Published online Jun 26, 2021. doi: 10.4252/wjsc.v13.i6.542
Peer-review started: February 25, 2021
First decision: April 20, 2021
Revised: May 2, 2021
Accepted: June 4, 2021
Article in press: June 4, 2021
Published online: June 26, 2021
Processing time: 120 Days and 12.5 Hours
Aberrant epigenetic alterations play a decisive role in cancer initiation and propagation via the regulation of key tumor suppressor genes and oncogenes or by modulation of essential signaling pathways. Autophagy is a highly regulated mechanism required for the recycling and degradation of surplus and damaged cytoplasmic constituents in a lysosome dependent manner. In cancer, autophagy has a divergent role. For instance, autophagy elicits tumor promoting functions by facilitating metabolic adaption and plasticity in cancer stem cells (CSCs) and cancer cells. Moreover, autophagy exerts pro-survival mechanisms to these cancerous cells by influencing survival, dormancy, immunosurveillance, invasion, metastasis, and resistance to anti-cancer therapies. In addition, recent studies have demonstrated that various tumor suppressor genes and oncogenes involved in autophagy, are tightly regulated via different epigenetic modifications, such as DNA methylation, histone modifications and non-coding RNAs. The impact of epigenetic regulation of autophagy in cancer cells and CSCs is not well-understood. Therefore, uncovering the complex mechanism of epigenetic regulation of autophagy provides an opportunity to improve and discover novel cancer therapeutics. Subsequently, this would aid in improving clinical outcome for cancer patients. In this review, we provide a comprehensive overview of the existing knowledge available on epigenetic regulation of autophagy and its importance in the maintenance and homeostasis of CSCs and cancer cells.
Core Tip: Cancer stem cells are a distinct population in the tumor bulk with enhanced self-renewal capability. Autophagy primarily exerts oncogenic activity and adaptive signals during cancer progression. Similarly, epigenetic modifications display a crucial role in tumor initiation and cancer development through its regulation of tumor suppressor genes and oncogenes. Emerging studies report epigenetic modifications regulate autophagy and metabolic pathways promoting tumor growth, elicit immunosuppressive activity and contribute to therapy resistance. Therefore, understanding this complex signaling patterns can theoretically lead to a more efficient and targeted cancer treatment.
- Citation: Mandhair HK, Novak U, Radpour R. Epigenetic regulation of autophagy: A key modification in cancer cells and cancer stem cells. World J Stem Cells 2021; 13(6): 542-567
- URL: https://www.wjgnet.com/1948-0210/full/v13/i6/542.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i6.542
Autophagy has been described to be a “self-eating” function. Autophagy is a tightly regulated catabolic process involved in the degradation of damaged organelles and misfolded proteins. The generated intermediate metabolites, such as free fatty acids, serve as an energy supply for cellular components, thus, supporting cellular homeostasis and differentiation[1]. Autophagy is activated by a multitude of environmental factors, including hypoxia, nutrient availability, DNA damage, oxidative stress, inflammation, and infections[2-6]. Defective autophagy has been associated to several pathological conditions, including inflammatory disease and cancer[7]. In cancer, autophagy has a context dependent role in disease initiation and propagation[8].
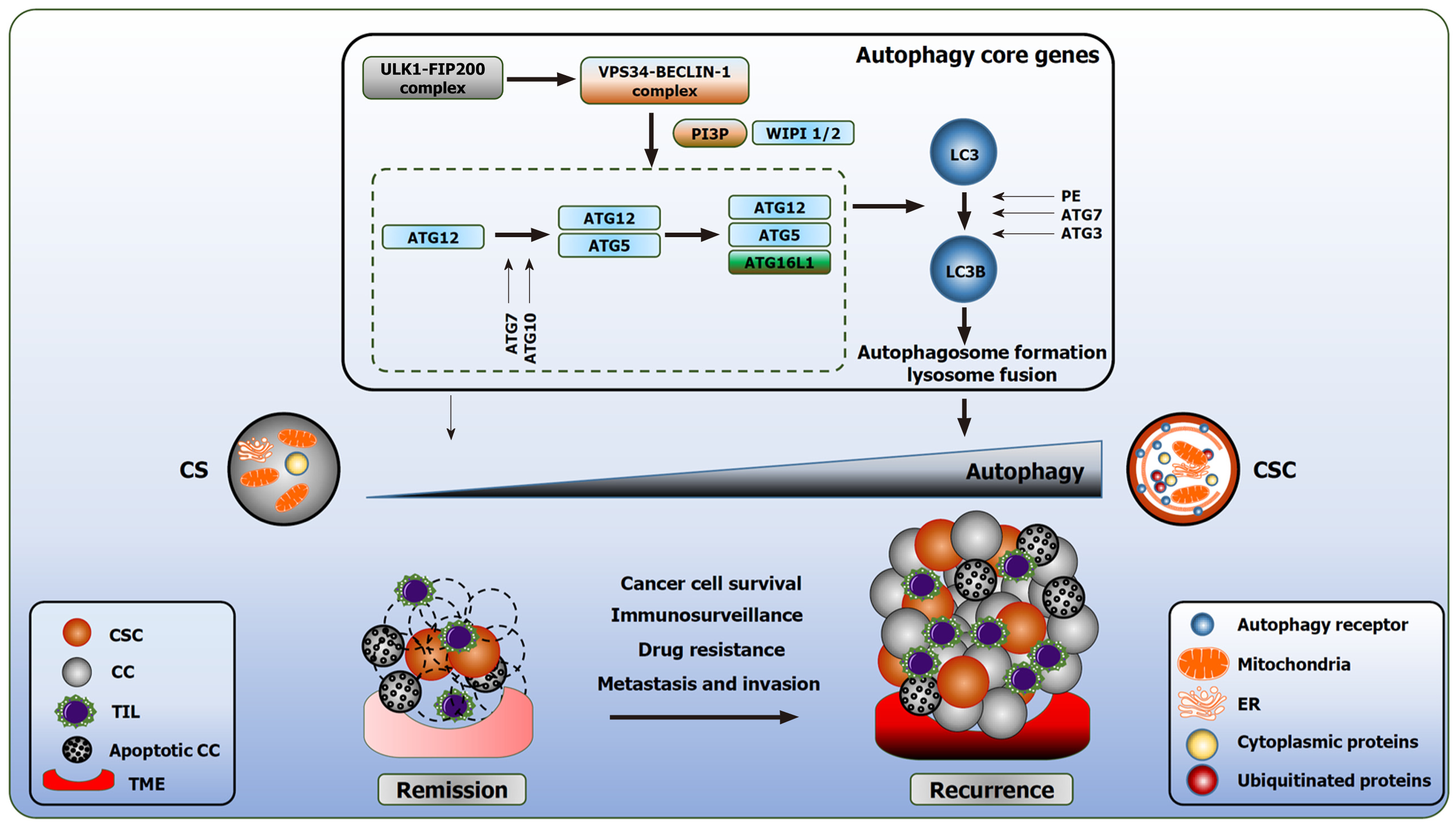
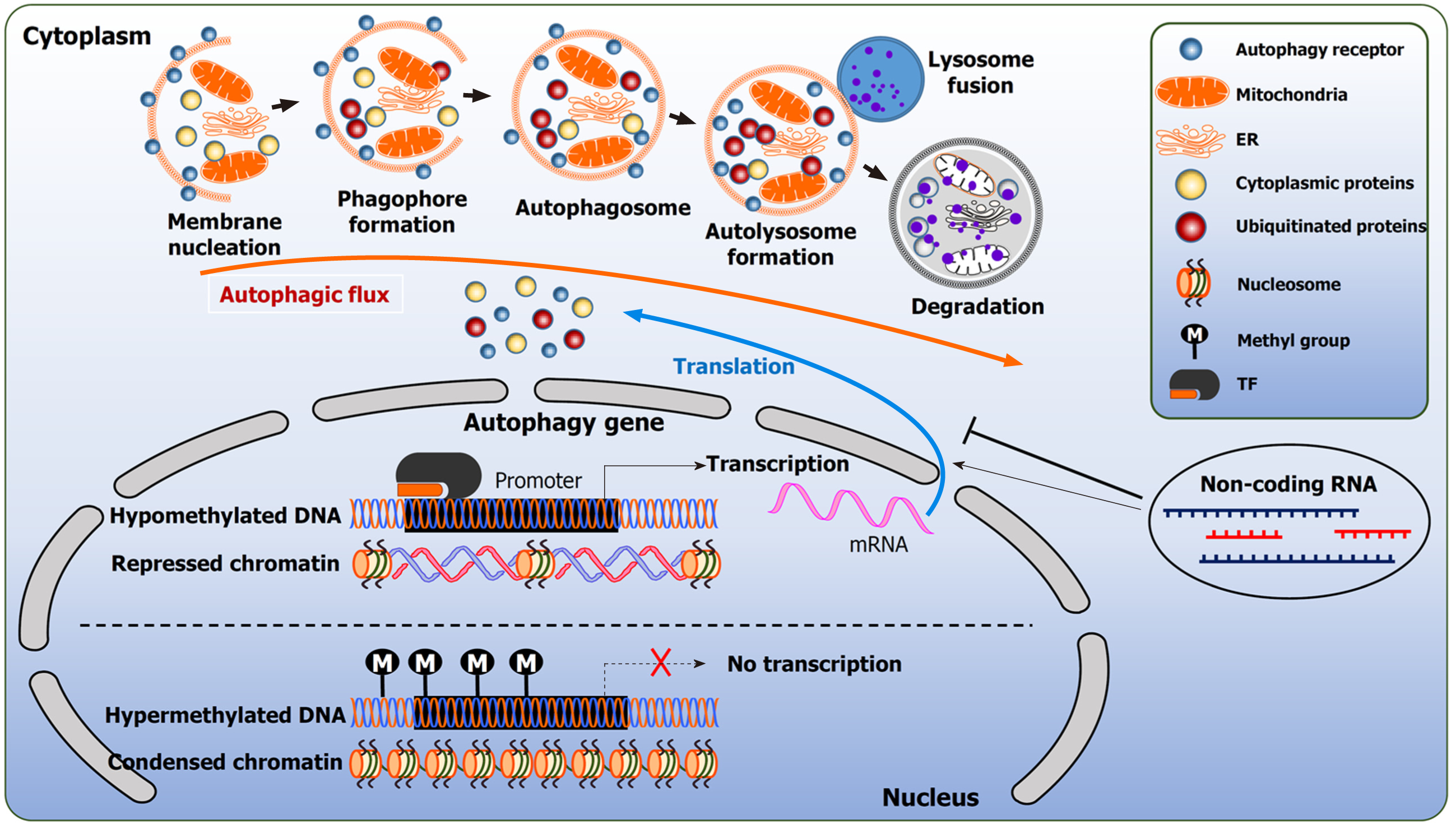
The orchestrated events of autophagy lead to the lysosome fusion for degradation. Three distinct forms of autophagy exist: microautophagy, chaperone mediated autophagy (CMA), and macroautophagy. Microautophagy is a poorly understood process. In mammalian cells, microautophagy is involved in the direct internalization of the cytosolic substrates through indentation of the lysosomal membrane. This resembles the formation of the late endosomes multivesicular bodies[9]. CMA is a form of selective autophagy. CMA targets substrates encoded with a specific pentapeptide sequence (KFERQ-like motifs). Cytosolic chaperones recognize these proteins and bind to the sequence. This interaction promotes the translocation of the cargo protein to the lysosomal membrane and bind to lysosomal associated membrane protein 2A (LAMP2A). This interaction will eventually facilitate degradation[10]. In contrast, macroautophagy (herein referred to as autophagy) is involved in the clearance of bulk cargo. In this instance, double membraned vesicles called autophagosomes, sequester their cytoplasmic cargo and fuse with the lysosome for the breakdown of the intracellular components. The biogenesis of the autophagosomes is a hallmark of autophagy[11,12]. The formation of the autophagosomes proceeds in multiple stages: initiation, elongation, and maturation. Thereafter, the autophagosome fuses with the lysosomes (Figure 1).
A consensus of studies indicate that the autophagosome membrane originates from the mitochondria and the endoplasmic reticulum (ER)[13]. However, emerging studies implicate additional cellular compartments that act as autophagy contact sites, such as the plasma membrane, Golgi and recycling endosomes[14-16]. These sites contribute to the expansion of the nascent autophagosome. The process of autophagy is governed by autophagy related genes (ATGs).
Nutrient sensing and amino acid availability are finely regulated by mammalian target of rapamycin (mTOR) and 5’ adenosine monophosphate activated protein kinase (AMPK). It is generally assumed that under glucose deprivation, the mTOR pathway is inhibited; whereas, increased amino acid availability and the promotion of cellular anabolism inhibits autophagy by activating mTOR[17,18]. Both pathways converge on unc-51-like kinase 1 (ULK1). Under nutrient rich conditions, the ULK1 complex is bound to mTOR and remains inactive[17,18].
The initiation of autophagy requires the activation of the ULK1 complex consisting of ULK2, FAK family kinase interacting protein of 200 kDa (FIP200), ATG13 and ATG101. This is followed by translocation to the ER and the phosphorylation of class III phosphatidylinositol-3-kinase vacuole protein sorting (VPS) 34 (VPS34/PI3KC3) complex, composed of VPS15, Beclin-1 (BECN1) and ATG14. This complex is also referred to as the BECN1 complex. The activation of these complexes generates a reservoir of phosphatidyl-inositol-3-phosphate (PI3P)[19]. ATG9 positive vesicles on ER contribute to the autophagosome nucleation. PI3P enriched membranes recruit effector proteins, such as WD-repeat domain phosphoinositide-interacting protein-2 (WIPI-2) and double FYVE-containing protein 1 (DFCP1)[20,21].
Furthermore, WIPI-2 promotes the expansion of the phagophore which assists in the recruitment of two conjugation systems[22]. The first conjugation complex is the covalent conjugation of ATG12-ATG5-ATG16L proteins by ATG7 and ATG10. The second conjugation system functions as an E3-like ligase, mediated by ATG12 and ATG5; assisting in the attachment of ATG8 family member microtubule associated proteins 1A/1B light chain (LC3) to phosphatidylethanolamine. The membrane bound LC3 matures and expands the autophagosome. Prior to the closure of the matured autophagosome, the ATG proteins dissociate from the autophagosome membrane, leaving the lipidated LC3 (LC3B protein, MAP1LC3B gene encoding) inside the autophagosome[23] (Figure 1).
Proteins comprising an LC3-interacting region interact with LC3 and serve as cargo receptors to target defined structures. Cargo receptors like sequestisome-1 (SQSTM1, also known as p62) and neighbor of BRCA1 facilitate the degradation of misfolded and ubiquitin-positive proteins[24]. LC3B and SQSTM1 are referred as the gold standard of measuring autophagy[25].
The formation of the autophagosome without the hierarchical activity of the core autophagy proteins is referred to as non-canonical autophagy. Limited information is currently available characterizing these alternative mechanisms[8].
Transcription factor EB (TFEB) plays a crucial role in lysosome biogenesis and autophagy by modulating the coordinated lysosomal expression and regulation (CLEAR) gene network[26]. TFEB belongs to the microphthalmia family of basic helix-loop-helix-leucine-zipper (bHLH-Zip) transcription factors (MiT family), including, transcription factor E3 (TFE3) and transcription factor EC[27]. These transcriptional factors are commonly dysregulated in cancer[27,28]. Nutrient sufficient conditions promote the phosphorylation at serine amino acids 142 and 211 in TFEB or at Serine 321 in TFE3 mediated by mTOR or extracellular signal regulated kinase-2 (ERK2). These proteins then translocate into the cytosol by 14-3-3 proteins and remain inactive[29-32]. In contrast, under starvation, lysosomal calcium is released, activating calcineurin, which triggers TFEB dephosphorylation, and nuclear translocation[33,34]. TFEB binding has been found to be enhanced under starved conditions as the promoters of autophagy core genes contain TFEB binding sites, including, UVRAG, WIPI, MAP1LC3B, SQSTM1, VPS11, VPS18 and ATG9B[35]. In contrast, zinc finger transcription factor (ZKSCAN3) has been identified as a master transcriptional repressor of autophagy[36]. Bladder cancer cells (UM-UC13) and colon cancer cells (RKO) transiently transfected with streptavidin flag tagged ZKSCAN3 vector was treated with Rapamycin (mTOR inhibitor) this downregulated LC3B protein expression. Thus, indicating the mTOR-TFEB/MiT family-ZKSCAN3 transcriptional axis is tightly regulating autophagy[37].
Nuclear factor kappa-B (NF-κB) is a crucial signaling pathway and exerts predominately pro-survival regulation of several biological functions, for example, immune responses, inflammation, cellular proliferation, differentiation, and anti-apoptotic functions. To the contrary, NF-κB activation facilitated apoptosis by upregulating BAX in breast cancer cells[38,39]. Indeed, this action required the nuclear translocation of RELA/p65 to initiate the relocalization of nucleophosmin to the cytoplasm. In consequence, this stimulated the mitochondrial localization of BAX, independent of NF-κB transcriptional activity[40]. These findings reveal a context dependent role for NF-κB.
Emerging studies report a reciprocal crosstalk between NF-κB and autophagy. Notably, under nutrient deprived conditions, the expression of autophagic genes Lc3, Atg5 and Becn1 were found to be increased in an IKK dependent phosphorylation of the p85α regulatory subunit of PI3K[41,42], which led to Akt and mTOR inhibition[42]. In contrast, in PTEN null prostate cancer cells, IKKα mediated mTOR activation resulted in autophagy suppression[43]. Interestingly, prolonged starvation promoted the accumulation of non-canonical NF-κB p52. These findings suggest the IKK complex is an essential mediator of autophagy and participates in the regulation of ATGs[41].
Furthermore, loss of IKKα in pancreatic acinar cells resulted in the accumulation of ubiquitinated proteins aggregating SQSTM1, with subsequent autophagy impairment and ER stress[44]. Moreover, knockdown of SQSTM1 in IKKα deficient pancreatic acinar cells ameliorated pancreatitis, reduced oxidative stress and ER stress markers[44]. These findings demonstrate a crucial interaction between IKKα, autophagy and ER. Interestingly, RELA/p65 regulates BECN1 transcription as it can bind to BECN1 promotor in T cells and induce autophagy[45]. Indeed, human T cell leukemia virus type 1 (HTLV-1) transformed T cells expressing retroviral oncoprotein TAX required BECN1, ATG5 and PI3KC3 to maintain constitutive activation of IκB kinase (IKK)/NF-κB and Stat3[46].
In mantle cell lymphoma (MCL), it has been reported that transglutaminase TG2/NF-κB activation stimulated interleukin 6 (IL-6) dependent autophagy for cytoprotection and tumorigenesis. ATG5KO in SP53 and JeKo cell lines proved to inhibit these signaling patterns, whilst demonstrating impaired autophagic structures, such as autophagosomes and autolysosomes, reduced proliferation rate, decreased chemoresistance, and increased apoptosis[47]. As expected, increased TG2, p50 and p65 levels were observed in MCL patients and correlated with poor prognosis[47]. These findings suggest therapeutically targeting TG2/NF-κB/IL-6 and autophagy may prove to be beneficial for MCL patients. Similar findings were reported in amino acid and serum deprived conditions in HeLa cells. Silencing BECN1 and ATG5 or BECN1 and VPS34 decreased STAT3 phosphorylation and IL-6 as compared to the control[48].
NF-κB activation in mouse model of Ras induced lung adenocarcinoma requires SQSTM1. Sqstm1-/- mice significantly reduced Ras transformed cells in colony formation assay and tumor burden. Furthermore, genetic ablation of Sqstm1 impaired NF-κB activation as Ras is necessary to stimulate IKK through the poly ubiquitination of tumor necrosis factor receptor associated factor 6[49]. As consequence, increased c-Jun NH2-terminal kinase (JNK) phosphorylation in the knockdowns promoted the reduction of reactive oxygen species (ROS) scavenger FHC. This study identified SQSTM1 as a crucial mediator of Ras induced transformed cells. In squamous cell carcinoma and melanoma cells, Chloroquine (CQ; lysosomotropic agent) treatment induced NF-κB activation, and in turn, increased the expression of hypoxia inducible factor 1-alpha (HIF-1α), and IL-8. Additionally, ATG5 and ATG7 knockdown in Mel624 melanoma cells decreased NF-κB activation and increased SQSTM1 protein, though decreased expression LC3B protein, indicating the loss of autophagosome formation. SQSTM1 or JNK knockdown impaired CQ induced IKK phosphorylation, NF-κB activation and SQSTM1[50]. It can be postulated that NF-κB signaling pathway regulates SQSTM1 levels via a positive feedback mechanism. However, SQSTM1 knockdown or NF-κB inhibition augmented CQ cytotoxicity leading to apoptosis in cancer cells[50]. To the contrary, NF-κB inhibition in macrophages due to IKKβ ablation or pharmacological IKKβ inhibitors, can enhance IL-1β secretion and mitochondrial damage by reducing SQSTM1 levels. NF-κB activation and SQSTM1 is capable of countering excessive inflammatory by suppressing NLR family pyrin domain containing 3 inflammasome activation[51]. In this instance, NF-κB activation mediates an anti-inflammatory response.
Of note, IKK complex is degraded by autophagy and inhibits NF-κB signaling. For instance, Bortezomib (proteasomal inhibitor) promoted the accumulation of poly ubiquitinated proteins in diffuse large b cell lymphoma (DLBCL) cell lines. This led to CHOP accumulation- an indicator of ER stress and LC3B dependent autophagy[52]. CQ treatment in DLBCL cell lines significantly reduced Bortezomib induced IκBα degradation and DNA binding activity of NF-κB/cREL and NF-κB nuclear translocation. Moreover, immunofluorescence data revealed accumulation of IκBα/SQSTM1 aggregation. Furthermore, the synergistic effect of CQ on Bortezomib promoted caspase 3 activation preceding apoptosis. These findings were confirmed in primary DLBCL and follicular lymphoma cells[52].
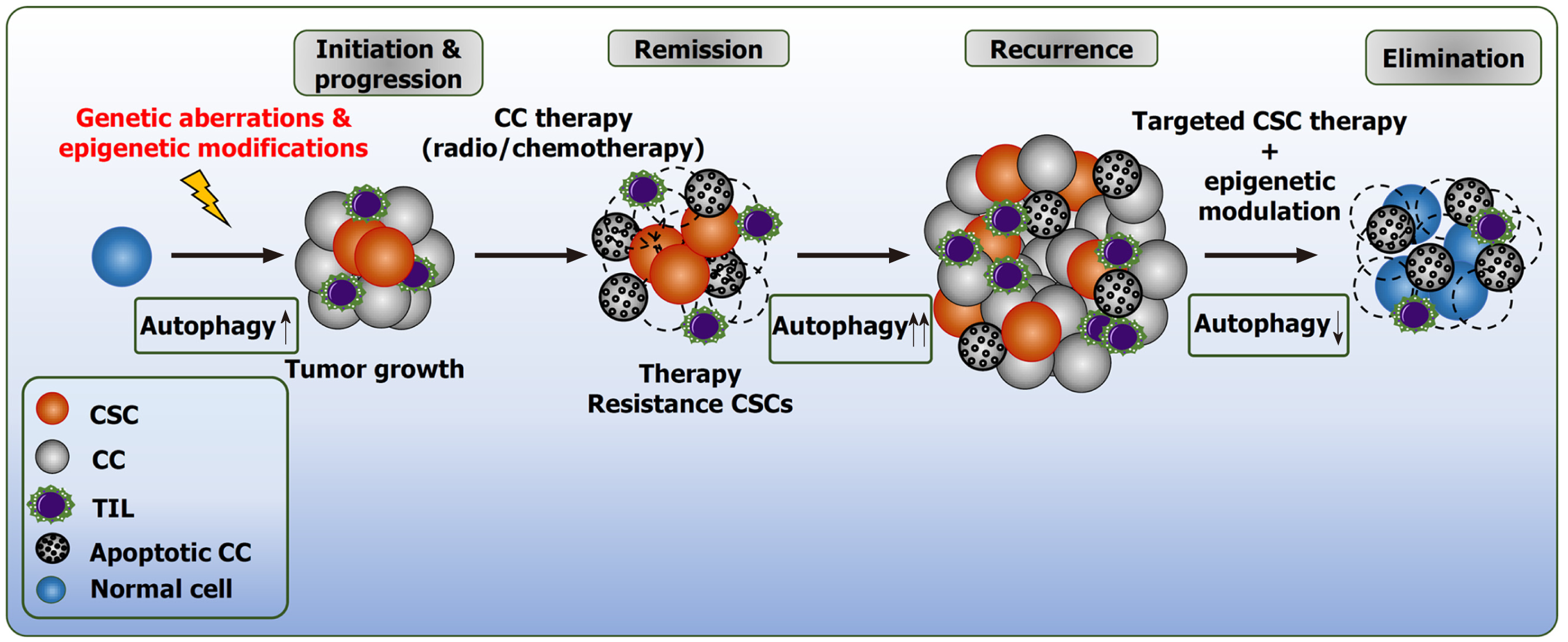
Tumorigenic potential in neoplasms is defined by phenotypical and functional heterogeneity. The intra-tumoral heterogeneity is a hallmark in cancer initiation, chemotherapeutic resistance and, in turn, negatively influences the clinical outcome for cancer patients[53]. Multiple factors contribute to this diversity, including, genetic mutations, pathologic epigenetic alterations, tumor microenvironment (TME) and the presence of cancer stem cells (CSCs; also known as tumor initiating cells)[53-55]. CSCs exhibit stem cell-properties with enhanced capabilities to escape immune response, self-renew, proliferate and metastasize[53]. In CSCs, the acquisition of genetic mutations and atypical epigenetic modifications are key underlying mechanisms involved in immunosurveillance and therapeutic resistance[56]. Overall, these factors grant CSCs resilience to chemotherapeutics and radiation[56-58]. The presence of CSCs have been detected in hematological malignancies[59-61], as well as in multiple solid cancers, including, glioblastoma[62], pancreatic[63], breast[64], ovarian[65] and liver[66].
Autophagy is a bimodal process with a context dependent role in tumorigenesis (Figure 1). In the early stage of tumor formation, autophagy is regarded as a longevity and elicits tumor suppressive functions by fostering the clearance of damaged mitochondria, preserving cellular integrity by limiting genotoxic stress and tissue damage, and decreasing inflammation[67]. During advanced stages of tumorigenesis and neoplastic transformation, autophagy deserts the above role and executes oncogenic activity by providing adaptive responses towards extracellular stimuli, including oxidative stress, hypoxia, and nutrient deprivation. Autophagy provides CSCs with recycled bioenergetic substrates for growth, supports migration and invasion by modulating the focal adhesion molecules dependent on ATG5 and FIP200[68]. In addition, autophagy stimulates the secretion of pro migratory cytokines through Rho family of small GTPases CDC42, for example, IL-6[69]. For further details, we would like to refer to our previous review deciphering the divergent roles of autophagy in CSCs and cancer cells[8].
Liu et al[66] reported, PIK3C3 governs the stemness and expansion of CD133+ liver CSCs independent of LC3B. Notably, PIK3C3 silencing reduced the protein expression of CD133 and NANOG. Overexpression of PIK3C3 increased the number of sphere formation in xenograft model treated with VPS34-IN-1 (PI3KC3 inhibitor), while reducing the proportion of CD133+ CSCs, as wells as the tumor formation capability[66]. Lung CSC stemness is dependent on TP53 signaling. TP53 knockdown prevented autophagy inhibition when ATG5 is silenced, suggesting that autophagy requires TP53 to sustain lung stemness[70]. HIF genes are transcriptionally active under oxygen sensing, such as hypoxia. Hypoxia promotes the transcription of pluripotent stem cell inducing transcription factors NANOG, SOX2, OCT4, KLF4, MYC in numerous cancer models[71,72]. In addition, primary prostate tumors expressing increased NANOG, OCT4 and HIF1α markers correlated with increased prostate tumor stage[71]. The leukemia stem cells (LSCs) in acute myeloid leukemia (AML) are dependent on ATG5 expression, an essential protein for basal autophagy. ATG5 knockdown or 3-Methyladenine (3-MA, autophagy inhibitor) demonstrated less proliferative capacity of LSCs and an increased proportion of cells in G0/G1 phase in comparison to G2[73]. Breast CSCs expressing CD44+/CD24- exhibit stem cell like properties through amplified expression of OCT4, NANOG and SQSTM1 genes. Xenograft models with depleted SQSTM1, abolished CSCs frequency and tumor growth[74]. The role of autophagy in epithelial-mesenchymal transition (EMT) is complex. CD44+/CD24-breast cancer stem-like phenotype is regulated by ATG5 gene. ATG5 knockdown and CQ treatment suppressed Vimentin (an invasion marker) in response to transforming growth factor1-β (TGF-1β) and in parallel increased CD24 transcription, and disrupted invasion[75]. On the other hand, death-effector domain-containing DNA-binding protein (DEDD) abrogated EMT transcriptional factors (SNAIL and TWIST) by inducing autophagy through PI3KC3/BECN1 complex and resulted to their degradation. Additionally, DEDD acted as a tumor suppressor by inhibiting tumor development and metastasis in breast cancer[76].
Primary DLBCL tumors expressing high BECN1 with low B cell lymphoma-2 (Bcl-2) correlated with the presence of LC3. This association led to favorable clinical outcome of patients[77,78]. Conversely, in gastric cancer, BECN1, LC3 and SQSTM1 substantially correlated with lymph node and hepatic metastasis and invasion. Unlike the previous studies, these indicators correlated with poor clinical outcome for patients with early-stage disease[79]. Similar findings were observed in patients with non-small cell lung cancer (NSCLC)[80]. Autophagy deficiency in triple negative breast cancer (TNBC) cells suppressing the trafficking of CD3+/CD28+ T cells within tumors in vivo. It can be speculated that autophagy deficiency results to T cell mediated immunosuppression. Furthermore, in TNBC patients, a negative correlation was identified with CD8+ T cell tumor infiltration and LC3B expression[81]. Moreover, downregulation of ATG7 has been reported in TNBC patients, and this correlated with a poor survival outcome. Corresponding in vitro findings demonstrated ATG7 overexpression impaired proliferation, migration and decreased EMT proteins (e.g., N-cadherin, SMA, Vimentin, SNAIL and SLUG) and upregulated E-cadherin, through abrogation of aerobic glycolysis metabolism[82].
Notably, autophagy repression improves antigen presentation by augmenting CD8+ T cell proliferation and function by attenuating tumor growth in vivo[83]. CQ treatment with dual immune-checkpoint therapy (anti-PD1 and anti-CTLA-4 antibodies) led to enhanced anti-tumoral activity by elevating the immune response. Therefore, it can be elucidated that pronounced autophagy degrades MHC-I to promote immune evasion[83].
Interestingly, autophagosomes containing cytoplasmic cargo and tumor specific antigens that fail to fuse with the lysosome are released into the extracellular milieu by cells under stressful conditions, including, hypoxia[84]; this is termed as tumor cell-released autophagosomes (TRAPs)[85,86]. In colorectal cancer and invasive melanomas, abundance of autophagosomes were reported and were associated with tumor cell proliferation, malignancy, and poor clinical outcome[87,88]. TRAPs harvested from supernatant of tumor cells or malignant effusions or ascites of cancer patients expressed LC3B positive autophagosomes accompanied with HMGB1 expression[84,86]. HMGB1 is a pro autophagic protein that directly interacts with BECN1 by displacing Bcl-2[89]. TRAPs promoted B cell differentiation into IL-10 producing regulatory B cells (B regs)[86]. TRAPs were reported to polarize monocytes to M2- like phenotype and enhance programmed death ligand-1 (PD-L1), CD163 and IL-10 levels with poor HLA-DR (MHC-II cell surface receptor) expression[90]. TRAPs elicit further immunosuppressive functions by diminishing CD4+ and CD8+ T cell proliferation and suppress interferon-gamma secretion; thus, promoting tumor growth and metastasis[86,90-92].
Epigenetics is the chemical and physical modification of DNA and chromatin, and these changes result in the regulation of gene expression without altering DNA sequences. Epigenetics mediate the gene expression via DNA methylation, histone modifications and non-coding RNAs (ncRNAs) that modifies the accessibility of the chromatin or changes the expression of different genes[93]. Epigenetic modifications are stimulated by individual genetic background or environmental factors, and therefore, can influence the occurrence of pathological conditions, including, cancer[93]. As a consequence, detrimental alterations in the epigenome can be the cause, mediator or consequence of genomic instabilities and contribute to cancer initiation and progression[94,95]. The underlying epigenetic signature in cancer cells is also referred to as “epimutation”, and similar to a gene mutation, can lead to unco
DNA methylation is the covalent binding of a methyl group to the 5'-position of cytosine, resulting to the formation of 5'-methylcytosine (5mC). It is catalyzed by DNA methyltransferase (DNMT) enzymes, which transfer the methyl groups from S-adenosyl methionine[93]. Methylation predominately affects cytosine nucleotide, as it is located next to the guanine on the 5'-side of the sequence, cytosine-p-guanine (CpG). DNA sections with high frequency sequences of CpG sites (so-called CpG islands) are found in the promoter region of several genes[96,97].
One of the epigenetic characteristics of cancer is genome-wide DNA hy
DNA methylation patterns are plastic. Depending on the degree of cell differentiation; type and age, they vary among individuals and cell types. DNA methylation analysis of tumors provides information concerning the transcriptional regulation and repression of gene expressions with tumor biological relevance[94,95,101]. Accumulating studies demonstrate that promoter hypermethylation of individual tumor entities assign as diagnostic, prognostic, or predictive biomarkers[94,95,98,99,102,103].
The second major mechanism of epigenetic regulation is histone modification, a process that controls gene expression patterns by changing the chromatin structure, making the DNA and the genes encoded on it accessible to the transcription apparatus[104,105]. Histones are nuclear proteins that associate with DNA in the nucleus and help condense it into chromatin structure. The smallest packaging unit of the compressed DNA is named a nucleosome, composed of two of each histone protein H2A, H2B, H3 and H4. The remaining histone H1 links the individual nucleosomes[105]. Histones consist of a globular center and flexible terminal arms (“histone tails”). In addition to the histone nuclei, the amino acids in these arms in particular can be chemically modified[105]. Beside methyl groups, other chemical tags, such as acetyl or phosphate residues or the addition of ubiquitin and similar smaller proteins are attached to histones. The result is variable patterns and a regular histone code that is interpreted differently by the cell's genetic apparatus[104].
The following modifications are frequently observed: H3K27ac (acetylation of H3 to lysine 27), H3K4me1, H3K4me3, H3K36me3, H3K27me3 and H3K9me3 (methyl group(s) to lysines)[106]. For instance, specific acetylation of histone H3 (H3K9ac) leads to accessibility of the chromatin and increased in the gene expression. In contrast, the methylation of the amino acid lysine in histone 3 (H3K27me2 or H3K27me3) results in compression of the chromatin with subsequently reduced transcription of the affected gene loci[106,107]. By determining these histone modifications, different chromatin states of region can be defined[107]. Histone modifications can be subjected to tightening or loose packaging under pathological conditions, including, cancer[108,109]. Histones are modified by specific enzymes. Therefore, chromatin-modifying enzymes are ideal targets for the development of specific inhibitors to modulate atypical histone modifications. Different histone deacetylase (HDAC) inhibitors (HDACis) have been approved and are currently effective drug targets in oncology.
ncRNAs are additional epigenetic regulators[93,110]. This group includes long ncRNAs (lncRNAs), comprising of at least 200 nucleotides and mainly regulate the expression of target genes. They do this by forming mRNA-riboprotein complexes with proteins. These complexes are bound to specific sites in the genome and modify those regions[110]. In comparison, short ncRNAs, such as microRNAs (miRNAs), consisting of 17-25 nucleotides regulate the expression at the post-transcriptional level[111]. They bind to the untranslated mRNA region of a target gene and suppress mRNA translation through degradation. Alternatively, gene expression is activated by an RNA interference mechanism (RNAi), using the RNA-induced silencing complex[102,111]. Therefore, lncRNAs and miRNAs effect a complex fine-tuning of the gene products on various molecular levels and play crucial role in carcinogenesis[112].
Autophagy has been implicated in cancer as an entity governing cancer progression, invasion, and metastasis. Additionally, multiple studies have recognized the contributory role of DNA methylation, histone modifications and ncRNAs in cancer. Recent accumulating reports have unveiled the convergence of autophagy and epigenetics in CSCs and cancer cells (Figure 2).
Autophagy associated genes display oncogenic function due to DNA hypomethylation, consequently leading to tumor progression. In ovarian CSCs, hypomethylation of ATG4A and histone cluster 1 H2B family member N (HIST1H2BN) were identified. Moreover, patients that harbor these genetic characteristics were found to have a poor clinical outcomes and survival[113]. Overexpression of ATG4A in SKOV3 and CP70 ovarian carcinoma cells demonstrated the tumorigenic functions of AT4A. For example, transcription factors associated to the regulation of human embryonic stem cells (ESCs) pluripotency, were found to be enhanced, such as, SOX2, NANOG, OCT4 and CD44[113]. These findings highlight the function of ATG4 promoter hypomethylation in ovarian cancer and a rational to target DNA methylation in these patients as a therapeutic opportunity[113]. Zhu et al[114] reported overexpression ATG7 promoted demethylation of ubiquitin specific peptidase (USP28) mediated through TET methylcytosine dioxygenase 1 (TET1), leading to increased USP28 expression; resulting to accumulation of CD44 protein that contributed to the invasion and lung metastasis of bladder CSCs.
Likewise, promoter hypomethylation of extracellular leucine rich repeat and fibronectin type III domain containing 2 (ELFEN2) was reported in patients with an astrocytoma, which correlated with increased ELFEN2 expression. Similar associations were found in glioma patients. ELFEN2 is a putative oncogene and elicits tumorigenic behavior by promoting autophagy via increasing the expressions of BECN1, ATG7, ATG3 and LC3B proteins[115]. In lung adenocarcinoma, the promoter of MAP1LC3A was found to be hypomethylated and contributed to resistance to epidermal growth factor receptor-tyrosine kinase inhibitors by promoting cytoprotective autophagy[116]. Aberrant DNA methylation has been described to modulate the TME. For example, hypomethylation of PIK3R5 was identified in inducible pluripotent stem cells conditioned with media of Lewis lung carcinoma[117].
Chen et al[118] reported the anti-tumoral role of autophagy in esophageal squamous cell carcinoma (ESCC). Hypomethylation of phospholipase C epsilon 1 (PLCE1) in primary ESCC tumors elicited poor clinical prognosis. PLCE1 triggers tumorigenesis through autophagy suppression and downregulation of P53 activity and MDM2 ubiquitination resulting in P53 degradation. PLCE1 silencing induced autophagy and subsequently attenuated tumor cell proliferation through P53[118]. Moreover, Caveolin-1 (CAV1) has been associated with glucose metabolism. In primary colorectal tumors and various corresponding cell lines, an abnormal overexpression of CAV1 due to promoter hypomethylation was demonstrated. CAV1 silencing led to the promotion of autophagy through AMPK and P53 dependent cell cycle arrest[119].
Promoter hypermethylation is an important causative factor in repressing tumor suppressor genes; for example, hypermethylation of BECN1 gene. In primary sporadic breast tumors, monoallelic loss of BECN1 was found in 45% of tumors and this loss was accompanied with significant promoter hypermethylation[120]. Equally, ATG2B, ATG4D, ATG9A and ATG9B promoter hypermethylation was identified in specimens of invasive ductal carcinoma. In autophagy, these genes are relevant. For instance, ATG2 homologs act as peripheral membrane proteins and are associated to cellular nucleation. ATG4D is part of the ATG4 family and is associated in regulating the ATG8-LC3 conjugation system. ATG9 protein is a functional orthologue that interacts with the phagophore[121]. Genome-wide methylation analysis and bisulfite sequencing reported low levels of ULK2 transcripts due to hypermethylation in glioblastoma[122]. In NSCLC, promoter methylation of transcription factor 21 is associated with repressed autophagy; this negatively correlated with tumor stage, metastasis, and invasion[123]. Methylation analysis revealed silencing of MAP1LC3Av1 caused by Helicobacter pylori infection in non-cancerous and cancerous gastric mucosae cells, which led to impaired autophagy[124]. Equally, MAP1LC3Av1, not MAP1LC3B, was frequently inactivated in ESCC due to demethylation and overexpression of MAP1LC3Av1 in those cells and exhibited anti-tumoral activity, such as decreasing the tumor volume and weight in vivo[125].
In gastric cancer, promoter hypermethylation of tumor suppresser gene KLOTHE was identified. Overexpression of KLOTHE engaged in autophagy induction by increasing LC3-I/II ratio and decreased the protein phosphorylation of insulin growth factor-1 receptor, insulin receptor substrate-1, PI3K, Akt and mTOR signaling, as well as apoptosis in gastric cancer cells[126]. Hypermethylation of BCL2/Adenovirus E1B 19KDa Protein-Interacting Protein 3 (BNIP3) promoter has been reported in human colorectal cancer cells. Treatment with demethylating agents, such as 5-aza-2’-deoxycytidine (DAC) is capable of restoring this BNIP3 via KRAS dependency and MAPK kinase activation[127]. GABARAP family members were differentially expressed in human breast cancer biopsies, suggesting global aberrant DNA methylation. Grade III lymph node-positive breast cancer tissues strongly correlated with the downregulation of GABARAPL1[128]. It was determined that nicotinamide N-methyl transferase (NNMT) negatively regulates autophagy. NNMT knockdown enhanced liver tumor growth under nutrient deprived conditions through PP2A methylation and decreased the ULK1 activity augmenting protective autophagy[129]. ATG5 promoter was hypermethylated in melanoma and was associated with suppressed basal autophagy, hence, promoting oncogene induced cell proliferation in primary epidermal melanocytes[130].
Several findings report core autophagy-related genes could be silenced via histone deacetylations[131]. In human and mouse CSCs, HDAC enzyme activity has been suggested to function as a pluripotent factor. Pharmacological inhibition or knockdown of HDAC6, inhibited CSCs proliferation and reduced the protein levels of POU5F1, NANOG and SOX2 (pluripotent factors) in human NT2/D1 and murine P19 embryonic carcinoma CSCs[132]. HDAC6 silencing led to the activation of autophagy with increased proteins levels of ATG5, ATG7 and decreased SQSTM1. ATG7 and ATG12 knockdown NT2/D1 decreased HDAC6 protein levels and promoted differentiation. In comparison, HDAC6 silencing, downregulated autophagy and promoted apoptosis in differentiated breast cancer cells[132]. These findings are indicative of the discriminatory role of HDAC6 in the maintenance of CSCs, as well as differentiated cancer cells. Similarly, glioma CSCs expressing increased levels of HDAC6 contributed to their stemness[132,133]. Chemotherapy and radiotherapy resistance is often mediated by the stemness characteristic of CSCs and is an important prognostic factor in various tumors. Yang et al[133] indicated HDAC6 inhibition rendered the transcription of SHH signaling pathway, decreased glioma CSCs neurosphere formation and protein expression of SOX2 and BMIL1, suggesting the induction of cell differentiation. Subsequently, HDAC6 knockdown resulted to radiosensitivity in glioma CSCs[133]. HDAC6 silencing achieved radio sensitization through the activation of BECN1; however, autophagy inhibition through 3-MA countered this phenomenon[134]. It can be proposed that HDAC6 promotes radio resistance by suppressing BECN1.
A study on neuroblastoma cohort indicated that ATG4D positively correlated with HDAC10 expression. HDAC10highexpression was correlated with significantly poor survival outcome of patients. In addition, HDAC10 overexpression in neuroblastoma cells promoted Doxorubicin resistance in neuroblastoma cells through HSC70/HSP70 interaction via its deacetylation function[135]. SIRT6 (Sirtuin family member of NAD dependent deacetylase) was reported to be overexpressed in primary ESCC samples. SIRT6 initiated LC3B mediated autophagic flux in ESCC cells by interacting with ULK1 and inhibited mTOR. In parallel, SIRT6 promoted cellular proliferation and participated in regulating the G2M phase. These observations support the potential oncogenic role of SIRT6 and its role in activating autophagy[136]. HDAC1 suppression led to tumor growth regression by inciting mitotic defects and caspase-independent of autophagic cell death via LC3B in hepatocellular carcinoma (HCC)[137]. Similarly, overexpression of HDAC8 is prevalent in oral squamous cell carcinoma and HDAC8 silencing led to anti-proliferative effects and cell death mediated through caspase 9, 3 and 7. The administration of CQ with silenced HDAC8 substantially reduced cellular viability (as compared to HDAC8 knockdown without CQ)[138]. In salivary mucoepidermoid carcinoma cells, HDAC7 silencing attenuated cellular proliferation and c-MYC expression and triggered G2/M phase cell cycle arrest mediated through P27. This stimulated apoptosis and autophagy[139].
To the contrary, HDAC activity has been implicated in positive regulation of autophagy in differentiated cancer cells. It has been reported that HDAC6 dependent autophagy compensated for the impaired ubiquitin-proteosome pathway[140]. Ectopic overexpression of HDAC6 in hepatocellular carcinoma cell line Hep3B reduced cell growth and proliferation without inducing pro-apoptotic proteins. Notably, HDAC6 activated autophagic cell death. Xenograft mouse model demonstrated similar findings and determined that autophagy cell death required the activation of BECN1 and JNK[141].
The Bromodomain and extra-terminal domain (BET) family are epigenetic regulators that preferentially bind to acetylated histones. Proteomic analysis revealed binding of BET proteins caused them to localize by the chromosome recruiting positive transcription elongation factor b (P-TEFb). Transcriptional kinase cyclin dependent kinase-9 (CDK9) and regulatory subunits CyclinT1, T2 or K bind to BRD4 resulting in the phosphorylation of pol II (RNA polymerase II), which results in gene transcription[142]. The BET family is composed of four members: BRD2, BRD3, BRD4 and BRDT[143]. BRD4 has a prominent role in G1 phase in the cell cycle[144]. Colocalization of BRD4 and P-TEFB was identified in late mitotic to early G1 phase. This interaction promoted the recruitment of P-TEFb to mitotic chromosomes to stimulate gene transcription relating to growth and trigger progression to S phase[145].
Impairment of histone acetylation results in aberrant gene expression. For example, BRD4 overexpression has been attributed to enhanced transcription of MYC[146]. In colon cancer cell lines and primary tumors, BRD4 is frequently aberrantly hyper
BET inhibitor JQ1 and genetic silencing of BRD4 in pancreatic ductal adenocarcinoma (PDAC) KP-4 cells led to an increase in LC3B and WIPI expression and autophagic flux, suggesting the formation of autophagosomes and upregulation of autophagosome-lysosome fusion protein[152]. BRD4 is a negative repressor of autophagy; its knockdown upregulated the autophagy genes BECN1, VMP1 (vacuole membrane protein-1), PIK3C3, ATG2A, ATG9B and MAP1LC3B, the autophagy cargo proteins SQSTM1 and OPTN (optineurin), as well as the autophagosome-lysosome fusion genes PLEKHM1 and TECPR1. Consistent findings were observed in overexpression studies, whilst the addition of JQ1 countered these findings. BRD4 knockdown promoted an upregulation in the lysosome biogenesis and function genes and at protein levels: LAMP1, LAMP2, acid sphingomyelinase (ASM), a-glucosidase (GAA), and heavy chain of mature cathepsin B (CTSB HC) and cathepsin D (CTSD HC). Furthermore, silencing studies confirmed that the BRD4-NUT axis is capable of transcriptionally regulating autophagy independently of the MiT family (TFEB, TFE3 and MITF)[152].
As discussed previously, starvation induced autophagy acquires the activation of AMPK and the direct phosphorylation of ULK1 and inhibition of mTOR. ATG7 is crucial in starvation induced autophagy for autophagosome formation, recycling of amino acids, mitochondria integrity and the clearance of ubiquitin-positive aggregates[153]. The role of AMPK, mTOR and ULK1 has gained much attention in numerous solid cancers[146,154-158]. Treatment of AML cell lines and primary CD34+ enriched LSCs with the Bet inhibitor JQ1 led to the downregulation of c-MYC protein[159]. Autophagy activation was preferentially observed in JQ1-resistant AML primary cells and in selected LSC cell lines KG1 and KG1a. AMPK (pThr172)/ULK1 (pSer555) pathway was found to induce autophagy independent of mTOR, thereby conferring resistance to JQ1 mediated apoptosis[160]. AMPK provides metabolic adaption in cancer cells in vitro and xenograft models through maintenance of ATP and NADH homeostasis[161]. AMPK deletion in MLL-AF9 (mixed lineage leukemia-AF-9 genes) suppressed disease propagation and depleted the LSCs in the hypoxic environment of the bone marrow[159]. Sakamaki et al[152] suggests AMPK and SIRT1 (Sirtuin-1) function as nutrient sensing mechanisms with the ability to directly interact with BRD4 to govern the transcription of autophagy genes. As such, nutrient deprivation would initiate the dissociation of BRD4 from the autophagy gene promoters, thus, inducing de-repression of autophagy gene transcription and cell survival[152].
Interestingly, the BR4 inhibitor 9f induced ATG5 dependent autophagy associated cell death in breast cancer cells by preventing the interaction between BRD4-AMPK. Furthermore, ATG5 silencing led to LC3B lipidation and accumulation of SQSTM1; however, this did not disrupt AMPK activation. These results indicate that 9f modulates autophagy through ATG5 by using the AMPK-mTOR-ULK1 pathway[155]. ATG5 silencing in bladder cancer cells diminished anti-proliferative ability of BRD4 inhibitor JQ1. In addition, AMPKα knockdown elicited similar results. Collectively, these findings suggest ATG5 dependent autophagy is induced by JQ1, utilizing the LKB1-AMPK-mTOR axis[157]. It was reported that inactivation of Akt (Ser473)-mTOR (Ser2448) contributed to cellular resistance to JQ1 in ovarian cancer cells and overexpression of AKT1 reversed the resistant phenotype[146]. To the contrary, Akt inhibitors are thought to overcome BET inhibitor resistance in primary prostate cancer cells harboring mutated Speckle Type POZ Protein[162].
G9a (also known as EHMT2) is a histone methyltransferase (KMT) enzyme targeting the lysine. Specifically, this enzyme mediates the histone H3K9 mono-methylation and demethylation at histone 3 lysine 9 (H3K9me1 and H3K9me2). Functionally, this promotes the recruitment of additional epigenetic regulators and repressors of transcription[163]. Gene silencing usually requires the methylation of H3K9. G9a silencing led to the formation of vacuole like structures in the pancreatic cancer cell line SU86.86. These findings indicate that G9a regulates the MAP1LC3B and WIPI1 promoters, as well as, diabetes and obesity regulated (DOR) gene promoters. Starvation induced autophagy led to the reduction of H3K9me2 and an increased H3K9ac[164]. Treatment of MCF-7 breast cancer cells with the G9a inhibitor BIX0124 led to the recruitment of NF-κB on the BECN1 promoter and elevated the intracellular ROS. These events reduced the levels of H3K9me2, resulting in an open chromatin structure. This increased the upregulation of BECN1 and promoted autophagy. Breast tumor samples with high G9a and low BECN1 expression exhibited a poor prognosis[165]. It can be postulated that BECN1 is a tumor suppressor governed by G9a. Immunohistochemistry data of paired lung adenocarcinoma and lung squamous cell carcinoma samples revealed a significant higher expression of G9a correlating with metastasis and a poor prognosis of patients[166]. In comparison, low expressions of H3K9me2 and G9a could predict a better prognosis for patients with gastric cancer[167].
Autophagy is an essential pro-survival mechanism and provides adaptive responses. The inhibition of G9a elicits autophagy. mTOR is an integral part of nutrient and energy sensing. G9a inhibitor BIX01294 administration in HeLa, SHEP1 and U2OS cell lines induced LC3B. Interestingly, BIX01294 treatment decreased the phosphorylation of ribosome protein S6 kinase (S6K), an essential mTOR substrate[168]. RHEB overexpression studies in bladder transitional cancer cells attenuated autophagy and autophagic cell death capacity of BIX01294, indicating G9a inhibition is mTOR mediated[169]. Similarly, GA001, an G9 antagonist, induced autophagy in breast cancer cells via the AMPK-mTOR-ULK1 pathway[170]. Ding et al[168] suggests that G9a mediates H3K9 methylation, serving as a potential sensor between amino acid availability, cellular growth and proliferation functioning by the activation of transcription factor 4 (ATF4). ATF4 is part of the unfolded protein response triggered by metabolic stress[171]. Glioblastoma cell lines: A172 and U87MG, treated with BIX01294 and knockdown of G9a, revealed activation of LC3B dependent autophagy. Inhibition of G9a, activated Akt/HIF1α expression. Tumor cells treated with BIX01294 exhibited elevated LC3B and PKM2 protein levels resulting in activation of autophagy[172].
Hypoxic stress has shown to increase H3K9me2 and decrease in acetylated H3K9, in multiple cancer cell lines. Additionally, hypoxia mimetics similarly enhanced the global expression of H3K9me2, G9a expression and activity. Hypoxic stress decreased the mRNA levels of MIH1 (involved in mismatch repair) and DHFR (dihydrofolate reductase) genes and increased H3K9me2 levels in their promoter regions[173]. Hypoxia induced autophagy has been implicated in CSCs of different tumor types, including breast and glioma, and this correlated with poor clinical outcome[174,175]. Ablation of BECN1, ATG5 and ATG7 has been reported to enhance cell death in hypoxia condition[176]. Kaempferol (flavonoid, HDACi) was found to mediate autophagy in gastric cancer cells by increased protein expression of LC3B, BECN1 and ATG5 and reduced levels of SQSTM1[131]. Kaempferol induced autophagy by targeting G9a expression. G9a knockdown and Kaempferol co-treated experiments indicated a reduction in G9a binding to LC3B promoter. However, 3-MA rescued this effect by repressing LC3B and cell death[131]. It has been proposed that inhibition of HDAC-G9a pathway may potentiate anti-tumoral activity in cancer cells[177].
Transcriptome analysis detected upregulation of gallbladder cancer drug resistant-associated IncRNA1 (GBCDRInc1) in gallbladder cancer tissues, and this increase is implicated in chemoresistance of gallbladder cancer cells[178]. Phosphoglycerate kinase 1 (PGK1) was found to directly interact with GBCDRInc1 by preventing its ubiquitination and breakdown of PGK1, resulting to the formation of ATG5-ATG12 complexes. GBCRlnc1 knockdown models treated with CQ reduced the autophagic activity and enhanced sensitivity to Doxorubicin in resistant gallbladder cancer cells in vitro and in vivo[178]. In colorectal cancer, the expression of LncRNA-H19Highis associated with poor recurrent free survival. H19 is associated to 5’Fluorouracil (5-FU) chemoresistance mediated by increased autophagy induction via SIRT1[179]. LncRNA MALAT-1 is upregulated in DLBCL compared to normal B lymphocytes. Silencing of MALAT-1 decreased lymphoma proliferation and invasion, enhanced cell cycle arrest and apoptosis. MALAT-1 knockdown promoted the generation of autophagosomes by increasing the protein levels of LC3 I/II along with SQSTM1 expressions to induce autophagy. MALAT-1 silencing in xenograft model significantly reduced tumor volume and weight[180].
Several studies have shown the controversial role of miRNA (miR) in the context of autophagy, tumorigenesis and chemoresistance of cancer cells. Indeed, miR-1251-5p levels were significantly elevated in advanced stages of primary ovarian tumors. MiR-1251-5p elicited oncogenic behavior through hyperproliferation, mediating cell cycle and initiating the LC3B dependent autophagy by targeting the tubulin binding cofactor CC (TBCC) in ovarian cancer[181]. Metastatic breast cancer invading lymphatic nodes, expressed increased miR-224-5p levels which correlated with low levels of LC3B protein and increased SQSTM1, suggesting the suppression of autophagy in a SMAD4 dependent manner[182]. SMAD4 protein is a crucial mediator of TGF-β[183]. Interactions between acute promyelocytic leukemia cells and bone marrow stromal cells activate NF-κB signaling, resulting in a negative regulation of miR-23a-5p. Consequently, increased levels of the autophagic proteins (for example BECN1, ATG5-ATG12 complex and LC3B), indicated the induction of cytoprotective autophagy. MiR-23a-5p overexpression led to Arsenic trioxide (APO) and Daunorubicin (DNR) sensitivity[184]. Autophagy inhibition with adjuvant ATO treatment re-established chemotherapy sensitivity in leukemic cells[184]. Invasion and migration of glioma cells is dependent on P72 expression, the downregulation of BECN1 and autophagy, causing an increase in miR-34-5p and miR-5195-3P expression[185].
Similarly, glioma stem cells are reliant on MIR93 (miR-93) for cell growth and sphere formation in vitro by repressing BECN1, ATG5, ATG4B and SQSTM1 proteins[186]. ATG7 gene overexpression facilitated in the degradation of the forkhead transcription factor FOXO4a mediated through autophagy. Subsequently, repressing miR-145 transcription and further reducing its binding to 3’UTR (3’ untranslated region) of PD-L1, thus promoting PD-L1 expression. These events enhance the stem like property, tumorigenesis, and invasive features of bladder cancer cells[187]. Similarly, in cervical and lung cancer, MiR7-3HG targeted the 3’UTR of AMBRA1 mRNA promoting the downregulation of AMBRA1, acting as oncogenesis and MYC phosphorylation, leading to autophagy blockade[188].
Notably, certain tumor suppressor miRNAs elicit anti-tumoral activity through the regulation of autophagy. For instance, miR-1262 was detected in gastric cardia adenocarcinoma[189]. ULK1 gene expression was negatively regulated with the expression of miR-1262. Functional assays, such as, proliferation and cell cycle analysis, colony formation and wound healing elucidated the tumor suppressive function of miR-1262[189]. MiR-101 negatively regulates basal and Rapamycin-induced autophagy in breast cancer cells by targeting ATG4D, RAB5A and STMN1 genes[190]. Likewise, miR-137 overexpression inhibited ATG5 dependent autophagy in pancreatic cells by sensitizing the cells to the anti-tumoral activity of Doxorubicin in vitro and in vivo[191]. MiR-130a downregulated DICER1 and ATG2B mRNA expressions in chronic lymphocytic leukemia. This led to a reduction in the autophagosome generation due to autophagy inhibition and promoting apoptosis[192]. Consistent with the previous findings, autophagy inhibition is essential in treating AML by targeting HMGB1[193]. Increased MiR-32a levels accompanied by low HMGB1 expression, inhibited all-trans retinoic acid and induced autophagy in AML cells via stimulating LC3 Lipidation[193]. MiR-224-3p overexpression repressed glioblastoma cell proliferation and ablated hypoxia stimulated protective autophagy through targeting ATG5 and FIP200 genes[194].
Epigenetic therapeutics are promising targets to modify autophagy and to reactivate repressed tumor suppressor genes in different tumor types (Figure 3). Epigenetic abnormalities have been identified in several cancers modulating ATGs (Table 1). Inhibition of DNMTs and HDACs have been clinically developed to achieve the above objective.
| Epigenetic | Type of epigenetic modification | Cancer model | Genes | Autophagy modulation | Ref. |
| DNA methylation | Hypermethylation | Breast cancer | ATG2B, ATG4D, ATG9A, ATG9B, Beclin-1, ARHI | Repressed | Li et al[120], Zhang et al[121] and Yu et al[203] |
| Hypermethylation | Colorectal cancer | BTG1, PCDH17, BTG1, BTG3, MAP1LC3Av1 | Repressed | Muhammad et al[124], Zhao et al[204], Hu et al[205] and Gou et al[206] | |
| Hypermethylation | Glioma and Glioblastoma | Ulk2, ANKDD1A | Repressed | Shukla et al[122] and Feng et al[207] | |
| Hypomethylation | Glioblastoma | ELFN2 | Activated | Liu et al[115] | |
| Hypermethylation | Hepatocellular carcinoma | BCLB | Repressed | Liu et al[208] | |
| Hypermethylation | Liver cancer | PP2A | Activated | Shin et al[129] | |
| Hypermethylation | Lung cancer | TCF21, TUSC3 | Repressed | Chen et al[123] and Peng et al[209] | |
| Hypomethylation | Lung cancer | LC3A | Activated | Nihira et al[116] | |
| Hypermethylation | Medulloblastoma | ATG16L1 | Repressed | Cruzeiro et al[210] | |
| Hypermethylation | Melanoma | ATG5 | Repressed | Liu et al[130] | |
| Hypermethylation | Ovarian cancer | ARHI | Repressed | Yu et al[203] | |
| Hypomethylation | Ovarian cancer | ATG4A | Activated | Liao et al[113] | |
| Histone modification | Histone methylation or acetylation | Breast cancer | EHMT2, Beclin-1 | Repressed | Park et al[165] and Sun et al[211] |
| Histone methylation | Bladder cancer | SMYD3 | Activated | Shen et al[212] | |
| Histone acetylation | Colorectal cancer | FOXO1 | Activated | Zhao et al[213] | |
| Histone demethylation | Gastric cancer | KDM2B | Repressed | Zhao et al[214] | |
| Histone demethylation or deacetylation | Glioma | KDM4A, SIRT3 | Repressed | Wang et al[215] and Qiao et al[216] | |
| Histone deacetylation | Hepatocellular carcinoma | HDAC6 | Activated | Jung et al[141] | |
| Histone deacetylation | Neuroblastoma | HDAC10 | Activated | Oehme et al[135] | |
| Histone methylation | Neuroblastoma | G9a | Repressed | Ke et al[217] | |
| Histone deacetylation | Prostate cancer | SIRT1 | Activated | Powell et al[218] | |
| Histone deacetylation | Salivary mucoepidermoid carcinoma | HDAC7 | Activated | Ahn and Yoon[139] |
It is reported that DAC treatment and additional administration of Panobinostat or valproic acid (HDAC inhibitors) downregulated oncogenic MYC expression and epigenetic modifiers, such as lysine demethylase KDM2B (demethylase for H3K36me2/ H3K4me3) and histone-lysine methyltransferase SUV39H1, leading to anti-leukemic activity in AML. Moreover, genes associated with metabolism were enriched under the combination therapy[195]. Monotherapy of DAC at low doses ablated clonogenicity of primary leukemic cells. Combined therapy of DAC and Azacitidine (DNMT inhibitor), decreased tumorigenicity in a xenograft model of breast cancer and in human primary breast cancer cells. Additionally, human breast CSCs displayed decreased self-renewal capacity in mammospheres[196]. In colorectal cancer, HDAC1 inhibitors, such as valproic acid and suberoylanilide hydroxamic acid, increased the expression of UVRAG (component of BECN1 complex). Increased UVRAG levels attenuated 5-FU mediated toxicity in colorectal cancer cells. HDAC1 inhibition potentiated cell death via DNA damage[197]. The novel HDAC8 inhibitor (HMC) elicited pro-apoptotic functions by activating ATG5 and LC3B autophagy proteins in MCF-7 breast cancer cells. Co-treatment of HMC with 3-MA or CQ autophagy inhibitors partially countered HMC-induced cell death, suggesting autophagy elicited a protective role[198].
Trichostatin A (HDAC inhibitor) and valproic acid promoted autophagy and apoptosis in pancreatic cancer cells[199]. CM-272 (G9a/DNMT dual methyltransferase inhibitor) elicited immunogenic cell death and apoptosis in human bladder cancer. Furthermore, CM-272 decreased proliferation, inhibited cell cycle progression and induced autophagy; this correlated with a decrease in H3K9me2 and 5-methylcytosine. In vivo model demonstrated CM-272 enhanced the response to anti-PDL1 and attenuated tumorigenesis in PIK3CA mutated bladder cancer cells. DMNT1 inhibition enhanced MHC-I in breast cancer leading to the recruitment and activation of CD8+ T cells[200].
LncRNA-HOTAIR elicited anti-tumoral activity in chondrosarcoma by upregulating miR-454-3p leading to STAT3 activation and elevation of ATG12 protein[201]. Combination treatment of valproic acid and Temsirolimus (mTOR inhibi
Abnormal epigenetic alterations have been implicated in cancer initiation, development, and therapy resistance. Epigenetic mechanisms, such as DNA methylation, histone modification or ncRNAs, can regulate crucial cellular processes like autophagy. In aggressive tumors, epigenetic changes of autophagy can deliberately influence immunosurveillance, maintenance, therapy resistance and invasion. Therefore, understanding the underlying mechanisms involved in epigenetic regulation of autophagy can enhance cytotoxic effects, and thus eliminate tumor cell resistance and prevent disease reoccurrence. Moreover, the application of epigenetic modulators, such as demethylating agents or HDAC inhibitors not only aim to normalize atypical epigenetic patterns on DNA sequences or histones but provide a newer therapeutic opportunity to regulate autophagy in malignant cells. Preclinical and small cohort studies have provided evidence that this approach can be effective and improve cancer prognosis in patients. In hindsight, a challenge lies in using epigenetic modulators on a defined section of the genome. For instance, clinically approved DNA methylation inhibitors or HDACis act genome wide. Currently, patient-specific modification of target genes, using CRISPR/Cas9-based epigenome editors are being developed. It is therefore imperative to identify and validate novel therapeutic approaches to directly target epigenetic changes of autophagy-dependent genes or pathways in resistant cancer cells and CSCs, as this will potentially improve personalized cancer therapy and clinical outcome for cancer patients.
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country/Territory of origin: Switzerland
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cao ZF, Morenikeji OB, Witte KE, Wu QN S-Editor: Gao CC L-Editor: Filipodia P-Editor: Zhang YL
| 1. | Riffelmacher T, Clarke A, Richter FC, Stranks A, Pandey S, Danielli S, Hublitz P, Yu Z, Johnson E, Schwerd T, McCullagh J, Uhlig H, Jacobsen SEW, Simon AK. Autophagy-Dependent Generation of Free Fatty Acids Is Critical for Normal Neutrophil Differentiation. Immunity 2017; 47: 466-480. e5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 229] [Article Influence: 28.6] [Reference Citation Analysis (1)] |
| 2. | Cosin-Roger J, Simmen S, Melhem H, Atrott K, Frey-Wagner I, Hausmann M, de Vallière C, Spalinger MR, Spielmann P, Wenger RH, Zeitz J, Vavricka SR, Rogler G, Ruiz PA. Hypoxia ameliorates intestinal inflammation through NLRP3/mTOR downregulation and autophagy activation. Nat Commun. 2017;8:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 245] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 3. | Thomas M, Davis T, Loos B, Sishi B, Huisamen B, Strijdom H, Engelbrecht AM. Autophagy is essential for the maintenance of amino acids and ATP levels during acute amino acid starvation in MDAMB231 cells. Cell Biochem Funct. 2018;36:65-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 4. | Wang Y, Zhang N, Zhang L, Li R, Fu W, Ma K, Li X, Wang L, Wang J, Zhang H, Gu W, Zhu WG, Zhao Y. Autophagy Regulates Chromatin Ubiquitination in DNA Damage Response through Elimination of SQSTM1/p62. Mol Cell. 2016;63:34-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 169] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 5. | Bartolini D, Dallaglio K, Torquato P, Piroddi M, Galli F. Nrf2-p62 autophagy pathway and its response to oxidative stress in hepatocellular carcinoma. Transl Res. 2018;193:54-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 6. | Liu K, Hong D, Zhang F, Li X, He M, Han X, Zhang G, Xu G, Stonehouse NJ, Jiang Z, An W, Guo L. MicroRNA-106a Inhibits Autophagy Process and Antimicrobial Responses by Targeting ULK1, ATG7, and ATG16L1 During Mycobacterial Infection. Front Immunol. 2020;11:610021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 7. | Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-1075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5421] [Cited by in RCA: 5293] [Article Influence: 311.4] [Reference Citation Analysis (0)] |
| 8. | Mandhair HK, Arambasic M, Novak U, Radpour R. Molecular modulation of autophagy: New venture to target resistant cancer stem cells. World J Stem Cells. 2020;12:303-322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (1)] |
| 9. | Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131-139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 715] [Cited by in RCA: 673] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 10. | Hao Y, Kacal M, Ouchida AT, Zhang B, Norberg E, Vakifahmetoglu-Norberg H. Targetome analysis of chaperone-mediated autophagy in cancer cells. Autophagy. 2019;15:1558-1571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 11. | Nakatogawa H. Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol. 2020;21:439-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 577] [Article Influence: 115.4] [Reference Citation Analysis (0)] |
| 12. | Djavaheri-Mergny M, Giuriato S, Tschan MP, Humbert M. Therapeutic Modulation of Autophagy in Leukaemia and Lymphoma. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 13. | Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1168] [Cited by in RCA: 1348] [Article Influence: 112.3] [Reference Citation Analysis (0)] |
| 14. | Nascimbeni AC, Giordano F, Dupont N, Grasso D, Vaccaro MI, Codogno P, Morel E. ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. EMBO J. 2017;36:2018-2033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 164] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 15. | Gui X, Yang H, Li T, Tan X, Shi P, Li M, Du F, Chen ZJ. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567:262-266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 393] [Cited by in RCA: 878] [Article Influence: 146.3] [Reference Citation Analysis (0)] |
| 16. | Imai K, Hao F, Fujita N, Tsuji Y, Oe Y, Araki Y, Hamasaki M, Noda T, Yoshimori T. Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J Cell Sci. 2016;129:3781-3791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 111] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 17. | Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214-226. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3114] [Cited by in RCA: 2998] [Article Influence: 176.4] [Reference Citation Analysis (0)] |
| 18. | Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2835] [Cited by in RCA: 3009] [Article Influence: 136.8] [Reference Citation Analysis (0)] |
| 19. | Su H, Yang F, Wang Q, Shen Q, Huang J, Peng C, Zhang Y, Wan W, Wong CCL, Sun Q, Wang F, Zhou T, Liu W. VPS34 Acetylation Controls Its Lipid Kinase Activity and the Initiation of Canonical and Non-canonical Autophagy. Mol Cell 2017; 67: 907-921. e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 115] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 20. | Karanasios E, Walker SA, Okkenhaug H, Manifava M, Hummel E, Zimmermann H, Ahmed Q, Domart MC, Collinson L, Ktistakis NT. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun. 2016;7:12420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 230] [Cited by in RCA: 237] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 21. | Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell. 2012;23:1860-1873. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 448] [Cited by in RCA: 440] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 22. | Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 Links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55:238-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 617] [Cited by in RCA: 674] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 23. | Fracchiolla D, Chang C, Hurley JH, Martens S. A PI3K-WIPI2 positive feedback loop allosterically activates LC3 Lipidation in autophagy. J Cell Biol. 2020;219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 24. | Turco E, Witt M, Abert C, Bock-Bierbaum T, Su MY, Trapannone R, Sztacho M, Danieli A, Shi X, Zaffagnini G, Gamper A, Schuschnig M, Fracchiolla D, Bernklau D, Romanov J, Hartl M, Hurley JH, Daumke O, Martens S. FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates. Mol Cell 2019; 74: 330-346. e11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 245] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 25. | Gottlieb RA, Andres AM, Sin J, Taylor DP. Untangling autophagy measurements: all fluxed up. Circ Res. 2015;116:504-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 129] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 26. | He R, Wang M, Zhao C, Shen M, Yu Y, He L, Zhao Y, Chen H, Shi X, Zhou M, Pan S, Liu Y, Guo X, Li X, Qin R. TFEB-driven autophagy potentiates TGF-β induced migration in pancreatic cancer cells. J Exp Clin Cancer Res. 2019;38:340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 27. | Slade L, Pulinilkunnil T. The MiTF/TFE Family of Transcription Factors: Master Regulators of Organelle Signaling, Metabolism, and Stress Adaptation. Mol Cancer Res. 2017;15:1637-1643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 28. | Steingrímsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 541] [Cited by in RCA: 599] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 29. | Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 756] [Cited by in RCA: 1027] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 30. | Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 821] [Cited by in RCA: 1014] [Article Influence: 78.0] [Reference Citation Analysis (0)] |
| 31. | Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095-1108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1414] [Cited by in RCA: 1512] [Article Influence: 116.3] [Reference Citation Analysis (0)] |
| 32. | Martina JA, Diab HI, Brady OA, Puertollano R. TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 2016;35:479-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 243] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 33. | Yang TT, Yu RY, Agadir A, Gao GJ, Campos-Gonzalez R, Tournier C, Chow CW. Integration of protein kinases mTOR and extracellular signal-regulated kinase 5 in regulating nucleocytoplasmic localization of NFATc4. Mol Cell Biol. 2008;28:3489-3501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 34. | Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, Settembre C, Wang W, Gao Q, Xu H, Sandri M, Rizzuto R, De Matteis MA, Ballabio A. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17:288-299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 773] [Cited by in RCA: 1073] [Article Influence: 107.3] [Reference Citation Analysis (0)] |
| 35. | Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429-1433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2700] [Cited by in RCA: 2554] [Article Influence: 182.4] [Reference Citation Analysis (0)] |
| 36. | Pan H, Yan Y, Liu C, Finkel T. The role of ZKSCAN3 in the transcriptional regulation of autophagy. Autophagy. 2017;13:1235-1238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 37. | Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM, Boyd DD. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50:16-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 218] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 38. | Fan P, Tyagi AK, Agboke FA, Mathur R, Pokharel N, Jordan VC. Modulation of nuclear factor-kappa B activation by the endoplasmic reticulum stress sensor PERK to mediate estrogen-induced apoptosis in breast cancer cells. Cell Death Discov. 2018;4:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 39. | Mohan S, Abdelwahab SI, Kamalidehghan B, Syam S, May KS, Harmal NS, Shafifiyaz N, Hadi AH, Hashim NM, Rahmani M, Taha MM, Cheah SC, Zajmi A. Involvement of NF-κB and Bcl2/Bax signaling pathways in the apoptosis of MCF7 cells induced by a xanthone compound Pyranocycloartobiloxanthone A. Phytomedicine. 2012;19:1007-1015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 113] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 40. | Khandelwal N, Simpson J, Taylor G, Rafique S, Whitehouse A, Hiscox J, Stark LA. Nucleolar NF-κB/RelA mediates apoptosis by causing cytoplasmic relocalization of nucleophosmin. Cell Death Differ. 2011;18:1889-1903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 108] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 41. | Comb WC, Cogswell P, Sitcheran R, Baldwin AS. IKK-dependent, NF-κB-independent control of autophagic gene expression. Oncogene. 2011;30:1727-1732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 87] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 42. | Comb WC, Hutti JE, Cogswell P, Cantley LC, Baldwin AS. p85α SH2 domain phosphorylation by IKK promotes feedback inhibition of PI3K and Akt in response to cellular starvation. Mol Cell. 2012;45:719-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 43. | Dan HC, Adli M, Baldwin AS. Regulation of mammalian target of rapamycin activity in PTEN-inactive prostate cancer cells by I kappa B kinase alpha. Cancer Res. 2007;67:6263-6269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 44. | Li N, Wu X, Holzer RG, Lee JH, Todoric J, Park EJ, Ogata H, Gukovskaya AS, Gukovsky I, Pizzo DP, VandenBerg S, Tarin D, Atay C, Arkan MC, Deerinck TJ, Moscat J, Diaz-Meco M, Dawson D, Erkan M, Kleeff J, Karin M. Loss of acinar cell IKKα triggers spontaneous pancreatitis in mice. J Clin Invest. 2013;123:2231-2243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 45. | Copetti T, Bertoli C, Dalla E, Demarchi F, Schneider C. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol. 2009;29:2594-2608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 211] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 46. | Chen L, Liu D, Zhang Y, Zhang H, Cheng H. The autophagy molecule Beclin 1 maintains persistent activity of NF-κB and Stat3 in HTLV-1-transformed T lymphocytes. Biochem Biophys Res Commun. 2015;465:739-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 47. | Zhang H, Chen Z, Miranda RN, Medeiros LJ, McCarty N. TG2 and NF-κB Signaling Coordinates the Survival of Mantle Cell Lymphoma Cells via IL6-Mediated Autophagy. Cancer Res. 2016;76:6410-6423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 48. | Yoon S, Woo SU, Kang JH, Kim K, Shin HJ, Gwak HS, Park S, Chwae YJ. NF-κB and STAT3 cooperatively induce IL6 in starved cancer cells. Oncogene. 2012;31:3467-3481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 107] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 49. | Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 465] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 50. | Yang S, Qiang L, Sample A, Shah P, He YY. NF-κB Signaling Activation Induced by Chloroquine Requires Autophagosome, p62 Protein, and c-Jun N-terminal Kinase (JNK) Signaling and Promotes Tumor Cell Resistance. J Biol Chem. 2017;292:3379-3388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 51. | Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, He F, Boassa D, Perkins G, Ali SR, McGeough MD, Ellisman MH, Seki E, Gustafsson AB, Hoffman HM, Diaz-Meco MT, Moscat J, Karin M. NF-κB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell. 2016;164:896-910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 835] [Cited by in RCA: 928] [Article Influence: 103.1] [Reference Citation Analysis (0)] |
| 52. | Jia L, Gopinathan G, Sukumar JT, Gribben JG. Blocking autophagy prevents bortezomib-induced NF-κB activation by reducing I-κBα degradation in lymphoma cells. PLoS One. 2012;7:e32584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 53. | Radpour R, Forouharkhou F. Single-cell analysis of tumors: Creating new value for molecular biomarker discovery of cancer stem cells and tumor-infiltrating immune cells. World J Stem Cells. 2018;10:160-171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 54. | Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4428] [Cited by in RCA: 4184] [Article Influence: 85.4] [Reference Citation Analysis (1)] |
| 55. | Radpour R. Tracing and targeting cancer stem cells: New venture for personalized molecular cancer therapy. World J Stem Cells. 2017;9:169-178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 56. | Miao Y, Yang H, Levorse J, Yuan S, Polak L, Sribour M, Singh B, Rosenblum MD, Fuchs E. Adaptive Immune Resistance Emerges from Tumor-Initiating Stem Cells. Cell 2019; 177: 1172-1186. e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 220] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 57. | Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 883] [Cited by in RCA: 1184] [Article Influence: 131.6] [Reference Citation Analysis (0)] |
| 58. | Forster S, Radpour R. Molecular Immunotherapy: Promising Approach to Treat Metastatic Colorectal Cancer by Targeting Resistant Cancer Cells or Cancer Stem Cells. Front Oncol. 2020;10:569017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 59. | Song S, Li Y, Zhang K, Zhang X, Huang Y, Xu M, Li S, Guan X, Yang T, Liu Z, Jiang J, Luo Y, Lan Y. Cancer Stem Cells of Diffuse Large B Cell Lymphoma Are Not Enriched in the CD45+CD19- cells but in the ALDHhigh Cells. J Cancer. 2020;11:142-152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 60. | Radpour R, Riether C, Simillion C, Höpner S, Bruggmann R, Ochsenbein AF. CD8+ T cells expand stem and progenitor cells in favorable but not adverse risk acute myeloid leukemia. Leukemia. 2019;33:2379-2392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 61. | Riether C, Radpour R, Kallen NM, Bürgin DT, Bachmann C, Schürch CM, Lüthi U, Arambasic M, Hoppe S, Albers CE, Baerlocher GM, Ochsenbein AF. Metoclopramide treatment blocks CD93-signaling-mediated self-renewal of chronic myeloid leukemia stem cells. Cell Rep. 2021;34:108663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 62. | Wang X, Prager BC, Wu Q, Kim LJY, Gimple RC, Shi Y, Yang K, Morton AR, Zhou W, Zhu Z, Obara EAA, Miller TE, Song A, Lai S, Hubert CG, Jin X, Huang Z, Fang X, Dixit D, Tao W, Zhai K, Chen C, Dong Z, Zhang G, Dombrowski SM, Hamerlik P, Mack SC, Bao S, Rich JN. Reciprocal Signaling between Glioblastoma Stem Cells and Differentiated Tumor Cells Promotes Malignant Progression. Cell Stem Cell 2018; 22: 514-528. e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 187] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 63. | Wang VM, Ferreira RMM, Almagro J, Evan T, Legrave N, Zaw Thin M, Frith D, Carvalho J, Barry DJ, Snijders AP, Herbert E, Nye EL, MacRae JI, Behrens A. CD9 identifies pancreatic cancer stem cells and modulates glutamine metabolism to fuel tumour growth. Nat Cell Biol. 2019;21:1425-1435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 64. | Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983-3988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7830] [Cited by in RCA: 7730] [Article Influence: 351.4] [Reference Citation Analysis (0)] |
| 65. | Parte SC, Batra SK, Kakar SS. Characterization of stem cell and cancer stem cell populations in ovary and ovarian tumors. J Ovarian Res. 2018;11:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 66. | Liu F, Wu X, Qian Y, Jiang X, Wang Y, Gao J. PIK3C3 regulates the expansion of liver CSCs and PIK3C3 inhibition counteracts liver cancer stem cell activity induced by PI3K inhibitor. Cell Death Dis. 2020;11:427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 67. | Clarke AJ, Simon AK. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat Rev Immunol. 2019;19:170-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 265] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 68. | Assar EA, Tumbarello DA. Loss of the Essential Autophagy Regulators FIP200 or Atg5 Leads to Distinct Effects on Focal Adhesion Composition and Organization. Front Cell Dev Biol. 2020;8:733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 69. | Razidlo GL, Burton KM, McNiven MA. Interleukin-6 promotes pancreatic cancer cell migration by rapidly activating the small GTPase CDC42. J Biol Chem. 2018;293:11143-11153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 70. | Wang J, Liu D, Sun Z, Ye T, Li J, Zeng B, Zhao Q, Rosie Xing H. Autophagy augments the self-renewal of lung cancer stem cells by the degradation of ubiquitinated p53. Cell Death Dis. 2021;12:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 71. | Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, Hubaud A, Stadler B, Choi M, Bar M, Tewari M, Liu A, Vessella R, Rostomily R, Born D, Horwitz M, Ware C, Blau CA, Cleary MA, Rich JN, Ruohola-Baker H. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640-4652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 418] [Cited by in RCA: 418] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 72. | Yan Y, Liu F, Han L, Zhao L, Chen J, Olopade OI, He M, Wei M. HIF-2α promotes conversion to a stem cell phenotype and induces chemoresistance in breast cancer cells by activating Wnt and Notch pathways. J Exp Clin Cancer Res. 2018;37:256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 133] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 73. | Li Y, Jiang Y, Cheng J, Ma J, Li Q, Pang T. ATG5 regulates mesenchymal stem cells differentiation and mediates chemosensitivity in acute myeloid leukemia. Biochem Biophys Res Commun. 2020;525:398-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 74. | Xu LZ, Li SS, Zhou W, Kang ZJ, Zhang QX, Kamran M, Xu J, Liang DP, Wang CL, Hou ZJ, Wan XB, Wang HJ, Lam EW, Zhao ZW, Liu Q. p62/SQSTM1 enhances breast cancer stem-like properties by stabilizing MYC mRNA. Oncogene. 2017;36:304-317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 75. | Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, Martin-Castillo B, Vellon L, Menendez JA. Autophagy positively regulates the CD44(+) CD24(-/Low) breast cancer stem-like phenotype. Cell Cycle. 2011;10:3871-3885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 155] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 76. | Lv Q, Wang W, Xue J, Hua F, Mu R, Lin H, Yan J, Lv X, Chen X, Hu ZW. DEDD interacts with PI3KC3 to activate autophagy and attenuate epithelial-mesenchymal transition in human breast cancer. Cancer Res. 2012;72:3238-3250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 144] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 77. | Nicotra G, Mercalli F, Peracchio C, Castino R, Follo C, Valente G, Isidoro C. Autophagy-active beclin-1 correlates with favourable clinical outcome in non-Hodgkin lymphomas. Mod Pathol. 2010;23:937-950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 78. | Huang JJ, Zhu YJ, Lin TY, Jiang WQ, Huang HQ, Li ZM. Beclin 1 expression predicts favorable clinical outcome in patients with diffuse large B-cell lymphoma treated with R-CHOP. Hum Pathol. 2011;42:1459-1466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 79. | Masuda GO, Yashiro M, Kitayama K, Miki Y, Kasashima H, Kinoshita H, Morisaki T, Fukuoka T, Hasegawa T, Sakurai K, Toyokawa T, Kubo N, Tanaka H, Muguruma K, Masaichi O, Hirakawa K. Clinicopathological Correlations of Autophagy-related Proteins LC3, Beclin 1 and p62 in Gastric Cancer. Anticancer Res. 2016;36:129-136. [PubMed] |
| 80. | Wang X, Du Z, Li L, Shi M, Yu Y. Beclin 1 and p62 expression in non-small cell lung cancer: relation with malignant behaviors and clinical outcome. Int J Clin Exp Pathol. 2015;8:10644-10652. [PubMed] |
| 81. | Li ZL, Zhang HL, Huang Y, Huang JH, Sun P, Zhou NN, Chen YH, Mai J, Wang Y, Yu Y, Zhou LH, Li X, Yang D, Peng XD, Feng GK, Tang J, Zhu XF, Deng R. Autophagy deficiency promotes triple-negative breast cancer resistance to T cell-mediated cytotoxicity by blocking tenascin-C degradation. Nat Commun. 2020;11:3806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 123] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 82. | Li M, Liu J, Li S, Feng Y, Yi F, Wang L, Wei S, Cao L. Autophagy-related 7 modulates tumor progression in triple-negative breast cancer. Lab Invest. 2019;99:1266-1274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 83. | Yamamoto K, Venida A, Yano J, Biancur DE, Kakiuchi M, Gupta S, Sohn ASW, Mukhopadhyay S, Lin EY, Parker SJ, Banh RS, Paulo JA, Wen KW, Debnath J, Kim GE, Mancias JD, Fearon DT, Perera RM, Kimmelman AC. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature. 2020;581:100-105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 335] [Cited by in RCA: 834] [Article Influence: 166.8] [Reference Citation Analysis (0)] |
| 84. | Zhang Y, Pan N, Sheng Y, Zhou M, Wen Z, Chen Y, Huang F, Wang LX. Hypoxia enhances IL-10-producing B cell generation through upregulating high-mobility group B1 on tumor cell-released autophagosomes. Immunol Lett. 2019;216:36-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 85. | Yi Y, Zhou Z, Shu S, Fang Y, Twitty C, Hilton TL, Aung S, Urba WJ, Fox BA, Hu HM, Li Y. Autophagy-assisted antigen cross-presentation: Autophagosome as the argo of shared tumor-specific antigens and DAMPs. Oncoimmunology. 2012;1:976-978. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 86. | Zhou M, Wen Z, Cheng F, Ma J, Li W, Ren H, Sheng Y, Dong H, Lu L, Hu HM, Wang LX. Tumor-released autophagosomes induce IL-10-producing B cells with suppressive activity on T lymphocytes via TLR2-MyD88-NF-κB signal pathway. Oncoimmunology. 2016;5:e1180485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 87. | Lazova R, Camp RL, Klump V, Siddiqui SF, Amaravadi RK, Pawelek JM. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res. 2012;18:370-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 255] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 88. | Katheder NS, Khezri R, O'Farrell F, Schultz SW, Jain A, Rahman MM, Schink KO, Theodossiou TA, Johansen T, Juhász G, Bilder D, Brech A, Stenmark H, Rusten TE. Microenvironmental autophagy promotes tumour growth. Nature. 2017;541:417-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 358] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 89. | Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ 3rd, Lotze MT. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881-892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 646] [Cited by in RCA: 782] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 90. | Wen ZF, Liu H, Gao R, Zhou M, Ma J, Zhang Y, Zhao J, Chen Y, Zhang T, Huang F, Pan N, Zhang J, Fox BA, Hu HM, Wang LX. Tumor cell-released autophagosomes (TRAPs) promote immunosuppression through induction of M2-like macrophages with increased expression of PD-L1. J Immunother Cancer. 2018;6:151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 129] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 91. | Chen YQ, Li PC, Pan N, Gao R, Wen ZF, Zhang TY, Huang F, Wu FY, Ou XL, Zhang JP, Zhu XJ, Hu HM, Chen K, Cai YL, Wang LX. Tumor-released autophagosomes induces CD4+ T cell-mediated immunosuppression via a TLR2-IL-6 cascade. J Immunother Cancer. 2019;7:178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 92. | Gao R, Ma J, Wen Z, Yang P, Zhao J, Xue M, Chen Y, Aldarouish M, Hu HM, Zhu XJ, Pan N, Wang LX. Tumor cell-released autophagosomes (TRAP) enhance apoptosis and immunosuppressive functions of neutrophils. Oncoimmunology. 2018;7:e1438108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 93. | Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1920] [Cited by in RCA: 2277] [Article Influence: 175.2] [Reference Citation Analysis (0)] |
| 94. | Aghagolzadeh P, Radpour R. New trends in molecular and cellular biomarker discovery for colorectal cancer. World J Gastroenterol. 2016;22:5678-5693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 51] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (2)] |
| 95. | Radpour R, Barekati Z, Kohler C, Holzgreve W, Zhong XY. New trends in molecular biomarker discovery for breast cancer. Genet Test Mol Biomarkers. 2009;13:565-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 96. | Radpour R, Kohler C, Haghighi MM, Fan AX, Holzgreve W, Zhong XY. Methylation profiles of 22 candidate genes in breast cancer using high-throughput MALDI-TOF mass array. Oncogene. 2009;28:2969-2978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 97. | Radpour R, Haghighi MM, Fan AX, Torbati PM, Hahn S, Holzgreve W, Zhong XY. High-throughput hacking of the methylation patterns in breast cancer by in vitro transcription and thymidine-specific cleavage mass array on MALDI-TOF silico-chip. Mol Cancer Res. 2008;6:1702-1709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 98. | Radpour R, Barekati Z, Haghighi MM, Kohler C, Asadollahi R, Torbati PM, Holzgreve W, Zhong XY. Correlation of telomere length shortening with promoter methylation profile of p16/Rb and p53/p21 pathways in breast cancer. Mod Pathol. 2010;23:763-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 99. | Weissmann C, Weber H. The interferon genes. Prog Nucleic Acid Res Mol Biol. 1986;33:251-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 100. | Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930-935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4860] [Cited by in RCA: 4389] [Article Influence: 274.3] [Reference Citation Analysis (0)] |
| 101. | Radpour R, Sikora M, Grussenmeyer T, Kohler C, Barekati Z, Holzgreve W, Lefkovits I, Zhong XY. Simultaneous isolation of DNA, RNA, and proteins for genetic, epigenetic, transcriptomic, and proteomic analysis. J Proteome Res. 2009;8:5264-5274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 102. | Jaggi B, Poon SS, MacAulay C, Palcic B. Imaging system for morphometric assessment of absorption or fluorescence in stained cells. Cytometry. 1988;9:566-572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 103. | Dellon AL, Mackinnon SE. Musculoaponeurotic variations along the course of the median nerve in the proximal forearm. J Hand Surg Br. 1987;12:359-363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 52] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 104. | Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 559] [Cited by in RCA: 581] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 105. | Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6680] [Cited by in RCA: 6767] [Article Influence: 241.7] [Reference Citation Analysis (0)] |
| 106. | Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1676] [Cited by in RCA: 1642] [Article Influence: 126.3] [Reference Citation Analysis (0)] |
| 107. | Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016-1028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1143] [Cited by in RCA: 1368] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 108. | Mancarella D, Plass C. Epigenetic signatures in cancer: proper controls, current challenges and the potential for clinical translation. Genome Med. 2021;13:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 109. | Radpour R, Stucki M, Riether C, Ochsenbein AF. Epigenetic Silencing of Immune-Checkpoint Receptors in Bone Marrow- Infiltrating T Cells in Acute Myeloid Leukemia. Front Oncol. 2021;11:663406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 110. | Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3151] [Cited by in RCA: 3684] [Article Influence: 263.1] [Reference Citation Analysis (0)] |
| 111. | Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2370] [Cited by in RCA: 2257] [Article Influence: 161.2] [Reference Citation Analysis (0)] |
| 112. | Zhang P, Brinton LT, Williams K, Sher S, Orwick S, Tzung-Huei L, Mims AS, Coss CC, Kulp SK, Youssef Y, Chan WK, Mitchell S, Mustonen A, Cannon M, Phillips H, Lehman AM, Kauffman T, Beaver L, Canfield D, Grieselhuber NR, Alinari L, Sampath D, Yan P, Byrd JC, Blachly JS, Lapalombella R. Targeting DNA Damage Repair Functions of Two Histone Deacetylases, HDAC8 and SIRT6, Sensitizes Acute Myeloid Leukemia to NAMPT Inhibition. Clin Cancer Res. 2021;27:2352-2366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 113. | Liao YP, Chen LY, Huang RL, Su PH, Chan MW, Chang CC, Yu MH, Wang PH, Yen MS, Nephew KP, Lai HC. Hypomethylation signature of tumor-initiating cells predicts poor prognosis of ovarian cancer patients. Hum Mol Genet. 2014;23:1894-1906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 114. | Zhu J, Huang G, Hua X, Li Y, Yan H, Che X, Tian Z, Liufu H, Huang C, Li J, Xu J, Dai W, Huang H. CD44s is a crucial ATG7 downstream regulator for stem-like property, invasion, and lung metastasis of human bladder cancer (BC) cells. Oncogene. 2019;38:3301-3315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 115. | Liu C, Fu H, Liu X, Lei Q, Zhang Y, She X, Liu Q, Sun Y, Li G, Wu M. LINC00470 Coordinates the Epigenetic Regulation of ELFN2 to Distract GBM Cell Autophagy. Mol Ther. 2018;26:2267-2281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 116. | Nihira K, Miki Y, Iida S, Narumi S, Ono K, Iwabuchi E, Ise K, Mori K, Saito M, Ebina M, Sato I, Maemondo M, Yamada-Okabe H, Kondo T, Sasano H. An activation of LC3A-mediated autophagy contributes to de novo and acquired resistance to EGFR tyrosine kinase inhibitors in lung adenocarcinoma. J Pathol. 2014;234:277-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 117. | Oo AKK, Calle AS, Nair N, Mahmud H, Vaidyanath A, Yamauchi J, Khayrani AC, Du J, Alam MJ, Seno A, Mizutani A, Murakami H, Iwasaki Y, Chen L, Kasai T, Seno M. Up-Regulation of PI 3-Kinases and the Activation of PI3K-Akt Signaling Pathway in Cancer Stem-Like Cells Through DNA Hypomethylation Mediated by the Cancer Microenvironment. Transl Oncol. 2018;11:653-663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 118. | Chen Y, Xin H, Peng H, Shi Q, Li M, Yu J, Tian Y, Han X, Chen X, Zheng Y, Li J, Yang Z, Yang L, Hu J, Huang X, Liu Z, Zhou H, Cui X, Li F. Hypomethylation-Linked Activation of PLCE1 Impedes Autophagy and Promotes Tumorigenesis through MDM2-Mediated Ubiquitination and Destabilization of p53. Cancer Res. 2020;80:2175-2189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 119. | Ha TK, Her NG, Lee MG, Ryu BK, Lee JH, Han J, Jeong SI, Kang MJ, Kim NH, Kim HJ, Chi SG. Caveolin-1 increases aerobic glycolysis in colorectal cancers by stimulating HMGA1-mediated GLUT3 transcription. Cancer Res. 2012;72:4097-4109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 120. | Li Z, Chen B, Wu Y, Jin F, Xia Y, Liu X. Genetic and epigenetic silencing of the beclin 1 gene in sporadic breast tumors. BMC Cancer. 2010;10:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 107] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 121. | Zhang X, Li C, Wang D, Chen Q, Li CL, Li HJ. Aberrant methylation of ATG2B, ATG4D, ATG9A and ATG9B CpG island promoter is associated with decreased mRNA expression in sporadic breast carcinoma. Gene. 2016;590:285-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 122. | Shukla S, Patric IR, Patil V, Shwetha SD, Hegde AS, Chandramouli BA, Arivazhagan A, Santosh V, Somasundaram K. Methylation silencing of ULK2, an autophagy gene, is essential for astrocyte transformation and tumor growth. J Biol Chem. 2014;289:22306-22318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 123. | Chen B, Zeng C, Ye Y, Wu D, Mu Z, Liu J, Xie Y, Wu H. Promoter methylation of TCF21 may repress autophagy in the progression of lung cancer. J Cell Commun Signal. 2018;12:423-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 124. | Muhammad JS, Nanjo S, Ando T, Yamashita S, Maekita T, Ushijima T, Tabuchi Y, Sugiyama T. Autophagy impairment by Helicobacter pylori-induced methylation silencing of MAP1LC3Av1 promotes gastric carcinogenesis. Int J Cancer. 2017;140:2272-2283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 125. | Bai H, Inoue J, Kawano T, Inazawa J. A transcriptional variant of the LC3A gene is involved in autophagy and frequently inactivated in human cancers. Oncogene. 2012;31:4397-4408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 126. | Xie B, Zhou J, Shu G, Liu DC, Chen J, Yuan L. Restoration of klotho gene expression induces apoptosis and autophagy in gastric cancer cells: tumor suppressive role of klotho in gastric cancer. Cancer Cell Int. 2013;13:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 127. | Swiderek E, Kalas W, Wysokinska E, Pawlak A, Rak J, Strzadala L. The interplay between epigenetic silencing, oncogenic KRas and HIF-1 regulatory pathways in control of BNIP3 expression in human colorectal cancer cells. Biochem Biophys Res Commun. 2013;441:707-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 128. | Hervouet E, Claude-Taupin A, Gauthier T, Perez V, Fraichard A, Adami P, Despouy G, Monnien F, Algros MP, Jouvenot M, Delage-Mourroux R, Boyer-Guittaut M. The autophagy GABARAPL1 gene is epigenetically regulated in breast cancer models. BMC Cancer. 2015;15:729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 129. | Shin JH, Park CW, Yoon G, Hong SM, Choi KY. NNMT depletion contributes to liver cancer cell survival by enhancing autophagy under nutrient starvation. Oncogenesis. 2018;7:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 130. | Liu H, He Z, von Rütte T, Yousefi S, Hunger RE, Simon HU. Down-regulation of autophagy-related protein 5 (ATG5) contributes to the pathogenesis of early-stage cutaneous melanoma. Sci Transl Med. 2013;5:202ra123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 137] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 131. | Kim TW, Lee SY, Kim M, Cheon C, Ko SG. Kaempferol induces autophagic cell death via IRE1-JNK-CHOP pathway and inhibition of G9a in gastric cancer cells. Cell Death Dis. 2018;9:875. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 229] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 132. | Sharif T, Martell E, Dai C, Ghassemi-Rad MS, Hanes MR, Murphy PJ, Margam NN, Parmar HB, Giacomantonio CA, Duncan R, Lee PWK, Gujar S. HDAC6 differentially regulates autophagy in stem-like vs differentiated cancer cells. Autophagy. 2019;15:686-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 133. | Yang W, Liu Y, Gao R, Yu H, Sun T. HDAC6 inhibition induces glioma stem cells differentiation and enhances cellular radiation sensitivity through the SHH/Gli1 signaling pathway. Cancer Lett. 2018;415:164-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 134. | Marampon F, Megiorni F, Camero S, Crescioli C, McDowell HP, Sferra R, Vetuschi A, Pompili S, Ventura L, De Felice F, Tombolini V, Dominici C, Maggio R, Festuccia C, Gravina GL. HDAC4 and HDAC6 sustain DNA double strand break repair and stem-like phenotype by promoting radioresistance in glioblastoma cells. Cancer Lett. 2017;397:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 135. | Oehme I, Linke JP, Böck BC, Milde T, Lodrini M, Hartenstein B, Wiegand I, Eckert C, Roth W, Kool M, Kaden S, Gröne HJ, Schulte JH, Lindner S, Hamacher-Brady A, Brady NR, Deubzer HE, Witt O. Histone deacetylase 10 promotes autophagy-mediated cell survival. Proc Natl Acad Sci USA. 2013;110:E2592-E2601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 167] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 136. | Huang N, Liu Z, Zhu J, Cui Z, Li Y, Yu Y, Sun F, Pan Q, Yang Q. Sirtuin 6 plays an oncogenic role and induces cell autophagy in esophageal cancer cells. Tumour Biol. 2017;39:1010428317708532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 137. | Xie HJ, Noh JH, Kim JK, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Lee JY, Park H, Nam SW. HDAC1 inactivation induces mitotic defect and caspase-independent autophagic cell death in liver cancer. PLoS One. 2012;7:e34265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 138. | Ahn MY, Yoon JH. Histone deacetylase 8 as a novel therapeutic target in oral squamous cell carcinoma. Oncol Rep. 2017;37:540-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 139. | Ahn MY, Yoon JH. Histone deacetylase 7 silencing induces apoptosis and autophagy in salivary mucoepidermoid carcinoma cells. J Oral Pathol Med. 2017;46:276-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 140. | Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 973] [Cited by in RCA: 954] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 141. | Jung KH, Noh JH, Kim JK, Eun JW, Bae HJ, Chang YG, Kim MG, Park WS, Lee JY, Lee SY, Chu IS, Nam SW. Histone deacetylase 6 functions as a tumor suppressor by activating c-Jun NH2-terminal kinase-mediated beclin 1-dependent autophagic cell death in liver cancer. Hepatology. 2012;56:644-657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 142. | Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 909] [Cited by in RCA: 1037] [Article Influence: 51.9] [Reference Citation Analysis (0)] |
| 143. | Venkataraman S, Alimova I, Balakrishnan I, Harris P, Birks DK, Griesinger A, Amani V, Cristiano B, Remke M, Taylor MD, Handler M, Foreman NK, Vibhakar R. Inhibition of BRD4 attenuates tumor cell self-renewal and suppresses stem cell signaling in MYC driven medulloblastoma. Oncotarget. 2014;5:2355-2371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 92] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 144. | Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, Natsume H, Yao H, Ozato K. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J Biol Chem. 2008;283:9040-9048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 190] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 145. | Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 303] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 146. | Luan W, Pang Y, Li R, Wei X, Jiao X, Shi J, Yu J, Mao H, Liu P. Akt/mTOR-Mediated Autophagy Confers Resistance To BET Inhibitor JQ1 In Ovarian Cancer. Onco Targets Ther. 2019;12:8063-8074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 147. | Rodriguez RM, Huidobro C, Urdinguio RG, Mangas C, Soldevilla B, Domínguez G, Bonilla F, Fernandez AF, Fraga MF. Aberrant epigenetic regulation of bromodomain BRD4 in human colon cancer. J Mol Med (Berl). 2012;90:587-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 148. | Liu J, Duan Z, Guo W, Zeng L, Wu Y, Chen Y, Tai F, Wang Y, Lin Y, Zhang Q, He Y, Deng J, Stewart RL, Wang C, Lin PC, Ghaffari S, Evers BM, Liu S, Zhou MM, Zhou BP, Shi J. Targeting the BRD4/FOXO3a/CDK6 axis sensitizes AKT inhibition in luminal breast cancer. Nat Commun. 2018;9:5200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 149. | Shafran JS, Andrieu GP, Györffy B, Denis GV. BRD4 Regulates Metastatic Potential of Castration-Resistant Prostate Cancer through AHNAK. Mol Cancer Res. 2019;17:1627-1638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 150. | Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1395] [Cited by in RCA: 1577] [Article Influence: 112.6] [Reference Citation Analysis (0)] |
| 151. | Spriano F, Stathis A, Bertoni F. Targeting BET bromodomain proteins in cancer: The example of lymphomas. Pharmacol Ther. 2020;215:107631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 152. | Sakamaki JI, Wilkinson S, Hahn M, Tasdemir N, O'Prey J, Clark W, Hedley A, Nixon C, Long JS, New M, Van Acker T, Tooze SA, Lowe SW, Dikic I, Ryan KM. Bromodomain Protein BRD4 Is a Transcriptional Repressor of Autophagy and Lysosomal Function. Mol Cell 2017; 66: 517-532. e9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 193] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 153. | Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425-434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1755] [Cited by in RCA: 1926] [Article Influence: 96.3] [Reference Citation Analysis (0)] |
| 154. | Yi Y, Chen D, Ao J, Zhang W, Yi J, Ren X, Fei J, Li F, Niu M, Chen H, Luo Y, Luo Z, Xiao ZJ. Transcriptional suppression of AMPKα1 promotes breast cancer metastasis upon oncogene activation. Proc Natl Acad Sci USA. 2020;117:8013-8021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 155. | Ouyang L, Zhang L, Liu J, Fu L, Yao D, Zhao Y, Zhang S, Wang G, He G, Liu B. Discovery of a Small-Molecule Bromodomain-Containing Protein 4 (BRD4) Inhibitor That Induces AMP-Activated Protein Kinase-Modulated Autophagy-Associated Cell Death in Breast Cancer. J Med Chem. 2017;60:9990-10012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 156. | da Motta LL, Ledaki I, Purshouse K, Haider S, De Bastiani MA, Baban D, Morotti M, Steers G, Wigfield S, Bridges E, Li JL, Knapp S, Ebner D, Klamt F, Harris AL, McIntyre A. The BET inhibitor JQ1 selectively impairs tumour response to hypoxia and downregulates CA9 and angiogenesis in triple negative breast cancer. Oncogene. 2017;36:122-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 119] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 157. | Li F, Yang C, Zhang HB, Ma J, Jia J, Tang X, Zeng J, Chong T, Wang X, He D, Guo P. BET inhibitor JQ1 suppresses cell proliferation via inducing autophagy and activating LKB1/AMPK in bladder cancer cells. Cancer Med. 2019;8:4792-4805. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 158. | Zhang P, Wang D, Zhao Y, Ren S, Gao K, Ye Z, Wang S, Pan CW, Zhu Y, Yan Y, Yang Y, Wu D, He Y, Zhang J, Lu D, Liu X, Yu L, Zhao S, Li Y, Lin D, Wang Y, Wang L, Chen Y, Sun Y, Wang C, Huang H. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat Med. 2017;23:1055-1062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 230] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 159. | Saito Y, Chapple RH, Lin A, Kitano A, Nakada D. AMPK Protects Leukemia-Initiating Cells in Myeloid Leukemias from Metabolic Stress in the Bone Marrow. Cell Stem Cell. 2015;17:585-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 204] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 160. | Jang JE, Eom JI, Jeung HK, Cheong JW, Lee JY, Kim JS, Min YH. AMPK-ULK1-Mediated Autophagy Confers Resistance to BET Inhibitor JQ1 in Acute Myeloid Leukemia Stem Cells. Clin Cancer Res. 2017;23:2781-2794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 101] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 161. | Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 809] [Cited by in RCA: 928] [Article Influence: 71.4] [Reference Citation Analysis (0)] |
| 162. | Xu M, Xu L, Wang Y, Dai G, Xue B, Liu YY, Zhu J. BRD4 inhibition sensitizes renal cell carcinoma cells to the PI3K/mTOR dual inhibitor VS-5584. Aging (Albany NY). 2020;12:19147-19158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 163. | Tian YF, Wang HC, Luo CW, Hung WC, Lin YH, Chen TY, Li CF, Lin CY, Pan MR. Preprogramming therapeutic response of PI3K/mTOR dual inhibitor via the regulation of EHMT2 and p27 in pancreatic cancer. Am J Cancer Res. 2018;8:1812-1822. [PubMed] |
| 164. | Artal-Martinez de Narvajas A, Gomez TS, Zhang JS, Mann AO, Taoda Y, Gorman JA, Herreros-Villanueva M, Gress TM, Ellenrieder V, Bujanda L, Kim DH, Kozikowski AP, Koenig A, Billadeau DD. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013;33:3983-3993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 173] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 165. | Park SE, Yi HJ, Suh N, Park YY, Koh JY, Jeong SY, Cho DH, Kim CS, Hwang JJ. Inhibition of EHMT2/G9a epigenetically increases the transcription of Beclin-1 via an increase in ROS and activation of NF-κB. Oncotarget. 2016;7:39796-39808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 166. | Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, Shiah SG, Chen PS, Jeng YM, Cheng TY, Lai TC, Chang JS, Jan YH, Chien MH, Yang CJ, Huang MS, Hsiao M, Kuo ML. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010;70:7830-7840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 284] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 167. | Chen P, Qian Q, Zhu Z, Shen X, Yu S, Yu Z, Sun R, Li Y, Guo D, Fan H. Increased expression of EHMT2 associated with H3K9me2 Level contributes to the poor prognosis of gastric cancer. Oncol Lett. 2020;20:1734-1742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 168. | Ding J, Li T, Wang X, Zhao E, Choi JH, Yang L, Zha Y, Dong Z, Huang S, Asara JM, Cui H, Ding HF. The histone H3 methyltransferase G9A epigenetically activates the serine-glycine synthesis pathway to sustain cancer cell survival and proliferation. Cell Metab. 2013;18:896-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 190] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 169. | Li F, Zeng J, Gao Y, Guan Z, Ma Z, Shi Q, Du C, Jia J, Xu S, Wang X, Chang L, He D, Guo P. G9a Inhibition Induces Autophagic Cell Death via AMPK/mTOR Pathway in Bladder Transitional Cell Carcinoma. PLoS One. 2015;10:e0138390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 170. | Zhang J, Yao D, Jiang Y, Huang J, Yang S, Wang J. Synthesis and biological evaluation of benzimidazole derivatives as the G9a Histone Methyltransferase inhibitors that induce autophagy and apoptosis of breast cancer cells. Bioorg Chem. 2017;72:168-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 171. | Cubillos-Ruiz JR, Bettigole SE, Glimcher LH. Tumorigenic and Immunosuppressive Effects of Endoplasmic Reticulum Stress in Cancer. Cell. 2017;168:692-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 672] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 172. | Ahmad F, Dixit D, Joshi SD, Sen E. G9a inhibition induced PKM2 regulates autophagic responses. Int J Biochem Cell Biol. 2016;78:87-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 173. | Chen H, Yan Y, Davidson TL, Shinkai Y, Costa M. Hypoxic stress induces dimethylated histone H3 Lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 2006;66:9009-9016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 173] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 174. | Kitajima S, Lee KL, Fujioka M, Sun W, You J, Chia GS, Wanibuchi H, Tomita S, Araki M, Kato H, Poellinger L. Hypoxia-inducible factor-2 alpha up-regulates CD70 under hypoxia and enhances anchorage-independent growth and aggressiveness in cancer cells. Oncotarget. 2018;9:19123-19135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 175. | Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, Hjelmeland AB, Rich JN. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1113] [Cited by in RCA: 1036] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 176. | Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouysségur J, Mazure NM. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570-2581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 971] [Cited by in RCA: 1177] [Article Influence: 73.6] [Reference Citation Analysis (0)] |
| 177. | Nakajima NI, Niimi A, Isono M, Oike T, Sato H, Nakano T, Shibata A. Inhibition of the HDAC/Suv39/G9a pathway restores the expression of DNA damage-dependent major histocompatibility complex class I-related chain A and B in cancer cells. Oncol Rep. 2017;38:693-702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 178. | Cai Q, Wang S, Jin L, Weng M, Zhou D, Wang J, Tang Z, Quan Z. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol Cancer. 2019;18:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 150] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 179. | Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang R, Jin Y, Zou C, Chen Y, Wang G, Gao X, Wang X. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9:1149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 155] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 180. | Li LJ, Chai Y, Guo XJ, Chu SL, Zhang LS. The effects of the long non-coding RNA MALAT-1 regulated autophagy-related signaling pathway on chemotherapy resistance in diffuse large B-cell lymphoma. Biomed Pharmacother. 2017;89:939-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 181. | Shao Y, Liu X, Meng J, Zhang X, Ma Z, Yang G. MicroRNA-1251-5p Promotes Carcinogenesis and Autophagy via Targeting the Tumor Suppressor TBCC in Ovarian Cancer Cells. Mol Ther. 2019;27:1653-1664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 182. | Cheng Y, Li Z, Xie J, Wang P, Zhu J, Li Y, Wang Y. MiRNA-224-5p inhibits autophagy in breast cancer cells via targeting Smad4. Biochem Biophys Res Commun. 2018;506:793-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 183. | Luo K. Signaling Cross Talk between TGF-β/Smad and Other Signaling Pathways. Cold Spring Harb Perspect Biol. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 437] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 184. | Ganesan S, Palani HK, Lakshmanan V, Balasundaram N, Alex AA, David S, Venkatraman A, Korula A, George B, Balasubramanian P, Palakodeti D, Vyas N, Mathews V. Stromal cells downregulate miR-23a-5p to activate protective autophagy in acute myeloid leukemia. Cell Death Dis. 2019;10:736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 185. | Zhang Z, Tian H, Miao Y, Feng X, Li Y, Wang H, Song X. Upregulation of p72 Enhances Malignant Migration and Invasion of Glioma Cells by Repressing Beclin1 Expression. Biochemistry (Mosc). 2016;81:574-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 186. | Huang T, Wan X, Alvarez AA, James CD, Song X, Yang Y, Sastry N, Nakano I, Sulman EP, Hu B, Cheng SY. MIR93 (microRNA -93) regulates tumorigenicity and therapy response of glioblastoma by targeting autophagy. Autophagy. 2019;15:1100-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 187. | Zhu J, Li Y, Luo Y, Xu J, Liufu H, Tian Z, Huang C, Li J. A Feedback Loop Formed by ATG7/Autophagy, FOXO3a/miR-145 and PD-L1 Regulates Stem-Like Properties and Invasion in Human Bladder Cancer. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 188. | Capizzi M, Strappazzon F, Cianfanelli V, Papaleo E, Cecconi F. MIR7-3HG, a MYC-dependent modulator of cell proliferation, inhibits autophagy by a regulatory loop involving AMBRA1. Autophagy. 2017;13:554-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 189. | Zheng Y, Xie M, Zhang N, Liu J, Song Y, Zhou L, Yang M. miR-1262 suppresses gastric cardia adenocarcinoma via targeting oncogene ULK1. J Cancer. 2021;12:1231-1239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 190. | Frankel LB, Wen J, Lees M, Høyer-Hansen M, Farkas T, Krogh A, Jäättelä M, Lund AH. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011;30:4628-4641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 294] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 191. | Wang ZC, Huang FZ, Xu HB, Sun JC, Wang CF. MicroRNA-137 inhibits autophagy and chemosensitizes pancreatic cancer cells by targeting ATG5. Int J Biochem Cell Biol. 2019;111:63-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 192. | Kovaleva V, Mora R, Park YJ, Plass C, Chiramel AI, Bartenschlager R, Döhner H, Stilgenbauer S, Pscherer A, Lichter P, Seiffert M. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012;72:1763-1772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 154] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 193. | Liu L, Ren W, Chen K. MiR-34a Promotes Apoptosis and Inhibits Autophagy by Targeting HMGB1 in Acute Myeloid Leukemia Cells. Cell Physiol Biochem. 2017;41:1981-1992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 194. | Guo X, Xue H, Guo X, Gao X, Xu S, Yan S, Han X, Li T, Shen J, Li G. MiR224-3p inhibits hypoxia-induced autophagy by targeting autophagy-related genes in human glioblastoma cells. Oncotarget. 2015;6:41620-41637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 195. | Blagitko-Dorfs N, Schlosser P, Greve G, Pfeifer D, Meier R, Baude A, Brocks D, Plass C, Lübbert M. Combination treatment of acute myeloid leukemia cells with DNMT and HDAC inhibitors: predominant synergistic gene downregulation associated with gene body demethylation. Leukemia. 2019;33:945-956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 196. | Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, Shin JJ, Harbom KM, Beaty R, Pappou E, Harris J, Yen RW, Ahuja N, Brock MV, Stearns V, Feller-Kopman D, Yarmus LB, Lin YC, Welm AL, Issa JP, Minn I, Matsui W, Jang YY, Sharkis SJ, Baylin SB, Zahnow CA. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 477] [Cited by in RCA: 499] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 197. | Jo YK, Park NY, Shin JH, Jo DS, Bae JE, Choi ES, Maeng S, Jeon HB, Roh SA, Chang JW, Kim JC, Cho DH. Up-regulation of UVRAG by HDAC1 Inhibition Attenuates 5FU-induced Cell Death in HCT116 Colorectal Cancer Cells. Anticancer Res. 2018;38:271-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 198. | Chiu CF, Chin HK, Huang WJ, Bai LY, Huang HY, Weng JR. Induction of Apoptosis and Autophagy in Breast Cancer Cells by a Novel HDAC8 Inhibitor. Biomolecules. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 199. | Gilardini Montani MS, Granato M, Santoni C, Del Porto P, Merendino N, D'Orazi G, Faggioni A, Cirone M. Histone deacetylase inhibitors VPA and TSA induce apoptosis and autophagy in pancreatic cancer cells. Cell Oncol (Dordr). 2017;40:167-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 200. | Luo N, Nixon MJ, Gonzalez-Ericsson PI, Sanchez V, Opalenik SR, Li H, Zahnow CA, Nickels ML, Liu F, Tantawy MN, Sanders ME, Manning HC, Balko JM. DNA methyltransferase inhibition upregulates MHC-I to potentiate cytotoxic T lymphocyte responses in breast cancer. Nat Commun. 2018;9:248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 188] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 201. | Bao X, Ren T, Huang Y, Sun K, Wang S, Liu K, Zheng B, Guo W. Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth. Cell Death Dis. 2017;8:e2605. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 202. | Dong LH, Cheng S, Zheng Z, Wang L, Shen Y, Shen ZX, Chen SJ, Zhao WL. Histone deacetylase inhibitor potentiated the ability of MTOR inhibitor to induce autophagic cell death in Burkitt leukemia/Lymphoma. J Hematol Oncol. 2013;6:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 203. | Yu Y, Luo R, Lu Z, Wei Feng W, Badgwell D, Issa JP, Rosen DG, Liu J, Bast RC Jr. Biochemistry and biology of ARHI (DIRAS3), an imprinted tumor suppressor gene whose expression is lost in ovarian and breast cancers. Methods Enzymol. 2006;407:455-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 51] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 204. | Zhao S, Chen SR, Yang XF, Shen DF, Takano Y, Su RJ, Zheng HC. BTG1 might be employed as a biomarker for carcinogenesis and a target for gene therapy in colorectal cancers. Oncotarget. 2017;8:7502-7520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 205. | Hu X, Sui X, Li L, Huang X, Rong R, Su X, Shi Q, Mo L, Shu X, Kuang Y, Tao Q, He C. Protocadherin 17 acts as a tumour suppressor inducing tumour cell apoptosis and autophagy, and is frequently methylated in gastric and colorectal cancers. J Pathol. 2013;229:62-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 206. | Gou WF, Yang XF, Shen DF, Zhao S, Liu YP, Sun HZ, Takano Y, Su RJ, Luo JS, Zheng HC. The roles of BTG3 expression in gastric cancer: a potential marker for carcinogenesis and a target molecule for gene therapy. Oncotarget. 2015;6:19841-19867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 207. | Feng J, Zhang Y, She X, Sun Y, Fan L, Ren X, Fu H, Liu C, Li P, Zhao C, Liu Q, Li G, Wu M. Hypermethylated gene ANKDD1A is a candidate tumor suppressor that interacts with FIH1 and decreases HIF1α stability to inhibit cell autophagy in the glioblastoma multiforme hypoxia microenvironment. Oncogene. 2019;38:103-119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 208. | Liu X, Hu X, Kuang Y, Yan P, Li L, Li C, Tao Q, Cai X. BCLB, methylated in hepatocellular carcinoma, is a starvation stress sensor that induces apoptosis and autophagy through the AMPK-mTOR signaling cascade. Cancer Lett. 2017;395:63-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 209. | Peng Y, Cao J, Yao XY, Wang JX, Zhong MZ, Gan PP, Li JH. TUSC3 induces autophagy in human non-small cell lung cancer cells through Wnt/β-catenin signaling. Oncotarget. 2017;8:52960-52974. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 210. | Cruzeiro GAV, Dos Reis MB, Silveira VS, Lira RCP, Carlotti CG Jr, Neder L, Oliveira RS, Yunes JA, Brandalise SR, Aguiar S, Eterovic AK, Tone LG, Scrideli CA, Valera ET. HIF1A is Overexpressed in Medulloblastoma and its Inhibition Reduces Proliferation and Increases EPAS1 and ATG16L1 Methylation. Curr Cancer Drug Targets. 2018;18:287-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 211. | Sun T, Li X, Zhang P, Chen WD, Zhang HL, Li DD, Deng R, Qian XJ, Jiao L, Ji J, Li YT, Wu RY, Yu Y, Feng GK, Zhu XF. Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun. 2015;6:7215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 175] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 212. | Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y, Fan Y. Upregulated SMYD3 promotes bladder cancer progression by targeting BCLAF1 and activating autophagy. Tumour Biol. 2016;37:7371-7381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 213. | Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, Zhu WG. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 492] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 214. | Zhao E, Tang C, Jiang X, Weng X, Zhong X, Zhang D, Hou J, Wang F, Huang M, Cui H. Inhibition of cell proliferation and induction of autophagy by KDM2B/FBXL10 knockdown in gastric cancer cells. Cell Signal. 2017;36:222-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 215. | Wang B, Fan X, Ma C, Lei H, Long Q, Chai Y. Downregulation of KDM4A Suppresses the Survival of Glioma Cells by Promoting Autophagy. J Mol Neurosci. 2016;60:137-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 216. | Qiao A, Wang K, Yuan Y, Guan Y, Ren X, Li L, Chen X, Li F, Chen AF, Zhou J, Yang JM, Cheng Y. Sirt3-mediated mitophagy protects tumor cells against apoptosis under hypoxia. Oncotarget. 2016;7:43390-43400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 217. | Ke XX, Zhang D, Zhu S, Xia Q, Xiang Z, Cui H. Inhibition of H3K9 methyltransferase G9a repressed cell proliferation and induced autophagy in neuroblastoma cells. PLoS One. 2014;9:e106962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 218. | Powell MJ, Casimiro MC, Cordon-Cardo C, He X, Yeow WS, Wang C, McCue PA, McBurney MW, Pestell RG. Disruption of a Sirt1-dependent autophagy checkpoint in the prostate results in prostatic intraepithelial neoplasia lesion formation. Cancer Res. 2011;71:964-975. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 4.3] [Reference Citation Analysis (0)] |