Copyright
©The Author(s) 2021.
World J Stem Cells. Mar 26, 2021; 13(3): 221-235
Published online Mar 26, 2021. doi: 10.4252/wjsc.v13.i3.221
Published online Mar 26, 2021. doi: 10.4252/wjsc.v13.i3.221
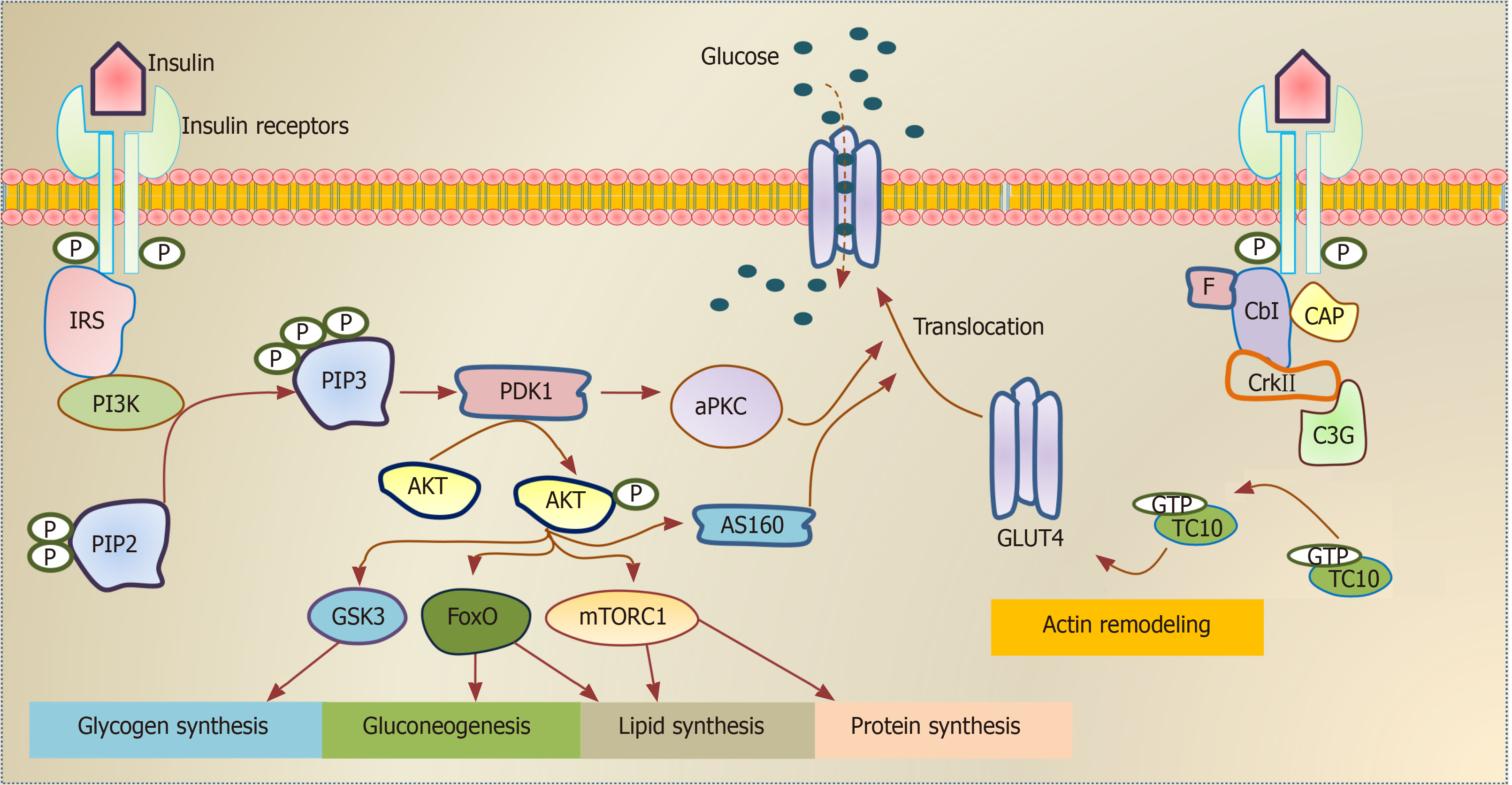
Figure 1 Schematic illustration of insulin signaling pathways.
Insulin binding activates the insulin receptor (INSR), which enables the recruitment of insulin receptor substrate isoforms and subsequent activation of the phosphatidylinositol 3-kinase (PI3K). The downstream event of PI3K enhances glucose uptake by translocation of glucose transporter proteins over cell membrane, enhances glycogen, lipid and protein synthesis and regulates lipolysis and gluconeogenesis. Alternative pathway for glucose transporter type 4 (GLUT-4) translocation by insulin stimulation. Insulin binding activates the INSR, which enables F binding to Cbl associated protein phosphorylates Cbl and recruit CrkII/C3G complex. This complex converts guanosine diphosphate into guanosine triphosphate (GTP) on TC10. The stimulated GTP containing TC10 involved in GLUT-4 translocation by actin remodeling on GLUT-4. INSR: Insulin receptor; IRS: Insulin receptor substrate; PI3K: Phosphatidylinositol 3-kinase; PIP2: Phosphatidylinositol 4,5 bisphosphate; PIP3: Phosphatidylinositol 3,4,5 trisphosphate; PDK1: Phosphatidylinositide dependent protein kinase 1; aPKC: Atypical protein kinase C; GSK3: Glycogen synthase kinase 3; FoxO: Forkhead box O; mTORC: mTOR complex; AS160: Akt substrate 160kDa; F: Flotillin; CAP: Cbl associated protein; C3G: Guanine nucleotide exchange factor; GTP: Guanosine triphosphate; GDP: Guanosine diphosphate; GLUT-4: Glucose transporter type 4.
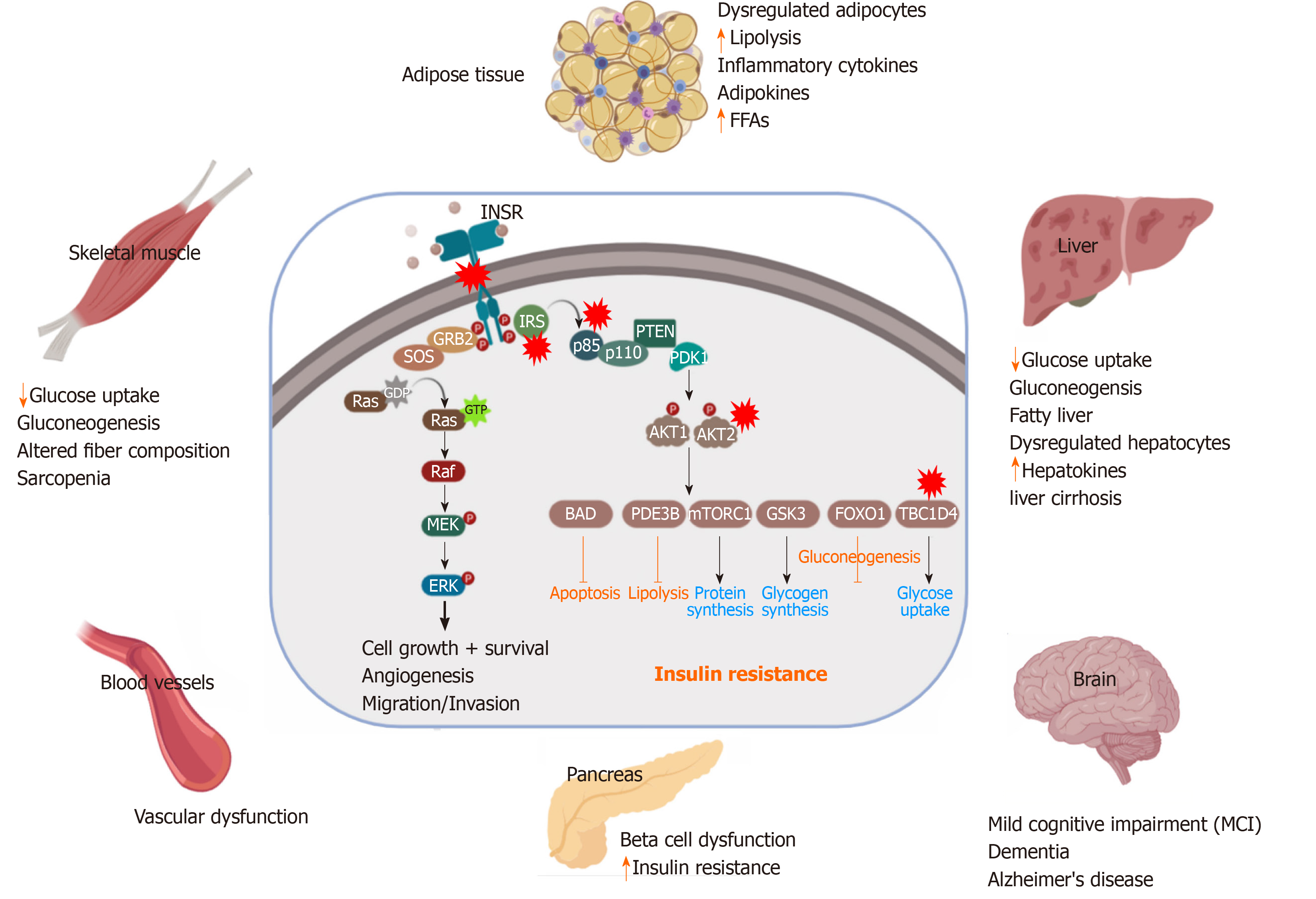
Figure 2 Pathological effect and consequence of insulin resistance.
Genetic alterations and mutations in the insulin signaling pathway can lead to insulin resistance (IR) in the insulin-target tissues. The red stars indicate the reported genetically defective molecules, such as insulin receptor, insulin receptor substrates, p85, AKT and TBC1D4. In response to IR, the insulin target tissues (adipose tissue, skeletal muscle, liver, pancreas, brain and blood vessels) show activation of the catabolic processes and accumulation of toxic metabolic byproducts and the inflammatory cytokines, leading to pancreatic β-cell dysfunction and other metabolic disorders. INSR: Insulin receptor; IRS: Insulin receptor substrate; PDK1: Phosphatidylinositide dependent protein kinase 1; GSK3: Glycogen synthase kinase 3; FoxO: Forkhead box O; mTORC: mTOR complex; GTP: Guanosine triphosphate; FFAs: Free fatty acids.
Figure 3 Using human induced pluripotent stem cells to study insulin resistance.
A schematic diagram showed the possibility of using human induced pluripotent stem cells (hiPSCs) to study insulin resistance (IR) with special focus on the genetic factors. Somatic cells are reprogrammed into pluripotency to generate induced pluripotent stem cells (iPSCs) carrying the genetic signatures of insulin sensitive and IR individuals. The generated iPSCs can be differentiated in vitro into the main insulin-target cells, including hepatocyte, adipocyte, and skeletal myotubes as well as insulin producing β-cells. Genome editing tools can be used to correct specific mutations in the generated iPSCs to establish the isogenic iPSC control. Studying those cells can help in understanding the signaling pathways involved in the development of IR and type 2 diabetes. Also, these hiPSC-based models can be used for drug screening. iPSCs: Induced pluripotent stem cells.
- Citation: Elsayed AK, Vimalraj S, Nandakumar M, Abdelalim EM. Insulin resistance in diabetes: The promise of using induced pluripotent stem cell technology. World J Stem Cells 2021; 13(3): 221-235
- URL: https://www.wjgnet.com/1948-0210/full/v13/i3/221.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i3.221