Published online Dec 26, 2020. doi: 10.4252/wjsc.v12.i12.1439
Peer-review started: March 30, 2020
First decision: September 21, 2020
Revised: October 7, 2020
Accepted: October 27, 2020
Article in press: October 27, 2020
Published online: December 26, 2020
Processing time: 272 Days and 19.5 Hours
Cancer stem-like cells (CSCs) with potential of self-renewal drive tumorigenesis. Brain tumor microenvironment (TME) has been identified as a critical regulator of malignancy progression. Many researchers are searching new ways to characterize tumors with the goal of predicting how they respond to treatment. Here, we describe the striking parallels between normal stem cells and CSCs. We review the microenvironmental aspects of brain tumors, in particular composition and vital roles of immune cells infiltrating glioma and medulloblastoma. By highlighting that CSCs cooperate with TME via various cellular communication approaches, we discuss the recent advances in therapeutic strategies targeting the components of TME. Identification of the complex and interconnected factors can facilitate the development of promising treatments for these deadly malignancies.
Core Tip: To better understand the effects of interplaying between cancer stem-like cells (CSCs) and tumor microenvironment (TME) on brain tumor progression, we review the distinct characters of CSCs and the mechanisms regarding how TME regulates CSC self-renewal. Moreover, we emphasize the valuable application of sing-cell RNA sequencing technology in the cancer research.
- Citation: Liu HL, Wang YN, Feng SY. Brain tumors: Cancer stem-like cells interact with tumor microenvironment. World J Stem Cells 2020; 12(12): 1439-1454
- URL: https://www.wjgnet.com/1948-0210/full/v12/i12/1439.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v12.i12.1439
Brain tumors respond poorly to existing therapies, leading to the poor prognoses. The recurrence of malignant brain tumors accounts for a majority of mortality and points a significant challenge for conventional treatment modalities[1,2]. The available therapies including operation and chemoradiotherapy mainly focus on the bulk of tumor cells, however, they are not able to impair the subpopulation that has been identified as cancer stem-like cells (CSCs)[3]. Actually, CSCs present the strong self-proliferation and diverse differentiation capability, which induces tumor progression and therapeutic resistance[4,5]. However, it is noticeable that CSCs cannot maintain the stem-like properties by relying on themselves. They need to interact with tumor micro-environment (TME) to insist the stemness and protect themselves against the chemotherapeutic elements and radiation[6]. Even when spreading along the circulation, CSCs recruit microenvironmental components, forming a cluster of metastasis niche or generating a pre-metastatic niche at the faraway organs before arriving. Previously, although bulk tumor sequence analysis can interrogate the genetic status with average expression profiles, it provides limited insight into the specific cell type, especially the immune heterogenicity[7]. Importantly, it is difficult to identify the mechanisms of TME activities at the different stages of brain tumors through traditional methods. To address these challenges, sing-cell RNA sequencing (scRNA-seq) has characterized cancer and immune cell types at high resolution, which figures out the oncogenic signaling, proliferation, and complement/immune response[8,9]. Therefore, to better understand the effects of interplaying between CSCs and TME on brain tumor progression, we review the distinct characters of CSCs and the mechanisms regarding how TME regulates CSC self-renewal. Moreover, we emphasize the valuable application of scRNA-seq technology in the cancer research.
Stem cells can perpetuate themselves via self-renewal and generate the particular mature cells via differentiation. Nevertheless, the rigorous ways to purification and identification of the somatic stem cells have been under debate in some conditions[10]. Neural stem cells (NSCs), an undifferentiated cell type originating in the central nervous system (CNS), have the potential to give rise to offspring cells differentiating to multiple lineages, including neuronal and non-neuronal populations[11]. NSCs have been isolated from mice and humans using the traditional methods as well as confirmed by the advanced scRNA-seq analysis[12,13]. The pathways mediating the stemness of NSCs, such as Notch, WNT/β-catenin, and Hedgehog signaling, are also critical in glioma stem-like cells to drive tumorigenicity[14,15]. Recently, novel therapeutic approaches are established with the goal of not only reducing the tumor burden but also targeting CSCs involved[16].
The differentiation and self-proliferation potential of NSCs and their clinical applications have been discovered in many studies[17]. For example, the stem cell biology can provide new insights into cancer research and treatment. The intermediate filament protein, nestin, serves as a biomarker for stem cells and has been further identified to label the CSCs[18]. We are now studying the critical role of nestin in Hedgehog signaling and revealing the proper interaction between Bergmann glia of the cerebellum and granular neuronal precursors induced by nestin. Based on these findings, our team continuously demonstrated that nestin drives sonic hedgehog (SHH)-medulloblastoma and uncovered Gli3 as a therapeutic target to treat these malignancies[19]. Furthermore, the study was expanded to TME, indicating that tumor-associated astrocytes (TAAs)-derived SHH drives nestin expression in medullo-blastoma cells through a smoothened-dependent mechanism[20]. Therefore, the similarity between stem cells and CSCs sheds light on targeting the stemness associated profiles to control cancer progression.
Heterogeneity has been phenotypically and functionally identified among all the malignancies[21]. Importantly, the multiple differentiation of CSCs and micro-environmental influences provide a model for generating phenotypic and functional heterogeneity beyond the clonal evolution[22]. The CSC models have been well established for cancer research, which differentiate into progenies with limited proliferation potential. Some cancers including medulloblastoma, neuroblastoma, and hierarchically organized cancers can arise from normal stem cells or restricted progenitors through mutations[23,24]. Although CSCs may not address the cell of origin, this population can significantly affect the microenvironment construction. CSCs are enough “clever” to order the other populations, such as fibroblasts, astrocytes, and microglia/macrophages, to activate for serving themselves via cytokine or exosome secretion. Phenotypic and functional heterogeneity always occurs in cancer-associated fibroblasts (CAFs)[25]. Differences in subtypes and functions of CAF behaviors lead to microenvironmental heterogeneity, which is mediated by augmented expression of proteolytic enzymes, deposition of extracellular matrix, and pathogenic angiogenesis derived from CSCs. In addition to the existing components, CSCs can contribute to tumor heterogeneity via differentiating to various kinds of stroma cells as occasion requires[26]. Especially in resistance to chemoradiotherapies or recurrences, the tumor cells undergo de-differentiation to stem-like status and then differentiate to the stromal populations to override the wicked conditions[27]. To better understand the heterogeneity of cancers, the typical flow cytometry and advanced sing-cell multi-omics sequencing are the most popular technologies. The application of flow cytometry makes it possible to harvest the distinct subpopulations of malignant and non-malignant cells based on the well-known markers[28]. Using this approach, the different kinds of cells can be separated for the following culture and experiments.
Trans-differentiation and de-differentiation have been discussed extensively in many studies. Similar to normal stem cells, CSCs can trans-differentiate into other cell lineages in addition to the original lineage arising from tumors[29]. Trans-differentiation of CSCs provides a possible therapeutic target to control recurrence even though this molecular basis has not yet been fully recovered. A cell or tissue from one differentiated state changes to another. The de-differentiated state is unnatural and unstable, which sometimes may present during trans-differentiation[30]. It is supposed that the cell de-differentiates to immature status and then differentiates to the other lineage[31]. However, this routine seems to consume more energy than the direct trans-differentiation, which is mediated or affected by microenvironmental factors. CAFs have been reported to be abundant in gliomas, breast, prostate, and pancreatic cancers. The production of TGF-β1, TGF-β2, PDGF, IL6, and bFGF and protein kinase C in cancer cells play crucial roles in tumor-induced trans-differentiation of surrounding fibroblasts[32]. Furthermore, TGF-β1 or TGF-β2 actually makes sense to the full-extent trans-differentiation, whereas the others, such as PDGF, bFGF, or IL6 (each alone), induce only partial trans-differentiation[33]. In addition to cytokines, the cancer cell-derived exosomes contain abundant and diverse signaling factors particularly under hypoxic conditions[32], which interact with CAFs, astrocytes, and immune cells to mediate trans-differentiation.
Having identified the functions of factors on inducing trans-differentiation, the cell fusion is another approach involved in cell fate reprogramming. Somatic cells are fused with the embryonic stem cells, thereby exposing them to the reprogramming milieu of stem cells[34,35]. The method has been confirmed in cancers using both murine and human cells. We are interested in the interaction between CSCs and microglia. We have found that microglia phagocytosed the oligodendrocyte progenitor cell like malignant cells, therefore forming de-differentiated microglia presenting both parental cell features including self-proliferation and proinflammation characters. However, the molecular mechanism regarding this two-lineage fusion induced de-differentiation remains unclear. Thus, it is important for malignant cells to achieve the ability to reprogram host body cells into stroma cells and to modulate their microenvironment and receive positive feedback for growth and drug resistance[36].
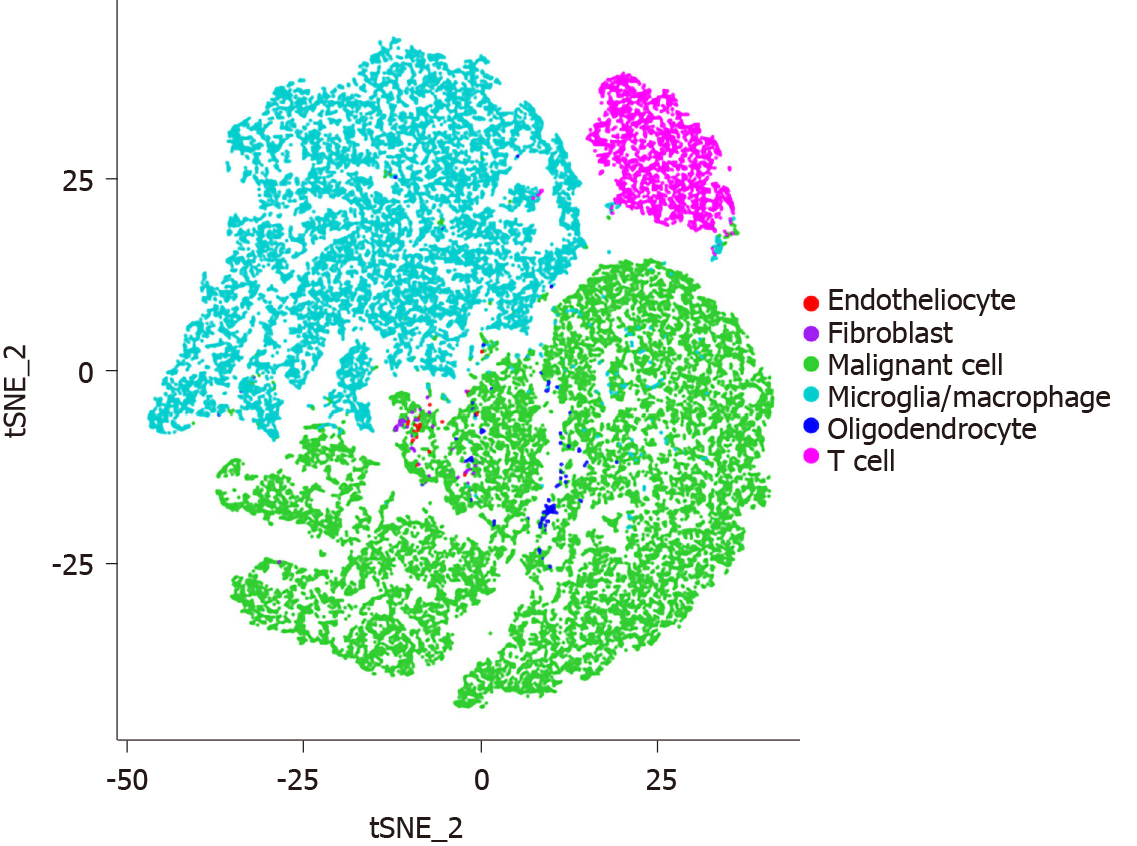
The flow cytometry sorting programs are restricted to the well-established cell surface markers, thus resulting in incapable identification of new subpopulations. Additionally, a cluster of cells rather than single cells in one unit are obtained after sorting[28,37]. On the other hand, it is difficult to accurately analyze heterogeneous populations and status due to technical limitations of marker-based approaches. Over the past decade, the powerful scRNA-seq technology has been applied to overcome the limitations and provide an unbiased view of cell-to-cell variability with gene signatures of each subgroup[38] (Figure 1). Both microfluidic and barcoding approaches are most commonly utilized to assay the transcriptomes from tens of thousands of single cells[39]. Due to the exponential increase in the amounts of single-cell transcriptomic data, it is also necessary to develop computational tools to achieve the meaningful findings. To analyze the cancer heterogeneity, two bioinformatic approaches in scRNA-seq data have been developed: (1) The discrete cluster indicators for cell subtypes and status are labelled in a discrete latent variable approach; and (2) the continuous pseudo-time for differentiation trajectories is constructed in a latent variable approach[9]. However, the double droplets and limited throughput resolution are still the major challenges.
Glioma is the most common primary malignant brain tumor, accounting for almost 40% of primary CNS tumors, of which glioblastoma is the leading cause of mortality[40]. Unfortunately, in spite of significant advances in diagnostic and therapeutic approaches, the median survival of glioblastoma patients remains about 14.2 mo. This could be attributed to the existing classic treatment producing limited efficacy on CSCs[41]. Although many studies discuss pathways driving tumor initiation and progression, epigenetic reprogramming increases oncogenic potential of CSCs, which can lead to tumor growth or therapeutic resistance[42]. Our previous study revealed that the tumor-specific maternal embryonic leucine zipper kinase (MELK) activity was essential for the EZH2/NF-κB interaction via enhancing the methy-ltransferase activity and maintained the stemness[43]. NF-κB as the downstream of the MELK/EZH2 complex opens another exciting pathway to better understand the mechanism of tumorigenesis, beyond the well-established Rel/NF-κB interaction[44]. Activation of NF-κB involves a series of sequential events including cooperation with TME via activating the inflammation associated transcriptions. On the other hand, immune microenvironment also contributes to the glioma stem-like property insistence. Zhang et al[45] reported that C-C motif ligand 8 (CCL8) was a tumor-associated macrophage (TAM) associated factor to mediate glioblastoma stemness via ERK1/2 signaling and targeting CCL8 could provide an insight strategy for glioma treatment.
Medulloblastoma constitutes the most common malignant brain tumor in childhood[46]. Despite the advanced therapeutic strategies, the 5-year survival rate in high-risk group is only about 40% and about half of patients suffer from metastasizing along the neuraxis[47,48]. Recurrent or disseminated medulloblastoma accounts for the majority of pediatric brain tumor-related mortality[49]. Previously, medulloblastoma stem-like cells (MBSCs) have been identified to drive tumorigenesis and recurrence with the potential of self-renewal and resistance to chemoradiotherapy[50]. Among the primary medulloblastoma, MBSCs maintain stemness via activation of key pathways, such as Notch, WNT/β-catenin, and JAK2/STAT3 signaling[51]. Nestin-expressing medulloblastoma cells are the source of medulloblastoma proliferation. MBSCs show restricted capacity to maintain stemness when undergoing metastasis, which requires the efficient cooperation of MBSC niche to protect stem-like properties. Astrocytes, the most abundance of glial cells, are reactivated to play a critical role in supporting tumor growth and inducing protection from chemotherapy[20,52]. We have found the elevated proportion of TAAs in disseminated medulloblastoma compared with primary medulloblastoma. MBSC enrichment in recurrent medulloblastoma was attributed to an increased level of C-C motif ligand 2 (CCL2) released by TAAs undergoing necroptosis[53]. Noticeably, no specific markers for MBSCs have been identified until now, which restricts their purification. CD133, CD15, CD34, and nestin are usually used to label or collect the MBSCs. However, the usage of CD133 to mark MBSCs has its specific drawbacks as follows: The percentage of CD133+ cells is less than that of CD15+ in medulloblastoma, which shows greater potential in labeling the CSCs. Only one marker is chosen to identify MBSCs with less meaningful results probably coming from two indexes, such as CD133+CD15, CD15+CD34, or CD133+nestin[5,23,54].
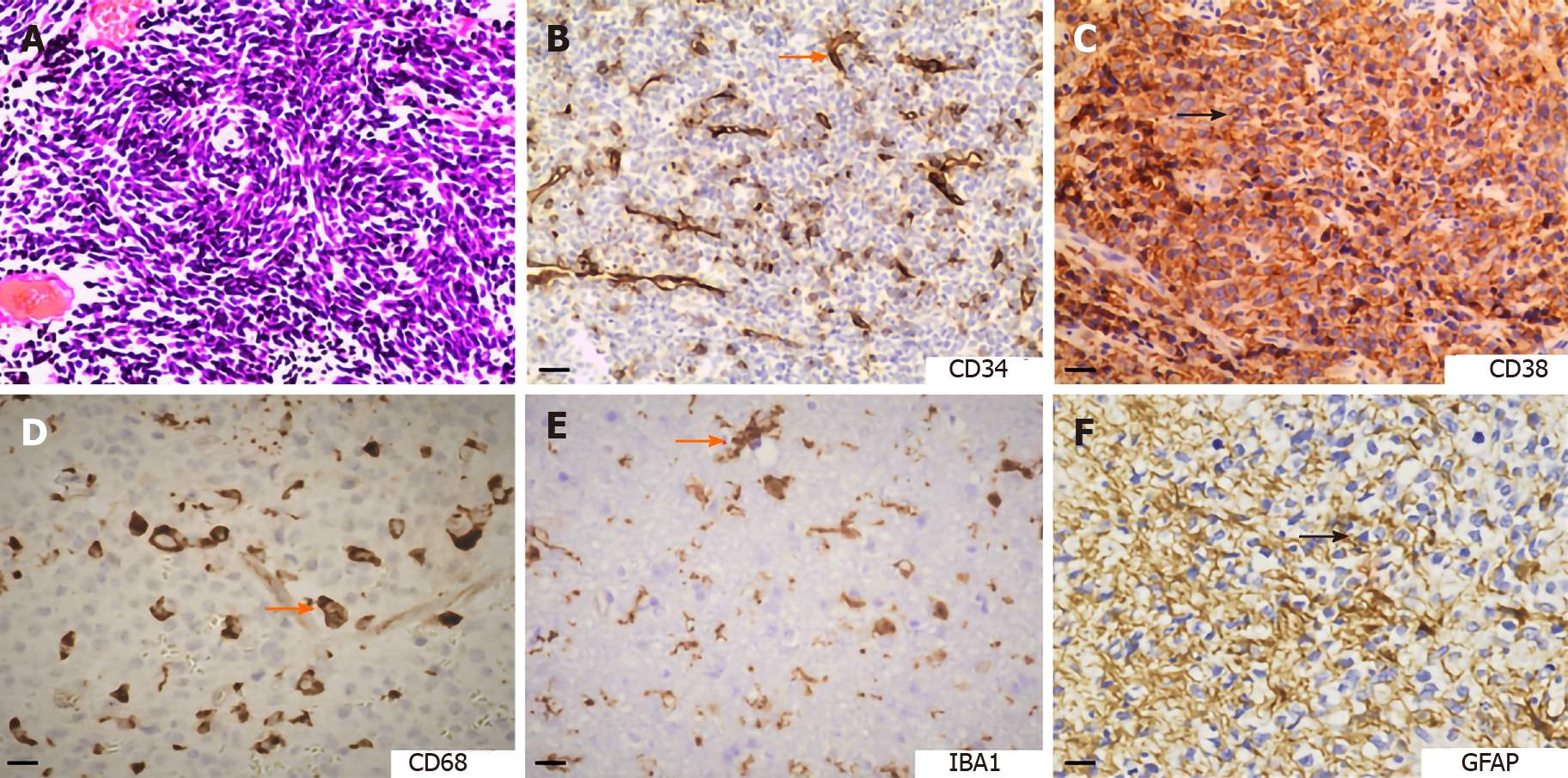
TME contains many non-malignant cells in addition to cancer cells, including immune cells, endothelial cells, pericytes, fibroblasts, and others[21]. Especially, the astrocytes, microglia, and neurons are special tissue-resident populations in the CNS. The unique properties of the CNS require a specific framework to generate the TME-targeted interventions. As shown in Figure 2, the expression of markers for vessels, immune cells, and astroglia stroma suggested the complex compositions of malignant brain tumors. TME has been emerging as a crucial mediator for tumor progression in both primary and metastatic brain malignancies. Brain tumors respond poorly to current therapies, in which TME has been recognized to play critical roles[55]. The CSCs may be considered as the cell origin for tumor recurrence and TME construction[56]. However, the tumor-associated parenchymal cells also importantly function in controlling the course of pathogenesis. We focus on glioma and medulloblastoma to review how brain TME regulates tumor progression and therapeutic response via interacting with CSCs.
Microglia and macrophage: The normal brain has been considered to be “immune privileged” in the whole body, which must be sheltered from immune cell entrance. The activated immune cells produce inflammatory factors that are cytotoxic to cause neurodegeneration. When dissociating the brain tumor tissues into single cells, the majority of cells are TAMs including the blood-derived macrophages and resident microglia accounting for about 35%[57]. Some studies focus on defining context-specific microglia/macrophage activation and phenotype as a measure of functional diversity. Microglia/macrophage activation is classified into the pro-inflammatory M1 state and anti-inflammatory M2 state[57,58]. TAMs exist along a linear M1-to-M2 phenotypic continuum. TAMs tend to be pro-tumorigenic and accumulate gradually with higher tumor grade[59], which produce high levels of pro-inflammatory cytokines promoting tumor proliferation and stemness maintenance. However, macrophage infiltration is considered as a double-edged sword, exerting both tumor-promoting and anti-tumor effects[60]. To support the role of macrophage-mediated inflammation in cancer induction, a previous study has discovered that genetic ablation of STAT3, an anti-inflammatory transcription factor, in macrophages resulted in a chronic inflammatory response in the colon that was sufficient to induce invasive adenocarcinoma. Additionally, loss of IL10 that acts through STAT3 enhanced carcinogen-induced tumorigenesis in the intestine[61]. Macrophage infiltration varies relying on the pathologic type or process. The immune function of macrophages can be suppressed when they are located in the glioma microenvironment[57,62].
Microglia, the resident myeloid cell population in the CNS and a major component of brain immune system, play an essential role in neuronal homeostasis and regulate multiple pathogeneses of disorders, such as neurodegenerative diseases and brain tumors[63]. Recently, accumulated research has reported that microglia distinguish from macrophages. Under homeostatic conditions, microglia originate from hematopoietic stem cells in the yolk sac but not from bone marrow[64]. Although microglia share some traits with monocyte-derived macrophages, they express numbers of special genes at high levels, which may be affected by CNS environment[65]. In glioma, microglia are reactivated within or in close proximity to masses up to half of TME, which shape the TME via releasing a wide range of cytokines for tumor proliferation and invasion[66]. Therefore, targeting the microglia has represented a novel therapeutic approach to this malignancy.
Previously, Venteicher et al[67] detected a continuum model characterizing transition from microglia-like state to macrophage-like state in IDH1/2 mutant gliomas. Müller et al[9] clearly separated microglial TAMs and monocyte-derived TAMs by ontogeny in IDH1/2 wild-type glioblastoma. Both studies provided evidence that microglia vary across different grades and subtypes of gliomas. However, the underlying molecular basis involved in reactivated microglia transition and interaction with glioma cells remains poorly understood. By scRNA-seq analysis of > 50000 single cells isolated from gliomas, we are now focusing a microglial subtype associated with high-grade glioma possessing inflammasome mediated proinflammation and stem-like features, which shapes cytokine microenvironment and promotes oncogenesis. Further analysis in our study also depicts TGF-β1 derived from IDH1/2 wild-type glioblastoma cells with SETD2-deficiency is required for high-grade glioma associated microglia activation.
Previous studies on the differences between brain-resident microglia and blood-derived macrophages have been confounded by a lack of specific markers. Recently, the scRNA-seq study has suggested that CD11b+/CX3CR1+/P2RY12+ population should be the murine microglia[68]. Based on this finding, we are purifying microglia from gliomas by utilizing CD11b and CX3CR1 markers in the scRNA-seq or fluorescent-activated cell sorting experiments. However, the expression of P2RY12 is dramatically compromised in microglia derived from glioblastoma, indicating the specific microglia activation driven by IDH1/2 wild-type cancer cells. Although much evidence supports that microglia/macrophage activation is classified into M1 and M2 state, recent scRNA-seq analysis indicates that macrophages simultaneously express gene profiles of both M1 and M2 phenotypes in an injury mouse model, raising the concern that this classification may not accurately reflect the microglia activation features[60]. Consistent with this finding, Keren-Shaul et al[69] described a novel microglia type associated with neurodegenerative diseases by using single-cell transcripts. Similarly, we established an experimental paradigm by analyzing the scRNA-seq data and histopathological staining to identify a special cohort of microglia associated with high grade glioma. We discovered the genetic program encoding a large number of risk factors, corresponding to the need for proinflammatory response and self-proliferation.
Lymphocytes: Lymphocytes consist of T cells, B cells, and natural killer cells. Mature T cells are released to peripheral lymphoid organs where they can be primed by engaging with antigen presenting cells[70]. In patients, whether further adaptation to specific microenvironment occurs during anti-tumor immunity remains poorly understood. Glioblastoma is a highly immunosuppressive brain tumor because of their T cell paucity[71,72]. Recently, Garris et al[70] revealed a specific mechanism regarding escaping immunosurveillance in brain tumors by trapping T cells in bone marrows via the deficiency of S1P receptor on T cells. The interaction between different cell types within TME also produces obvious effects on immunosuppression. The brain-dependent immune suppression is apparently mediated by microglial cells through TGF-β1/TβRI signaling. Pharmacological obligation of TGF-β1/TβRI signaling can partially reverse the immune suppression but cannot contribute to prolonging the survival of mice, which is due to the lack of sufficient T cells in brain tumors[73]. Additionally, tumor associated CD8+ T cells exhibit proliferation and differentiation potential within the brain, leading to enhanced retention[74]. The researchers have detected that cooperation with brain TME reduced the population of CD8+ T cells in human glioma samples. CD8+ T cells in TME consist of two distinct populations of stem-like and terminally differentiated ones. The stem-like subset gives rise to more terminally differentiated, effector-expressing daughter cells. Similar to this finding, we are also interested in a subpopulation of microglia associated with high-grade glioma, which presents the stem-like property after phagocytosing the oligodendrocyte precursor cell (OPC)-like malignant cells[75]. Therefore, these studies critically suggest that the local microenvironment can modify T cell effector functions during anti-tumor immunity[76]. It is currently under clinical investigation that enhancing T cell activation is induced by co-stimulations through the usage of checkpoint inhibitors among the patients with brain primary or metastatic tumors. In mouse glioma models, inhibition of CTLA-4, a checkpoint molecule, leads to a prolonged survival and activity enhancement of CD4+ helper T cell[77,78]. The standard care treatment is recommended to combine with these advanced clinical studies in recurrent stages. For instance, combination of temozolomide with this treatment regimen reveals an even more pronounced effect on prognosis[79].
Astrocytes as the specialized glial cells distribute ubiquitously throughout the CNS, which play critical roles in providing neurotransmitters and cholesterol, constructing microcirculation, producing energy metabolites, and maintaining homeostasis[52]. Antibodies against glial fibrillary acidic protein (GFAP), S100β, astrocyte cell surface antigen 2, and brain lipid binding protein are often used to detect astrocytes in immunohistochemistry assays[80,81]. Specifically, astrocytes in the cerebellum are identified as Bergmann glial cells supporting proliferation and migration of granular neuronal precursors. In pathological status, such as trauma or tumor growth, astrocytes are activated with upregulated expression levels of GFAP and S100β and enhanced proliferative capability. TAAs release many cytokines to potentially develop a supportive TME for tumor growth and aggressiveness[20]. This program is linked to the significantly elevated expression of malignancy associated genes in cancer cells, which have been proposed to protect against chemoradiotherapy. In a previous study, reactive astrocytes have been reported to mediate glioblastoma invasion through hyperactivation of matrix metalloproteinase 2[82]. Another study has demonstrated that reactive astrocytes expressing SHH were highly concentrated in the perivascular regions of glioblastoma[83]. Consistent with this finding, our work discovered that TAAs, enriched in medulloblastoma, expressed and secreted SHH to promote medulloblastoma cell proliferation. Genetic ablation of TAAs dramatically inhibited nestin expression in medulloblastoma cells, resulting in reduced tumor growth[20]. Furthermore, we found a higher proportion of TAAs in recurrent or disseminated medulloblastoma and TAAs within recurrent TME underwent necroptosis, releasing CCL2 to interplay with MBSCs[53]. The fact that the CSCs are enriched more dramatically in relapsed tumors can be attributed to the dynamic variation of microenvironmental components.
Fibrosis is a common pathophysiological response to chronic injury in many tissues. The processes of wound healing and tissue remodeling are protective mechanisms activated in response to stress and injury with the goal of maintaining functional integrity of systems[84]. Additionally, fibrosis is the marker of chronic inflammation, which results from deregulation of normal healing and exposure to chronic injury. Chronic inflammation has been identified within TME, especially after receiving chemoradiotherapy[85]. Stromal fibroblasts activated by tumor cells in TME have been reported to function in angiogenesis development and metastasis formation. Fibroblasts can be activated at all stages of tumor progression and their structural and functional influences on the process work through cytokine secretion[33]. The growth factors, chemokines and extracellular matrix derived from CAFs facilitate the angiogenic recruitment of endothelial cells and pericytes. Fibroblasts are therefore a key determinant in malignancy progression and represent an important target for cancer therapies. It is hypothesized that both CSCs and CAFs cooperate with and support each other relying on the communicating messenger or reside preferentially at the tumor–stroma interfaces[86]. To develop the favorable niche for CSC self-proliferation, CAFs can also interact with other cells in TME, such as immune and endothelial cells[87]. Choi et al[88] reported that CAFs promoted cell adhesion to human brain microvascular endothelial cells via upregulating expression of integrin α5β1 and αvβ3, c-MET, and α2,6-siayltransferase. A similar role of CAFs within brain tumors has also been suggested. When coculturing human brain-derived fibroblasts and glioblastoma cells, the production and hyperactivation of matrix metalloproteinase 2/9 have been shown to be involved in tumor migration[89].
In addition to glial cells, neurons as a key regulator of CNS development and plasticity are the highly specialized cells contributing to tumor initiation and progression. Recently, accumulated research suggests that glioma arises from neural stem/precursor cells, specifically oligodendrocyte precursor cells (OPCs), pre-OPCs, or earlier neural precursor cells[90]. It is known that the proliferation of neuronal cells and OPCs is stimulated by neurons via the mitogenic signals, which recalls our understanding of neuronal activity as important components of TME. The active neurons influence the proliferation, differentiation, and invasion of glioma cells[91]. A previous study has reported that upregulation of neuroligin-3 in post-synaptic neurons promoted proliferation of cancer cells of patient-derived xenograft glioblastoma models. The mechanism involved PI3K/Akt signaling activation induced by neuronal upregulation of neuroligin-3, which subsequently elevated the expression of FOS and feedforward-upregulation of neuroligin-3 gene expression to enhance the cancerous proliferative activity[92]. In addition to proliferation, the neuron activity affords convenient condition to malignant cell spreading. Wang et al[93] demonstrated that CSCs were preferentially located along neuronal white matter tracts presenting a demyelinated phenotype at the invasive frontiers of glioblastoma. The Notch-induced Sox9 promoted the elevated expression of Sox2 and the methylation level of the Notch1 promoter was attenuated by the upregulation of Sox2 to reinforce Notch1 expression in CD133+/Notch1+ CSCs. Inhibition of Notch signaling attenuated the white-matter-tract tropism of CSCs. For the metastases, the neoplasms could mimic neurons by activating neurotransmitter signaling via the critical elements, for example, upregulating the expression of GABA receptors and transporters[91]. Collectively, these studies suggest that neuronal-specific processes regarding the synaptic transmission can promote brain tumor progression, which warrants further investigation to generate the indispensable roles of neurons within TME.
Communication between cancer cells and non-malignant cells within TME is a two-way process involving a wide variety of stroma cells and a diverse range of mechanisms. Cells communicate in the direct and indirect manners[94]. The essential components of cell-cell communication include the cellular junctions (chemical synapses, pannexins, connexins, and ion channels), anchoring junctions (adherence, focal adhesions, and desmosomes), and tight junctions, as well as cytokines (inflammatory factors and growth factors), exosomes, extracellular matrix, extracellular microRNAs, and different transmembrane adhesion proteins (cadherins and integrins)[94-96]. Sharing information via cellular communication is mediated by different mechanisms: The direct cell-cell communication involves intracrine /autocrine and adjacent communication with nearby cells, which themselves are also regulated by other distinct patterns; and the indirect intercellular communication involves local communication over short distances (paracrine and synaptic signaling) and long distances via hormones (endocrine)[97]. Here, we give some examples including cytokines, exosomes, and matrix.
The function of inflammation in cancer development has been established well. Cytokines, low-molecular-weight proteins mainly derived from immune and stromal cells, regulate proliferation, differentiation, migration, activation, and death[98]. Within the chronic inflammatory TME, they induce malignancy transformation and affect immunotherapy based on the balance of pro- and anti-inflammatory process, relative concentrations, associated receptor expression, and surrounding cell conditions[99]. Targeting the cytokines have delivered promising prospects on cancer therapy. Our study has revealed that inhibition of CCL2/CCR2 blocked the communication between MBSCs and TAAs and compromised the disseminated medulloblastoma stemness[53].
Exosomes, transporting all the main biomolecules, perform intercellular transfer of components locally and systemically[100]. Exosomes have emerged as new influencers in tumor progression by acting both tumor cells and tumor-associated cells. Exosomes derived from glioblastoma have been reported to induce the tumor-promoting transformation of NSCs[101].
Brain extracellular matrix constituents of the normal brain parenchyma, such as heparan sulfate proteoglycans and hyaluronic acid, are mainly concentrated in neural stem cell niches, modifying normal stem cell homeostasis[102]. Dramatically increased production of heparan sulfate proteoglycans in gliomas has been identified as a reservoir for heparin-binding angiogenic growth factors[103].
An understanding of the contribution of TME will allow us to truly tailor therapeutic strategy for each patient. Current standard treatment for glioblastoma, for example, is resection followed by radiation and temozolomide chemotherapy[104] as well as ifosfamide, carboplatin, vincristine, and teniposide chemotherapy for medu-lloblastoma[105]. Adverse effects range in severity between individuals, such as the loss of blood-brain barrier integrity, cytokine deregulation, cognitive dysfunction, and changes in neuronal integrity. Clinical evaluation of benefit vs risk in a quantifiable manner should be considered to minimize additional unnecessary harm. Recently, several approaches to target the TME of brain tumors are ongoing in preclinical and clinical studies. Among them, targeting the vasculature through anti-angiogenic reagents, such as bevacizumab and apatinib, is relatively successful in glioblastoma patients because of highly distributed vessels[106,107]. We have also found that apatinib exhibits efficient effects on the glioblastoma resistant to temozolomide (Figure 3). Immune checkpoint inhibitors are popular with treatment of both primary and metastatic brain tumors, such as nivolumab and/or ipilimumab vs bevacizumab in glioblastoma (NCT02017717)[108] and ipilimumab with nivolumab/fotemustine in brain metastasis (NCT02460068)[109]. The success of immune checkpoint inhibitors utilized in various kinds of cancers is an excellent example of addressing a TME-mediated resistance mechanism to obtain prognostic benefits. Another promising immunotherapy involves the development of T cells engineered to target proteins on the surfaces of cancer cells[110]. Chimeric antigen receptor T-cells are constructed to target a number of different tumorous antigens. As TME provides a safe haven for cancer cells, strategies to mobilize cells from tumor niche will render the malignant cells more sensitive to therapy. However, as the engineered T cells remain to be subject to suppression by microenvironmental factors, it is necessary to illustrate the mechanisms regarding the efficacy of novel agents. In addition to targeting the TME components, oncologists should pay more attention to CSCs that have been identified as the root of tumor recurrence. Some agents targeting stemness associated genes, such as the Notch, Hedgehog, and WNT signaling, are underway in many cancers[111]. We are now trying to treat medulloblastoma using LY3039478, an oral Notch inhibitor[112], indicating a promising prospect (Figure 3).
Accumulating laboratory works support the thrilling concept that brain tumors rely on the interplay between CSCs and TME during progression. CSCs govern the surrounding components to maintain stem-like properties and other cells create a more permissive niche via production of extracellular substrates facilitating tumor growth and invasion. Identifying the cellular and extracellular dependent relationships unique to TME can provide exceptional opportunities to develop effective treatments targeting these symbiotic associations that support brain tumor progression.
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hann HW, Huang AHC, Tanabe S S-Editor: Chen XF L-Editor: Wang TQ P-Editor: Ma YJ
| 1. | Yung WK. Imaging endpoints in brain tumor clinical trials: proceedings of the January 30, 2014 Workshop. Introduction. Neuro Oncol. 2014;16 Suppl 7:vii1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 2. | Aref D, Croul S. Medulloblastoma: recurrence and metastasis. CNS Oncol. 2013;2:377-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 3. | Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 842] [Cited by in RCA: 874] [Article Influence: 67.2] [Reference Citation Analysis (0)] |
| 4. | Peitzsch C, Tyutyunnykova A, Pantel K, Dubrovska A. Cancer stem cells: The root of tumor recurrence and metastases. Semin Cancer Biol. 2017;44:10-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 291] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 5. | Garg N, Bakhshinyan D, Venugopal C, Mahendram S, Rosa DA, Vijayakumar T, Manoranjan B, Hallett R, McFarlane N, Delaney KH, Kwiecien JM, Arpin CC, Lai PS, Gómez-Biagi RF, Ali AM, de Araujo ED, Ajani OA, Hassell JA, Gunning PT, Singh SK. CD133+ brain tumor-initiating cells are dependent on STAT3 signaling to drive medulloblastoma recurrence. Oncogene. 2017;36:606-617. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 6. | Kenda Suster N, Virant-Klun I. Presence and role of stem cells in ovarian cancer. World J Stem Cells. 2019;11:383-397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 7. | Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1349] [Cited by in RCA: 2279] [Article Influence: 284.9] [Reference Citation Analysis (0)] |
| 8. | Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1265] [Cited by in RCA: 1299] [Article Influence: 118.1] [Reference Citation Analysis (0)] |
| 9. | Müller S, Kohanbash G, Liu SJ, Alvarado B, Carrera D, Bhaduri A, Watchmaker PB, Yagnik G, Di Lullo E, Malatesta M, Amankulor NM, Kriegstein AR, Lim DA, Aghi M, Okada H, Diaz A. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 2017;18:234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 308] [Cited by in RCA: 465] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 10. | Abazova N, Krijgsveld J. Advances in stem cell proteomics. Curr Opin Genet Dev. 2017;46:149-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 11. | Strzyz P. Proteins clog neural stem cell activation. Nat Rev Mol Cell Biol. 2018;19:346-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 12. | Shang Z, Chen D, Wang Q, Wang S, Deng Q, Wu L, Liu C, Ding X, Wang S, Zhong J, Zhang D, Cai X, Zhu S, Yang H, Liu L, Fink JL, Chen F, Liu X, Gao Z, Xu X. Single-cell RNA-seq reveals dynamic transcriptome profiling in human early neural differentiation. Gigascience. 2018;7:giy117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Monaco S, Baur K, Hellwig A, Hölzl-Wenig G, Mandl C, Ciccolini F. A Flow Cytometry-Based Approach for the Isolation and Characterization of Neural Stem Cell Primary Cilia. Front Cell Neurosci. 2018;12:519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 14. | Bajaj J, Diaz E, Reya T. Stem cells in cancer initiation and progression. J Cell Biol. 2020;219:e201911053. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 15. | Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6844] [Cited by in RCA: 6917] [Article Influence: 288.2] [Reference Citation Analysis (0)] |
| 16. | Worley JR, Parker GC. Effects of environmental stressors on stem cells. World J Stem Cells. 2019;11:565-577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 17. | Tuazon JP, Castelli V, Lee JY, Desideri GB, Stuppia L, Cimini AM, Borlongan CV. Neural Stem Cells. Adv Exp Med Biol. 2019;1201:79-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (1)] |
| 18. | Lendahl U, Zimmerman LB, McKay RD. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2348] [Cited by in RCA: 2438] [Article Influence: 69.7] [Reference Citation Analysis (0)] |
| 19. | Li P, Lee EH, Du F, Gordon RE, Yuelling LW, Liu Y, Ng JM, Zhang H, Wu J, Korshunov A, Pfister SM, Curran T, Yang ZJ. Nestin Mediates Hedgehog Pathway Tumorigenesis. Cancer Res. 2016;76:5573-5583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 20. | Liu Y, Yuelling LW, Wang Y, Du F, Gordon RE, O'Brien JA, Ng JMY, Robins S, Lee EH, Liu H, Curran T, Yang ZJ. Astrocytes Promote Medulloblastoma Progression through Hedgehog Secretion. Cancer Res. 2017;77:6692-6703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 21. | Quail DF, Joyce JA. The Microenvironmental Landscape of Brain Tumors. Cancer Cell. 2017;31:326-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 887] [Cited by in RCA: 1229] [Article Influence: 153.6] [Reference Citation Analysis (0)] |
| 22. | McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell. 2017;168:613-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1799] [Cited by in RCA: 1841] [Article Influence: 230.1] [Reference Citation Analysis (0)] |
| 23. | Li P, Du F, Yuelling LW, Lin T, Muradimova RE, Tricarico R, Wang J, Enikolopov G, Bellacosa A, Wechsler-Reya RJ, Yang ZJ. A population of Nestin-expressing progenitors in the cerebellum exhibits increased tumorigenicity. Nat Neurosci. 2013;16:1737-1744. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 24. | Boeva V, Louis-Brennetot C, Peltier A, Durand S, Pierre-Eugène C, Raynal V, Etchevers HC, Thomas S, Lermine A, Daudigeos-Dubus E, Geoerger B, Orth MF, Grünewald TGP, Diaz E, Ducos B, Surdez D, Carcaboso AM, Medvedeva I, Deller T, Combaret V, Lapouble E, Pierron G, Grossetête-Lalami S, Baulande S, Schleiermacher G, Barillot E, Rohrer H, Delattre O, Janoueix-Lerosey I. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat Genet. 2017;49:1408-1413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 332] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 25. | Su S, Chen J, Yao H, Liu J, Yu S, Lao L, Wang M, Luo M, Xing Y, Chen F, Huang D, Zhao J, Yang L, Liao D, Su F, Li M, Liu Q, Song E. CD10+GPR77+ Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell. 2018;172:841-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 879] [Article Influence: 125.6] [Reference Citation Analysis (0)] |
| 26. | Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer. 2017;16:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 331] [Cited by in RCA: 565] [Article Influence: 70.6] [Reference Citation Analysis (0)] |
| 27. | Bai X, Ni J, Beretov J, Graham P, Li Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat Rev. 2018;69:152-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 227] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 28. | Montante S, Brinkman RR. Flow cytometry data analysis: Recent tools and algorithms. Int J Lab Hematol. 2019;41 Suppl 1:56-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 29. | Brown DV, Mantamadiotis T. Insights into the next generation of cancer stem cell research. Front Biosci. 19:1015-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 30. | Dall'Agnese A, Caputo L, Nicoletti C, di Iulio J, Schmitt A, Gatto S, Diao Y, Ye Z, Forcato M, Perera R, Bicciato S, Telenti A, Ren B, Puri PL. Transcription Factor-Directed Re-wiring of Chromatin Architecture for Somatic Cell Nuclear Reprogramming toward trans-Differentiation. Mol Cell. 2019;76:453-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 31. | Gong L, Cao L, Shen Z, Shao L, Gao S, Zhang C, Lu J, Li W. Materials for Neural Differentiation, Trans-Differentiation, and Modeling of Neurological Disease. Adv Mater. 2018;30:e1705684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 32. | Heneberg P. Paracrine tumor signaling induces transdifferentiation of surrounding fibroblasts. Crit Rev Oncol Hematol. 2016;97:303-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 33. | Malanchi I, Santamaria-Martínez A, Susanto E, Peng H, Lehr HA, Delaloye JF, Huelsken J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481:85-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 943] [Cited by in RCA: 1075] [Article Influence: 76.8] [Reference Citation Analysis (0)] |
| 34. | Wang R, Chen S, Li C, Ng KT, Kong CW, Cheng J, Cheng SH, Li RA, Lo CM, Man K, Sun D. Fusion with stem cell makes the hepatocellular carcinoma cells similar to liver tumor-initiating cells. BMC Cancer. 2016;16:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 35. | Imai H, Kusakabe KT, Kiso Y, Hattori S, Kai C, Ono E, Kano K. Induction of pluripotency in mammalian fibroblasts by cell fusion with mouse embryonic stem cells. Biochem Biophys Res Commun. 2020;521:24-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 36. | Friedl P. Cell fusion: new mechanisms of plasticity in cancer? Lancet Oncol. 2005;6:916-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 37. | Pozarowski P, Darzynkiewicz Z. Analysis of cell cycle by flow cytometry. Methods Mol Biol. 2004;281:301-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 230] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 38. | Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, Vijayaraghavan R, Wong J, Chen A, Sheng X, Kaper F, Shen R, Ronaghi M, Fan JB, Wang W, Chun J, Zhang K. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586-1590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 626] [Cited by in RCA: 655] [Article Influence: 72.8] [Reference Citation Analysis (0)] |
| 39. | Freytag S, Tian L, Lönnstedt I, Ng M, Bahlo M. Comparison of clustering tools in R for medium-sized 10x Genomics single-cell RNA-sequencing data. F1000Res. 2018;7:1297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 40. | Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10993] [Cited by in RCA: 10867] [Article Influence: 1207.4] [Reference Citation Analysis (0)] |
| 41. | Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4467] [Cited by in RCA: 4760] [Article Influence: 250.5] [Reference Citation Analysis (0)] |
| 42. | Hu B, Wang Q, Wang YA, Hua S, Sauvé CG, Ong D, Lan ZD, Chang Q, Ho YW, Monasterio MM, Lu X, Zhong Y, Zhang J, Deng P, Tan Z, Wang G, Liao WT, Corley LJ, Yan H, Zhang J, You Y, Liu N, Cai L, Finocchiaro G, Phillips JJ, Berger MS, Spring DJ, Hu J, Sulman EP, Fuller GN, Chin L, Verhaak RGW, DePinho RA. Epigenetic Activation of WNT5A Drives Glioblastoma Stem Cell Differentiation and Invasive Growth. Cell. 2016;167:1281-1295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 209] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 43. | Liu H, Sun Y, Qi X, Gordon RE, O'Brien JA, Yuan H, Zhang J, Wang Z, Zhang M, Song Y, Yu C, Gu C. EZH2 Phosphorylation Promotes Self-Renewal of Glioma Stem-Like Cells Through NF-κB Methylation. Front Oncol. 2019;9:641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 44. | Siggers T, Gilmore TD, Barron B, Penvose A. Characterizing the DNA binding site specificity of NF-κB with protein-binding microarrays (PBMs). Methods Mol Biol. 2015;1280:609-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 45. | Zhang X, Chen L, Dang WQ, Cao MF, Xiao JF, Lv SQ, Jiang WJ, Yao XH, Lu HM, Miao JY, Wang Y, Yu SC, Ping YF, Liu XD, Cui YH, Zhang X, Bian XW. CCL8 secreted by tumor-associated macrophages promotes invasion and stemness of glioblastoma cells via ERK1/2 signaling. Lab Invest. 2020;100:619-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 120] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 46. | Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, Garzia L, Torchia J, Nor C, Morrissy AS, Agnihotri S, Thompson YY, Kuzan-Fischer CM, Farooq H, Isaev K, Daniels C, Cho BK, Kim SK, Wang KC, Lee JY, Grajkowska WA, Perek-Polnik M, Vasiljevic A, Faure-Conter C, Jouvet A, Giannini C, Nageswara Rao AA, Li KKW, Ng HK, Eberhart CG, Pollack IF, Hamilton RL, Gillespie GY, Olson JM, Leary S, Weiss WA, Lach B, Chambless LB, Thompson RC, Cooper MK, Vibhakar R, Hauser P, van Veelen MC, Kros JM, French PJ, Ra YS, Kumabe T, López-Aguilar E, Zitterbart K, Sterba J, Finocchiaro G, Massimino M, Van Meir EG, Osuka S, Shofuda T, Klekner A, Zollo M, Leonard JR, Rubin JB, Jabado N, Albrecht S, Mora J, Van Meter TE, Jung S, Moore AS, Hallahan AR, Chan JA, Tirapelli DPC, Carlotti CG, Fouladi M, Pimentel J, Faria CC, Saad AG, Massimi L, Liau LM, Wheeler H, Nakamura H, Elbabaa SK, Perezpeña-Diazconti M, Chico Ponce de León F, Robinson S, Zapotocky M, Lassaletta A, Huang A, Hawkins CE, Tabori U, Bouffet E, Bartels U, Dirks PB, Rutka JT, Bader GD, Reimand J, Goldenberg A, Ramaswamy V, Taylor MD. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell. 2017;31:737-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 810] [Cited by in RCA: 841] [Article Influence: 105.1] [Reference Citation Analysis (0)] |
| 47. | Liu H, Zhang J, Liu Y, Sun Y, Li C, Gu C, Wang H, Zhang H, Yu C, Zhang M. Neuraxis Metastases Of Primary Central Nervous System Tumors: A Review Of Clinicopathological And Radiographic Characters Of 198 Cases In A Single Center. Cancer Manag Res. 2019;11:9829-9841. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 48. | Liu H, Zhang X, Zhang M, Zhang J, Ning W, Yue A, Zhao R, Sun Y, Yu C. Skull bone tumor: a review of clinicopathological and neuroimaging characteristics of 426 cases at a single center. Cancer Commun. 39:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 49. | Northcott PA, Robinson GW, Kratz CP, Mabbott DJ, Pomeroy SL, Clifford SC, Rutkowski S, Ellison DW, Malkin D, Taylor MD, Gajjar A, Pfister SM. Medulloblastoma. Nat Rev Dis Primers. 2019;5:11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 420] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 50. | Liu H, Sun Q, Sun Y, Zhang J, Yuan H, Pang S, Qi X, Wang H, Zhang M, Zhang H, Yu C, Gu C. MELK and EZH2 Cooperate to Regulate Medulloblastoma Cancer Stem-like Cell Proliferation and Differentiation. Mol Cancer Res. 2017;15:1275-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 51. | Ramaswamy V, Taylor MD. Medulloblastoma: From Myth to Molecular. J Clin Oncol. 2017;35:2355-2363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 52. | Seifert G, Schilling K, Steinhäuser C. Astrocyte dysfunction in neurological disorders: a molecular perspective. Nat Rev Neurosci. 2006;7:194-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 581] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 53. | Liu H, Sun Y, O'Brien JA, Franco-Barraza J, Qi X, Yuan H, Jin W, Zhang J, Gu C, Zhao Z, Yu C, Feng S, Yu X. Necroptotic astrocytes contribute to maintaining stemness of disseminated medulloblastoma through CCL2 secretion. Neuro Oncol. 2020;22:625-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 54. | Read TA, Fogarty MP, Markant SL, McLendon RE, Wei Z, Ellison DW, Febbo PG, Wechsler-Reya RJ. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell. 2009;15:135-147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 287] [Cited by in RCA: 270] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 55. | Celià-Terrassa T, Kang Y. Metastatic niche functions and therapeutic opportunities. Nat Cell Biol. 2018;20:868-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 128] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 56. | Thomas TM, Yu JS. Metabolic regulation of glioma stem-like cells in the tumor micro-environment. Cancer Lett. 2017;408:174-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 57. | Gieryng A, Pszczolkowska D, Walentynowicz KA, Rajan WD, Kaminska B. Immune microenvironment of gliomas. Lab Invest. 2017;97:498-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 400] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 58. | Herz J, Filiano AJ, Smith A, Yogev N, Kipnis J. Myeloid Cells in the Central Nervous System. Immunity. 2017;46:943-956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 264] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 59. | Szulzewsky F, Pelz A, Feng X, Synowitz M, Markovic D, Langmann T, Holtman IR, Wang X, Eggen BJ, Boddeke HW, Hambardzumyan D, Wolf SA, Kettenmann H. Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS One. 2015;10:e0116644. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 320] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 60. | Van Hove H, Martens L, Scheyltjens I, De Vlaminck K, Pombo Antunes AR, De Prijck S, Vandamme N, De Schepper S, Van Isterdael G, Scott CL, Aerts J, Berx G, Boeckxstaens GE, Vandenbroucke RE, Vereecke L, Moechars D, Guilliams M, Van Ginderachter JA, Saeys Y, Movahedi K. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci. 2019;22:1021-1035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 671] [Article Influence: 111.8] [Reference Citation Analysis (0)] |
| 61. | Wang Y, Shen Y, Wang S, Shen Q, Zhou X. The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett. 2018;415:117-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 270] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 62. | Ghosh A, Bhattacharya M, Sarkar P, Acharya S, Chaudhuri S. T11 target structure exerts effector function by activating immune cells in CNS against glioma where cytokine modulation provide favorable microenvironment. Indian J Exp Biol. 2010;48:879-888. [PubMed] |
| 63. | Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15:300-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 870] [Cited by in RCA: 1011] [Article Influence: 91.9] [Reference Citation Analysis (0)] |
| 64. | Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19:20-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 786] [Cited by in RCA: 1227] [Article Influence: 136.3] [Reference Citation Analysis (0)] |
| 65. | Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O'Connor C, Fitzpatrick C, Pasillas MP, Pena M, Adair A, Gonda DD, Levy ML, Ransohoff RM, Gage FH, Glass CK. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356:eaal3222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 630] [Cited by in RCA: 841] [Article Influence: 105.1] [Reference Citation Analysis (0)] |
| 66. | Bettinger I, Thanos S, Paulus W. Microglia promote glioma migration. Acta Neuropathol. 2002;103:351-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 131] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 67. | Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, Hovestadt V, Escalante LE, Shaw ML, Rodman C, Gillespie SM, Dionne D, Luo CC, Ravichandran H, Mylvaganam R, Mount C, Onozato ML, Nahed BV, Wakimoto H, Curry WT, Iafrate AJ, Rivera MN, Frosch MP, Golub TR, Brastianos PK, Getz G, Patel AP, Monje M, Cahill DP, Rozenblatt-Rosen O, Louis DN, Bernstein BE, Regev A, Suvà ML. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science. 2017;355:eaai8478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 523] [Cited by in RCA: 691] [Article Influence: 86.4] [Reference Citation Analysis (0)] |
| 68. | Masuda T, Sankowski R, Staszewski O, Böttcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, Coenen VA, Reinacher PC, Michel A, Sure U, Gold R, Grün D, Priller J, Stadelmann C, Prinz M. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature. 2019;566:388-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 547] [Cited by in RCA: 870] [Article Influence: 145.0] [Reference Citation Analysis (0)] |
| 69. | Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, Itzkovitz S, Colonna M, Schwartz M, Amit I. A Unique Microglia Type Associated with Restricting Development of Alzheimer's Disease. Cell. 2017;169:1276-1290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2095] [Cited by in RCA: 3443] [Article Influence: 430.4] [Reference Citation Analysis (1)] |
| 70. | Garris CS, Blaho VA, Hla T, Han MH. Sphingosine-1-phosphate receptor 1 signalling in T cells: trafficking and beyond. Immunology. 2014;142:347-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 124] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 71. | Vella G, Bergers G. Where Have All the T Cells Gone? Immunity. 2018;49:592-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 72. | With GBM, T Cells May Be Stuck in Bone Marrow. Cancer Discov. 2018;8:1203-1204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 73. | Ooi YC, Tran P, Ung N, Thill K, Trang A, Fong BM, Nagasawa DT, Lim M, Yang I. The role of regulatory T-cells in glioma immunology. Clin Neurol Neurosurg. 2014;119:125-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 74. | Jansen CS, Prokhnevska N, Master VA, Sanda MG, Carlisle JW, Bilen MA, Cardenas M, Wilkinson S, Lake R, Sowalsky AG, Valanparambil RM, Hudson WH, McGuire D, Melnick K, Khan AI, Kim K, Chang YM, Kim A, Filson CP, Alemozaffar M, Osunkoya AO, Mullane P, Ellis C, Akondy R, Im SJ, Kamphorst AO, Reyes A, Liu Y, Kissick H. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature. 2019;576:465-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 572] [Article Influence: 95.3] [Reference Citation Analysis (0)] |
| 75. | Kucharova K, Stallcup WB. NG2-proteoglycan-dependent contributions of oligodendrocyte progenitors and myeloid cells to myelin damage and repair. J Neuroinflammation. 2015;12:161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 76. | Khan AB, Carpenter B, Santos E Sousa P, Pospori C, Khorshed R, Griffin J, Velica P, Zech M, Ghorashian S, Forrest C, Thomas S, Gonzalez Anton S, Ahmadi M, Holler A, Flutter B, Ramirez-Ortiz Z, Means TK, Bennett CL, Stauss H, Morris E, Lo Celso C, Chakraverty R. Redirection to the bone marrow improves T cell persistence and antitumor functions. J Clin Invest. 2018;128:2010-2024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 77. | Bagley SJ, Desai AS, Linette GP, June CH, O'Rourke DM. CAR T-cell therapy for glioblastoma: recent clinical advances and future challenges. Neuro Oncol. 2018;20:1429-1438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 219] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 78. | Rodriguez A, Brown C, Badie B. Chimeric antigen receptor T-cell therapy for glioblastoma. Transl Res. 2017;187:93-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 79. | Kleijn A, van den Bossche W, Haefner ES, Belcaid Z, Burghoorn-Maas C, Kloezeman JJ, Pas SD, Leenstra S, Debets R, de Vrij J, Dirven CMF, Lamfers MLM. The Sequence of Delta24-RGD and TMZ Administration in Malignant Glioma Affects the Role of CD8+T Cell Anti-tumor Activity. Mol Ther Oncolytics. 2017;5:11-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 80. | Hol EM, Pekny M. Glial fibrillary acidic protein (GFAP) and the astrocyte intermediate filament system in diseases of the central nervous system. Curr Opin Cell Biol. 2015;32:121-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 609] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 81. | Ghandour MS, Labourdette G, Vincendon G, Gombos G. A biochemical and immunohistological study of S100 protein in developing rat cerebellum. Dev Neurosci. 1981;4:98-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 82. | Kegelman TP, Wu B, Das SK, Talukdar S, Beckta JM, Hu B, Emdad L, Valerie K, Sarkar D, Furnari FB, Cavenee WK, Wei J, Purves A, De SK, Pellecchia M, Fisher PB. Inhibition of radiation-induced glioblastoma invasion by genetic and pharmacological targeting of MDA-9/Syntenin. Proc Natl Acad Sci USA. 2017;114:370-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 79] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 83. | Morgenroth A, Vogg AT, Ermert K, Zlatopolskiy B, Mottaghy FM. Hedgehog signaling sensitizes glioma stem cells to endogenous nano-irradiation. Oncotarget. 2014;5:5483-5493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 84. | Buechler MB, Turley SJ. A short field guide to fibroblast function in immunity. Semin Immunol. 2018;35:48-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 85. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47141] [Article Influence: 3367.2] [Reference Citation Analysis (5)] |
| 86. | Liu J, Chen S, Wang W, Ning BF, Chen F, Shen W, Ding J, Chen W, Xie WF, Zhang X. Cancer-associated fibroblasts promote hepatocellular carcinoma metastasis through chemokine-activated hedgehog and TGF-β pathways. Cancer Lett. 2016;379:49-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 180] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 87. | Valenti G, Quinn HM, Heynen GJJE, Lan L, Holland JD, Vogel R, Wulf-Goldenberg A, Birchmeier W. Cancer Stem Cells Regulate Cancer-Associated Fibroblasts via Activation of Hedgehog Signaling in Mammary Gland Tumors. Cancer Res. 2017;77:2134-2147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 130] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 88. | Choi YP, Lee JH, Gao MQ, Kim BG, Kang S, Kim SH, Cho NH. Cancer-associated fibroblast promote transmigration through endothelial brain cells in three-dimensional in vitro models. Int J Cancer. 2014;135:2024-2033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 89. | Clavreul A, Guette C, Faguer R, Tétaud C, Boissard A, Lemaire L, Rousseau A, Avril T, Henry C, Coqueret O, Menei P. Glioblastoma-associated stromal cells (GASCs) from histologically normal surgical margins have a myofibroblast phenotype and angiogenic properties. J Pathol. 2014;233:74-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 90. | Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, Dewitt J, Gritsch S, Perez EM, Gonzalez Castro LN, Lan X, Druck N, Rodman C, Dionne D, Kaplan A, Bertalan MS, Small J, Pelton K, Becker S, Bonal D, Nguyen QD, Servis RL, Fung JM, Mylvaganam R, Mayr L, Gojo J, Haberler C, Geyeregger R, Czech T, Slavc I, Nahed BV, Curry WT, Carter BS, Wakimoto H, Brastianos PK, Batchelor TT, Stemmer-Rachamimov A, Martinez-Lage M, Frosch MP, Stamenkovic I, Riggi N, Rheinbay E, Monje M, Rozenblatt-Rosen O, Cahill DP, Patel AP, Hunter T, Verma IM, Ligon KL, Louis DN, Regev A, Bernstein BE, Tirosh I, Suvà ML. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell. 2019;178:835-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 769] [Cited by in RCA: 1611] [Article Influence: 268.5] [Reference Citation Analysis (0)] |
| 91. | Johung T, Monje M. Neuronal activity in the glioma microenvironment. Curr Opin Neurobiol. 2017;47:156-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 92. | Venkatesh HS, Tam LT, Woo PJ, Lennon J, Nagaraja S, Gillespie SM, Ni J, Duveau DY, Morris PJ, Zhao JJ, Thomas CJ, Monje M. Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature. 2017;549:533-537. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 380] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 93. | Wang J, Xu SL, Duan JJ, Yi L, Guo YF, Shi Y, Li L, Yang ZY, Liao XM, Cai J, Zhang YQ, Xiao HL, Yin L, Wu H, Zhang JN, Lv SQ, Yang QK, Yang XJ, Jiang T, Zhang X, Bian XW, Yu SC. Invasion of white matter tracts by glioma stem cells is regulated by a NOTCH1-SOX2 positive-feedback loop. Nat Neurosci. 2019;22:91-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 118] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 94. | Brücher BL, Jamall IS. Cell-cell communication in the tumor microenvironment, carcinogenesis, and anticancer treatment. Cell Physiol Biochem. 2014;34:213-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 148] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 95. | Maia J, Caja S, Strano Moraes MC, Couto N, Costa-Silva B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front Cell Dev Biol. 2018;6:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 346] [Cited by in RCA: 506] [Article Influence: 72.3] [Reference Citation Analysis (0)] |
| 96. | Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 311] [Cited by in RCA: 589] [Article Influence: 84.1] [Reference Citation Analysis (0)] |
| 97. | Birbrair A. Stem Cell Microenvironments and Beyond. Adv Exp Med Biol. 2017;1041:1-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 98. | Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17:559-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 964] [Cited by in RCA: 1542] [Article Influence: 192.8] [Reference Citation Analysis (0)] |
| 99. | Landskron G, De la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. 2014;2014:149185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1033] [Cited by in RCA: 1233] [Article Influence: 112.1] [Reference Citation Analysis (0)] |
| 100. | Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 944] [Cited by in RCA: 1418] [Article Influence: 157.6] [Reference Citation Analysis (0)] |
| 101. | Wang J, Liu J, Sun G, Meng H, Wang J, Guan Y, Yin Y, Zhao Z, Dong X, Yin S, Li H, Cheng Y, Wu H, Wu A, Yu X, Chen L. Glioblastoma extracellular vesicles induce the tumour-promoting transformation of neural stem cells. Cancer Lett. 2019;466:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 102. | Reinhard J, Brösicke N, Theocharidis U, Faissner A. The extracellular matrix niche microenvironment of neural and cancer stem cells in the brain. Int J Biochem Cell Biol. 2016;81:174-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 103. | Harris NC, Achen MG. The proteolytic activation of angiogenic and lymphangiogenic growth factors in cancer--its potential relevance for therapeutics and diagnostics. Curr Med Chem. 2014;21:1821-1842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 104. | Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987-996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14033] [Cited by in RCA: 15825] [Article Influence: 791.3] [Reference Citation Analysis (0)] |
| 105. | Weller M, van den Bent M, Tonn JC, Stupp R, Preusser M, Cohen-Jonathan-Moyal E, Henriksson R, Le Rhun E, Balana C, Chinot O, Bendszus M, Reijneveld JC, Dhermain F, French P, Marosi C, Watts C, Oberg I, Pilkington G, Baumert BG, Taphoorn MJB, Hegi M, Westphal M, Reifenberger G, Soffietti R, Wick W; European Association for Neuro-Oncology (EANO) Task Force on Gliomas. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18:e315-e329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 639] [Cited by in RCA: 776] [Article Influence: 97.0] [Reference Citation Analysis (0)] |
| 106. | Diaz RJ, Ali S, Qadir MG, De La Fuente MI, Ivan ME, Komotar RJ. The role of bevacizumab in the treatment of glioblastoma. J Neurooncol. 2017;133:455-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 159] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 107. | Wang Y, Meng X, Zhou S, Zhu Y, Xu J, Tao R. Apatinib Plus Temozolomide for Recurrent Glioblastoma: An Uncontrolled, Open-Label Study. Onco Targets Ther. 2019;12:10579-10585. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 108. | Preusser M, Lim M, Hafler DA, Reardon DA, Sampson JH. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat Rev Neurol. 2015;11:504-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 302] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 109. | Johnson DB, Sullivan RJ, Menzies AM. Immune checkpoint inhibitors in challenging populations. Cancer. 2017;123:1904-1911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 261] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 110. | Labanieh L, Majzner RG, Mackall CL. Programming CAR-T cells to kill cancer. Nat Biomed Eng. 2018;2:377-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 298] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 111. | Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang SX, Ivy SP. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12:445-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 916] [Cited by in RCA: 996] [Article Influence: 99.6] [Reference Citation Analysis (2)] |
| 112. | Massard C, Azaro A, Soria JC, Lassen U, Le Tourneau C, Sarker D, Smith C, Ohnmacht U, Oakley G, Patel BKR, Yuen ESM, Benhadji KA, Rodon J. First-in-human study of LY3039478, an oral Notch signaling inhibitor in advanced or metastatic cancer. Ann Oncol. 2018;29:1911-1917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 111] [Article Influence: 18.5] [Reference Citation Analysis (0)] |