Published online Nov 26, 2020. doi: 10.4252/wjsc.v12.i11.1327
Peer-review started: July 10, 2020
First decision: August 9, 2020
Revised: August 21, 2020
Accepted: September 15, 2020
Article in press: September 15, 2020
Published online: November 26, 2020
Processing time: 138 Days and 16.5 Hours
Tooth enamel, a highly mineralized tissue covering the outermost area of teeth, is always damaged by dental caries or trauma. Tooth enamel rarely repairs or renews itself, due to the loss of ameloblasts and dental epithelial stem cells (DESCs) once the tooth erupts. Unlike human teeth, mouse incisors grow continuously due to the presence of DESCs that generate enamel-producing ameloblasts and other supporting dental epithelial lineages. The ready accessibility of mouse DESCs and wide availability of related transgenic mouse lines make mouse incisors an excellent model to examine the identity and heterogeneity of dental epithelial stem/progenitor cells; explore the regulatory mechanisms underlying enamel formation; and help answer the open question regarding the therapeutic development of enamel engineering. In the present review, we update the current understanding about the identification of DESCs in mouse incisors and summarize the regulatory mechanisms of enamel formation driven by DESCs. The roles of DESCs during homeostasis and repair are also discussed, which should improve our knowledge regarding enamel tissue engineering.
Core Tip: In the present review, we update the current understanding about the identification of dental epithelial stem cells (DESCs) in mouse incisors and summarize the regulatory mechanisms of enamel formation driven by DESCs. The roles of DESCs during homeostasis and repair are also discussed, which should improve our knowledge regarding enamel tissue engineering.
- Citation: Gan L, Liu Y, Cui DX, Pan Y, Wan M. New insight into dental epithelial stem cells: Identification, regulation, and function in tooth homeostasis and repair. World J Stem Cells 2020; 12(11): 1327-1340
- URL: https://www.wjgnet.com/1948-0210/full/v12/i11/1327.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v12.i11.1327
Tooth enamel, the most highly mineralized tissue in the human body, consists of hydroxyapatite organized into enamel rods and inter-rods which are interwoven, therefore serving as a protective covering for the tooth crown. During enamel formation, inner enamel epithelial cells in the enamel organ differentiate into enamel-forming ameloblasts, which secrete enamel matrix and create an extracellular environment for mineralization[1-4]. These cells commit apoptosis once the enamel formation is accomplished and the tooth erupts into the oral cavity[5]. The loss of ameloblasts and neighboring environment renders enamel an acellular and nonvital tissue that, when insulted by dental caries or trauma, is incapable of repair or renewal. To restore the missing enamel tissue, current treatment is limited to using acid-etching techniques and artificial materials such as resin, amalgam, and porcelain, which are not perfect due to frequent microleakage, limited life span, or inherent inability to fully restore its function[6-8].
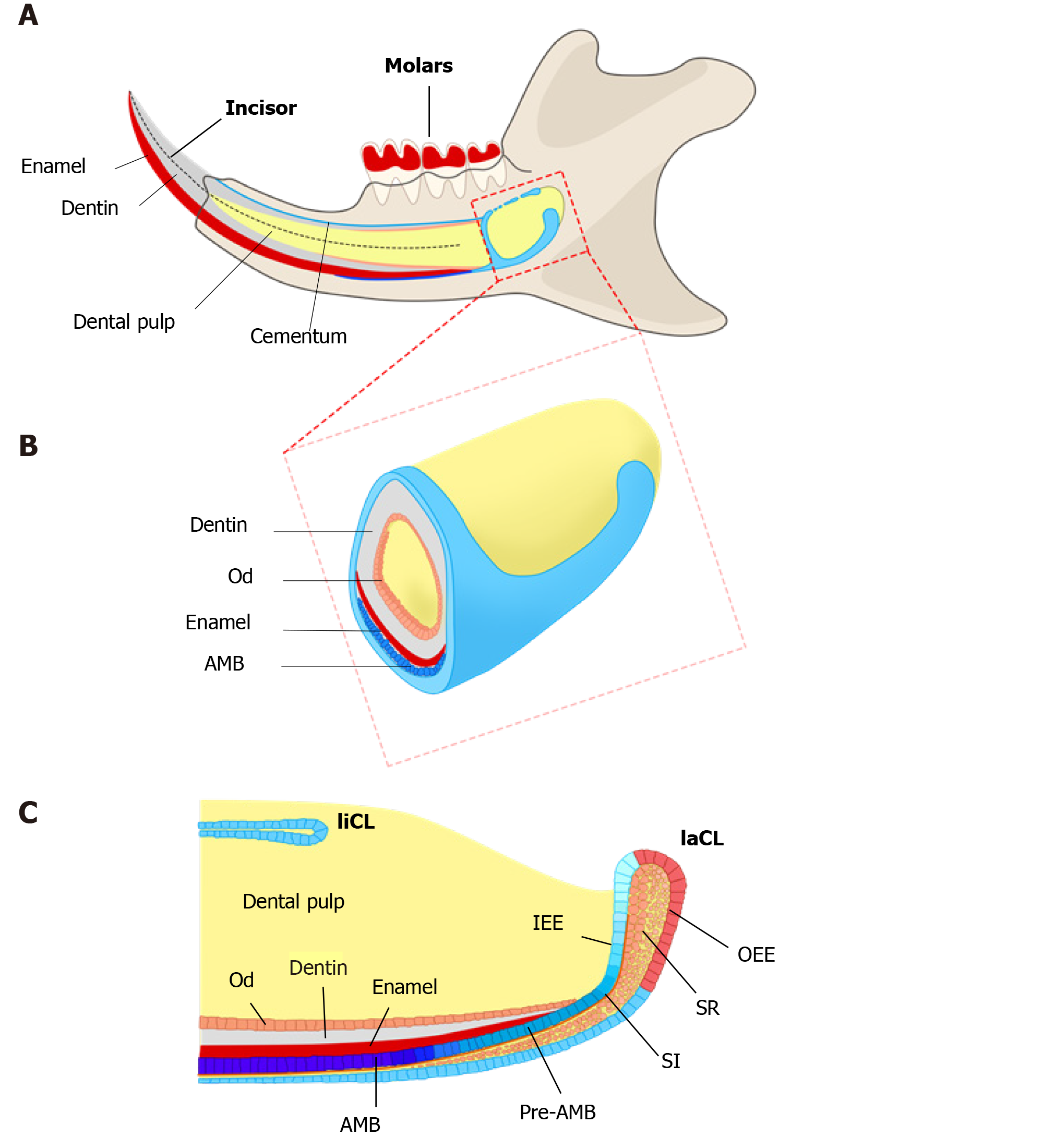
One potential remedy for this is to construct natural enamel. Enamel–dentin complex structure has been detected on polyglycolic acid fiber mesh using dissociated porcine third molar tooth germ cells, suggesting tissue engineering as an alternative strategy to regenerate enamel[9]. The classic tissue engineering relies on three elements, including stem cells, suitable scaffolds, and bioactive molecules to initiate a sequence of events inducing tissue formation[10,11]. However, unlike successful implementation of tissue engineering in other dental tissues, such as dentin and pulp regeneration, enamel tissue engineering is hindered since ameloblasts and dental epithelial stem cells (DESCs) are lost when the tooth erupts[5]. Consequently, the generation of potent and viable DESCs would be a major step toward promising enamel tissue engineering[12]. Interestingly, nature has provided us a good example, the rodent incisor, which grows continuously throughout the animal’s life[13]. Harada et al[14] have identified DESCs at the proximal end of mouse incisors, in a structure named the labial cervical loop (laCL). These cells can self-renew and differentiate into enamel-secreting ameloblasts and the other supporting dental epithelial lineages[15-18]. Owing to the ready accessibility of mouse DESCs and the wide availability of related transgenic mouse lines, mouse incisors serve as an ideal system to explore the identity and heterogeneity of dental epithelial stem/progenitor cells. The continuous replenishment of enamel tissue is fueled by the DESCs, making mouse incisors an excellent model to uncover the regulatory mechanisms underlying enamel formation[19] (Figure 1). Moreover, studying how the homeostasis and repair are maintained by DESCs in mouse incisors can help us answer the open question regarding the therapeutic development of enamel engineering.
In the present review, we update the current understanding about the identification of DESCs in mouse incisors and summarize the regulatory mechanisms of enamel formation driven by DESCs. The roles of DESCs during homeostasis and repair are also discussed, which could improve our knowledge regarding enamel tissue engineering.
In the classical model of tissue supported by stem cells, rare slow-cycling stem cells, which divide infrequently, contribute to the production of active-cycling transit-amplifying (TA) cells, which then generate all the related lineages[20-23]. Therefore, the stem cells in the adult tissue are always identified by their characteristics, for instance, their quiescent feature and their potential of uni- or multilineage differentiation[21].
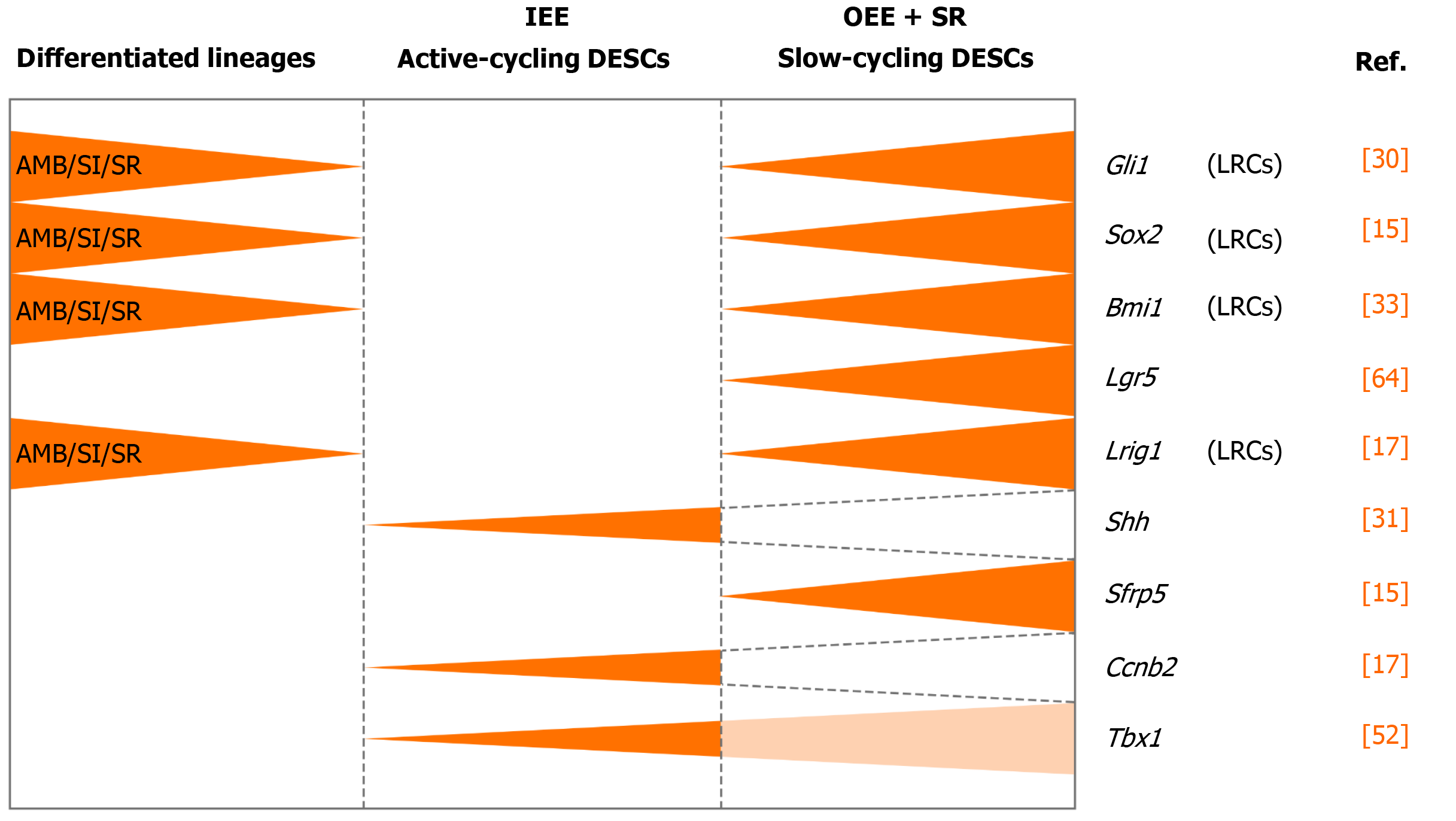
A credible approach to locate the stem cell niche in adult tissue is to take the benefit of their quiescent property by positioning the distribution of label-retaining cells (LRCs)[21]. LRCs are capable of retaining DNA synthesis labels, including tritiated thymidine (3H-thymidine) and 5-bromo-2’-deoxyuridine (BrdU), after a long-term chase[24,25]. Active-cycling cells, such as TA cells, remove the DNA label following cell divisions within a short time. On the contrary, the quiescent stem cells retain the label during a long chasing period owing to their infrequent division[21-23]. LRCs have been confirmed as stem cells in various tissues and organs, such as the epidermis, intestine, and mammary gland[26-28]. With one administration of 3H-thymidine, an early study showed proliferating cells in the adult rat enamel organ, which exited the cell cycle and migrated distally. After 32 d, the cells retaining the label were detected in the outer enamel epithelium (OEE) and underlying stellate reticulum (SR) of laCL, suggesting the existence of DESCs at the proximal end of the laCL[29,30]. To detect LRCs in vivo, the neonatal mice were pulsed with BrdU, to label the cycling cells in the dental epithelium at the time of tissue expansion and subsequently to identify the LRCs that rarely divide and retain the label till adulthood. The distribution of BrdU+ LRCs in the OEE and underlying SR confirmed the presence of slow-cycling cells in laCL[14,31]. Later, this assumption was demonstrated with K5Tta; H2B-GFP mice, in which the expression of doxycycline-repressible H2B-GFP is regulated by the keratin 5 promoter[32]. All dental epithelial lineages uniformly express green fluorescent protein (GFP) in the absence of doxycycline administration. Following 2 mo administration of doxycycline, H2B-GFP-retaining cells were observed in the same compartments, suggesting that putative DESCs were present in these compartments and might have contributed to the continuous growth of the incisor enamel[33,34].
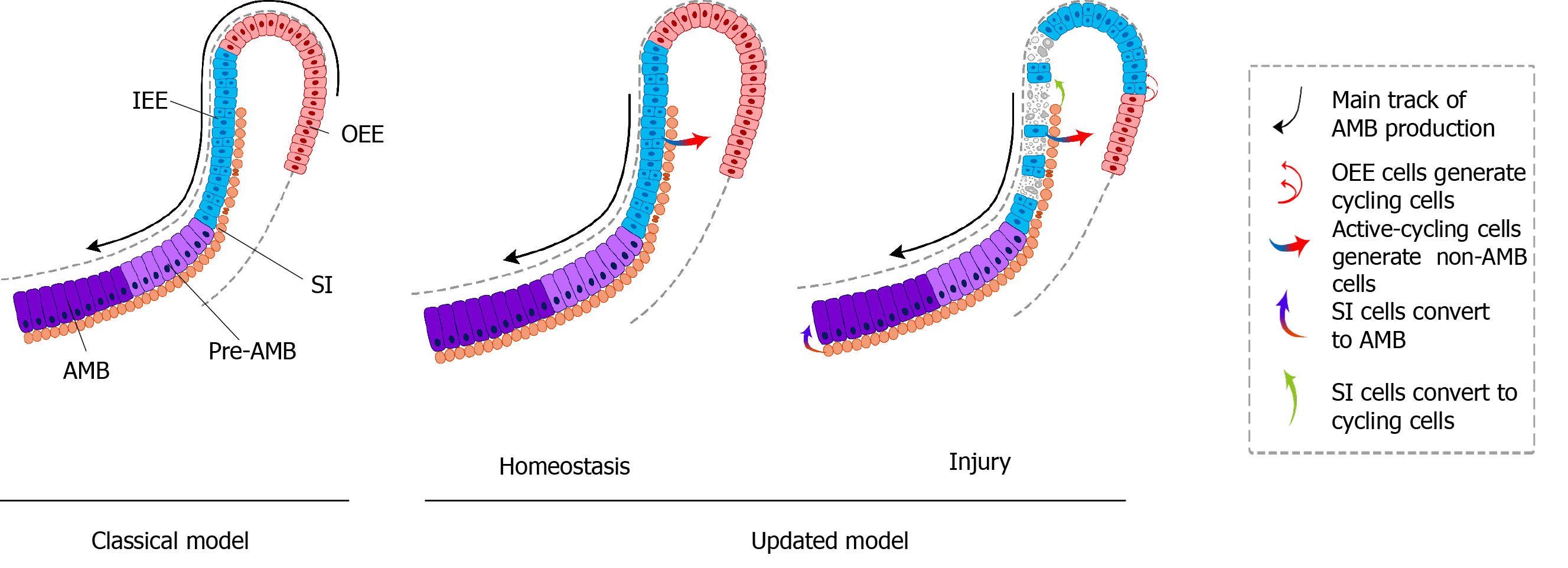
To further assess the uni- or multilineage differentiation potential of putative stem cells, the cells and their progeny are permanently genetically marked and chased using lineage tracing, an essential strategy in the identification of stem cells in adult mammalian tissue[21,35]. Genetic lineage tracing in mice is preferably achieved with the Cre-loxP system. In this system, the Cre specifically activates the reporter in cells upon the control of the tissue- or cell-specific promoter, by excising the loxP-STOP-loxP sequence. With inducible recombination (Cre recombinase is fused to estrogen receptor), the Cre recombinase activity can be manipulated temporally and spatially with tamoxifen[36-38]. The application of inducible Cre for lineage tracing has provided maximum information about all the progeny of the stem cells in postnatal tissue. With CreER controlled by Gli1 promoter (GliCreER), Gli1+ cells residing in the LRCs region (OEE and underlying SR) have been demonstrated to be capable of generating functional ameloblasts and supporting epithelial lineages, such as stratum intermedium (SI) cells[31]. The distribution of Bmi1+ cells in laCL is consistent with that of LRCs and Gli1+ cells. Similarly, Bmi1+ cells also give rise to enamel-producing ameloblasts and neighboring SI cells[33]. Unlike Gli1 and Bmi1, Sox2 has a broader expression domain, which expands both distally and proximally in the laCL. Genetic lineage tracing in vivo has revealed that Sox2+ cells could generate all the mouse incisor epithelial lineages[15]. Later, Lrig1 has been proposed as a putative stem cell marker by gene coexpression module analysis. Moreover, Lrig1+ cells have been shown to contribute to dental epithelial lineages via in vivo lineage tracing[17]. These previous studies have constructed the classical model of enamel renewal, in which slow-cycling DESCs, residing in the OEE and underlying SR of the laCL, regularly move to the inner enamel epithelium (IEE) and generate active-cycling TA cells[16]. These active-cycling cells migrate distally, exit the cell cycle, and differentiate into enamel-producing ameloblasts. However, identification of cell types, properties, and cellular relationships remains unknown in this classical model. Furthermore, the latest study has shown that the expression of the putative DESCs markers, including Bmi1, Gli1, and Sox2, are broader than we expected, since they have been detected in the IEE as well, which unravels the limitation of the currently available genetic strategies (Figure 2).
The previous studies mentioned above characterized stem cells based on the known properties, which would inevitably introduce researchers’ preconceptions, such as significant signaling pathways certified in the putative stem cell niche[39]. Therefore, unbiased technology is desired to identify cells and characterize cells with new clusters of markers. Single-cell sequencing, in particular, mRNA sequencing from single cells (scRNA-seq), contributes to the unbiased profiling of cells from tissues and organs[40-42]. Generally, single cells are isolated and assigned to a specific barcode; thus, gathered mRNA can be sequenced and reassigned to its cellular origin. According to the transcriptomes, cells are divided into groups with unsupervised clustering[43]. This scRNA-seq shows a series of advantages in uncovering heterogeneity within the population which was considered to be homogeneous, discovering novel and rare cell types, and raveling the relationships between cell types.
Sharir et al[18] performed scRNA-seq of sorted dental epithelial cells to resolve the cellular heterogeneity and lineage dynamics of the adult incisor epithelium in an unbiased manner. Single cells from mouse incisors were generated and sequenced. The high-dimensional, whole-transcriptome data were visualized using SPRING[18], which is suitable for analyzing differentiation trajectories as maintaining relationships of cells with the similar transcriptome. Mouse incisor epithelial cells are divided into three groups, including cycling cells (class 1), ameloblast lineages (class 2), and nonameloblast epithelial cells (class 3)[17]. With the expression of cell-cycle markers like Cdc20 and Ccnb2, class 1 cells have the majority of dividing and cycling cells, located in the IEE and adjacent SI region. Moreover, class 1 cells maintain active self-renewal as their transcriptomes have shown successive phases of the cell cycle, and several cells return to their original state at the end of the cell cycle. Signatures reflecting classes 2 and 3 populations have also been observed in class 1 cell populations, suggesting that progenitors are cycling with upregulated expression of differentiation genes. Combined with differentiation trajectories and kinetics experiments, class 1 houses progenitor cells considered as the root, which produces cells of classes 2 and 3[18]. Therefore, distinct from the classical views, an updated dynamic model of stem cells in mouse incisors reveals that the IEE (class 1) possesses active cycling stem cells that differentiate into both the functional ameloblasts and the surrounding nonameloblast epithelial lineages (classes 2 and 3).
The classic model of DESCs in mouse incisors is similar to that of hematopoietic stem cells. The homeostasis of mouse incisor enamel is fueled by the quiescent stem cells in the OEE and underlying SR of the laCL[13-17,19]. These slow-cycling cells produce TA cells in the IEE that undergo limited divisions before terminal differentiation (Figure 3A). Even though scientists stick to this concept for a long time, the concept is still incapable of accounting for the great demand of ameloblasts in daily production of enamel. In this model, DESCs were identified with LRCs and lineage tracing of specific stem cell marker candidates, including Sox2, Bmi1, Gli1, and Lrig1. On the contrary, the novel model is based on scRNA-seq, an unbiased method. The dental epithelial cells in laCL can be divided into three different groups. The daily production of enamel is supported by a group of actively cycling progenitor cells in the IEE, which are responsible for the production of ameloblasts and nonameloblast epithelial lineages, including the OEE and underlying SR, which are considered as DESCs in the classic model[18] (Figure 3B). Previously established stem cell markers, including Sox2, Bmi1, Gli1, and Lrig1, are not expressed specifically in any groups of the updated model.
The proliferation and differentiation of DESCs are regulated by genetic regulatory signals in a temporospatial manner[39]. Surrounding dental mesenchymal cells provide essential signals for maintenance and differentiation of DESCs[44]. To date, several signaling pathways have been found to be important for the steady state of the DESCs niche and subsequent differentiation in mouse incisors[16,19]. Fibroblast growth factor (FGF) signaling was first put forward. Fgf3, Fgf9, and Fgf10 are key mesenchymal signals for stimulating the proliferation of DESCs in the developing cervical loops[30,45-48]. Transforming growth factor (TGF)-β signaling participates in DESC maintenance and TA cell proliferation by regulating the activity of FGF signaling[47,48]. The expression of Fgf3 and Fgf10 and the number of BrdU+ LRCs are markedly reduced in the laCL of mice with mutation of the Alk5 gene, which is responsible for encoding the TGF-β type I receptor[48]. The reduction of Fgf3/10 expression by TGF-β type II receptor (Tgfbr2) deletion in mesenchyme-promoted differentiation of DESCs results in wavy mineralized structure formation[49]. Sprouty genes, which encode the intracellular antagonists of receptor tyrosine kinase signaling, ensure lineage differentiation of DESCs through the FGF signaling pathway[50]. Once Sprouty genes are deleted, the inhibitory signal is removed, leading to increased sensitivity to Fgf3/10 expression in both liCL and laCL as well as the adjacent mesenchyme[51]. The inhibitory effect of Sprouty protein on FGF signaling functions is mediated by changing the expression of TBX1 and BCL11B indirectly in the Spry4-/- mice[52,53]. The removal of BCL11B affects the differentiation and proliferation of DESCs, reduces the size of the laCL, and shortens the zone of ameloblast progenitors[53]. Conversely, an allele of BCL11B promotes the proliferation of TA cells, thus ensuring a stable number of DESCs[54]. Cao et al[55] have demonstrated that TBX1 regulates DESCs by suppressing the transcriptional activity of Pitx2 which is in connection with a cell cycling inhibitor p21. The downstream of FGF signaling has been explored by Goodwin et al[56]. Ras signaling is activated after FGFs bind to receptor tyrosine kinases (RTKs) and then regulate the proliferative activity of DESCs through the mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K) pathways. Further evidence about the role of MAPK and PI3K in amelogenesis has been proposed through a mouse incisor model of Ras dysregulation[57].
Another important signaling pathway is Hedgehog (Hh), which is essential for maintaining epithelial cell size, proliferation, and polarization[58]. Runx2 mutation results in downregulated expression of Shh in the dental epithelium[59]. It has been shown that Runx gene and its binding protein core binding factor β gene (Cbfb) modulate the continuous proliferation and differentiation of DESCs by activating FGF signaling loops and maintaining the expression of Shh mRNA[60]. The BMP–Smad4 signaling cascade inhibits the activity of Shh–Gli1 signaling to maintain Sox2+ DESCs in the CL region of mouse molars. Conversely, loss of Smad4 prolongs maintenance of the CL and affects cell expansion and differentiation[61]. Ptch1 and Ptch2 are binding receptors of Hh ligands and have distinct functional roles. Ptch1 transduces Hh signaling to maintain Sox2+ stem cells, whereas Ptch2 with Desert hedgehog negatively regulates P-cadherin expression, suggesting that Hh signaling contributes to the maintenance and differentiation of DESCs simultaneously[62]. Several studies have revealed that the Notch signaling pathway is required for survival of DESCs and the formation of ameloblasts[63]. Multiple genes of this pathway are expressed in the CL, including Delta-like 1 (Dll1) and Jagged 2 (Jag2) genes encoding the Notch ligands, as well as lunatic fringe (Lfng) encoding transferase that modifies Notch receptors[63-65]. The notch responsive gene Hes1 is expressed in the SR. When dissected CL is cocultured with the Notch signaling inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT), the size of the CL is reduced because of increased apoptosis and reduced proliferation of DESCs[63]. Jag2 and Lfng genes are regulated by FGF and bone morphogenetic protein (BMP) signaling. Deletion of Jag2 results in abnormal ameloblast differentiation[65]. The number of SI cells was increased when CL-derived dental epithelial cells were cultured with Jagged 1 protein and overexpressed the Notch1 internal domain. Differentiation of SI cells was inhibited when Jagged1 was neutralized with specific antibody, suggesting that Notch signaling regulates SI cells, which function as a reserve progenitor pool[66]. Elimination of Notch1+ cells disrupted the repair process of injured epithelium and obstructed the regeneration of damaged dental epithelium[18]. The essential role of Hippo signaling has been suggested in coordinating the proliferation and differentiation of DESCs[67]. The effectors of the evolutionarily conserved Hippo signaling pathway, Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ), are expressed in TA cells[67]. It has been reported that YAP/TAZ induce the ITG43–FAK–CDC42 signaling axis in the TA zone and activate mammalian target of rapamycin (mTOR) signaling by controlling Rheb expression, maintaining TA cell proliferation and survival, and inhibiting precocious differentiation[67].
Intriguingly, there is neither expression of the Wnt-responsive gene Axin2 nor expression of Wnt pathway mediators or inhibitors in the region of putative stem cells. These signs indicate that DESCs are not modulated directly by Wnt/β-catenin signaling[68]. However, Wnt/β-catenin can inhibit expression of Fgf10, a key antiapoptotic signal, to maintain a proper rate of apoptosis in DESCs[69]. And Lgr5, a Wnt signaling target gene and stem cell marker, is expressed in the putative stem cell region, the SR region underlying the OEE. These Lgr5+ cells are identified as slow-cycling stem cells and a subpopulation of Sox2+ DESCs[70,71]. A recent study reports that Runx1 regulates the Lgr5-expressing epithelial stem cells and differentiation of ameloblast progenitors in the developing incisors. The Runx1–Lgr5 axis is partially modulated by signal transducer and activator of transcription 3 (STAT3) phosphorylation in the CL of growing incisors. Runx1 deficiency results in downregulated expression of Lgr5 and Sox2 and underdevelopment of the CL[70].
Apart from signaling pathways, stem cell markers, like Bmi1 and Sox2, are functional in the maintenance and differentiation of DESCs. The deletion of Bmi1 decreased the number of stem cells, disorganized gene expression, and impaired enamel production. Knockdown of Ink4a/Arf partially rescued Bmi1-null phenotypes. Ink4a/Arf is one critical target gene of Bmi1, which encodes the cell cycle inhibitors. Bmi1 also targets Hox genes, which maintain the undifferentiated state of stem cells and prevent inappropriate differentiation[33]. Conditional removal of Sox2 during incisor renewal resulted in the morphological change of laCL and slowed down incisor growth[72]. Conditional overexpression of lymphoid enhancer-binding factor (Lef-1) in the dental epithelium increased cell proliferation, created a new stem cell compartment in the laCL, and rescued tooth arrest resulting from deletion of Sox2[72]. These results show that Lef-1 regulates maintenance of DESCs and enamel formation, but the underlying mechanism remains unresolved.
In addition to transcription factors, epigenetics also regulates the gene expression in mammalian development without alterations in the DNA sequence[73-75]. Epigenetic mechanisms involve DNA methylation, modification of histone tails, and gene regulation by noncoding RNAs and miRNAs[76]. Although various studies have shown the epigenetic regulation in tooth development and regeneration, there are only a few reports about the epigenetic effects in DESCs in mouse incisors.
Several studies have demonstrated that miRNAs play a role in enamel formation and the renewal and differentiation of DESCs in mouse incisors. Conditional deletion of Dicer1, an essential factor for mature miRNA formation, led to the complete loss of miRNA genesis, resulting in ectopic formation of the CL[77]. This was the result of increased proliferation of incisor epithelial stem cells and impaired differentiation of ameloblasts. Furthermore, microarray analysis unraveled that the distinct expression pattern of miRNAs in different compartments of the dental epithelium, including the laCL, lingual CL, and ameloblasts, suggests the potential role of miRNAs in the self-renewal and differentiation of DESCs[77]. miR-200c, one of the differentially expressed miRNAs, participates in differentiation by repressing noggin, an antagonist of BMP signaling. Noggin induces expression of E-cadherin and Amelogenin to maintain cell adhesion and promote ameloblast differentiation, respectively[74]. BMP signaling is simultaneously the upstream regulator of miRNA-200c, and a positive-feedback loop has been shown between miR-200c and BMP signaling. Another upstream regulatory pathway has also been identified in which endogenous Pitx2 interacts with the miR-200c/141 promoter to activate miR-200c[75]. Also, miR-200a-3p is activated by Pitx2 and targets the BMP antagonist Bmper to further regulate BMP signaling[74]. Recently, miR-1 expressed at the CL of the dental epithelium was revealed by an in situ hybridization assay and its expression was inversely correlated with its target connexin (Cx) 43. Deletion of miR-1 induced DESCs to express Cx43, which regulated cell proliferation during DESC differentiation, by formation of cell–cell gap junctions and hemichannels at the plasma membrane[78].
Interactions among different cell populations and between cells and the extracellular matrix are indispensable for the homeostasis of the stem cell niche in the dental epithelium[79]. For instance, integrin β3 is required for the formation and maintenance of the CL and proliferation of DESCs. The knockdown of the CD61 gene results in a reduction of the CL size with downregulated expression of Lgr5 and Notch1[80]. E-cadherin, a protein for cell–cell adhesion, regulates proliferation in TA cells and controls differentiation of DESCs[81]. Prominin-1(Prom1/CD133), an essential protein for ciliary kinetics, regulates DESCs to respond appropriately to extracellular signals. Conditional removal of Prom1 impairs ciliary dynamics and the positive effects of SHH treatment, leading to the destruction of stem cell activation and homeostasis in mouse incisor tooth epithelium[82].
Finally, DESCs are regulated by other factors, such as follistatin, activin, heparin-binding molecule midkine, and heparin-binding growth-associated molecule[45,83]. Exogenous retinoic acid (RA) has negative effects on DESCs via inhibition of Fgf10. It has been demonstrated that supplementation of FGF10 in incisor cultures blocks RA’s negative effects to antagonize apoptosis and increase proliferation of DESCs in the laCL[84].
The renewal and differentiation of DESCs are critical drivers of continuously growing mouse incisors[16]. The heterogeneity in DESCs has been discovered, hinting that different stem cell populations play a distinct function in mouse incisors[19]. Numerous studies have found that renewal of mouse incisors requires a balance of proliferation and differentiation of DESCs, which is controlled tightly by a complex regulated network, to maintain proper lineage ratios (Figure 3).
The renewal process of incisor enamel has been studied by constantly developing investigation methods. Continuous renewal of mouse incisors was initially observed by cutting the erupted enamel[85,86]. Sequential 3H-thymidine tracing showed the growth rate of rat incisors to be approximately 365 μm/d[13]. It was confirmed that proper incisor growth requires proliferative dividing cells in an early experiment in animals treated with mitotic arrest agents[87]. The discovery of stem cell marker candidates and genetic lineage tracing helped researchers to propose a classical model of renewal of mouse incisors. It is thought that quiescent stem cells residing in the OEE generate actively proliferating TA cells, which migrate distally and differentiate into ameloblasts that are responsible for enamel formation[16,19]. How the slowly cycling DESCs meet the daily requirement of ameloblasts has been an open question for a long time. To address this, Sharir et al[18] established a novel model of mouse incisor DESCs, in which actively cycling dental epithelial progenitor cells generate both the functional ameloblasts and the surrounding nonameloblast epithelial cell populations, which are subsequently responsible for the homeostasis of mouse incisor enamel (See session ‘Updated model’ for details)[18,19]. Even though this study identified the DESCs responsible for the homeostasis, the mechanism responsible for the committed differentiation is still absent. More studies are needed to determine the underlying mechanisms by which incisor epithelial self-renewal and cell lineage distribution remain stable under physiological conditions.
DESCs also support damaged incisor epithelium regeneration after injury (Figure 3). The number of Sox2 and Lgr5 transcripts decreased significantly and the spherical shape of the laCL was lost after transient deletion of Sox2. Some Sox2 transcripts could be detected and the laCL shape was restored after 5 d. During recovery, the percentage of EdU+ cells was significantly increased in the central and proximal sections of the SR. These findings demonstrated that the Sox2+ DESC population could be regenerated quickly from the SR[87].
A recent study has demonstrated the migration and plasticity of DESCs during recovery. In the IEE of mouse incisors, the proliferative cells eliminated by 5-fluorouracil (5-FU) treatment were supplemented by the burgeoning proliferating cell population after 3 d. After 10 d, abnormal ameloblast organization and disorganized enamel matrix resulting from 5-FU treatment both recovered to normal. The function and dynamic changes of DESCs were further analyzed by scRNA-seq during injury repair. After cytotoxic injury, the number of cycling cells was increased with expanded expression domains of Ccnb1 and Birc5, but the numbers of pre-ameloblasts and ameloblasts were significantly decreased with the distal shift. Expression of Sfrp5 and Cldn10 was upregulated and expanded towards the proliferating regions, suggesting an increased nonameloblast population. Further study has demonstrated that Notch1+ SI cells are induced to differentiate into ameloblasts and critical for tissue recovery. These findings have shown that dental epithelium regeneration after injury is driven by recruiting more progenitors or nonmitotic pre-ameloblasts to divide, shortening the cell cycle, and delaying onset of ameloblast differentiation. Therefore, DESCs play an essential role in an appropriate and rapid response to tissue damage[18].
Whole tooth regeneration based on epithelial and mesenchymal interaction through simulating tooth development is a promising strategy for replacing lost teeth. The odontogenic potential could be retained in epithelial and mesenchymal cells isolated from the tooth germ of early development. Tooth-like structures can be produced in vitro based on individual dental epithelial cells and mesenchymal cells from mouse embryos[88]. Bioengineered tooth germs, which are reconstructed by epithelial and mesenchymal cells, have generated functional teeth when they were placed into the alveolar socket of adult mice[89]. In addition to the mouse regenerative model, a recent study, using an allogeneic cell reassociation approach, achieved whole-tooth regeneration in minipig jawbone in situ[90].
To resolve the problem of a source of DESCs for tooth engineering, DESCs from mouse incisors have been demonstrated as an excellent tool. Besides, several studies have attempted to induce available stem cells to form dental epithelial cells. It has been reported that human tooth germ stem cells can differentiate into epithelial cell types, but not functional ameloblasts[91]. Mouse induced pluripotent stem cells (miPSCs) can differentiate into dental epithelial-like cells in serum-free culture conditions, with the addition of neurotrophin-4. These cells derived from miPSCs can express dental epithelial surface marker CD49f and ameloblast-specific markers[92]. As possible alternative sources for the human dental epithelium, human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) may be options due to their potency of multilineage differentiation[93,94]. Based on the vital role of interactions between dental epithelial and ectodermal mesenchymal cells in dental development, both hESCs and hiPSCs are induced to differentiate into epithelial-like stem cells by the HERS/ERM cell line[95]. Newly ex-vivo-formed differentiated hESCs express special epithelial stem cell markers including E-cadherin, ABCG2, Bmi-1, p63, and p75[94]. Even though rare ameloblasts, enamel, or dentin-enamel tissue were detected in this study, some progress has been made on how to obtain DESCs.
Ameloblast cell lines are indispensable for enamel formation and regeneration, because they secrete amelogenin, which is an essential constituent of enamel[96,97]. The generation of ameloblasts is still an obstacle. Although several mouse ameloblast-like cell lines, such as ALC and LS8, have been established, they do not generate enamel[98,99]. Human gingival epithelial cells have been a source of ameloblast-like cells induced by BMPs and TGF-β. It has been reported that there are 20 ameloblast-specific genes as cell surface markers, which will contribute to the isolation of human ameloblast-like cells[100].
To replace enamel defects due to caries or trauma, dentists use several artificial materials, which do not completely resemble the mechanical, physical, and esthetic features of the lost enamel[101]. Enamel regeneration has been considered as an alternative clinical strategy. Our understanding of identification, regulation, and role of DESCs has been strengthened by studying continuously growing mouse incisor models. By scRNA-seq, the heterogeneity of DESCs in the laCL has been identified and a novel mouse incisor model distinct from early evidence has been established. The updated understanding of the regulation and role of DESCs in tissue homeostasis and repair contributes to the therapeutic development of enamel engineering. Despite all this progress with DESCs in recent years, enamel regeneration still faces various challenges, which have been outlined in two recent conferences[97,102]. The difficulties include the acellular structure, high mineralization, essential epigenetic regulation during mineralization, unique migration of ameloblasts during crystal formation, and ultimate organization with prismatic and interprismatic structures of natural enamel. Several issues remain to be addressed before clinical application, such as the combination of regenerated enamel with natural teeth and the control of shape, size, color, and time. Although there are limitations in enamel tissue engineering, the exciting progress with DESCs provides researchers with novel insight into stem cell-based tooth engineering, and consequently, to pave the way for future treatments.
Manuscript source: Invited manuscript
Specialty type: Cell biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cenciarelli C, Mitsiadis T S-Editor: Gong ZM L-Editor: Wang TQ P-Editor: Wang LL
| 1. | Bartlett JD. Dental enamel development: proteinases and their enamel matrix substrates. ISRN Dent. 2013;2013:684607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 2. | Honda MJ, Sumita Y, Kagami H, Ueda M. Histological and immunohistochemical studies of tissue engineered odontogenesis. Arch Histol Cytol. 2005;68:89-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 67] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 3. | Fincham AG, Moradian-Oldak J, Simmer JP. The structural biology of the developing dental enamel matrix. J Struct Biol. 1999;126:270-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 423] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 4. | Fukumoto S, Yamada Y. Review: extracellular matrix regulates tooth morphogenesis. Connect Tissue Res. 2005;46:220-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 5. | Kaneko H, Ogiuchi H, Shimono M. Cell death during tooth eruption in the rat: surrounding tissues of the crown. Anat Embryol (Berl). 1997;195:427-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 6. | Arrow P. Dental enamel defects, caries experience and oral health-related quality of life: a cohort study. Aust Dent J. 2017;62:165-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 7. | Tirlet G, Crescenzo H, Crescenzo D, Bazos P. Ceramic adhesive restorations and biomimetic dentistry: tissue preservation and adhesion. Int J Esthet Dent. 2014;9:354-369. [PubMed] |
| 8. | BaniHani A, Deery C, Toumba J, Munyombwe T, Duggal M. The impact of dental caries and its treatment by conventional or biological approaches on the oral health-related quality of life of children and carers. Int J Paediatr Dent. 2018;28:266-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 9. | Young CS, Terada S, Vacanti JP, Honda M, Bartlett JD, Yelick PC. Tissue engineering of complex tooth structures on biodegradable polymer scaffolds. J Dent Res. 2002;81:695-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 297] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 10. | Zhang W, Vázquez B, Yelick PC. Bioengineered post-natal recombinant tooth bud models. J Tissue Eng Regen Med. 2017;11:658-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Bakhshandeh B, Zarrintaj P, Oftadeh MO, Keramati F, Fouladiha H, Sohrabi-Jahromi S, Ziraksaz Z. Tissue engineering; strategies, tissues, and biomaterials. Biotechnol Genet Eng Rev. 2017;33:144-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 12. | Yu T, Volponi AA, Babb R, An Z, Sharpe PT. Stem Cells in Tooth Development, Growth, Repair, and Regeneration. Curr Top Dev Biol. 2015;115:187-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 13. | Smith CE, Warshawsky H. Cellular renewal in the enamel organ and the odontoblast layer of the rat incisor as followed by radioautography using 3H-thymidine. Anat Rec. 1975;183:523-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 145] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 14. | Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147:105-120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 402] [Cited by in RCA: 394] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 15. | Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, Klein OD, Thesleff I, Michon F. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev Cell. 2012;23:317-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 188] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 16. | Kuang-Hsien Hu J, Mushegyan V, Klein OD. On the cutting edge of organ renewal: Identification, regulation, and evolution of incisor stem cells. Genesis. 2014;52:79-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 17. | Seidel K, Marangoni P, Tang C, Houshmand B, Du W, Maas RL, Murray S, Oldham MC, Klein OD. Resolving stem and progenitor cells in the adult mouse incisor through gene co-expression analysis. Elife. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 18. | Sharir A, Marangoni P, Zilionis R, Wan M, Wald T, Hu JK, Kawaguchi K, Castillo-Azofeifa D, Epstein L, Harrington K, Pagella P, Mitsiadis T, Siebel CW, Klein AM, Klein OD. A large pool of actively cycling progenitors orchestrates self-renewal and injury repair of an ectodermal appendage. Nat Cell Biol. 2019;21:1102-1112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 19. | Yu T, Klein OD. Molecular and cellular mechanisms of tooth development, homeostasis and repair. Development. 2020;147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 159] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 20. | Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, Dela Cruz-Racelis J, Fuchs E. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155-169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 671] [Cited by in RCA: 626] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 21. | Snippert HJ, Clevers H. Tracking adult stem cells. EMBO Rep. 2011;12:113-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 140] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 22. | Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 2009;137:811-819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 312] [Cited by in RCA: 283] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 23. | Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 954] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 24. | Goodlad RA. Quantification of epithelial cell proliferation, cell dynamics, and cell kinetics in vivo. Wiley Interdiscip Rev Dev Biol. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 25. | Maurer HR. Potential pitfalls of [3H]thymidine techniques to measure cell proliferation. Cell Tissue Kinet. 1981;14:111-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 26. | Sada A, Jacob F, Leung E, Wang S, White BS, Shalloway D, Tumbar T. Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin. Nat Cell Biol. 2016;18:619-631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 151] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 27. | Li N, Nakauka-Ddamba A, Tobias J, Jensen ST, Lengner CJ. Mouse Label-Retaining Cells Are Molecularly and Functionally Distinct From Reserve Intestinal Stem Cells. Gastroenterology 2016; 151: 298-310. e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 28. | Boras-Granic K, Dann P, Wysolmerski JJ. Embryonic cells contribute directly to the quiescent stem cell population in the adult mouse mammary gland. Breast Cancer Res. 2014;16:487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 29. | Smith CE. Cell turnover in the odontogenic organ of the rat incisor as visualized by graphic reconstructions following a single injection of 3H-thymidine. Am J Anat. 1980;158:321-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 30. | Samperiz MM, Blumen G, Merzel J. Effect of vinblastine on the cell cycle and migration of ameloblasts of mouse incisors as shown by autoradiography using 3H-thymidine. Cell Tissue Kinet. 1985;18:493-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 31. | Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, Cao T, Carano RA, Curran T, Schober M, Fuchs E, Joyner A, Martin GR, de Sauvage FJ, Klein OD. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development. 2010;137:3753-3761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 147] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 32. | Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1649] [Cited by in RCA: 1556] [Article Influence: 74.1] [Reference Citation Analysis (0)] |
| 33. | Biehs B, Hu JK, Strauli NB, Sangiorgi E, Jung H, Heber RP, Ho S, Goodwin AF, Dasen JS, Capecchi MR, Klein OD. BMI1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat Cell Biol. 2013;15:846-852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 125] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 34. | Zhao H, Feng J, Seidel K, Shi S, Klein O, Sharpe P, Chai Y. Secretion of shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor. Cell Stem Cell. 2014;14:160-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 352] [Cited by in RCA: 345] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 35. | He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 402] [Cited by in RCA: 407] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 36. | Joyner AL, Zervas M. Genetic inducible fate mapping in mouse: establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn. 2006;235:2376-2385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 139] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 37. | Wu SS, Lee JH, Koo BK. Lineage Tracing: Computational Reconstruction Goes Beyond the Limit of Imaging. Mol Cells. 2019;42:104-112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 26] [Reference Citation Analysis (0)] |
| 38. | Yang H, Adam RC, Ge Y, Hua ZL, Fuchs E. Epithelial-Mesenchymal Micro-niches Govern Stem Cell Lineage Choices. Cell 2017; 169: 483-496. e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 200] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 39. | Simons BD, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145:851-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 363] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 40. | Svensson V, Vento-Tormo R, Teichmann SA. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc. 2018;13:599-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 579] [Article Influence: 82.7] [Reference Citation Analysis (0)] |
| 41. | Haque A, Engel J, Teichmann SA, Lönnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 589] [Cited by in RCA: 668] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 42. | Clevers H, Watt FM. Defining Adult Stem Cells by Function, not by Phenotype. Annu Rev Biochem. 2018;87:1015-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 154] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 43. | Kiselev VY, Andrews TS, Hemberg M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev Genet. 2019;20:273-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 612] [Article Influence: 102.0] [Reference Citation Analysis (0)] |
| 44. | Tummers M, Thesleff I. The importance of signal pathway modulation in all aspects of tooth development. J Exp Zool B Mol Dev Evol. 2009;312B:309-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 178] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 45. | Wang XP, Suomalainen M, Jorgez CJ, Matzuk MM, Werner S, Thesleff I. Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev Cell. 2004;7:719-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 152] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 46. | Harada H, Toyono T, Toyoshima K, Ohuchi H. FGF10 maintains stem cell population during mouse incisor development. Connect Tissue Res. 2002;43:201-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 47. | Yokohama-Tamaki T, Ohshima H, Fujiwara N, Takada Y, Ichimori Y, Wakisaka S, Ohuchi H, Harada H. Cessation of Fgf10 signaling, resulting in a defective dental epithelial stem cell compartment, leads to the transition from crown to root formation. Development. 2006;133:1359-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 99] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 48. | Zhao H, Li S, Han D, Kaartinen V, Chai Y. Alk5-mediated transforming growth factor β signaling acts upstream of fibroblast growth factor 10 to regulate the proliferation and maintenance of dental epithelial stem cells. Mol Cell Biol. 2011;31:2079-2089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 49. | Yang G, Zhou J, Teng Y, Xie J, Lin J, Guo X, Gao Y, He M, Yang X, Wang S. Mesenchymal TGF-β signaling orchestrates dental epithelial stem cell homeostasis through Wnt signaling. Stem Cells. 2014;32:2939-2948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 50. | Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, Boran T, Peterkova R, Martin GR. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377-385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 141] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 51. | Boran T, Peterkova R, Lesot H, Lyons DB, Peterka M, Klein OD. Temporal analysis of ectopic enamel production in incisors from sprouty mutant mice. J Exp Zool B Mol Dev Evol. 2009;312B:473-485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 52. | Catón J, Luder HU, Zoupa M, Bradman M, Bluteau G, Tucker AS, Klein O, Mitsiadis TA. Enamel-free teeth: Tbx1 deletion affects amelogenesis in rodent incisors. Dev Biol. 2009;328:493-505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 45] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 53. | Kyrylkova K, Kyryachenko S, Biehs B, Klein O, Kioussi C, Leid M. BCL11B regulates epithelial proliferation and asymmetric development of the mouse mandibular incisor. PLoS One. 2012;7:e37670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 54. | Katsuragi Y, Anraku J, Nakatomi M, Ida-Yonemochi H, Obata M, Mishima Y, Sakuraba Y, Gondo Y, Kodama Y, Nishikawa A, Takagi R, Ohshima H, Kominami R. Bcl11b transcription factor plays a role in the maintenance of the ameloblast-progenitors in mouse adult maxillary incisors. Mech Dev. 2013;130:482-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 55. | Cao H, Florez S, Amen M, Huynh T, Skobe Z, Baldini A, Amendt BA. Tbx1 regulates progenitor cell proliferation in the dental epithelium by modulating Pitx2 activation of p21. Dev Biol. 2010;347:289-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 56. | Goodwin AF, Tidyman WE, Jheon AH, Sharir A, Zheng X, Charles C, Fagin JA, McMahon M, Diekwisch TG, Ganss B, Rauen KA, Klein OD. Abnormal Ras signaling in Costello syndrome (CS) negatively regulates enamel formation. Hum Mol Genet. 2014;23:682-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 57. | Zheng X, Goodwin AF, Tian H, Jheon AH, Klein OD. Ras Signaling Regulates Stem Cells and Amelogenesis in the Mouse Incisor. J Dent Res. 2017;96:1438-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 58. | Gritli-Linde A, Bei M, Maas R, Zhang XM, Linde A, McMahon AP. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development. 2002;129:5323-5337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 214] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 59. | Wang XP, Aberg T, James MJ, Levanon D, Groner Y, Thesleff I. Runx2 (Cbfa1) inhibits Shh signaling in the lower but not upper molars of mouse embryos and prevents the budding of putative successional teeth. J Dent Res. 2005;84:138-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 60. | Kurosaka H, Islam MN, Kuremoto K, Hayano S, Nakamura M, Kawanabe N, Yanagita T, Rice DP, Harada H, Taniuchi I, Yamashiro T. Core binding factor beta functions in the maintenance of stem cells and orchestrates continuous proliferation and differentiation in mouse incisors. Stem Cells. 2011;29:1792-1803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 61. | Li J, Feng J, Liu Y, Ho TV, Grimes W, Ho HA, Park S, Wang S, Chai Y. BMP-SHH signaling network controls epithelial stem cell fate via regulation of its niche in the developing tooth. Dev Cell. 2015;33:125-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 108] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 62. | Binder M, Chmielarz P, Mckinnon PJ, Biggs LC, Thesleff I, Balic A. Functionally Distinctive Ptch Receptors Establish Multimodal Hedgehog Signaling in the Tooth Epithelial Stem Cell Niche. Stem Cells. 2019;37:1238-1248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 63. | Felszeghy S, Suomalainen M, Thesleff I. Notch signalling is required for the survival of epithelial stem cells in the continuously growing mouse incisor. Differentiation. 2010;80:241-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 64. | Harada H, Ichimori Y, Yokohama-Tamaki T, Ohshima H, Kawano S, Katsube K, Wakisaka S. Stratum intermedium lineage diverges from ameloblast lineage via Notch signaling. Biochem Biophys Res Commun. 2006;340:611-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 65. | Mitsiadis TA, Graf D, Luder H, Gridley T, Bluteau G. BMPs and FGFs target Notch signalling via jagged 2 to regulate tooth morphogenesis and cytodifferentiation. Development. 2010;137:3025-3035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 66. | Liang Y, Chen G, Yang Y, Li Z, Chen T, Sun W, Yu M, Pan K, Guo W, Tian W. Effect of canonical NF-κB signaling pathway on the differentiation of rat dental epithelial stem cells. Stem Cell Res Ther. 2019;10:139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 67. | Hu JK, Du W, Shelton SJ, Oldham MC, DiPersio CM, Klein OD. An FAK-YAP-mTOR Signaling Axis Regulates Stem Cell-Based Tissue Renewal in Mice. Cell Stem Cell 2017; 21: 91-106. e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 184] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 68. | Suomalainen M, Thesleff I. Patterns of Wnt pathway activity in the mouse incisor indicate absence of Wnt/beta-catenin signaling in the epithelial stem cells. Dev Dyn. 2010;239:364-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (1)] |
| 69. | Yang Z, Balic A, Michon F, Juuri E, Thesleff I. Mesenchymal Wnt/β-Catenin Signaling Controls Epithelial Stem Cell Homeostasis in Teeth by Inhibiting the Antiapoptotic Effect of Fgf10. Stem Cells. 2015;33:1670-1681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 70. | Sarper SE, Inubushi T, Kurosaka H, Ono Minagi H, Kuremoto KI, Sakai T, Taniuchi I, Yamashiro T. Runx1-Stat3 signaling regulates the epithelial stem cells in continuously growing incisors. Sci Rep. 2018;8:10906. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 71. | Chang JY, Wang C, Jin C, Yang C, Huang Y, Liu J, McKeehan WL, D'Souza RN, Wang F. Self-renewal and multilineage differentiation of mouse dental epithelial stem cells. Stem Cell Res. 2013;11:990-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 72. | Sun Z, Yu W, Sanz Navarro M, Sweat M, Eliason S, Sharp T, Liu H, Seidel K, Zhang L, Moreno M, Lynch T, Holton NE, Rogers L, Neff T, Goodheart MJ, Michon F, Klein OD, Chai Y, Dupuy A, Engelhardt JF, Chen Z, Amendt BA. Sox2 and Lef-1 interact with Pitx2 to regulate incisor development and stem cell renewal. Development. 2016;143:4115-4126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 73. | Yin K, Hacia JG, Zhong Z, Paine ML. Genome-wide analysis of miRNA and mRNA transcriptomes during amelogenesis. BMC Genomics. 2014;15:998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 74. | Sharp T, Wang J, Li X, Cao H, Gao S, Moreno M, Amendt BA. A pituitary homeobox 2 (Pitx2):microRNA-200a-3p:β-catenin pathway converts mesenchymal cells to amelogenin-expressing dental epithelial cells. J Biol Chem. 2014;289:27327-27341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 75. | Cao H, Jheon A, Li X, Sun Z, Wang J, Florez S, Zhang Z, McManus MT, Klein OD, Amendt BA. The Pitx2:miR-200c/141:noggin pathway regulates Bmp signaling and ameloblast differentiation. Development. 2013;140:3348-3359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 76. | Lin Y, Zheng L, Fan L, Kuang W, Guo R, Lin J, Wu J, Tan J. The Epigenetic Regulation in Tooth Development and Regeneration. Curr Stem Cell Res Ther. 2018;13:4-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 77. | Jheon AH, Li CY, Wen T, Michon F, Klein OD. Expression of microRNAs in the stem cell niche of the adult mouse incisor. PLoS One. 2011;6:e24536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 78. | Nakamura T, Iwamoto T, Nakamura HM, Shindo Y, Saito K, Yamada A, Yamada Y, Fukumoto S, Nakamura T. Regulation of miR-1-Mediated Connexin 43 Expression and Cell Proliferation in Dental Epithelial Cells. Front Cell Dev Biol. 2020;8:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 79. | Zhang X, Diekwisch TG, Luan X. Structure and function of ameloblastin as an extracellular matrix protein: adhesion, calcium binding, and CD63 interaction in human and mouse. Eur J Oral Sci. 2011;119 Suppl 1:270-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 80. | Yoshida T, Iwata T, Umemoto T, Shiratsuchi Y, Kawashima N, Sugiyama T, Yamato M, Okano T. Promotion of mouse ameloblast proliferation by Lgr5 mediated integrin signaling. J Cell Biochem. 2013;114:2138-2147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 81. | Li CY, Cha W, Luder HU, Charles RP, McMahon M, Mitsiadis TA, Klein OD. E-cadherin regulates the behavior and fate of epithelial stem cells and their progeny in the mouse incisor. Dev Biol. 2012;366:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 82. | Singer D, Thamm K, Zhuang H, Karbanová J, Gao Y, Walker JV, Jin H, Wu X, Coveney CR, Marangoni P, Lu D, Grayson PRC, Gulsen T, Liu KJ, Ardu S, Wann AK, Luo S, Zambon AC, Jetten AM, Tredwin C, Klein OD, Attanasio M, Carmeliet P, Huttner WB, Corbeil D, Hu B. Prominin-1 controls stem cell activation by orchestrating ciliary dynamics. EMBO J. 2019;38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 83. | Sonoda A, Iwamoto T, Nakamura T, Fukumoto E, Yoshizaki K, Yamada A, Arakaki M, Harada H, Nonaka K, Nakamura S, Yamada Y, Fukumoto S. Critical role of heparin binding domains of ameloblastin for dental epithelium cell adhesion and ameloblastoma proliferation. J Biol Chem. 2009;284:27176-27184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 84. | Xi J, He S, Wei C, Shen W, Liu J, Li K, Zhang Y, Yue J, Yang Z. Negative effects of retinoic acid on stem cell niche of mouse incisor. Stem Cell Res. 2016;17:489-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 85. | Thomson G. A Method of Crowning Incisor Teeth. Proc R Soc Med. 1915;8:77-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 86. | Warshawsky H, Smith CE. Morphological classification of rat incisor ameloblasts. Anat Rec. 1974;179:423-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 212] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 87. | Sanz-Navarro M, Seidel K, Sun Z, Bertonnier-Brouty L, Amendt BA, Klein OD, Michon F. Plasticity within the niche ensures the maintenance of a Sox2+ stem cell population in the mouse incisor. Development. 2018;145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 88. | Hu B, Nadiri A, Bopp-Küchler S, Perrin-Schmitt F, Lesot H. Dental Epithelial Histomorphogenesis in vitro. J Dent Res. 2005;84:521-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 57] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 89. | Ikeda E, Morita R, Nakao K, Ishida K, Nakamura T, Takano-Yamamoto T, Ogawa M, Mizuno M, Kasugai S, Tsuji T. Fully functional bioengineered tooth replacement as an organ replacement therapy. Proc Natl Acad Sci USA. 2009;106:13475-13480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 276] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 90. | Wu Z, Wang F, Fan Z, Wu T, He J, Wang J, Zhang C, Wang S. Whole-Tooth Regeneration by Allogeneic Cell Reassociation in Pig Jawbone. Tissue Eng Part A. 2019;25:1202-1212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 91. | Doğan A, Demirci S, Şahin F. In vitro differentiation of human tooth germ stem cells into endothelial- and epithelial-like cells. Cell Biol Int. 2015;39:94-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 92. | Abdullah AN, Miyauchi S, Onishi A, Tanimoto K, Kato K. Differentiation of mouse-induced pluripotent stem cells into dental epithelial-like cells in the absence of added serum. In Vitro Cell Dev Biol Anim. 2019;55:130-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 93. | Lee CH, Kim JH, Lee HJ, Jeon K, Lim H, Choi Hy, Lee ER, Park SH, Park JY, Hong S, Kim S, Cho SG. The generation of iPS cells using non-viral magnetic nanoparticle based transfection. Biomaterials. 2011;32:6683-6691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 94. | Zhu K, Li J, Lai H, Yang C, Guo C, Wang C. Reprogramming fibroblasts to pluripotency using arginine-terminated polyamidoamine nanoparticles based non-viral gene delivery system. Int J Nanomedicine. 2014;9:5837-5847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 95. | Kim GH, Yang J, Jeon DH, Kim JH, Chae GY, Jang M, Lee G. Differentiation and Establishment of Dental Epithelial-Like Stem Cells Derived from Human ESCs and iPSCs. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 96. | Pandya M, Diekwisch TGH. Enamel biomimetics-fiction or future of dentistry. Int J Oral Sci. 2019;11:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 97. | Kirkham J, Brookes SJ, Diekwisch TGH, Margolis HC, Berdal A, Hubbard MJ. Enamel Research: Priorities and Future Directions. Front Physiol. 2017;8:513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 98. | Huang Z, Sargeant TD, Hulvat JF, Mata A, Bringas P Jr, Koh CY, Stupp SI, Snead ML. Bioactive nanofibers instruct cells to proliferate and differentiate during enamel regeneration. J Bone Miner Res. 2008;23:1995-2006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 93] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 99. | Huang Z, Newcomb CJ, Zhou Y, Lei YP, Bringas P Jr, Stupp SI, Snead ML. The role of bioactive nanofibers in enamel regeneration mediated through integrin signals acting upon C/EBPα and c-Jun. Biomaterials. 2013;34:3303-3314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 100. | Hyun SY, Mun S, Kang KJ, Lim JC, Kim SY, Han K, Jang YJ. Amelogenic transcriptome profiling in ameloblast-like cells derived from adult gingival epithelial cells. Sci Rep. 2019;9:3736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 101. | Chatzistavrou X, Papagerakis S, Ma PX, Papagerakis P. Innovative approaches to regenerate enamel and dentin. Int J Dent. 2012;2012:856470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 102. | Klein OD, Duverger O, Shaw W, Lacruz RS, Joester D, Moradian-Oldak J, Pugach MK, Wright JT, Millar SE, Kulkarni AB, Bartlett JD, Diekwisch TG, DenBesten P, Simmer JP. Meeting report: a hard look at the state of enamel research. Int J Oral Sci. 2017;9:e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |