.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Philips CA, Zhang L S-Editor: Yan JP L-Editor: Webster JR P-Editor: Xing YX
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55826] [Article Influence: 7975.1] [Reference Citation Analysis (132)] |
| 2. | El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2881] [Cited by in RCA: 3088] [Article Influence: 220.6] [Reference Citation Analysis (0)] |
| 3. | Sangiovanni A, Prati GM, Fasani P, Ronchi G, Romeo R, Manini M, Del Ninno E, Morabito A, Colombo M. The natural history of compensated cirrhosis due to hepatitis C virus: A 17-year cohort study of 214 patients. Hepatology. 2006;43:1303-1310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 433] [Cited by in RCA: 444] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 4. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2508] [Article Influence: 192.9] [Reference Citation Analysis (2)] |
| 5. | Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology. 2019;156:477-491.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 754] [Cited by in RCA: 1216] [Article Influence: 202.7] [Reference Citation Analysis (1)] |
| 6. | Plaz Torres MC, Bodini G, Furnari M, Marabotto E, Zentilin P, Strazzabosco M, Giannini EG. Surveillance for Hepatocellular Carcinoma in Patients with Non-Alcoholic Fatty Liver Disease: Universal or Selective? Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 7. | European Association For The Study Of The Liver. European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4059] [Cited by in RCA: 4520] [Article Influence: 347.7] [Reference Citation Analysis (2)] |
| 8. | European Association for the Study of the Liver. European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5593] [Cited by in RCA: 6059] [Article Influence: 865.6] [Reference Citation Analysis (3)] |
| 9. | Llovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2645] [Cited by in RCA: 2876] [Article Influence: 110.6] [Reference Citation Analysis (1)] |
| 10. | Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Häussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J; SHARP Investigators Study Group. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9016] [Cited by in RCA: 10268] [Article Influence: 604.0] [Reference Citation Analysis (2)] |
| 11. | Chen S, Cao Q, Wen W, Wang H. Targeted therapy for hepatocellular carcinoma: Challenges and opportunities. Cancer Lett. 2019;460:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 178] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 12. | Greten TF, Lai CW, Li G, Staveley-O'Carroll KF. Targeted and Immune-Based Therapies for Hepatocellular Carcinoma. Gastroenterology. 2019;156:510-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 200] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 13. | Kole C, Charalampakis N, Tsakatikas S, Vailas M, Moris D, Gkotsis E, Kykalos S, Karamouzis MV, Schizas D. Immunotherapy for Hepatocellular Carcinoma: A 2021 Update. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 14. | Hudson K, Cross N, Jordan-Mahy N, Leyland R. The Extrinsic and Intrinsic Roles of PD-L1 and Its Receptor PD-1: Implications for Immunotherapy Treatment. Front Immunol. 2020;11:568931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 147] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 15. | Salmaninejad A, Khoramshahi V, Azani A, Soltaninejad E, Aslani S, Zamani MR, Zal M, Nesaei A, Hosseini SM. PD-1 and cancer: molecular mechanisms and polymorphisms. Immunogenetics. 2018;70:73-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 16. | Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27:409-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 601] [Article Influence: 60.1] [Reference Citation Analysis (0)] |
| 17. | El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, Kim TY, Choo SP, Trojan J, Welling TH Rd, Meyer T, Kang YK, Yeo W, Chopra A, Anderson J, Dela Cruz C, Lang L, Neely J, Tang H, Dastani HB, Melero I. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492-2502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3278] [Cited by in RCA: 3310] [Article Influence: 413.8] [Reference Citation Analysis (1)] |
| 18. | Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443-2454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8900] [Cited by in RCA: 9910] [Article Influence: 762.3] [Reference Citation Analysis (0)] |
| 19. | Zhu AX, Finn RS, Edeline J, Cattan S, Ogasawara S, Palmer D, Verslype C, Zagonel V, Fartoux L, Vogel A, Sarker D, Verset G, Chan SL, Knox J, Daniele B, Webber AL, Ebbinghaus SW, Ma J, Siegel AB, Cheng AL, Kudo M; KEYNOTE-224 investigators. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19:940-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1184] [Cited by in RCA: 1897] [Article Influence: 271.0] [Reference Citation Analysis (0)] |
| 20. | Sangro B, Gomez-Martin C, de la Mata M, Iñarrairaegui M, Garralda E, Barrera P, Riezu-Boj JI, Larrea E, Alfaro C, Sarobe P, Lasarte JJ, Pérez-Gracia JL, Melero I, Prieto J. A clinical trial of CTLA-4 blockade with tremelimumab in patients with hepatocellular carcinoma and chronic hepatitis C. J Hepatol. 2013;59:81-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 634] [Cited by in RCA: 750] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 21. | Duffy AG, Ulahannan SV, Makorova-Rusher O, Rahma O, Wedemeyer H, Pratt D, Davis JL, Hughes MS, Heller T, ElGindi M, Uppala A, Korangy F, Kleiner DE, Figg WD, Venzon D, Steinberg SM, Venkatesan AM, Krishnasamy V, Abi-Jaoudeh N, Levy E, Wood BJ, Greten TF. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma. J Hepatol. 2017;66:545-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 454] [Cited by in RCA: 640] [Article Influence: 80.0] [Reference Citation Analysis (0)] |
| 22. | Roderburg C, Berres ML, Wree A, Loosen SH, Luedde T, Trautwein C. Excellent Response to Anti-PD-1 Therapy in a Patient with Hepatocellular Carcinoma Intolerant to Sorafenib. Visc Med. 2019;35:43-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 23. | Zhang J, Zhao T, Xu C, Huang J, Yu H. The association between polymorphisms in the PDCD1 gene and the risk of cancer: A PRISMA-compliant meta-analysis. Medicine (Baltimore). 2016;95:e4423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Herbst RS, Giaccone G, de Marinis F, Reinmuth N, Vergnenegre A, Barrios CH, Morise M, Felip E, Andric Z, Geater S, Özgüroğlu M, Zou W, Sandler A, Enquist I, Komatsubara K, Deng Y, Kuriki H, Wen X, McCleland M, Mocci S, Jassem J, Spigel DR. Atezolizumab for First-Line Treatment of PD-L1-Selected Patients with NSCLC. N Engl J Med. 2020;383:1328-1339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 604] [Cited by in RCA: 1094] [Article Influence: 218.8] [Reference Citation Analysis (0)] |
| 25. | Horn L, Gettinger SN, Gordon MS, Herbst RS, Gandhi L, Felip E, Sequist LV, Spigel DR, Antonia SJ, Balmanoukian A, Cassier PA, Liu B, Kowanetz M, O'Hear C, Fassò M, Grossman W, Sandler A, Soria JC. Safety and clinical activity of atezolizumab monotherapy in metastatic non-small-cell lung cancer: final results from a phase I study. Eur J Cancer. 2018;101:201-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 26. | Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3965] [Cited by in RCA: 4183] [Article Influence: 380.3] [Reference Citation Analysis (0)] |
| 27. | Lee M, Ryoo BY, Hsu CH, Numata K, Stein S, Verret W, Hack S, Spahn J, Liu B, Abdullah H, He R, Lee KH. LBA39 - Randomised efficacy and safety results for atezolizumab (Atezo) + bevacizumab (Bev) in patients (pts) with previously untreated, unresectable Hepatocellular Carcinoma (HCC). Ann Oncol. 2019;30:v875. [RCA] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 28. | Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, Kudo M, Breder V, Merle P, Kaseb AO, Li D, Verret W, Xu DZ, Hernandez S, Liu J, Huang C, Mulla S, Wang Y, Lim HY, Zhu AX, Cheng AL; IMbrave150 Investigators. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N Engl J Med. 2020;382:1894-1905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2542] [Cited by in RCA: 4696] [Article Influence: 939.2] [Reference Citation Analysis (2)] |
| 29. | Sung PS, Jang JW, Lee J, Lee SK, Lee HL, Yang H, Nam HC, Lee SW, Bae SH, Choi JY, Han NI, Yoon SK. Real-World Outcomes of Nivolumab in Patients With Unresectable Hepatocellular Carcinoma in an Endemic Area of Hepatitis B Virus Infection. Front Oncol. 2020;10:1043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 30. | Siu EH, Chan AW, Chong CC, Chan SL, Lo KW, Cheung ST. Treatment of advanced hepatocellular carcinoma: immunotherapy from checkpoint blockade to potential of cellular treatment. Transl Gastroenterol Hepatol. 2018;3:89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 31. | Staron MM, Gray SM, Marshall HD, Parish IA, Chen JH, Perry CJ, Cui G, Li MO, Kaech SM. The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8(+) T cells during chronic infection. Immunity. 2014;41:802-814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 298] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 32. | Arasanz H, Gato-Cañas M, Zuazo M, Ibañez-Vea M, Breckpot K, Kochan G, Escors D. PD1 signal transduction pathways in T cells. Oncotarget. 2017;8:51936-51945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 192] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 33. | Chemnitz JM, Parry RV, Nichols KE, June CH, Riley JL. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol. 2004;173:945-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 775] [Cited by in RCA: 889] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 34. | Mizuno R, Sugiura D, Shimizu K, Maruhashi T, Watada M, Okazaki IM, Okazaki T. PD-1 Primarily Targets TCR Signal in the Inhibition of Functional T Cell Activation. Front Immunol. 2019;10:630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 35. | Sheppard KA, Fitz LJ, Lee JM, Benander C, George JA, Wooters J, Qiu Y, Jussif JM, Carter LL, Wood CR, Chaudhary D. PD-1 inhibits T-cell receptor induced phosphorylation of the ZAP70/CD3zeta signalosome and downstream signaling to PKCtheta. FEBS Lett. 2004;574:37-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 497] [Cited by in RCA: 615] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 36. | Dermani FK, Samadi P, Rahmani G, Kohlan AK, Najafi R. PD-1/PD-L1 immune checkpoint: Potential target for cancer therapy. J Cell Physiol. 2019;234:1313-1325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 333] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 37. | Lee SJ, Jang BC, Lee SW, Yang YI, Suh SI, Park YM, Oh S, Shin JG, Yao S, Chen L, Choi IH. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-γgamma-induced upregulation of B7-H1 (CD274). FEBS Lett. 2006;580:755-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 385] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 38. | Moreno-Cubero E, Larrubia JR. Specific CD8+ T cell response immunotherapy for hepatocellular carcinoma and viral hepatitis. World J Gastroenterol. 2016;22:6469-6483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 38] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 39. | Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, Iwai Y, Long AJ, Brown JA, Nunes R, Greenfield EA, Bourque K, Boussiotis VA, Carter LL, Carreno BM, Malenkovich N, Nishimura H, Okazaki T, Honjo T, Sharpe AH, Freeman GJ. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2:261-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2021] [Cited by in RCA: 2245] [Article Influence: 93.5] [Reference Citation Analysis (0)] |
| 40. | Hartley GP, Chow L, Ammons DT, Wheat WH, Dow SW. Programmed Cell Death Ligand 1 (PD-L1) Signaling Regulates Macrophage Proliferation and Activation. Cancer Immunol Res. 2018;6:1260-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 251] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 41. | Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms Controlling PD-L1 Expression in Cancer. Mol Cell. 2019;76:359-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 693] [Article Influence: 115.5] [Reference Citation Analysis (0)] |
| 42. | Youngblood B, Oestreich KJ, Ha SJ, Duraiswamy J, Akondy RS, West EE, Wei Z, Lu P, Austin JW, Riley JL, Boss JM, Ahmed R. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity. 2011;35:400-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 345] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 43. | Li H, Li X, Liu S, Guo L, Zhang B, Zhang J, Ye Q. Programmed cell death-1 (PD-1) checkpoint blockade in combination with a mammalian target of rapamycin inhibitor restrains hepatocellular carcinoma growth induced by hepatoma cell-intrinsic PD-1. Hepatology. 2017;66:1920-1933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 151] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 44. | Wang W, Li F, Mao Y, Zhou H, Sun J, Li R, Liu C, Chen W, Hua D, Zhang X. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet. 2013;132:641-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 45. | Zhang G, Li N, Li Z, Zhu Q, Li F, Yang C, Han Q, Lv Y, Zhou Z, Liu Z. microRNA-4717 differentially interacts with its polymorphic target in the PD1 3' untranslated region: A mechanism for regulating PD-1 expression and function in HBV-associated liver diseases. Oncotarget. 2015;6:18933-18944. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 46. | Xiao G, Jin LL, Liu CQ, Wang YC, Meng YM, Zhou ZG, Chen J, Yu XJ, Zhang YJ, Xu J, Zheng L. EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J Immunother Cancer. 2019;7:300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 47. | Wang H, Fu C, Du J, Wang H, He R, Yin X, Li H, Li X, Li K, Zheng L, Liu Z, Qiu Y. Enhanced histone H3 acetylation of the PD-L1 promoter via the COP1/c-Jun/HDAC3 axis is required for PD-L1 expression in drug-resistant cancer cells. J Exp Clin Cancer Res. 2020;39:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 48. | Coelho MA, de Carné Trécesson S, Rana S, Zecchin D, Moore C, Molina-Arcas M, East P, Spencer-Dene B, Nye E, Barnouin K, Snijders AP, Lai WS, Blackshear PJ, Downward J. Oncogenic RAS Signaling Promotes Tumor Immunoresistance by Stabilizing PD-L1 mRNA. Immunity. 2017;47:1083-1099.e6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 477] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 49. | Li H, Li CW, Li X, Ding Q, Guo L, Liu S, Liu C, Lai CC, Hsu JM, Dong Q, Xia W, Hsu JL, Yamaguchi H, Du Y, Lai YJ, Sun X, Koller PB, Ye Q, Hung MC. MET Inhibitors Promote Liver Tumor Evasion of the Immune Response by Stabilizing PDL1. Gastroenterology. 2019;156:1849-1861.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 50. | Zou J, Zhuang M, Yu X, Li N, Mao R, Wang Z, Wang J, Wang X, Zhou H, Zhang L, Shi Y. MYC inhibition increases PD-L1 expression induced by IFN-γ in hepatocellular carcinoma cells. Mol Immunol. 2018;101:203-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 51. | Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O'Donnell E, Chapuy B, Takeyama K, Neuberg D, Golub TR, Kutok JL, Shipp MA. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116:3268-3277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 893] [Cited by in RCA: 993] [Article Influence: 66.2] [Reference Citation Analysis (0)] |
| 52. | Kan G, Dong W. The expression of PD-L1 APE1 and P53 in hepatocellular carcinoma and its relationship to clinical pathology. Eur Rev Med Pharmacol Sci. 2015;19:3063-3071. [PubMed] |
| 53. | Tojyo I, Shintani Y, Nakanishi T, Okamoto K, Hiraishi Y, Fujita S, Enaka M, Sato F, Muragaki Y. PD-L1 expression correlated with p53 expression in oral squamous cell carcinoma. Maxillofac Plast Reconstr Surg. 2019;41:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 54. | Thiem A, Hesbacher S, Kneitz H, di Primio T, Heppt MV, Hermanns HM, Goebeler M, Meierjohann S, Houben R, Schrama D. IFN-γgamma-induced PD-L1 expression in melanoma depends on p53 expression. J Exp Clin Cancer Res. 2019;38:397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 117] [Article Influence: 19.5] [Reference Citation Analysis (1)] |
| 55. | Yan Y, Zheng L, Du Q, Yan B, Geller DA. Interferon regulatory factor 1 (IRF-1) and IRF-2 regulate PD-L1 expression in hepatocellular carcinoma (HCC) cells. Cancer Immunol Immunother. 2020;69:1891-1903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 78] [Article Influence: 15.6] [Reference Citation Analysis (1)] |
| 56. | Imai D, Yoshizumi T, Okano S, Itoh S, Ikegami T, Harada N, Aishima S, Oda Y, Maehara Y. IFN-γ Promotes Epithelial-Mesenchymal Transition and the Expression of PD-L1 in Pancreatic Cancer. J Surg Res. 2019;240:115-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 57. | Shrestha R, Prithviraj P, Anaka M, Bridle KR, Crawford DHG, Dhungel B, Steel JC, Jayachandran A. Monitoring Immune Checkpoint Regulators as Predictive Biomarkers in Hepatocellular Carcinoma. Front Oncol. 2018;8:269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 58. | Xu GL, Ni CF, Liang HS, Xu YH, Wang WS, Shen J, Li MM, Zhu XL. Upregulation of PD-L1 expression promotes epithelial-to-mesenchymal transition in sorafenib-resistant hepatocellular carcinoma cells. Gastroenterol Rep (Oxf). 2020;8:390-398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 59. | Huang CY, Wang Y, Luo GY, Han F, Li YQ, Zhou ZG, Xu GL. Relationship Between PD-L1 Expression and CD8+ T-cell Immune Responses in Hepatocellular Carcinoma. J Immunother. 2017;40:323-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 60. | Xiang J, Zhang N, Sun H, Su L, Zhang C, Xu H, Feng J, Wang M, Chen J, Liu L, Shan J, Shen J, Yang Z, Wang G, Zhou H, Prieto J, Ávila MA, Liu C, Qian C. Disruption of SIRT7 Increases the Efficacy of Checkpoint Inhibitor via MEF2D Regulation of Programmed Cell Death 1 Ligand 1 in Hepatocellular Carcinoma Cells. Gastroenterology. 2020;158:664-678.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 61. | Wen Q, Han T, Wang Z, Jiang S. Role and mechanism of programmed death-ligand 1 in hypoxia-induced liver cancer immune escape. Oncol Lett. 2020;19:2595-2601. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 62. | Guo R, Li Y, Wang Z, Bai H, Duan J, Wang S, Wang L, Wang J. Hypoxia-inducible factor-1α and nuclear factor-κB play important roles in regulating programmed cell death ligand 1 expression by epidermal growth factor receptor mutants in non-small-cell lung cancer cells. Cancer Sci. 2019;110:1665-1675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 63. | Dai X, Xue J, Hu J, Yang SL, Chen GG, Lai PBS, Yu C, Zeng C, Fang X, Pan X, Zhang T. Positive Expression of Programmed Death Ligand 1 in Peritumoral Liver Tissue is Associated with Poor Survival after Curative Resection of Hepatocellular Carcinoma. Transl Oncol. 2017;10:511-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 64. | Semaan A, Dietrich D, Bergheim D, Dietrich J, Kalff JC, Branchi V, Matthaei H, Kristiansen G, Fischer HP, Goltz D. CXCL12 expression and PD-L1 expression serve as prognostic biomarkers in HCC and are induced by hypoxia. Virchows Arch. 2017;470:185-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 65. | Wu Q, Zhou W, Yin S, Zhou Y, Chen T, Qian J, Su R, Hong L, Lu H, Zhang F, Xie H, Zhou L, Zheng S. Blocking Triggering Receptor Expressed on Myeloid Cells-1-Positive Tumor-Associated Macrophages Induced by Hypoxia Reverses Immunosuppression and Anti-Programmed Cell Death Ligand 1 Resistance in Liver Cancer. Hepatology. 2019;70:198-214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 211] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 66. | Long J, Qu T, Pan XF, Tang X, Wan HH, Qiu P, Xu YH. Expression of programmed death ligand-1 and programmed death 1 in hepatocellular carcinoma and its clinical significance. J Cancer Res Ther. 2018;14:S1188-S1192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 67. | Kim HD, Song GW, Park S, Jung MK, Kim MH, Kang HJ, Yoo C, Yi K, Kim KH, Eo S, Moon DB, Hong SM, Ju YS, Shin EC, Hwang S, Park SH. Association Between Expression Level of PD1 by Tumor-Infiltrating CD8+ T Cells and Features of Hepatocellular Carcinoma. Gastroenterology. 2018;155:1936-1950.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 202] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 68. | Ma J, Zheng B, Goswami S, Meng L, Zhang D, Cao C, Li T, Zhu F, Ma L, Zhang Z, Zhang S, Duan M, Chen Q, Gao Q, Zhang X. PD1Hi CD8+ T cells correlate with exhausted signature and poor clinical outcome in hepatocellular carcinoma. J Immunother Cancer. 2019;7:331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 238] [Cited by in RCA: 230] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 69. | Yun J, Yu G, Hu P, Chao Y, Li X, Chen X, Wei Q, Wang J. PD-1 expression is elevated in monocytes from hepatocellular carcinoma patients and contributes to CD8 T cell suppression. Immunol Res. 2020;68:436-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 70. | Guo M, Yuan F, Qi F, Sun J, Rao Q, Zhao Z, Huang P, Fang T, Yang B, Xia J. Expression and clinical significance of LAG-3, FGL1, PD-L1 and CD8+T cells in hepatocellular carcinoma using multiplex quantitative analysis. J Transl Med. 2020;18:306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 71. | Yarchoan M, Xing D, Luan L, Xu H, Sharma RB, Popovic A, Pawlik TM, Kim AK, Zhu Q, Jaffee EM, Taube JM, Anders RA. Characterization of the Immune Microenvironment in Hepatocellular Carcinoma. Clin Cancer Res. 2017;23:7333-7339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 72. | Peng H, Fu YX. The Inhibitory PVRL1/PVR/TIGIT Axis in Immune Therapy for Hepatocellular Carcinoma. Gastroenterology. 2020;159:434-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 73. | Chiu DK, Yuen VW, Cheu JW, Wei LL, Ting V, Fehlings M, Sumatoh H, Nardin A, Newell EW, Ng IO, Yau TC, Wong CM, Wong CC. Hepatocellular Carcinoma Cells Up-regulate PVRL1, Stabilizing PVR and Inhibiting the Cytotoxic T-Cell Response via TIGIT to Mediate Tumor Resistance to PD1 Inhibitors in Mice. Gastroenterology. 2020;159:609-623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 129] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 74. | Fabrizio FP, Trombetta D, Rossi A, Sparaneo A, Castellana S, Muscarella LA. Gene code CD274/PD-L1: from molecular basis toward cancer immunotherapy. Ther Adv Med Oncol. 2018;10:1758835918815598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 75. | Ribas A, Hu-Lieskovan S. What does PD-L1 positive or negative mean? J Exp Med. 2016;213:2835-2840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 266] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 76. | Chen CL, Pan QZ, Zhao JJ, Wang Y, Li YQ, Wang QJ, Pan K, Weng DS, Jiang SS, Tang Y, Zhang XF, Zhang HX, Zhou ZQ, Zeng YX, Xia JC. PD-L1 expression as a predictive biomarker for cytokine-induced killer cell immunotherapy in patients with hepatocellular carcinoma. Oncoimmunology. 2016;5:e1176653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 77. | Chen L, Huang X, Zhang W, Liu Y, Chen B, Xiang Y, Zhang R, Zhang M, Feng J, Liu S, Duan T, Chen X, Wang W, Pan T, Yan L, Jin T, Li G, Li Y, Xie T, Sui X. Correlation of PD-L1 and SOCS3 Co-expression with the Prognosis of Hepatocellular Carcinoma Patients. J Cancer. 2020;11:5440-5448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 78. | Jung HI, Jeong D, Ji S, Ahn TS, Bae SH, Chin S, Chung JC, Kim HC, Lee MS, Baek MJ. Overexpression of PD-L1 and PD-L2 Is Associated with Poor Prognosis in Patients with Hepatocellular Carcinoma. Cancer Res Treat. 2017;49:246-254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 170] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 79. | Umemoto Y, Okano S, Matsumoto Y, Nakagawara H, Matono R, Yoshiya S, Yamashita Y, Yoshizumi T, Ikegami T, Soejima Y, Harada M, Aishima S, Oda Y, Shirabe K, Maehara Y. Prognostic impact of programmed cell death 1 ligand 1 expression in human leukocyte antigen class I-positive hepatocellular carcinoma after curative hepatectomy. J Gastroenterol. 2015;50:65-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 80. | Qian Q, Wu C, Chen J, Wang W. Relationship between IL10 and PD-L1 in Liver Hepatocellular Carcinoma Tissue and Cell Lines. Biomed Res Int. 2020;2020:8910183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 81. | Li XS, Li JW, Li H, Jiang T. Prognostic value of programmed cell death ligand 1 (PD-L1) for hepatocellular carcinoma: a meta-analysis. Biosci Rep. 2020;40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 82. | Zeng Z, Shi F, Zhou L, Zhang MN, Chen Y, Chang XJ, Lu YY, Bai WL, Qu JH, Wang CP, Wang H, Lou M, Wang FS, Lv JY, Yang YP. Upregulation of circulating PD-L1/PD-1 is associated with poor post-cryoablation prognosis in patients with HBV-related hepatocellular carcinoma. PLoS One. 2011;6:e23621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 147] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 83. | Calderaro J, Rousseau B, Amaddeo G, Mercey M, Charpy C, Costentin C, Luciani A, Zafrani ES, Laurent A, Azoulay D, Lafdil F, Pawlotsky JM. Programmed death ligand 1 expression in hepatocellular carcinoma: Relationship With clinical and pathological features. Hepatology. 2016;64:2038-2046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 331] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 84. | Liao H, Chen W, Dai Y, Richardson JJ, Guo J, Yuan K, Zeng Y, Xie K. Expression of Programmed Cell Death-Ligands in Hepatocellular Carcinoma: Correlation With Immune Microenvironment and Survival Outcomes. Front Oncol. 2019;9:883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (1)] |
| 85. | Ihling C, Naughton B, Zhang Y, Rolfe PA, Frick-Krieger E, Terracciano LM, Dussault I. Observational Study of PD-L1, TGF-β, and Immune Cell Infiltrates in Hepatocellular Carcinoma. Front Med (Lausanne). 2019;6:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 86. | Gabrielson A, Wu Y, Wang H, Jiang J, Kallakury B, Gatalica Z, Reddy S, Kleiner D, Fishbein T, Johnson L, Island E, Satoskar R, Banovac F, Jha R, Kachhela J, Feng P, Zhang T, Tesfaye A, Prins P, Loffredo C, Marshall J, Weiner L, Atkins M, He AR. Intratumoral CD3 and CD8 T-cell Densities Associated with Relapse-Free Survival in HCC. Cancer Immunol Res. 2016;4:419-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 243] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 87. | An Y, Gao S, Zhao WC, Qiu BA, Xia NX, Zhang PJ, Fan ZP. Transforming growth factor-β and peripheral regulatory cells are negatively correlated with the overall survival of hepatocellular carcinoma. World J Gastroenterol. 2018;24:2733-2740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 12] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 88. | Huang CY, Wang H, Liao W, Han F, Li YQ, Chen SW, Lao XM. Transforming Growth Factor β is a Poor Prognostic Factor and Inhibits the Favorable Prognostic Value of CD8+ CTL in Human Hepatocellular Carcinoma. J Immunother. 2017;40:175-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 89. | Zhao T, Li C, Wu Y, Li B, Zhang B. Prognostic value of PD-L1 expression in tumor infiltrating immune cells in cancers: A meta-analysis. PLoS One. 2017;12:e0176822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 90. | Lu LC, Lee YH, Chang CJ, Shun CT, Fang CY, Shao YY, Liu TH, Cheng AL, Hsu CH. Increased Expression of Programmed Death-Ligand 1 in Infiltrating Immune Cells in Hepatocellular Carcinoma Tissues after Sorafenib Treatment. Liver Cancer. 2019;8:110-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 91. | Zong Z, Zou J, Mao R, Ma C, Li N, Wang J, Wang X, Zhou H, Zhang L, Shi Y. M1 Macrophages Induce PD-L1 Expression in Hepatocellular Carcinoma Cells Through IL-1β Signaling. Front Immunol. 2019;10:1643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 146] [Article Influence: 24.3] [Reference Citation Analysis (1)] |
| 92. | Zhu Y, Yang J, Xu D, Gao XM, Zhang Z, Hsu JL, Li CW, Lim SO, Sheng YY, Zhang Y, Li JH, Luo Q, Zheng Y, Zhao Y, Lu L, Jia HL, Hung MC, Dong QZ, Qin LX. Disruption of tumour-associated macrophage trafficking by the osteopontin-induced colony-stimulating factor-1 signalling sensitises hepatocellular carcinoma to anti-PD-L1 blockade. Gut. 2019;68:1653-1666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 304] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 93. | Zhou ZQ, Tong DN, Guan J, Tan HW, Zhao LD, Zhu Y, Yao J, Yang J, Zhang ZY. Follicular helper T cell exhaustion induced by PD-L1 expression in hepatocellular carcinoma results in impaired cytokine expression and B cell help, and is associated with advanced tumor stages. Am J Transl Res. 2016;8:2926-2936. [PubMed] |
| 94. | Xiao X, Lao XM, Chen MM, Liu RX, Wei Y, Ouyang FZ, Chen DP, Zhao XY, Zhao Q, Li XF, Liu CL, Zheng L, Kuang DM. PD-1hi Identifies a Novel Regulatory B-cell Population in Human Hepatoma That Promotes Disease Progression. Cancer Discov. 2016;6:546-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 256] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 95. | Wu M, Mei F, Liu W, Jiang J. Comprehensive characterization of tumor infiltrating natural killer cells and clinical significance in hepatocellular carcinoma based on gene expression profiles. Biomed Pharmacother. 2020;121:109637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 96. | Chen DP, Ning WR, Jiang ZZ, Peng ZP, Zhu LY, Zhuang SM, Kuang DM, Zheng L, Wu Y. Glycolytic activation of peritumoral monocytes fosters immune privilege via the PFKFB3-PD-L1 axis in human hepatocellular carcinoma. J Hepatol. 2019;71:333-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 127] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 97. | Yasuoka H, Asai A, Ohama H, Tsuchimoto Y, Fukunishi S, Higuchi K. Increased both PD-L1 and PD-L2 expressions on monocytes of patients with hepatocellular carcinoma was associated with a poor prognosis. Sci Rep. 2020;10:10377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 98. | Chang B, Huang T, Wei H, Shen L, Zhu D, He W, Chen Q, Zhang H, Li Y, Huang R, Li W, Wu P. The correlation and prognostic value of serum levels of soluble programmed death protein 1 (sPD-1) and soluble programmed death-ligand 1 (sPD-L1) in patients with hepatocellular carcinoma. Cancer Immunol Immunother. 2019;68:353-363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 99. | Nielsen C, Ohm-Laursen L, Barington T, Husby S, Lillevang ST. Alternative splice variants of the human PD-1 gene. Cell Immunol. 2005;235:109-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 180] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 100. | Chen Y, Wang Q, Shi B, Xu P, Hu Z, Bai L, Zhang X. Development of a sandwich ELISA for evaluating soluble PD-L1 (CD274) in human sera of different ages as well as supernatants of PD-L1+ cell lines. Cytokine. 2011;56:231-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 208] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 101. | Frigola X, Inman BA, Lohse CM, Krco CJ, Cheville JC, Thompson RH, Leibovich B, Blute ML, Dong H, Kwon ED. Identification of a soluble form of B7-H1 that retains immunosuppressive activity and is associated with aggressive renal cell carcinoma. Clin Cancer Res. 2011;17:1915-1923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 309] [Cited by in RCA: 287] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 102. | Finkelmeier F, Canli Ö, Tal A, Pleli T, Trojan J, Schmidt M, Kronenberger B, Zeuzem S, Piiper A, Greten FR, Waidmann O. High levels of the soluble programmed death-ligand (sPD-L1) identify hepatocellular carcinoma patients with a poor prognosis. Eur J Cancer. 2016;59:152-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 170] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 103. | Cheng HY, Kang PJ, Chuang YH, Wang YH, Jan MC, Wu CF, Lin CL, Liu CJ, Liaw YF, Lin SM, Chen PJ, Lee SD, Yu MW. Circulating programmed death-1 as a marker for sustained high hepatitis B viral load and risk of hepatocellular carcinoma. PLoS One. 2014;9:e95870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 104. | Elhag OA, Hu XJ, Wen-Ying Z, Li X, Yuan YZ, Deng LF, Liu DL, Liu YL, Hui G. Reconstructed adeno-associated virus with the extracellular domain of murine PD-1 induces antitumor immunity. Asian Pac J Cancer Prev. 2012;13:4031-4036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 105. | Shin SP, Seo HH, Shin JH, Park HB, Lim DP, Eom HS, Bae YS, Kim IH, Choi K, Lee SJ. Adenovirus expressing both thymidine kinase and soluble PD1 enhances antitumor immunity by strengthening CD8 T-cell response. Mol Ther. 2013;21:688-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 106. | Sica GL, Ramalingam SS. Assays for PD-L1 Expression: Do All Roads Lead to Rome? JAMA Oncol. 2017;3:1058-1059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 107. | Sangro B, Chan SL, Meyer T, Reig M, El-Khoueiry A, Galle PR. Diagnosis and management of toxicities of immune checkpoint inhibitors in hepatocellular carcinoma. J Hepatol. 2020;72:320-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 192] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 108. | Sangro B, Melero I, Wadhawan S, Finn RS, Abou-Alfa GK, Cheng AL, Yau T, Furuse J, Park JW, Boyd Z, Tang HT, Shen Y, Tschaika M, Neely J, El-Khoueiry A. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol. 2020;73:1460-1469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 309] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 109. | Ng HHM, Lee RY, Goh S, Tay ISY, Lim X, Lee B, Chew V, Li H, Tan B, Lim S, Lim JCT, Au B, Loh JJH, Saraf S, Connolly JE, Loh T, Leow WQ, Lee JJX, Toh HC, Malavasi F, Lee SY, Chow P, Newell EW, Choo SP, Tai D, Yeong J, Lim TKH. Immunohistochemical scoring of CD38 in the tumor microenvironment predicts responsiveness to anti-PD-1/PD-L1 immunotherapy in hepatocellular carcinoma. J Immunother Cancer. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 110. | Garnelo M, Tan A, Her Z, Yeong J, Lim CJ, Chen J, Lim KH, Weber A, Chow P, Chung A, Ooi LL, Toh HC, Heikenwalder M, Ng IO, Nardin A, Chen Q, Abastado JP, Chew V. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut. 2017;66:342-351. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 247] [Cited by in RCA: 360] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 111. | Romero D. Immunotherapy: PD-1 says goodbye, TIM-3 says hello. Nat Rev Clin Oncol. 2016;13:202-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 112. | Brown ZJ, Yu SJ, Heinrich B, Ma C, Fu Q, Sandhu M, Agdashian D, Zhang Q, Korangy F, Greten TF. Indoleamine 2,3-dioxygenase provides adaptive resistance to immune checkpoint inhibitors in hepatocellular carcinoma. Cancer Immunol Immunother. 2018;67:1305-1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 113. | Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, Gandhi L, Redig AJ, Rodig SJ, Asahina H, Jones RE, Kulkarni MM, Kuraguchi M, Palakurthi S, Fecci PE, Johnson BE, Janne PA, Engelman JA, Gangadharan SP, Costa DB, Freeman GJ, Bueno R, Hodi FS, Dranoff G, Wong KK, Hammerman PS. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7:10501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 870] [Cited by in RCA: 1196] [Article Influence: 132.9] [Reference Citation Analysis (0)] |
| 114. | Li F, Li N, Sang J, Fan X, Deng H, Zhang X, Han Q, Lv Y, Liu Z. Highly elevated soluble Tim-3 levels correlate with increased hepatocellular carcinoma risk and poor survival of hepatocellular carcinoma patients in chronic hepatitis B virus infection. Cancer Manag Res. 2018;10:941-951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 115. | Tampaki M, Ionas E, Hadziyannis E, Deutsch M, Malagari K, Koskinas J. Association of TIM-3 with BCLC Stage, Serum PD-L1 Detection, and Response to Transarterial Chemoembolization in Patients with Hepatocellular Carcinoma. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 116. | Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, Liu Z, Dong M, Hu X, Ouyang W, Peng J, Zhang Z. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 2017;169:1342-1356.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1011] [Cited by in RCA: 1503] [Article Influence: 187.9] [Reference Citation Analysis (0)] |
| 117. | Caruso S, O'Brien DR, Cleary SP, Roberts LR, Zucman-Rossi J. Genetics of Hepatocellular Carcinoma: Approaches to Explore Molecular Diversity. Hepatology. 2021;73 Suppl 1:14-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 118. | Zhu J, Zhang T, Li J, Lin J, Liang W, Huang W, Wan N, Jiang J. Association Between Tumor Mutation Burden (TMB) and Outcomes of Cancer Patients Treated With PD-1/PD-L1 Inhibitions: A Meta-Analysis. Front Pharmacol. 2019;10:673. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 119. | Forschner A, Battke F, Hadaschik D, Schulze M, Weißgraeber S, Han CT, Kopp M, Frick M, Klumpp B, Tietze N, Amaral T, Martus P, Sinnberg T, Eigentler T, Keim U, Garbe C, Döcker D, Biskup S. Tumor mutation burden and circulating tumor DNA in combined CTLA-4 and PD-1 antibody therapy in metastatic melanoma - results of a prospective biomarker study. J Immunother Cancer. 2019;7:180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 141] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 120. | Lee M, Samstein RM, Valero C, Chan TA, Morris LGT. Tumor mutational burden as a predictive biomarker for checkpoint inhibitor immunotherapy. Hum Vaccin Immunother. 2020;16:112-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 121. | Chen H, Chong W, Teng C, Yao Y, Wang X, Li X. The immune response-related mutational signatures and driver genes in non-small-cell lung cancer. Cancer Sci. 2019;110:2348-2356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 122. | Ang C, Klempner SJ, Ali SM, Madison R, Ross JS, Severson EA, Fabrizio D, Goodman A, Kurzrock R, Suh J, Millis SZ. Prevalence of established and emerging biomarkers of immune checkpoint inhibitor response in advanced hepatocellular carcinoma. Oncotarget. 2019;10:4018-4025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 131] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 123. | Lei J, Zhang D, Yao C, Ding S, Lu Z. Development of a Predictive Immune-Related Gene Signature Associated With Hepatocellular Carcinoma Patient Prognosis. Cancer Control. 2020;27:1073274820977114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 124. | Spahn S, Roessler D, Pompilia R, Gabernet G, Gladstone BP, Horger M, Biskup S, Feldhahn M, Nahnsen S, Hilke FJ, Scheiner B, Dufour JF, De Toni EN, Pinter M, Malek NP, Bitzer M. Clinical and Genetic Tumor Characteristics of Responding and Non-Responding Patients to PD-1 Inhibition in Hepatocellular Carcinoma. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 125. | Wong CN, Fessas P, Dominy K, Mauri FA, Kaneko T, Parcq PD, Khorashad J, Toniutto P, Goldin RD, Avellini C, Pinato DJ. Qualification of tumour mutational burden by targeted next-generation sequencing as a biomarker in hepatocellular carcinoma. Liver Int. 2021;41:192-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 126. | Goumard C, Desbois-Mouthon C, Wendum D, Calmel C, Merabtene F, Scatton O, Praz F. Low Levels of Microsatellite Instability at Simple Repeated Sequences Commonly Occur in Human Hepatocellular Carcinoma. Cancer Genomics Proteomics. 2017;14:329-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 127. | Hashemi M, Karami S, Sarabandi S, Moazeni-Roodi A, Małecki A, Ghavami S, Wiechec E. Association between PD-1 and PD-L1 Polymorphisms and the Risk of Cancer: A Meta-Analysis of Case-Control Studies. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 128. | Tang W, Wang Y, Jiang H, Liu P, Liu C, Gu H, Chen S, Kang M. Programmed death-1 (PD-1) rs2227981 C > T polymorphism is associated with cancer susceptibility: a meta-analysis. Int J Clin Exp Med. 2015;8:22278-22285. [PubMed] |
| 129. | Zheng L, Li D, Wang F, Wu H, Li X, Fu J, Chen X, Wang L, Liu Y, Wang S. Association between hepatitis B viral burden in chronic infection and a functional single nucleotide polymorphism of the PDCD1 gene. J Clin Immunol. 2010;30:855-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 130. | Peng H, Li QL, Hou SH, Hu J, Fan JH, Guo JJ. Association of genetic polymorphisms in CD8+ T cell inhibitory genes and susceptibility to and progression of chronic HBV infection. Infect Genet Evol. 2015;36:467-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 131. | Zhang G, Li Z, Han Q, Li N, Zhu Q, Li F, Lv Y, Chen J, Lou S, Liu Z. Altered TNF-α and IFN-γ levels associated with PD1 but not TNFA polymorphisms in patients with chronic HBV infection. Infect Genet Evol. 2011;11:1624-1630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 132. | Hou Z, Zhou Q, Lu M, Tan D, Xu X. A Programmed Cell Death-1 Haplotype is Associated with Clearance of Hepatitis B Virus. Ann Clin Lab Sci. 2017;47:334-343. [PubMed] |
| 133. | Zhang G, Liu Z, Duan S, Han Q, Li Z, Lv Y, Chen J, Lou S, Li N. Association of polymorphisms of programmed cell death-1 gene with chronic hepatitis B virus infection. Hum Immunol. 2010;71:1209-1213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 134. | Li Z, Li N, Zhu Q, Zhang G, Han Q, Zhang P, Xun M, Wang Y, Zeng X, Yang C, Liu Z. Genetic variations of PD1 and TIM3 are differentially and interactively associated with the development of cirrhosis and HCC in patients with chronic HBV infection. Infect Genet Evol. 2013;14:240-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 135. | Li Z, Li N, Li F, Zhou Z, Sang J, Chen Y, Han Q, Lv Y, Liu Z. Immune checkpoint proteins PD-1 and TIM-3 are both highly expressed in liver tissues and correlate with their gene polymorphisms in patients with HBV-related hepatocellular carcinoma. Medicine (Baltimore). 2016;95:e5749. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 82] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 136. | Dong W, Gong M, Shi Z, Xiao J, Zhang J, Peng J. Programmed Cell Death-1 Polymorphisms Decrease the Cancer Risk: A Meta-Analysis Involving Twelve Case-Control Studies. PLoS One. 2016;11:e0152448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 137. | Bayram S, Akkız H, Ülger Y, Bekar A, Akgöllü E, Yıldırım S. Lack of an association of programmed cell death-1 PD1.3 polymorphism with risk of hepatocellular carcinoma susceptibility in Turkish population: a case-control study. Gene. 2012;511:308-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 138. | Ülger Y, Bayram S, Sandıkçı MÜ, Akgöllü E, Bekar A. Relationship between programmed cell death-1 polymorphisms and clearance of hepatitis B virus. Int J Immunogenet. 2015;42:133-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 139. | Demirci AF, Demirtas CO, Eren F, Yilmaz D, Keklikkiran C, Ozdogan OC, Gunduz F. Evaluation of the Association between Programmed Cell Death-1 Gene Polymorphisms and Hepatocellular Carcinoma Susceptibility in Turkish Subjects. A Pilot Study. J Gastrointestin Liver Dis. 2020;29:617-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 140. | Darlay R, Eldafashi N, McStraw NM, Cordell H, Watson R, McCain MV, Mauricio-Muir J, Shukla R, Dufour J-F, Valenti L, Anstee Q, Reeves H. Exploring genetic variation of PDCD1, which encodes T-cell receptor PD-1, in patients with non-alcoholic fatty liver disease (NAFLD) and hepatocellular carcinoma (HCC). Gut. 2020;69:A4-A4. |
| 141. | Zou J, Wu D, Li T, Wang X, Liu Y, Tan S. Association of PD-L1 gene rs4143815 C>G polymorphism and human cancer susceptibility: A systematic review and meta-analysis. Pathol Res Pract. 2019;215:229-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 142. | Xie Q, Chen Z, Xia L, Zhao Q, Yu H, Yang Z. Correlations of PD-L1 gene polymorphisms with susceptibility and prognosis in hepatocellular carcinoma in a Chinese Han population. Gene. 2018;674:188-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 143. | Tao LH, Zhou XR, Li FC, Chen Q, Meng FY, Mao Y, Li R, Hua D, Zhang HJ, Wang WP, Chen WC. A polymorphism in the promoter region of PD-L1 serves as a binding-site for SP1 and is associated with PD-L1 overexpression and increased occurrence of gastric cancer. Cancer Immunol Immunother. 2017;66:309-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 144. | Huang C, Ge T, Xia C, Zhu W, Xu L, Wang Y, Wu F, Liu F, Zheng M, Chen Z. Association of rs10204525 genotype GG and rs2227982 CC combination in programmed cell death 1 with hepatitis B virus infection risk. Medicine (Baltimore). 2019;98:e16972. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 145. | Li F, Fan X, Wang X, Deng H, Zhang X, Zhang K, Li N, Han Q, Lv Y, Liu Z. Genetic association and interaction of PD1 and TIM3 polymorphisms in susceptibility of chronic hepatitis B virus infection and hepatocarcinogenesis. Discov Med. 2019;27:79-92. [PubMed] |
| 146. | Juran BD, Lazaridis KN. Update on the genetics and genomics of PBC. J Autoimmun. 2010;35:181-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 147. | Wagner M, Tupikowski K, Jasek M, Tomkiewicz A, Witkowicz A, Ptaszkowski K, Karpinski P, Zdrojowy R, Halon A, Karabon L. SNP-SNP Interaction in Genes Encoding PD-1/PD-L1 Axis as a Potential Risk Factor for Clear Cell Renal Cell Carcinoma. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 148. | Shi XL, Mancham S, Hansen BE, de Knegt RJ, de Jonge J, van der Laan LJ, Rivadeneira F, Metselaar HJ, Kwekkeboom J. Counter-regulation of rejection activity against human liver grafts by donor PD-L1 and recipient PD-1 interaction. J Hepatol. 2016;64:1274-1282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 67] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 149. | Wang X, Zhang D, Wang GQ, Duan AQ, Ruan X, Zhao T. Association between PD-L1 variants and PD-L1 expression: A pan-cancer analysis. J Clin Oncol. 2020;38:e13661. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 150. | Budczies J, Bockmayr M, Denkert C, Klauschen F, Gröschel S, Darb-Esfahani S, Pfarr N, Leichsenring J, Onozato ML, Lennerz JK, Dietel M, Fröhling S, Schirmacher P, Iafrate AJ, Weichert W, Stenzinger A. Pan-cancer analysis of copy number changes in programmed death-ligand 1 (PD-L1, CD274) - associations with gene expression, mutational load, and survival. Genes Chromosomes Cancer. 2016;55:626-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 77] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 151. | Ma LJ, Feng FL, Dong LQ, Zhang Z, Duan M, Liu LZ, Shi JY, Yang LX, Wang ZC, Zhang S, Ding ZB, Ke AW, Cao Y, Zhang XM, Zhou J, Fan J, Wang XY, Gao Q. Clinical significance of PD-1/PD-Ls gene amplification and overexpression in patients with hepatocellular carcinoma. Theranostics. 2018;8:5690-5702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 152. | Parfenov M, Pedamallu CS, Gehlenborg N, Freeman SS, Danilova L, Bristow CA, Lee S, Hadjipanayis AG, Ivanova EV, Wilkerson MD, Protopopov A, Yang L, Seth S, Song X, Tang J, Ren X, Zhang J, Pantazi A, Santoso N, Xu AW, Mahadeshwar H, Wheeler DA, Haddad RI, Jung J, Ojesina AI, Issaeva N, Yarbrough WG, Hayes DN, Grandis JR, El-Naggar AK, Meyerson M, Park PJ, Chin L, Seidman JG, Hammerman PS, Kucherlapati R; Cancer Genome Atlas Network. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci U S A. 2014;111:15544-15549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 304] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 153. | Hassounah NB, Malladi VS, Huang Y, Freeman SS, Beauchamp EM, Koyama S, Souders N, Martin S, Dranoff G, Wong KK, Pedamallu CS, Hammerman PS, Akbay EA. Identification and characterization of an alternative cancer-derived PD-L1 splice variant. Cancer Immunol Immunother. 2019;68:407-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 154. | Mahoney KM, Shukla SA, Patsoukis N, Chaudhri A, Browne EP, Arazi A, Eisenhaure TM, Pendergraft WF 3rd, Hua P, Pham HC, Bu X, Zhu B, Hacohen N, Fritsch EF, Boussiotis VA, Wu CJ, Freeman GJ. A secreted PD-L1 splice variant that covalently dimerizes and mediates immunosuppression. Cancer Immunol Immunother. 2019;68:421-432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 107] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 155. | Gong B, Kiyotani K, Sakata S, Nagano S, Kumehara S, Baba S, Besse B, Yanagitani N, Friboulet L, Nishio M, Takeuchi K, Kawamoto H, Fujita N, Katayama R. Secreted PD-L1 variants mediate resistance to PD-L1 blockade therapy in non-small cell lung cancer. J Exp Med. 2019;216:982-1000. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 202] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 156. | EDMONDSON HA, STEINER PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 157. | Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, Roberts LR, Demetris AJ, Sun Z, Nevens F, Roskams T, Thorgeirsson SS. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 748] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 158. | Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, Tang ZY, Wang XW. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451-1461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 545] [Cited by in RCA: 593] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 159. | Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 677] [Cited by in RCA: 691] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 160. | Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, Jeannot E, Hérault A, Saric J, Belghiti J, Franco D, Bioulac-Sage P, Laurent-Puig P, Zucman-Rossi J. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 827] [Cited by in RCA: 927] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 161. | Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, Watanabe G, Gabriel S, Friedman SL, Kumada H, Llovet JM, Golub TR. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385-7392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 956] [Cited by in RCA: 944] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 162. | Hirschfield H, Bian CB, Higashi T, Nakagawa S, Zeleke TZ, Nair VD, Fuchs BC, Hoshida Y. In vitro modeling of hepatocellular carcinoma molecular subtypes for anti-cancer drug assessment. Exp Mol Med. 2018;50:e419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 163. | Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, Castro de Moura M, Putra J, Camprecios G, Bassaganyas L, Akers N, Losic B, Waxman S, Thung SN, Mazzaferro V, Esteller M, Friedman SL, Schwartz M, Villanueva A, Llovet JM. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology. 2017;153:812-826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 690] [Cited by in RCA: 663] [Article Influence: 82.9] [Reference Citation Analysis (0)] |
| 164. | Zhang Q, Lou Y, Yang J, Wang J, Feng J, Zhao Y, Wang L, Huang X, Fu Q, Ye M, Zhang X, Chen Y, Ma C, Ge H, Wu J, Wei T, Chen Q, Yu C, Xiao Y, Feng X, Guo G, Liang T, Bai X. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut. 2019;68:2019-2031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 252] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 165. | Sukowati CHC. Heterogeneity of Hepatic Cancer Stem Cells. Adv Exp Med Biol. 2019;1139:59-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 166. | Yamashita T, Honda M, Nakamoto Y, Baba M, Nio K, Hara Y, Zeng SS, Hayashi T, Kondo M, Takatori H, Yamashita T, Mizukoshi E, Ikeda H, Zen Y, Takamura H, Wang XW, Kaneko S. Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology. 2013;57:1484-1497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 227] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 167. | Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6844] [Cited by in RCA: 6916] [Article Influence: 288.2] [Reference Citation Analysis (0)] |
| 168. | Holczbauer Á, Factor VM, Andersen JB, Marquardt JU, Kleiner DE, Raggi C, Kitade M, Seo D, Akita H, Durkin ME, Thorgeirsson SS. Modeling pathogenesis of primary liver cancer in lineage-specific mouse cell types. Gastroenterology. 2013;145:221-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 142] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 169. | Thorgeirsson SS. Stemness and reprogramming in liver cancer. Hepatology. 2016;63:1068-1070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 170. | Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer. 2015;15:653-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 236] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 171. | Chen Y, Yu D, Zhang H, He H, Zhang C, Zhao W, Shao RG. CD133(+)EpCAM(+) phenotype possesses more characteristics of tumor initiating cells in hepatocellular carcinoma Huh7 cells. Int J Biol Sci. 2012;8:992-1004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 172. | Kimura O, Kondo Y, Kogure T, Kakazu E, Ninomiya M, Iwata T, Morosawa T, Shimosegawa T. Expression of EpCAM increases in the hepatitis B related and the treatment-resistant hepatocellular carcinoma. Biomed Res Int. 2014;2014:172913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 173. | Ma S, Lee TK, Zheng BJ, Chan KW, Guan XY. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749-1758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 605] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 174. | Ma S. Biology and clinical implications of CD133(+) liver cancer stem cells. Exp Cell Res. 2013;319:126-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 175. | Yamashita M, Wada H, Eguchi H, Ogawa H, Yamada D, Noda T, Asaoka T, Kawamoto K, Gotoh K, Umeshita K, Doki Y, Mori M. A CD13 inhibitor, ubenimex, synergistically enhances the effects of anticancer drugs in hepatocellular carcinoma. Int J Oncol. 2016;49:89-98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 176. | Bort A, Sánchez BG, Mateos-Gómez PA, Vara-Ciruelos D, Rodríguez-Henche N, Díaz-Laviada I. Targeting AMP-activated kinase impacts hepatocellular cancer stem cells induced by long-term treatment with sorafenib. Mol Oncol. 2019;13:1311-1331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 177. | Kahraman DC, Kahraman T, Cetin-Atalay R. Targeting PI3K/Akt/mTOR Pathway Identifies Differential Expression and Functional Role of IL8 in Liver Cancer Stem Cell Enrichment. Mol Cancer Ther. 2019;18:2146-2157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 178. | Qiu Y, Shan W, Yang Y, Jin M, Dai Y, Yang H, Jiao R, Xia Y, Liu Q, Ju L, Huang G, Zhang J, Yang L, Li L, Li Y. Reversal of sorafenib resistance in hepatocellular carcinoma: epigenetically regulated disruption of 14-3-3η/hypoxia-inducible factor-1α. Cell Death Discov. 2019;5:120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 179. | Huang M, Chen C, Geng J, Han D, Wang T, Xie T, Wang L, Wang Y, Wang C, Lei Z, Chu X. Targeting KDM1A attenuates Wnt/β-catenin signaling pathway to eliminate sorafenib-resistant stem-like cells in hepatocellular carcinoma. Cancer Lett. 2017;398:12-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 90] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 180. | Xin HW, Ambe CM, Hari DM, Wiegand GW, Miller TC, Chen JQ, Anderson AJ, Ray S, Mullinax JE, Koizumi T, Langan RC, Burka D, Herrmann MA, Goldsmith PK, Stojadinovic A, Rudloff U, Thorgeirsson SS, Avital I. Label-retaining liver cancer cells are relatively resistant to sorafenib. Gut. 2013;62:1777-1786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 181. | Kahraman DC, Hanquet G, Jeanmart L, Lanners S, Šramel P, Boháč A, Cetin-Atalay R. Quinoides and VEGFR2 TKIs influence the fate of hepatocellular carcinoma and its cancer stem cells. Medchemcomm. 2017;8:81-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 182. | Wu Y, Chen M, Wu P, Chen C, Xu ZP, Gu W. Increased PD-L1 expression in breast and colon cancer stem cells. Clin Exp Pharmacol Physiol. 2017;44:602-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 85] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 183. | Castagnoli L, Cancila V, Cordoba-Romero SL, Faraci S, Talarico G, Belmonte B, Iorio MV, Milani M, Volpari T, Chiodoni C, Hidalgo-Miranda A, Tagliabue E, Tripodo C, Sangaletti S, Di Nicola M, Pupa SM. WNT signaling modulates PD-L1 expression in the stem cell compartment of triple-negative breast cancer. Oncogene. 2019;38:4047-4060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 151] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 184. | Mansour FA, Al-Mazrou A, Al-Mohanna F, Al-Alwan M, Ghebeh H. PD-L1 is overexpressed on breast cancer stem cells through notch3/mTOR axis. Oncoimmunology. 2020;9:1729299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 185. | Almozyan S, Colak D, Mansour F, Alaiya A, Al-Harazi O, Qattan A, Al-Mohanna F, Al-Alwan M, Ghebeh H. PD-L1 promotes OCT4 and Nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int J Cancer. 2017;141:1402-1412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 191] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 186. | Zhang C, Wang H, Wang X, Zhao C. CD44, a marker of cancer stem cells, is positively correlated with PD-L1 expression and immune cells infiltration in lung adenocarcinoma. Cancer Cell Int. 2020;20:583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 187. | Park JW, Um H, Yang H, Ko W, Kim DY, Kim HK. Proteogenomic analysis of NCC-S1M, a gastric cancer stem cell-like cell line that responds to anti-PD-1. Biochem Biophys Res Commun. 2017;484:631-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 188. | Komura N, Mabuchi S, Shimura K, Yokoi E, Kozasa K, Kuroda H, Takahashi R, Sasano T, Kawano M, Matsumoto Y, Kodama M, Hashimoto K, Sawada K, Kimura T. The role of myeloid-derived suppressor cells in increasing cancer stem-like cells and promoting PD-L1 expression in epithelial ovarian cancer. Cancer Immunol Immunother. 2020;69:2477-2499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 189. | Wei F, Zhang T, Deng SC, Wei JC, Yang P, Wang Q, Chen ZP, Li WL, Chen HC, Hu H, Cao J. PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Cancer Lett. 2019;450:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 129] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 190. | Hou YC, Chao YJ, Hsieh MH, Tung HL, Wang HC, Shan YS. Low CD8⁺ T Cell Infiltration and High PD-L1 Expression Are Associated with Level of CD44⁺/CD133⁺ Cancer Stem Cells and Predict an Unfavorable Prognosis in Pancreatic Cancer. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 191. | Lee Y, Shin JH, Longmire M, Wang H, Kohrt HE, Chang HY, Sunwoo JB. CD44+ Cells in Head and Neck Squamous Cell Carcinoma Suppress T-Cell-Mediated Immunity by Selective Constitutive and Inducible Expression of PD-L1. Clin Cancer Res. 2016;22:3571-3581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 180] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 192. | Koh YW, Han JH, Haam S. Expression of PD-L1, cancer stem cell and epithelial-mesenchymal transition phenotype in non-small cell lung cancer. Pathology. 2021;53:239-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 193. | Tamai K, Nakamura M, Mizuma M, Mochizuki M, Yokoyama M, Endo H, Yamaguchi K, Nakagawa T, Shiina M, Unno M, Muramoto K, Sato I, Satoh K, Sugamura K, Tanaka N. Suppressive expression of CD274 increases tumorigenesis and cancer stem cell phenotypes in cholangiocarcinoma. Cancer Sci. 2014;105:667-674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 194. | Nishida N, Sakai K, Morita M, Aoki T, Takita M, Hagiwara S, Komeda Y, Takenaka M, Minami Y, Ida H, Ueshima K, Nishio K, Kudo M. Association between Genetic and Immunological Background of Hepatocellular Carcinoma and Expression of Programmed Cell Death-1. Liver Cancer. 2020;9:426-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 195. | Zeng SS, Yamashita T, Kondo M, Nio K, Hayashi T, Hara Y, Nomura Y, Yoshida M, Oishi N, Ikeda H, Honda M, Kaneko S. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. J Hepatol. 2014;60:127-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 196. | Wei Y, Shi D, Liang Z, Liu Y, Li Y, Xing Y, Liu W, Ai Z, Zhuang J, Chen X, Gao Q, Jiang J. IL-17A secreted from lymphatic endothelial cells promotes tumorigenesis by upregulation of PD-L1 in hepatoma stem cells. J Hepatol. 2019;71:1206-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (1)] |
| 197. | Winograd P, Hou S, Court CM, Lee YT, Chen PJ, Zhu Y, Sadeghi S, Finn RS, Teng PC, Wang JJ, Zhang Z, Liu H, Busuttil RW, Tomlinson JS, Tseng HR, Agopian VG. Hepatocellular Carcinoma-Circulating Tumor Cells Expressing PD-L1 Are Prognostic and Potentially Associated With Response to Checkpoint Inhibitors. Hepatol Commun. 2020;4:1527-1540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 82] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 198. | Ruf B, Heinrich B, Greten TF. Immunobiology and immunotherapy of HCC: spotlight on innate and innate-like immune cells. Cell Mol Immunol. 2021;18:112-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 219] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 199. | Gao Y, Ruan B, Liu W, Wang J, Yang X, Zhang Z, Li X, Duan J, Zhang F, Ding R, Tao K, Dou K. Knockdown of CD44 inhibits the invasion and metastasis of hepatocellular carcinoma both in vitro and in vivo by reversing epithelial-mesenchymal transition. Oncotarget. 2015;6:7828-7837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 200. | Asai R, Tsuchiya H, Amisaki M, Makimoto K, Takenaga A, Sakabe T, Hoi S, Koyama S, Shiota G. CD44 standard isoform is involved in maintenance of cancer stem cells of a hepatocellular carcinoma cell line. Cancer Med. 2019;8:773-782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 201. | Koyama S, Tsuchiya H, Amisaki M, Sakaguchi H, Honjo S, Fujiwara Y, Shiota G. NEAT1 is Required for the Expression of the Liver Cancer Stem Cell Marker CD44. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 202. | Zhao Q, Zhou H, Liu Q, Cao Y, Wang G, Hu A, Ruan L, Wang S, Bo Q, Chen W, Hu C, Xu D, Tao F, Cao J, Ge Y, Yu Z, Li L, Wang H. Prognostic value of the expression of cancer stem cell-related markers CD133 and CD44 in hepatocellular carcinoma: From patients to patient-derived tumor xenograft models. Oncotarget. 2016;7:47431-47443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 203. | Zhu Z, Hao X, Yan M, Yao M, Ge C, Gu J, Li J. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int J Cancer. 2010;126:2067-2078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 226] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 204. | Yan Y, Zuo X, Wei D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Transl Med. 2015;4:1033-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 481] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 205. | Orian-Rousseau V, Sleeman J. CD44 is a multidomain signaling platform that integrates extracellular matrix cues with growth factor and cytokine signals. Adv Cancer Res. 2014;123:231-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 89] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 206. | Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4715] [Cited by in RCA: 6233] [Article Influence: 566.6] [Reference Citation Analysis (0)] |
| 207. | Sukowati CHC, Anfuso B, Fiore E, Ie SI, Raseni A, Vascotto F, Avellini C, Mazzolini G, Tiribelli C. Hyaluronic acid inhibition by 4-methylumbelliferone reduces the expression of cancer stem cells markers during hepatocarcinogenesis. Sci Rep. 2019;9:4026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 208. | Hsu JM, Xia W, Hsu YH, Chan LC, Yu WH, Cha JH, Chen CT, Liao HW, Kuo CW, Khoo KH, Hsu JL, Li CW, Lim SO, Chang SS, Chen YC, Ren GX, Hung MC. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat Commun. 2018;9:1908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 336] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 209. | Chen M, Sharma A, Lin Y, Wu Y, He Q, Gu Y, Xu ZP, Monteiro M, Gu W. Insluin and epithelial growth factor (EGF) promote programmed death ligand 1(PD-L1) production and transport in colon cancer stem cells. BMC Cancer. 2019;19:153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 210. | Sasidharan Nair V, Toor SM, Ali BR, Elkord E. Dual inhibition of STAT1 and STAT3 activation downregulates expression of PD-L1 in human breast cancer cells. Expert Opin Ther Targets. 2018;22:547-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 211. | Vasuri F, Nerpiti A, Zagnoni S, Ravaioli M, D'Errico A, Fiorentino M. Pd-ligand 1 is expressed in inflammatory cells but not in neoplastic cells in hepatocellular carcinoma: An immunohistochemical study. Acta Histochem. 2020;122:151468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 212. | Ma J, Li J, Qian M, Han W, Tian M, Li Z, Wang Z, He S, Wu K. PD-L1 expression and the prognostic significance in gastric cancer: a retrospective comparison of three PD-L1 antibody clones (SP142, 28-8 and E1L3N). Diagn Pathol. 2018;13:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 213. | Rimm DL, Han G, Taube JM, Yi ES, Bridge JA, Flieder DB, Homer R, West WW, Wu H, Roden AC, Fujimoto J, Yu H, Anders R, Kowalewski A, Rivard C, Rehman J, Batenchuk C, Burns V, Hirsch FR, Wistuba II. A Prospective, Multi-institutional, Pathologist-Based Assessment of 4 Immunohistochemistry Assays for PD-L1 Expression in Non-Small Cell Lung Cancer. JAMA Oncol. 2017;3:1051-1058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 497] [Cited by in RCA: 634] [Article Influence: 79.3] [Reference Citation Analysis (0)] |
| 214. | Smith J, Robida MD, Acosta K, Vennapusa B, Mistry A, Martin G, Yates A, Hnatyszyn HJ. Quantitative and qualitative characterization of Two PD-L1 clones: SP263 and E1L3N. Diagn Pathol. 2016;11:44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 215. | Ou DL, Lin YY, Hsu CL, Chen CW, Yu JS, Miaw SC, Hsu PN, Cheng AL, Hsu C. Development of a PD-L1-Expressing Orthotopic Liver Cancer Model: Implications for Immunotherapy for Hepatocellular Carcinoma. Liver Cancer. 2019;8:155-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |