Published online Nov 26, 2018. doi: 10.4252/wjsc.v10.i11.160
Peer-review started: August 30, 2018
First decision: October 16, 2018
Revised: October 18, 2018
Accepted: October 23, 2018
Article in press: October 23, 2018
Published online: November 26, 2018
Processing time: 88 Days and 17.6 Hours
Biomarker-driven individualized treatment in oncology has made tremendous progress through technological developments, new therapeutic modalities and a deeper understanding of the molecular biology for tumors, cancer stem cells and tumor-infiltrating immune cells. Recent technical developments have led to the establishment of a variety of cancer-related diagnostic, prognostic and predictive biomarkers. In this regard, different modern OMICs approaches were assessed in order to categorize and classify prognostically different forms of neoplasia. Despite those technical advancements, the extent of molecular heterogeneity at the individual cell level in human tumors remains largely uncharacterized. Each tumor consists of a mixture of heterogeneous cell types. Therefore, it is important to quantify the dynamic cellular variations in order to predict clinical parameters, such as a response to treatment and or potential for disease recurrence. Recently, single-cell based methods have been developed to characterize the heterogeneity in seemingly homogenous cancer cell populations prior to and during treatment. In this review, we highlight the recent advances for single-cell analysis and discuss the challenges and prospects for molecular characterization of cancer cells, cancer stem cells and tumor-infiltrating immune cells.
Core tip: Extensive heterogeneity in cancer cells negatively influences treatment efficacy and survival of patients. The existing molecular methods for biomarker discovery of cancer cells and cancer stem cells are often unsuited to capture the heterogeneous nature of cell populations. Recent advances in single-cell based profiling approaches allowed the detection of molecular changes in individual cancer cells. Therefore, single-cell analysis is leading to build a complete landscape of cell types within tumor cells and facilitating the study of complex molecular heterogeneity in cancer cell populations. This will improve the investigation of more specific biomarkers to identify and target cancer stem cells.
- Citation: Radpour R, Forouharkhou F. Single-cell analysis of tumors: Creating new value for molecular biomarker discovery of cancer stem cells and tumor-infiltrating immune cells. World J Stem Cells 2018; 10(11): 160-171
- URL: https://www.wjgnet.com/1948-0210/full/v10/i11/160.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v10.i11.160
In principle, three main classes of biomarkers are distinguished for cancer disease stratification: Diagnostic, prognostic and predictive biomarkers[1-4]. In oncology, the diagnostic biomarkers essentially serve to substantiate a specific entity association and suspected malignant disease-spreading pattern. Classical examples are the in situ immunophenotyping of a neoplasm such as lung cancer[5] by immunohistology as well as the specific representation of entity-defining molecules such as prostate-specific membrane antigen in prostate cancer[6].
By contrast, prognostic biomarkers have the function of predicting the natural course of a malignant disease. These include classical parameters such as clinical and pathological staging but also the collection of molecular factors, such as tumor specific genetic aberrations (chromosomal abnormalities, gene mutations, pathologic epigenetic changes or dysregulated genes/pathways) that may be associated with more aggressive disease progression. However, a prognostic biomarker has only a limited value for the patient, since mere knowledge about the prognosis of disease alone has little benefit[2,4,7].
The predictive biomarkers specifically describe the expected likelihood of a patient responding to an available therapy option based on the molecular properties of the tumor. This concept is currently used in the context of targeted drug-based tumor treatment with targeted drugs, e.g., with inhibitors of the epidermal growth factor receptor (EGFR) in the presence of a tissue-based EGFR mutation in lung carcinomas[8] or CD70-CD27 signaling in leukemia including acute myeloid leukemia (AML)[9] or chronic myeloid leukemia (CML)[10]. However, there are only a few approved tissue-based predictive biomarkers available. Predictive analytics using molecular imaging and blood-based technologies are still at the stage of development.
The boundaries between these biomarker types can be blurred. For example, a pathologic genetic alteration in different situations may represent a diagnostic, a prognostic, and a predictive biomarker. This is illustrated by the BRAF mutation, as it can support the early diagnosis of a thyroid carcinoma[11], prognostically define an unfavorable subtype of colorectal carcinoma[4] and predictably provide therapy with a BRAF-specific small molecule inhibitor (e.g., vemurafenib) in malignant melanoma[12].
Classical macroscopically assisted histomorphologic evaluation of a malignant tumor remains by far the most significant diagnostic, prognostic and in many respects predictive biomarker with the greatest impact on patient treatment. Nevertheless, in recent decades, a refinement of biomarker analysis by molecular methods has found its way into pathological diagnostics and shaped the new area of individualized medicine.
Cancer stem cells (CSCs) or tumor precursor cells are a minor fraction of cells within the bulk tumor population, which, because of their unique stem cell properties of relative quiescence and self-renewal, have been found to reconstitute and propagate the tumor and are considered to be essential for tumor neoplasm and metastasis[13]. The theory of CSCs was firstly postulated in the 1970s and was experimentally confirmed by the isolation of tumor-initiating cells in AML[14]. Furthermore, CSC has been demonstrated in a variety of solid tumors, such as tumors in brain, colorectal, hematopoietic malignancies (e.g., myeloid or lymphoid leukemia), head and neck, mammary glands, lung, liver, melanoma and also prostate carcinomas[9,10,15,16]. Heterogeneity is a major hallmark of tumor cells including CSCs. Each cancer cell clone is characterized by harboring different combinations of mutations or genetic alterations, and subsequently the processes of tumorigenesis occur differently based on the type of genetic lesions[17].
CSCs are often resistant against standard therapies such as irradiation, chemotherapy, cytotoxic drugs and probably also against immune attack. This may be due to different escape mechanisms of CSCs and/or due to protective mechanisms of the microenvironment. Unravelling the function of the CSCs has been one of the main challenges of cancer research[16,18].
Over the past few decade, a variety of biomarkers for a wide range of solid tumors and hematopoietic malignancies has been identified[2,4]. The technologies for biomarker analysis are developing rapidly. The next generation sequencing (NGS) technology promptly follows some of the technologies mentioned above, which will lead to further dynamization of the biomarker discovery in oncology. In addition, blood-based assays using circulating cell free DNA that move beyond the classic tumor marker determination will become more important for the monitoring of disease processes and resistance as well as the prediction of therapy outcome[16,19-21]. Mutational analysis in EGFR-mutated lung carcinoma prior to therapy with Osimertinib is an example of a blood-based assay that has already found way into the routine diagnostic pipelines[22]. Further assays are being developed to trace and target circulating tumor cells (mainly CSCs) in the blood, urine, cerebrospinal fluid and other body fluids. The goal must be to transfer molecular markers from tissue diagnostics into non-invasive molecular profiling approaches.
Targeted proteomics using tissue-based in situ methods such as immunohistology has been developed as an important biomarker analysis tool in oncology[23]. This approach is used in many areas of pathology including pathological oncology, and the predictive biomarker analysis still relies significantly on this method. Examples include the analysis of human epidermal growth factor receptor 2 (HER2) expression prior to treatment with HER2 inhibitors (e.g., trastuzumab) in gastric and breast carcinoma[24,25] as well as the stratifying assignment of treatment with immune checkpoint inhibitors in programmed death-ligand 1 (PD-L1)-positive advanced non-small cell lung cancer[26]. The development of multiplexable and quantitatively more precise proteomic methods promises new opportunities for biomarker discovery/analysis in the near future. These include using slice-based imaging mass spectrometry [e.g., matrix-assisted laser desorption ionization imaging mass spectrometry (MALDI-IMS)][27] or quantitative multiplex protein analysis using extract-based mass spectrometry (LC-MS)[28].
The earliest clinically relevant genomic studies on predictive biomarker analysis used in routine diagnostics were the application of fluorescence in situ hybridizations (FISH) to determine the gene copy number of ERBB2, the HER2 gene, in breast cancer, which could assign it to a positive or negative category for HER2 expression[2,29,30]. One of the first examples of large solid tumor profiling is mutation screening for KRAS and NRAS genes in metastatic colorectal carcinoma as a predictive biomarker for using the EGFR inhibitor panitumumab[4,31]. Today, numerous individual examinations of gene mutations or chromosomal aberrations (e.g., translocations or amplifications) are firmly anchored in the routine diagnostic of different tumors. Currently, new technologies such as massive parallel sequencing (MPS) have been priced into areas where routine diagnostic application has become possible. Those methods have already been adapted to high-throughput screening in routine applications[32]. Implementation of those high-throughput approaches has led to improvement of diagnosis and therapy of different cancer types[33,34].
The first introduced epigenetic biomarker into the routine diagnostic was investigation of promoter methylation of the MGMT gene using sequence-based techniques to predict response to treatment with temozolomide in glioblastoma[35]. However, newer epigenetic screening approaches, which are still in the process of diagnostic development, focus on the simultaneous investigation of DNA methylation in a large number of coding genes using array-based or high-throughput sequencing methods (e.g., Methyl-seq). Since it is postulated that pathologic methylation patterns in individual tumor entities are more stable and reproducible than transcriptome profiles, these technologies are currently being tested primarily in molecular entity assignment. Large studies have substantiated their overall suitability for cancer with unknown primary and for some rare tumor families but have not yet been implemented in the routine diagnostic pipelines[36-38].
The analysis of RNA expression signature within cancer cells and CSCs using quantitative polymerase chain reaction (qPCR), array-based capture, NanoString technology or massive parallel RNA sequencing (RNA-Seq) approaches has a long tradition in cancer molecular biomarker analysis. However, individual methods (e.g., quantitative polymerase chain reaction) could never prevail over immunohistology despite partially superior precision. Initially, the parallel analysis of RNA expression patterns was assessed with the hope that diagnostic assignments could be made in unclear cases (e.g., cancer with unknown primary)[39]. Despite their potential and some positive results, these applications could not establish themselves in the wide range of diagnostic services. In addition, many tumor entities have been used to develop predictors for the efficacy of conventional chemotherapies based on transcriptomic profiles. Some success in this context has been gene expression tests in breast cancer, which can be used as an additional decision-making aid in the therapy stratification of breast cancer patients for adjuvant chemotherapy[40]. However, these tests are currently not being used consistently in clinical care.
As indicated, tumors are a pool of heterogeneous cells including CSCs. Inter- or intra-tumor heterogeneity may completely render CSC biomarkers inapt. Seemingly homogenous cell populations that are enriched and purified by a set of well-known surface markers often hide exceptional heterogeneity. This is more pronounced in the hematological malignancies[16]. Such tumor heterogeneity can be the result of different genetically distinct clones within the tumor due to having various genetic lesions or dysregulation of markers via pathologic epigenetic regulations[2,4,41-46].
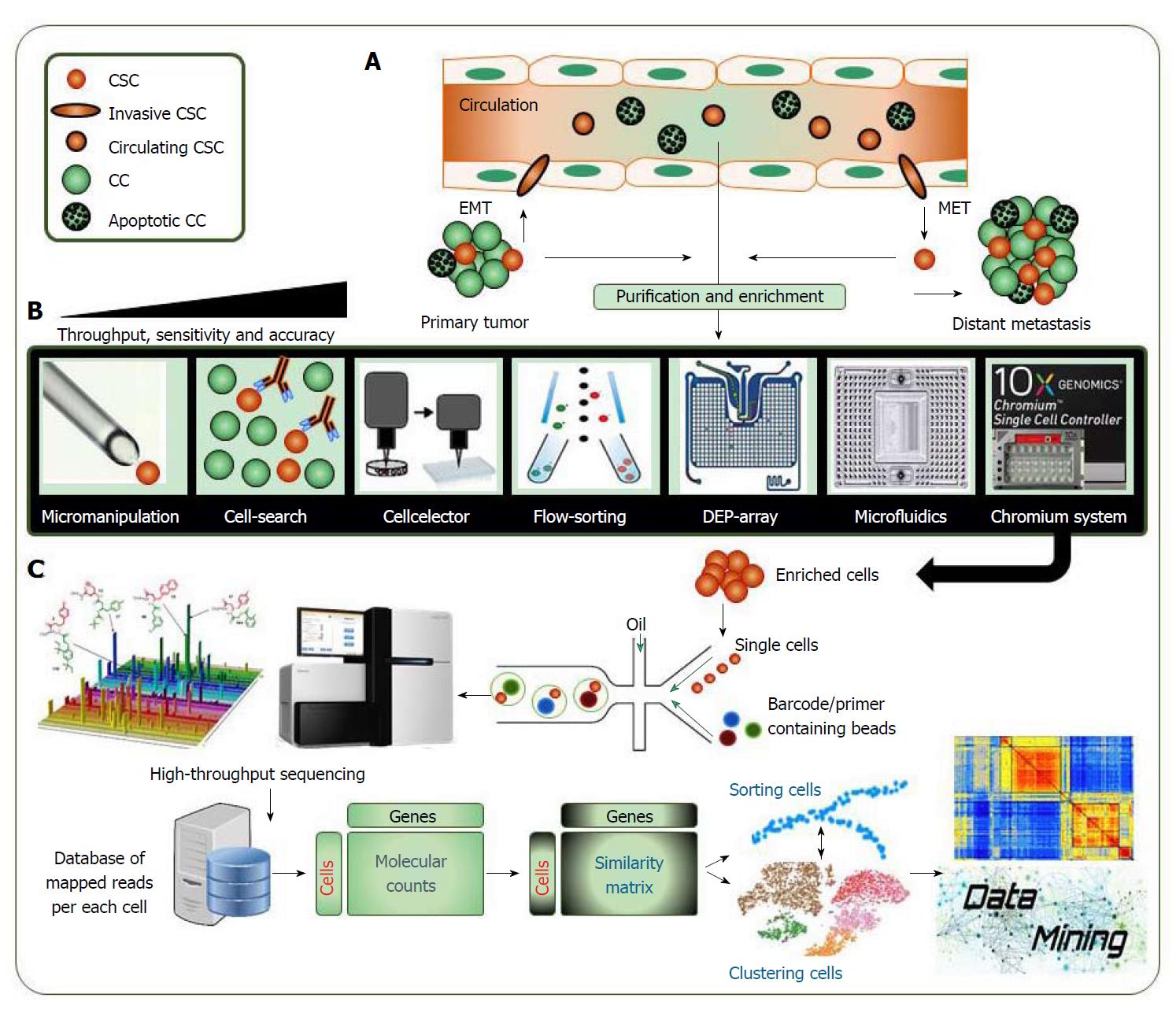
Different OMICs approaches have allowed for the discovery and characterization of a variety of cancer-related cell populations. However, those approaches are unsuited to capture the heterogeneous nature of cancer cell populations. Therefore, interest was shifted towards characterization of single-cells rather than cell populations. The technical advances that include single-cell imaging, genomics or transcriptomics assessed full characterization of different cell populations. The OMICs analysis is usually performed using samples of many cells. However, this type of analysis lacks the kind of detailed assessment needed for evaluating contribution of individual cells to the overall phenotype. In contrast, single-cell analysis allows comparing the captured OMICs data of thousands of individual cells (Figure 1). Applied methods for single-cell isolation have rapidly enhanced in the past few years from manual micromanipulation, cell-search antibody-based isolation or flow-sorting of cells to high-throughput isolation methods using dielectrophoresis (DEP) arrays, microfluidics, emulsion-based platforms or 10X genomics ChromiumTM single cell controller system. This technical advance could provide massive advantages by significantly increasing the throughput sensitivity and accuracy of employed approaches (Figure 1B).
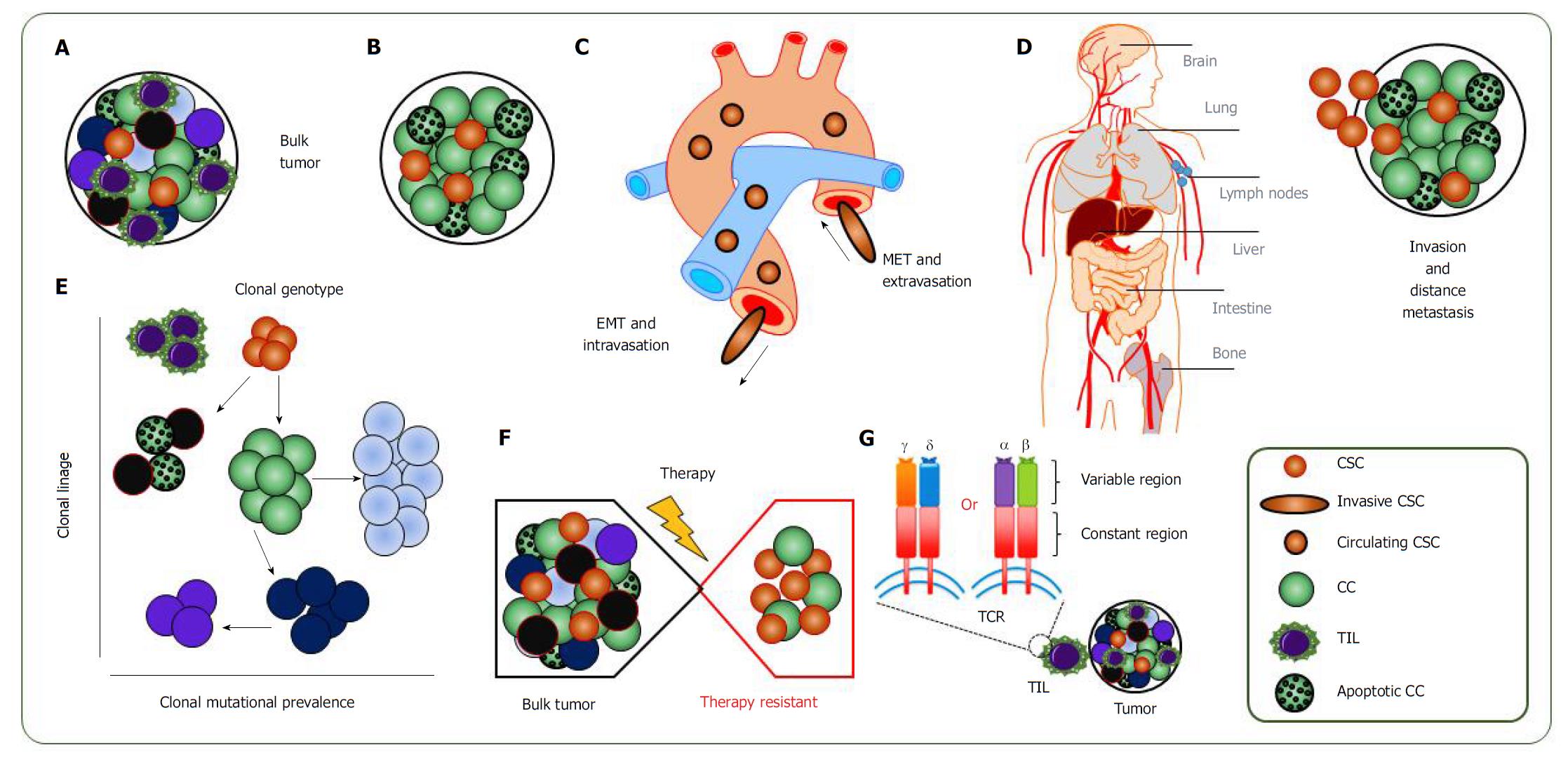
One of the prime reasons for using single-cell analysis is to evaluate heterogeneity in seemingly homogenous cell populations. Another reason is to detect small subpopulations that would otherwise be missed in bulk populations. In addition, by using single-cell analysis, it is possible to find CSCs and trace them in the circulation, investigate the clonal evolution and mutational rate of cancer cells, to study better the invasion and trace the metastatic dissemination and to understand the molecular mechanisms of therapy resistance of cancer cells and CSCs (Figure 2).
The first single-cell RNA-Seq study was published in 2009[47]. Since then the interest for the approach is growing[48,49]. Single-cell RNA sequencing is being used for identifying cellular intermediates during developmental processes. Different microfluidic systems have been proposed to isolate single cells and help in library preparation[50]. Several novel methods are available for single-cell analyses. Multiplexed error robust fluorescence (MERFISH), is a high-throughput method that uses sequential imaging with combinatorial labeling and multiplex single molecule FISH, allowing for robust detection of many genes at the same time in both tissues and cell culture conditions[51]. Another approach is quantitative hybridization chain reaction (qHCR), which uses probes harboring initiators for DNA interacting with fluorophore-labelled hairpins assembled into polymerase. Using this method, the mRNA expression of thousands of different genes can be captured simultaneously at a single-cell based resolution[52]. Single-cell linage tracking allows researchers to follow and trace the fate of individual cells over the time. This also includes the tracing of different cancer cells from primitive CSCs. Lineage tracing by nuclease-activated editing of ubiquitous sequences (LINNAEUS), is a novel method for cell type identification, characterization and massively parallel linage tracking. In this approach, a double strand break will be introduced to the cells using a CRISPR/Cas9 system, which upon repair, reacts as a unique heritable scar in the daughter cells in order to trace the cellular linages[53].
Pooled screenings rely on readouts that average properties of the cell population of interest. Although these approaches provide an assessment of gene function at the genome scale, they cannot identify the contribution of subpopulations to the bulk phenotype. Moreover, the consequences of distinct perturbations to the overall phenotype cannot be evaluated. To circumvent the problem, methods have been recently developed to study the impact of perturbations at the single-cell levels[54,55]. These approaches integrate parallel massive single-cell RNA-Seq and pooled screens to reconstruct the gene regulatory networks controlling particular biological processes. Perturb-Seq is a platform for multiplexed profiling of perturbations at the single-cell resolution[54]. Profiling the genomic perturbation and the transcriptome in the same cell provides a powerful means to simultaneously the function of multiple factors and their interactions.
Heterogeneity in the tumor cell population was recently evaluated in different forms of human cancers. In the ovarian cancer, single-cell analysis revealed two major subsets of cells characterized by stromal gene expression patterns [genes associated with epithelial-to-mesenchymal transition (EMT) and also extracellular matrix (ECM) genes] and epithelial gene expression signature (characterized by proliferation- and oxidative phosphorylation-related genes)[56]. Analysis of CSCs in CML, uncovered distinct molecular signatures of leukemia stem cells with a high level of heterogeneity in the seemingly homogenous cell populations of CSCs[57]. Single-cell whole exome sequencing (scWES), is a promising tool for detecting sub-clones and possibly leukemia stem cells in AML[58]. Furthermore, epigenetically distinct hematopoietic stem cell sub-populations have been detected by high-resolution single-cell DNA methylation analysis[59]. Single-cell sequencing of glioblastoma and glioma cells also detected a heterogeneous gene expression signature within the tumor population[60,61]. In breast cancer, regulatory networks influencing stemness, pluripotency, proliferation, differentiation and EMT have been identified using single-cell gene expression profiling. The analysis has shown that ALDH-CD44+CD24- and ALDH+ human mammary cells have mesenchymal-like and epithelial-like characteristics, respectively. At the single-cell level, these cells express high levels of stemness- and EMT-associated gene signatures. In contrast, both detected populations had some co-expressing ALDH+ and CD44+CD24- by flow cytometry[62]. Important findings using single-cell sequencing studies on variety of human primary tumors, including bladder, blood, brain, breast, colorectal, kidney, lung and ovarian cancer, are summarized in Table 1.
| Tumor type | Source | Platform | Major finding | Ref. |
| Bladder cancer | Squamous cell carcinoma | RNA-seq | Cellular heterogeneity in the gene expression affects the disease outcome | [73] |
| Muscle-invasive cell carcinoma | SNV-seq | Lineage-specific mutations are driving cancer initiation and progress | [74] | |
| Blood cancer | B-cell ALL | CNV-seq | CNVs were developed as an impact of environmental stressors, which was only detectable at single-cell level | [75] |
| Pediatric ALL | SNV-seq | Analysis revealed clonal somatic mutational prevalence at single-cell resolution | [76] | |
| Therapy resistant AML | RNA-seq | Identified molecular signature of resistant LSCs versus therapy-naive LSCs | [77] | |
| Secondary AML | SNV-seq | Genomic complexity was identified at single cells which was not seen at bulk leukemic populations | [78] | |
| CML | RNA-seq | Single-cell analysis uncovered molecular signature of LSCs | [57] | |
| JAK2 negative MPN | SNV-seq | Large genetic distances was observed between mono-clonal tumor cells | [79] | |
| JAK2V617F MPN | RNA-seq | Single-cell sequencing revealed the molecular networks driving self-renewal of CSCs | [80] | |
| Brain cancer | EGFR amplified GBM | CNV-seq | Heterogeneity in EGFR mutations among different tumor cells leading to variation in therapy response | [81] |
| GBM | RNA-seq | Heterogeneity in gene expression panthers was identified including EGFR gene | [82] | |
| Breast cancer | ER+ | CNV-seq | Showed clonal evolution of tumor cells at single-cell resolution | [83] |
| HER2+ | RNA-seq | 404 differentially expressed gene signature was identified in CSCs, which had a prognostic value | [84] | |
| MDA-MB-231 and CN34 cell lines | RNA-seq | Gene expression profiling identifies small sub-population with more metastatic potential, which was therapy resistant. | [85] | |
| TNBC | CNV-seq | Showed clonal evolution of tumor cells at single-cell level. Also, chemo-resistance evolution in TNBC was identified | [86,87] | |
| RNA-seq | ||||
| TNBC or ER+ HER2- | SNV-seq CNV-seq | ER+ HER2- tumors represented significantly less mutational rate compared to TNBC tumors | [88] | |
| Colorectal cancer | Colon tumor and adjacent normal cells | SNV-seq | Different mutational profiles were identified among tumors’ sub-populations | [89] |
| Colon tumor | CNV-seq | CSCs (EpCAMhighCD44+) and DTCs (EpCAMhighCD44-) had similar somatic CNV pattern, while they had regional differences | [90] | |
| Rectal tumor | CNV-seq | Multi-region single-cell analysis showed somatic copy number alterations are an early event in cancer development | [91] | |
| Kidney cancer | ccRCC primary carcinoma and paired metastasis | RNA-seq | Heterogeneity in the expression of targetable genes was identified. The finding highlights the necessity of multi-agent therapies | [92] |
| Lung cancer | NSCLC | RNA-seq | Characterization of tumor-infiltrating T cells revealed that inter-tissue effector T cells with a highly migratory nature | [93] |
| Clear cell renal cell carcinoma | SNV-seq | A complex mutational pattern was observed at single-cells compared to bulk tumors | [94] | |
| Adenocarcinoma PDX | RNA-seq | Single-cell sequencing identified KRAS+ drug resistant cell population within the tumor | [95] | |
| LC2/ad and LC2/ad-R cell lines | RNA-seq | Gene expression profiling identifies signature that is linked to therapy resistance | [96] | |
| Ovarian cancer | HGSOC | RNA-seq | Single-cell analysis could distinguish two major sub-populations within the tumor based on their gene expression signature | [56] |
Most immune cell types can be present in a tumor, and the fraction of immune cells can vary greatly across different tumors and patients[63]. T lymphocytes are among the most studied tumor-infiltrating immune cells, since they have the potential to recognize mutated protein epitopes displayed by human lymphocyte antigen (HLA) molecules on cancer cells and CSCs, thereby allowing immune recognition of the tumor. Different types of tumor-infiltrating lymphocytes (TILs) have different effects. For instance, CD4+ Tregs have been associated with poor survival and have been demonstrated to play an immune-suppressive role[64]. Conversely, CD8+ T cells can mediate cytolytic activity against cancer cells or CSCs. However, cancer cells, particularly CSCs, evade immune recognition and elimination by TILs via various mechanisms, including loss of antigen and the expression of immune inhibitory molecules. Tumor-infiltrating CD8+ T cell are often anergic, as characterized by their exhaustion phenotype[65]. Overall, it is clear that complex relationships govern the interactions between immune cells and cancer cells or CSCs.
During T cell development in the thymus, they gain the ability to recognize many different foreign antigens. This ability is assessed by the expression of highly polymorphic surface T cell receptors (TCRs). The enormous diversity of TCRs is resulted by random combinations of genes’ segments encoding TCR chains [including variable (V), diversity (D), and joining (J) segments][66]. Molecular profiling and characterization of TCRs in TILs could describe T cell dynamics in different tumors[67].
TILs are typically studied by immunohistochemistry or by flow cytometry, relying on a panel of antibodies targeting specific markers of immune cells. To complement this approach, gene expression of whole tumors can be used and expression of the immune cell type markers can inform us about the presence of the corresponding cell types[68]. One promising aspect of this approach is that it provides information about the whole transcriptome and is not restricted by the availability of antibodies; however, it is not capable of overcoming the extensive heterogeneity among TILs.
The next generation sequencing approach using genomic DNA (gDNA) as starting material was first used to characterize the TCR diversity in healthy individuals[69] and rapidly adapted to TCR profiling in tumor immunology[70]. However, the use of gDNA was more challenging due to the fact that non-productive TCR rearrangements were also sequenced. In addition, the presence of introns can introduce more technical biases. Therefore, RNA-seq was selected as a better approach. Upon introducing more advanced single-cell analysis approaches like microfluidics or 10 × genomics, there was promise to couple RNA-seq and TCR sequencing from the same cell, which has the great advantage to identify and characterize very rare T cell populations. A recent work using different single-cell analysis methods investigated the T cell repertoire according to their TCR variability in both mice and human Treg cells[71]. The results of this comprehensive TCR single-cell sequencing revealed that Tregs with some highly activated subpopulations could display a broad heterogeneity, while Treg sharing the same antigen recognition specificity were more transcriptionally similar than those with different TCR sequence.
The coupled profiling of TCRs sequencing and single-cell gene expression analysis from the same cell provides an unbiased classification of T cells based of their TCR signature, which is association of the transcriptional landscape of individual cell[72]. This approach will provide a powerful tool to study the potential impact of TILs on CSCs and will yield valuable insights to personalized immunotherapy of cancer patients.
The determination of diagnostic, prognostic and predictive biomarkers forms the basis of individualized patient treatment in oncology. As a biomarker, it is demanded to be reproducible, robust and quality-assured. As of today, the collection of specific biomarkers are not be able to define the complete subsequent of oncological therapy for cancer patients. This affects the efficacy of a treatment, the side effects that a patient is exposed to and the cost of therapy.
Despite some developments in the field of blood-based tests and molecular imaging, biomarker analysis in oncology continues to rely essentially on molecular tissue analysis. An exact molecular characterization of CSCs in the tumor requires the development of specific markers and suitable enrichment methods. New genomics, epigenomics, transcriptomics and proteomics methods as well as the introduction of novel single-cell based approached will result in an accelerating identification of specific oncological biomarkers.
Single-cell technologies are allowing for the detection of molecular changes in individual cancer cells. This can improve investigation of more specific biomarkers with unprecedented resolution leading to build a complete landscape of different cell types within tumors. Single-cell analysis of CSCs is challenging mainly due to their rarity and the small amount of total RNA in a single cell. Using a combination of different cellular enrichment strategies, such as flow cytometry for rare cell population like CSCs with the single-cell analyzing methods, will improve the resolution in profiling and characterization of CSCs. Likewise, the ability to amplify and sequence other RNA molecules, such as micro RNAs and long non-coding RNAs, will provide valuable information on gene regulation. New methods to simultaneously profile genomic DNA variants, DNA methylation and gene expression from the same cell coupled with potential proteomic analysis, could provide powerful tools for assessing the effects of genomic variation and gene expression profiles or epigenetic modifications on cancer cell heterogeneity. Particularly, from high-throughput single-cell based technologies, we can expect valuable insights regarding suitable associated biomarkers to identify and target CSCs. Furthermore, cancer immunotherapy may also benefit from single-cell methods that define the role of TILs within the CSCs and monitor the individual response to the immune-regulatory agents. This would be an important step towards individualized cancer management.
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country of origin: Switzerland
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Economescu M, Garg M, Van Seuningen IM, Wang YG S- Editor: Ma RY L- Editor: Filipodia E- Editor: Tan WW
| 1. | Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4510] [Cited by in RCA: 4130] [Article Influence: 172.1] [Reference Citation Analysis (0)] |
| 2. | Radpour R, Barekati Z, Kohler C, Holzgreve W, Zhong XY. New trends in molecular biomarker discovery for breast cancer. Genet Test Mol Biomarkers. 2009;13:565-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 3. | Zhang B, Barekati Z, Kohler C, Radpour R, Asadollahi R, Holzgreve W, Zhong XY. Proteomics and biomarkers for ovarian cancer diagnosis. Ann Clin Lab Sci. 2010;40:218-225. [PubMed] |
| 4. | Aghagolzadeh P, Radpour R. New trends in molecular and cellular biomarker discovery for colorectal cancer. World J Gastroenterol. 2016;22:5678-5693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 51] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (2)] |
| 5. | Warth A, Muley T, Herpel E, Meister M, Herth FJ, Schirmacher P, Weichert W, Hoffmann H, Schnabel PA. Large-scale comparative analyses of immunomarkers for diagnostic subtyping of non-small-cell lung cancer biopsies. Histopathology. 2012;61:1017-1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 89] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 6. | Maurer T, Eiber M, Schwaiger M, Gschwend JE. Current use of PSMA-PET in prostate cancer management. Nat Rev Urol. 2016;13:226-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 453] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 7. | Nalejska E, Mączyńska E, Lewandowska MA. Prognostic and predictive biomarkers: tools in personalized oncology. Mol Diagn Ther. 2014;18:273-284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 120] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 8. | Tan DS, Yom SS, Tsao MS, Pass HI, Kelly K, Peled N, Yung RC, Wistuba II, Yatabe Y, Unger M. The International Association for the Study of Lung Cancer Consensus Statement on Optimizing Management of EGFR Mutation-Positive Non-Small Cell Lung Cancer: Status in 2016. J Thorac Oncol. 2016;11:946-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 154] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 9. | Riether C, Schürch CM, Bührer ED, Hinterbrandner M, Huguenin AL, Hoepner S, Zlobec I, Pabst T, Radpour R, Ochsenbein AF. CD70/CD27 signaling promotes blast stemness and is a viable therapeutic target in acute myeloid leukemia. J Exp Med. 2017;214:359-380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 142] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 10. | Riether C, Schürch CM, Flury C, Hinterbrandner M, Drück L, Huguenin AL, Baerlocher GM, Radpour R, Ochsenbein AF. Tyrosine kinase inhibitor-induced CD70 expression mediates drug resistance in leukemia stem cells by activating Wnt signaling. Sci Transl Med. 2015;7:298ra119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 11. | Tennakoon TMPB, Rushdhi M, Ranasinghe ADCU, Dassanayake RS. Values of molecular markers in the differential diagnosis of thyroid abnormalities. J Cancer Res Clin Oncol. 2017;143:913-931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 12. | Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596-599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1536] [Cited by in RCA: 1439] [Article Influence: 95.9] [Reference Citation Analysis (0)] |
| 13. | Dawood S, Austin L, Cristofanilli M. Cancer stem cells: implications for cancer therapy. Oncology (Williston Park). 2014;28:1101-1107, 1110. [PubMed] |
| 14. | Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3316] [Cited by in RCA: 3387] [Article Influence: 109.3] [Reference Citation Analysis (0)] |
| 15. | Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2552] [Cited by in RCA: 2626] [Article Influence: 154.5] [Reference Citation Analysis (0)] |
| 16. | Radpour R. Tracing and targeting cancer stem cells: New venture for personalized molecular cancer therapy. World J Stem Cells. 2017;9:169-178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 17. | Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1549] [Cited by in RCA: 1802] [Article Influence: 150.2] [Reference Citation Analysis (0)] |
| 18. | Murone M, Radpour R, Attinger A, Chessex AV, Huguenin AL, Schürch CM, Banz Y, Sengupta S, Aguet M, Rigotti S. The Multi-kinase Inhibitor Debio 0617B Reduces Maintenance and Self-renewal of Primary Human AML CD34+ Stem/Progenitor Cells. Mol Cancer Ther. 2017;16:1497-1510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 19. | Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1634] [Cited by in RCA: 1767] [Article Influence: 220.9] [Reference Citation Analysis (0)] |
| 20. | Zachariah RR, Schmid S, Buerki N, Radpour R, Holzgreve W, Zhong X. Levels of circulating cell-free nuclear and mitochondrial DNA in benign and malignant ovarian tumors. Obstet Gynecol. 2008;112:843-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 21. | Zachariah R, Schmid S, Radpour R, Buerki N, Fan AX, Hahn S, Holzgreve W, Zhong XY. Circulating cell-free DNA as a potential biomarker for minimal and mild endometriosis. Reprod Biomed Online. 2009;18:407-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 22. | Mok TS, Wu Y-L, Ahn M-J, Garassino MC, Kim HR, Ramalingam SS, Shepherd FA, He Y, Akamatsu H, Theelen WS. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med. 2017;376:629-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2334] [Cited by in RCA: 2595] [Article Influence: 324.4] [Reference Citation Analysis (0)] |
| 23. | Prichard JW. Overview of automated immunohistochemistry. Arch Pathol Lab Med. 2014;138:1578-1582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 24. | Cameron D, Piccart-Gebhart MJ, Gelber RD, Procter M, Goldhirsch A, de Azambuja E, Castro G Jr, Untch M, Smith I, Gianni L, Baselga J, Al-Sakaff N, Lauer S, McFadden E, Leyland-Jones B, Bell R, Dowsett M, Jackisch C; Herceptin Adjuvant (HERA) Trial Study Team. 11 years’ follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive early breast cancer: final analysis of the HERceptin Adjuvant (HERA) trial. Lancet. 2017;389:1195-1205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 572] [Cited by in RCA: 714] [Article Influence: 89.3] [Reference Citation Analysis (0)] |
| 25. | Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5541] [Cited by in RCA: 5317] [Article Influence: 354.5] [Reference Citation Analysis (3)] |
| 26. | Hui R, Garon EB, Goldman JW, Leighl NB, Hellmann MD, Patnaik A, Gandhi L, Eder JP, Ahn MJ, Horn L. Pembrolizumab as first-line therapy for patients with PD-L1-positive advanced non-small cell lung cancer: a phase 1 trial. Ann Oncol. 2017;28:874-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 192] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 27. | Kriegsmann M, Casadonte R, Kriegsmann J, Dienemann H, Schirmacher P, Hendrik Kobarg J, Schwamborn K, Stenzinger A, Warth A, Weichert W. Reliable Entity Subtyping in Non-small Cell Lung Cancer by Matrix-assisted Laser Desorption/Ionization Imaging Mass Spectrometry on Formalin-fixed Paraffin-embedded Tissue Specimens. Mol Cell Proteomics. 2016;15:3081-3089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 28. | Panis C, Pizzatti L, Souza GF, Abdelhay E. Clinical proteomics in cancer: Where we are. Cancer Lett. 2016;382:231-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, Cameron D, Dowsett M, Barrios CH, Steger G, Huang CS, Andersson M, Inbar M, Lichinitser M, Láng I, Nitz U, Iwata H, Thomssen C, Lohrisch C, Suter TM, Rüschoff J, Suto T, Greatorex V, Ward C, Straehle C, McFadden E, Dolci MS, Gelber RD; Herceptin Adjuvant (HERA) Trial Study Team. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659-1672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3742] [Cited by in RCA: 3681] [Article Influence: 184.1] [Reference Citation Analysis (0)] |
| 30. | Radpour R, Sikora M, Grussenmeyer T, Kohler C, Barekati Z, Holzgreve W, Lefkovits I, Zhong XY. Simultaneous isolation of DNA, RNA, and proteins for genetic, epigenetic, transcriptomic, and proteomic analysis. J Proteome Res. 2009;8:5264-5274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626-1634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2504] [Cited by in RCA: 2403] [Article Influence: 141.4] [Reference Citation Analysis (0)] |
| 32. | Endris V, Stenzinger A, Pfarr N, Penzel R, Möbs M, Lenze D, Darb-Esfahani S, Hummel M, Sabine-Merkelbach-Bruse , Jung A. NGS-based BRCA1/2 mutation testing of high-grade serous ovarian cancer tissue: results and conclusions of the first international round robin trial. Virchows Arch. 2016;468:697-705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | Jesinghaus M, Pfarr N, Endris V, Kloor M, Volckmar AL, Brandt R, Herpel E, Muckenhuber A, Lasitschka F, Schirmacher P. Genotyping of colorectal cancer for cancer precision medicine: Results from the IPH Center for Molecular Pathology. Genes Chromosomes Cancer. 2016;55:505-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 34. | Haghighi MM, Radpour R, Mahmoudi T, Mohebbi SR, Vahedi M, Zali MR. Association between MTHFR polymorphism (C677T) with nonfamilial colorectal cancer. Oncol Res. 2009;18:57-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 35. | Wick W, Weller M, van den Bent M, Sanson M, Weiler M, von Deimling A, Plass C, Hegi M, Platten M, Reifenberger G. MGMT testing--the challenges for biomarker-based glioma treatment. Nat Rev Neurol. 2014;10:372-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 411] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 36. | Moran S, Martínez-Cardús A, Sayols S, Musulén E, Balañá C, Estival-Gonzalez A, Moutinho C, Heyn H, Diaz-Lagares A, de Moura MC. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol. 2016;17:1386-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 343] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 37. | Röhrich M, Koelsche C, Schrimpf D, Capper D, Sahm F, Kratz A, Reuss J, Hovestadt V, Jones DT, Bewerunge-Hudler M. Methylation-based classification of benign and malignant peripheral nerve sheath tumors. Acta Neuropathol. 2016;131:877-887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 139] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 38. | Radpour R, Barekati Z, Haghighi MM, Kohler C, Asadollahi R, Torbati PM, Holzgreve W, Zhong XY. Correlation of telomere length shortening with promoter methylation profile of p16/Rb and p53/p21 pathways in breast cancer. Mod Pathol. 2010;23:763-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 39. | Stenzinger A, Kriegsmann M, Weichert W. [The role of pathology in the diagnostics of CUP syndrome]. Radiologe. 2014;54:124-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 40. | Harris LN, Ismaila N, McShane LM, Andre F, Collyar DE, Gonzalez-Angulo AM, Hammond EH, Kuderer NM, Liu MC, Mennel RG. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2016;34:1134-1150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 594] [Cited by in RCA: 594] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 41. | Radpour R, Haghighi MM, Fan AX, Torbati PM, Hahn S, Holzgreve W, Zhong XY. High-throughput hacking of the methylation patterns in breast cancer by in vitro transcription and thymidine-specific cleavage mass array on MALDI-TOF silico-chip. Mol Cancer Res. 2008;6:1702-1709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 42. | Radpour R, Kohler C, Haghighi MM, Fan AX, Holzgreve W, Zhong XY. Methylation profiles of 22 candidate genes in breast cancer using high-throughput MALDI-TOF mass array. Oncogene. 2009;28:2969-2978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 43. | Barekati Z, Radpour R, Kohler C, Zhang B, Toniolo P, Lenner P, Lv Q, Zheng H, Zhong XY. Methylation profile of TP53 regulatory pathway and mtDNA alterations in breast cancer patients lacking TP53 mutations. Hum Mol Genet. 2010;19:2936-2946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 44. | Radpour R, Barekati Z, Kohler C, Schumacher MM, Grussenmeyer T, Jenoe P, Hartmann N, Moes S, Letzkus M, Bitzer J. Integrated epigenetics of human breast cancer: synoptic investigation of targeted genes, microRNAs and proteins upon demethylation treatment. PLoS One. 2011;6:e27355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 45. | Radpour R, Barekati Z, Kohler C, Lv Q, Bürki N, Diesch C, Bitzer J, Zheng H, Schmid S, Zhong XY. Hypermethylation of tumor suppressor genes involved in critical regulatory pathways for developing a blood-based test in breast cancer. PLoS One. 2011;6:e16080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 46. | Barekati Z, Radpour R, Lu Q, Bitzer J, Zheng H, Toniolo P, Lenner P, Zhong XY. Methylation signature of lymph node metastases in breast cancer patients. BMC Cancer. 2012;12:244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 52] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 47. | Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3043] [Cited by in RCA: 2445] [Article Influence: 152.8] [Reference Citation Analysis (0)] |
| 48. | Haque A, Engel J, Teichmann SA, Lönnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 589] [Cited by in RCA: 667] [Article Influence: 83.4] [Reference Citation Analysis (0)] |
| 49. | Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202-1214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5595] [Cited by in RCA: 4863] [Article Influence: 486.3] [Reference Citation Analysis (0)] |
| 50. | Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell. 2017;65:631-643.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 855] [Cited by in RCA: 960] [Article Influence: 120.0] [Reference Citation Analysis (0)] |
| 51. | Moffitt JR, Hao J, Bambah-Mukku D, Lu T, Dulac C, Zhuang X. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc Natl Acad Sci U S A. 2016;113:14456-14461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 219] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 52. | Trivedi V, Choi HMT, Fraser SE, Pierce NA. Multidimensional quantitative analysis of mRNA expression within intact vertebrate embryos. Development. 2018;145:pii: dev156869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 53. | Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N, Junker JP. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat Biotechnol. 2018;36:469-473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 363] [Article Influence: 51.9] [Reference Citation Analysis (0)] |
| 54. | Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016;167:1867-1882.e21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 768] [Cited by in RCA: 753] [Article Influence: 83.7] [Reference Citation Analysis (0)] |
| 55. | Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods. 2017;14:297-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 732] [Cited by in RCA: 694] [Article Influence: 86.8] [Reference Citation Analysis (0)] |
| 56. | Winterhoff BJ, Maile M, Mitra AK, Sebe A, Bazzaro M, Geller MA, Abrahante JE, Klein M, Hellweg R, Mullany SA. Single cell sequencing reveals heterogeneity within ovarian cancer epithelium and cancer associated stromal cells. Gynecol Oncol. 2017;144:598-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 57. | Giustacchini A, Thongjuea S, Barkas N, Woll PS, Povinelli BJ, Booth CAG, Sopp P, Norfo R, Rodriguez-Meira A, Ashley N. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat Med. 2017;23:692-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 310] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 58. | Walter C, Pozzorini C, Reinhardt K, Geffers R, Xu Z, Reinhardt D, von Neuhoff N, Hanenberg H. Single-cell whole exome and targeted sequencing in NPM1/FLT3 positive pediatric acute myeloid leukemia. Pediatr Blood Cancer. 2018;65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 59. | Hui T, Cao Q, Wegrzyn-Woltosz J, O’Neill K, Hammond CA, Knapp DJHF, Laks E, Moksa M, Aparicio S, Eaves CJ. High-Resolution Single-Cell DNA Methylation Measurements Reveal Epigenetically Distinct Hematopoietic Stem Cell Subpopulations. Stem Cell Reports. 2018;11:578-592. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 60. | Sen R, Dolgalev I, Bayin NS, Heguy A, Tsirigos A, Placantonakis DG. Single-Cell RNA Sequencing of Glioblastoma Cells. Methods Mol Biol. 2018;1741:151-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 61. | Johnson E, Dickerson KL, Connolly ID, Hayden Gephart M. Single-Cell RNA-Sequencing in Glioma. Curr Oncol Rep. 2018;20:42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 62. | Colacino JA, Azizi E, Brooks MD, Harouaka R, Fouladdel S, McDermott SP, Lee M, Hill D, Madden J, Boerner J. Heterogeneity of Human Breast Stem and Progenitor Cells as Revealed by Transcriptional Profiling. Stem Cell Reports. 2018;10:1596-1609. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 107] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 63. | Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3108] [Cited by in RCA: 3612] [Article Influence: 277.8] [Reference Citation Analysis (0)] |
| 64. | Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942-949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3582] [Cited by in RCA: 3878] [Article Influence: 184.7] [Reference Citation Analysis (0)] |
| 65. | Baitsch L, Baumgaertner P, Devêvre E, Raghav SK, Legat A, Barba L, Wieckowski S, Bouzourene H, Deplancke B, Romero P. Exhaustion of tumor-specific CD8+ T cells in metastases from melanoma patients. J Clin Invest. 2011;121:2350-2360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 573] [Cited by in RCA: 680] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 66. | Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 427] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 67. | Kirsch IR, Watanabe R, O’Malley JT, Williamson DW, Scott LL, Elco CP, Teague JE, Gehad A, Lowry EL, LeBoeuf NR. TCR sequencing facilitates diagnosis and identifies mature T cells as the cell of origin in CTCL. Sci Transl Med. 2015;7:308ra158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 159] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 68. | Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1792] [Cited by in RCA: 2952] [Article Influence: 246.0] [Reference Citation Analysis (0)] |
| 69. | Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099-4107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 824] [Cited by in RCA: 904] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 70. | Sherwood AM, Emerson RO, Scherer D, Habermann N, Buck K, Staffa J, Desmarais C, Halama N, Jaeger D, Schirmacher P. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol Immunother. 2013;62:1453-1461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 142] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 71. | Zemmour D, Zilionis R, Kiner E, Klein AM, Mathis D, Benoist C. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol. 2018;19:291-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 238] [Cited by in RCA: 289] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 72. | De Simone M, Rossetti G, Pagani M. Single Cell T Cell Receptor Sequencing: Techniques and Future Challenges. Front Immunol. 2018;9:1638. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 73. | Zhang X, Zhang M, Hou Y, Xu L, Li W, Zou Z, Liu C, Xu A, Wu S. Single-cell analyses of transcriptional heterogeneity in squamous cell carcinoma of urinary bladder. Oncotarget. 2016;7:66069-66076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 74. | Li Y, Xu X, Song L, Hou Y, Li Z, Tsang S, Li F, Im KM, Wu K, Wu H. Single-cell sequencing analysis characterizes common and cell-lineage-specific mutations in a muscle-invasive bladder cancer. Gigascience. 2012;1:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 75. | Bakker B, Taudt A, Belderbos ME, Porubsky D, Spierings DC, de Jong TV, Halsema N, Kazemier HG, Hoekstra-Wakker K, Bradley A. Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome Biol. 2016;17:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 157] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 76. | Gawad C, Koh W, Quake SR. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc Natl Acad Sci USA. 2014;111:17947-17952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 211] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 77. | Boyd AL, Aslostovar L, Reid J, Ye W, Tanasijevic B, Porras DP, Shapovalova Z, Almakadi M, Foley R, Leber B. Identification of Chemotherapy-Induced Leukemic-Regenerating Cells Reveals a Transient Vulnerability of Human AML Recurrence. Cancer Cell. 2018;34:483-498.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 78. | Hughes AE, Magrini V, Demeter R, Miller CA, Fulton R, Fulton LL, Eades WC, Elliott K, Heath S, Westervelt P. Clonal architecture of secondary acute myeloid leukemia defined by single-cell sequencing. PLoS Genet. 2014;10:e1004462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 108] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 79. | Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 428] [Cited by in RCA: 415] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 80. | Shepherd MS, Li J, Wilson NK, Oedekoven CA, Li J, Belmonte M, Fink J, Prick JCM, Pask DC, Hamilton TL. Single-cell approaches identify the molecular network driving malignant hematopoietic stem cell self-renewal. Blood. 2018;132:791-803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 81. | Francis JM, Zhang CZ, Maire CL, Jung J, Manzo VE, Adalsteinsson VA, Homer H, Haidar S, Blumenstiel B, Pedamallu CS. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014;4:956-971. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 230] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 82. | Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396-1401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2689] [Cited by in RCA: 3425] [Article Influence: 311.4] [Reference Citation Analysis (0)] |
| 83. | Baslan T, Kendall J, Ward B, Cox H, Leotta A, Rodgers L, Riggs M, D’Italia S, Sun G, Yong M. Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res. 2015;25:714-724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 105] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 84. | Lei B, Zhang XY, Zhou JP, Mu GN, Li YW, Zhang YX, Pang D. Transcriptome sequencing of HER2-positive breast cancer stem cells identifies potential prognostic marker. Tumour Biol. 2016;37:14757-14764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 85. | Nguyen A, Yoshida M, Goodarzi H, Tavazoie SF. Highly variable cancer subpopulations that exhibit enhanced transcriptome variability and metastatic fitness. Nat Commun. 2016;7:11246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 97] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 86. | Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, Tsai PC, Casasent A, Waters J, Zhang H. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet. 2016;48:1119-1130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 325] [Cited by in RCA: 345] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 87. | Kim C, Gao R, Sei E, Brandt R, Hartman J, Hatschek T, Crosetto N, Foukakis T, Navin NE. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell. 2018;173:879-893.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 756] [Cited by in RCA: 726] [Article Influence: 103.7] [Reference Citation Analysis (0)] |
| 88. | Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155-160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 761] [Cited by in RCA: 750] [Article Influence: 68.2] [Reference Citation Analysis (0)] |
| 89. | Yu C, Yu J, Yao X, Wu WK, Lu Y, Tang S, Li X, Bao L, Li X, Hou Y. Discovery of biclonal origin and a novel oncogene SLC12A5 in colon cancer by single-cell sequencing. Cell Res. 2014;24:701-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 90. | Liu M, Di J, Liu Y, Su Z, Jiang B, Wang Z, Su X. Comparison of EpCAMhighCD44+ cancer stem cells with EpCAMhighCD44- tumor cells in colon cancer by single-cell sequencing. Cancer Biol Ther. 2018;19:939-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 91. | Liu M, Liu Y, Di J, Su Z, Yang H, Jiang B, Wang Z, Zhuang M, Bai F, Su X. Multi-region and single-cell sequencing reveal variable genomic heterogeneity in rectal cancer. BMC Cancer. 2017;17:787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 92. | Kim KT, Lee HW, Lee HO, Song HJ, Jeong da E, Shin S, Kim H, Shin Y, Nam DH, Jeong BC. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol. 2016;17:80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 154] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 93. | Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, Kang B, Liu Z, Jin L, Xing R. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24:978-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 749] [Cited by in RCA: 1089] [Article Influence: 155.6] [Reference Citation Analysis (0)] |
| 94. | Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886-895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 504] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 95. | Kim KT, Lee HW, Lee HO, Kim SC, Seo YJ, Chung W, Eum HH, Nam DH, Kim J, Joo KM. Single-cell mRNA sequencing identifies subclonal heterogeneity in anti-cancer drug responses of lung adenocarcinoma cells. Genome Biol. 2015;16:127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 204] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 96. | Suzuki A, Matsushima K, Makinoshima H, Sugano S, Kohno T, Tsuchihara K, Suzuki Y. Single-cell analysis of lung adenocarcinoma cell lines reveals diverse expression patterns of individual cells invoked by a molecular target drug treatment. Genome Biol. 2015;16:66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |